Abstract
A number of viruses remodel the cellular gene expression landscape by globally accelerating messenger RNA (mRNA) degradation. Unlike the mammalian basal mRNA decay enzymes, which largely target mRNA from the 5′ and 3′ end, viruses instead use endonucleases that cleave their targets internally. This is hypothesized to more rapidly inactivate mRNA while maintaining selective power, potentially though the use of a targeting motif(s). Yet, how mRNA endonuclease specificity is achieved in mammalian cells remains largely unresolved. Here, we reveal key features underlying the biochemical mechanism of target recognition and cleavage by the SOX endonuclease encoded by Kaposi's sarcoma-associated herpesvirus (KSHV). Using purified KSHV SOX protein, we reconstituted the cleavage reaction in vitro and reveal that SOX displays robust, sequence-specific RNA binding to residues proximal to the cleavage site, which must be presented in a particular structural context. The strength of SOX binding dictates cleavage efficiency, providing an explanation for the breadth of mRNA susceptibility observed in cells. Importantly, we establish that cleavage site specificity does not require additional cellular cofactors, as had been previously proposed. Thus, viral endonucleases may use a combination of RNA sequence and structure to capture a broad set of mRNA targets while still preserving selectivity.
INTRODUCTION
Viral infection dramatically reshapes the gene expression landscape of the host cell. By changing overall messenger RNA (mRNA) abundance or translation, viruses can redirect host machinery towards viral gene expression while simultaneously dampening immune stimulatory signals (1–3). Suppression of host gene expression, termed host shutoff, can occur via a variety of mechanisms, but one common strategy is to accelerate degradation of mRNA (1–3). This occurs during infection with DNA viruses such as alphaherpesviruses, gammaherpesvirues, and vaccinia virus, as well as with RNA viruses such as influenza A virus and SARS and MERS coronaviruses (1,4,5). In the majority of these cases, a viral factor promotes endonucleolytic cleavage of target mRNAs. This strategy bypasses the normally rate limiting steps of deadenylation and decapping to effect rapid mRNA degradation by host exonucleases (1).
Virally encoded host shutoff endonucleases are usually specific for mRNA, yet broad-acting in that they target the majority of the mRNA population. This is exemplified by herpesviral nucleases, including the SOX endonuclease encoded by Kaposi's sarcoma-associated herpesvirus (KSHV), an oncogenic human gammaherpesvirus that causes Kaposi's sarcoma and B cell lymphoproliferative diseases (6,7). KSHV SOX is a member of the PD-(D/E)xK type II restriction endonuclease superfamily that possesses mechanistically distinct DNase and RNase activities (8–10). The RNase activity of the gammaherpesvirus SOX protein has been shown to play key roles in various aspects of the viral lifecycle, including immune evasion, cell type specific replication, and controlling the gene expression landscape of infected cells (11–14). However, the mechanism by which SOX targets mRNAs remains largely unknown.
Sequencing data indicate that within the mRNA pool there appears to be a range of SOX targeting efficiencies; some transcripts are efficiently cleaved in cells, while others are partially or fully refractory to cleavage (15–19). Additionally, SOX has been shown to cut within specific locations of mRNAs in cells, further emphasizing that there must be transcript features that confer selectivity (16,20). Indeed, a transcriptome-wide cleavage analysis indicated that SOX targeting is directed by a relatively degenerate motif, often containing an unpaired polyadenosine stretch shortly upstream of the cleavage site, which is located in a loop structure (20). Cleavage within an unpaired loop was confirmed in a recent crystal structure of SOX with RNA, although additional contacts that could confer sequence specificity were not observed (21).
Thus, a major outstanding question is how RNA sequence and/or structure contribute to SOX target recognition. In this context, it is unclear how sequence features surrounding the RNA cleavage site might impact SOX targeting, for example by changing its affinity for a given RNA or the efficiency with which cleavage occurs. To address these questions, we sought to reconstitute the SOX cleavage reaction in vitro using purified components. Using an RNA substrate that is efficiently cleaved by SOX in cells, we revealed that specific RNA sequences within and outside of the cleavage site significantly contribute to SOX binding efficiency and target processing. In particular, we found that the polyadenosine stretch adjacent to the cleavage site is critical for SOX binding, and we experimentally verified the importance of an open loop structure surrounding the cleavage site. Finally, we demonstrated that this in vitro system faithfully recapitulates the initial endonucleolytic cleavage event that is an essential component of mRNA target specificity in vivo. Collectively, our data reveal that specific sequence features potently impact SOX binding, and thus provide key insight into the breadth of SOX targeting efficiency observed across the transcriptome. More broadly, this information provides a framework for better understanding the target specificity of endonucleases, which play central roles in mammalian quality control processes and viral infection outcomes.
MATERIALS AND METHODS
Recombinant protein expression and purification
KSHV SOX was codon optimized for Sf9 expression and synthesized from GENEWIZ. SOX was then subcloned using restriction sites BamHI and SalI (New England BioLabs) into pFastBac HTD. This vector was modified to carry a GST affinity tag and PreScission protease cut site as described (22). All SOX mutants were generated using single primer site-directed mutagenesis (23). Sequences were validated using standard pGEX forward and reverse primers. Generation of viral bacmids and transfections were prepared as described in the Bac-to-Bac® Baculovirus Expression System (Thermo Fisher Scientific) manual. After transfection, Sf9 cells (Thermo Fisher Scientific) were grown for 96 h at 22°C using SF-900 SMF media (Gibco) substituted with 5% fetal bovine serum (FBS) and 1% antibiotic antimycotic (AA). Supernatant was transferred to a six-well tissue culture plate containing 1 ml of 2 × 10∧6 cells/well. Cells were incubated for 96hr to generate passage 1 (P1). The P1 supernatant was transferred to a flask containing 50 ml of 1 × 106 cells/ml and incubated for 96 h, a time point sufficient to yield 5 mg of SOX per 50 ml of cells. Protein expression was confirmed by western blot with an anti-GST antibody (GE Health Care Life Sciences).
Sf9 cell pellets were suspended in lysis buffer containing 600 mM NaCl, 5% glycerol, 0.5% Triton X-100, 3 mM DTT, 20 mM HEPES pH 7.0 with a cOmplete, EDTA-Free protease inhibitor Cocktail tablet (Roche). Cells were sonicated on ice using a macro trip for 30 s bursts with 1 min rests for 6 min at 50 A. Cell lysate was cleared using a pre-chilled (4°C) Sorvall LYNX 6000 Superspeed Centrifuge spun at 18 000 rpm for 45 min. The cleared lysate was incubated for 4 h at 4°C with rotation with 4 ml of a GST bead slurry (GE Healthcare Life Sciences) that had been pre-washed 3× with wash buffer (WB) containing 600 mM NaCl, 5% glycerol, 3 mM DTT, 20 mM HEPES pH 7.0. The bead-protein mixture was washed 3× with 15 ml of WB, then transferred to a 10 ml disposable column (Qiagen) and washed with an additional 50 ml of WB followed by 100 ml of low salt buffer (LSB) containing 250 mM NaCl, 5% glycerol, 3 mM DTT, 20 mM HEPES pH 7.0 with periodic resuspension to prevent compaction. SOX was then cleaved on column with PreScission protease (GE Healthcare Life Sciences) overnight at 4°C, and protein eluate was collected for a final volume of 10 ml in LSB.
Cleaved protein was concentrated to ∼1 ml using Amicon filter concentrator membrane cut off 30 kDa (EMD Millipore), then loaded onto a HiLoad Superdex S200 pg gel filtration column (GE Healthcare Life Sciences). Protein elutions were concentrated using an Amicon concentrator described above to 5 mg/ml and 25 μl aliquots were snap frozen in liquid N2 using nuclease-free 0.5 ml microfuge tubes (Ambion Life Technologies) and stored at –80°C.
RNA substrate preparation and end labeling
All RNA substrates (sequences in Supplementary Table S1) unless stated otherwise were synthesized by Dharmacon (GE Healthcare) with HPLC and page purification. RNAs were 5′ end labeled with γ-[32P]-ATP-6000 Ci/mmol 150 mCi/ml (Perkin Elmer) using T4 PNK (New England BioLabs). RNAs were 3′ end labeled with 5′-[32P]-pCp 3000 Ci/mMol 10 mCi/ml using T4 RNA ligase 1 (New England BioLabs). Labeled RNA substrates were purified using 20% urea–PAGE and were isolated from gel slices by incubating overnight at 8°C in a buffer containing 10 mM Tris–HCl, 1 mM EDTA pH 8.0. Eluted RNAs were ethanol precipitated and resuspended in RNase-free ddH2O.
Ribonuclease assays
k obs and Hill coefficients of SOX were determined from the cleavage kinetics of [32P]-labeled RNA substrates as previously described (23). Briefly, 1 μl (≤1 pM) of [32P]-labeled RNA was added to 9 μl of premixture containing 20 mM HEPES pH 7.1, 70 mM NaCl, 2 mM MgCl2, 1 mM TCEP, 1% glycerol, and increasing concentrations of purified SOX. Reactions were performed at room temperature under single turnover conditions, and quenched at the indicated time intervals with 8 μl stop solution (10 M urea, 0.1% SDS, 0.1 mM EDTA, 0.05% xylene cyanol, 0.05% bromophenol blue). Samples were resolved by 15% urea–PAGE, imaged using a Typhoon variable mode imager (GE Healthcare), and quantified using ImageQuant and GelQuant software packages (Molecular Devices). The data were plotted and fit to exponential curves using Prism 7 software package (GraphPad) to determine observed rate constants.
A FRET probe with excitation at 646 nM and emission at 662 nM (LIMD1 54 Flo) was purchased from Dharmacon (Supplementary Table S1). The RNA FRET probe was added at a final concentration of 100 nM to 9 μl of premixture containing 20 mM Hepes pH 7.1, 70 mM NaCl, 2 mM MgCl2, 1 mM TCEP, 1% glycerol with 2 μM of SOX (23). Terminator 5′ exonuclease (Lucigen) was added to reactions using a 1:1000 dilution of the enzyme. reactions were quenched at indicated time intervals with equal volumes of stop solution containing 95% formamide and 10 mM EDTA, then resolved using urea-PAGE and visualized using a Typhoon variable mode imager (GE Halthcare). The data were plotted using Prism 7 software package (GraphPad). All experiments were repeated >3 times and mean values were computed.
For assays designed to detect endonucleolytic cleavage intermediates, 1 μl of labeled RNA substrate was combined with 9 μl of reaction solution (20 mM HEPES pH 7.1, 70 mM NaCl, 0.200 mM CaCl, 0.700 mM MgCl2, 1% glycerol, 0.5 mM TCEP) in the presence or absence of 2 μM SOX for 10 min at room temperature. RNA was then ethanol precipitated, resuspended in 95% formamide solution containing 10 mM EDTA, and resolved on a 12% urea–PAGE analytical grade sequencing gel together with a ssRNA Decade ladder (Ambion Life Technologies) for 1.5 h at 22 W before imaging as described above.
In-line probing
The sequence surrounding the cut site in LIMD1 was inserted into a pBSSK (–) backbone using the BamHI and XbaI restriction sites. Mutations were introduced by the Quickchange site directed mutagenesis protocol (Agilent). The 100 nt sequence surrounding the GFP cut site was inserted using the BamHI and XhoI restriction site.
In-line probing was performed as described previously (24). Briefly, pBSSK(–) plasmids containing the indicated sequences (see Supplementary Table S2) were linearized by digestion with XhoI and ScaI for GFP or BlpI and SacI (NEB) for LIMD1, gel purified, phenol/chloroform extracted, and ethanol precipitated. The fragments were then used as templates for in vitro transcription with the HiScribe T7 High Yield RNA synthesis Kit (NEB) and afterwards subjected to Turbo DNase (Ambion by Life Technologies) treatment. RNA was resolved by 8% Urea PAGE, and full length transcripts were excised from the SYBR Gold stained gel (Thermo Fisher Scientific), eluted overnight in G50 buffer (20 mM Tris HCl pH 7.5, 300 mM NaOAc, 2 mM EDTA, 0.2% SDS), phenol/chloroform extracted, and ethanol precipitated. The RNA (∼40 pmol) was dephosphorylated using shrimp alkaline phosphatase (rSAP, NEB), labeled with 1 μl [γ32P] ATP (150 mCi/ml) using USB Optikinase (Affymetrix), then gel purified as described above and dissolved in 20 μl of nuclease free water. For the in-line probing reaction, 1 μl RNA (≥20 000 cpm) was incubated in 2× reaction buffer (100 mM Tris-HCl pH 8.3, 40 mM MgCl2, 200 mM KCl) at room temperature for 24 or 48 h. The reaction was quenched with 2× loading buffer (10 M urea, 1.5 mM EDTA pH 8.0). To generate ladders, 1 μl of the purified RNA was separately subjected to hydrolysis using the Next Magnesium RNA Fragmentation module (–OH) or RNase T1 digestion (T1) (NEB). Reactions were resolved by 8% urea–PAGE, exposed on a phoshorimager screen, and scanned using the Storm 820 imaging system (GE Healthcare). Deduced RNA structures were drawn using the RNA secondary structure visualization tool forna (Vienna RNA Web Services).
Electrophoretic mobility shift assays (EMSA)
RNA probes used in EMSA experiments were radiolabeled using the protocol described for ribonuclease activity assays. Reactions were incubated at RT for 30 min in buffer containing 20 mM HEPES pH 8.0, 30 mM KCl, 5 mM CaCl2, 0.01% Tween-20, 0.5 TCEP, 0.2 mg/ml BSA (Sigma-Aldrich), 40 μg/ml of yeast tRNA (Ambion Thermo Fisher), and the indicated amount of purified SOX protein. Calcium Chloride was used in these binding assays to prevent substrate processing and stabilize RNA–protein interactions. Reactions volumes were kept at 10 μl and stopped with 3 μl 7× EMSA loading dye (70 mM HEPES pH 8.0, 420 mM KCl, 35% glycerol). Reactions were resolved by 8% native PAGE, and gels were imaged on a Typhoon multivariable imager (GE Healthcare) and quantified using GelQuant software package (Molecular Dynamics).
SOX RNA footprinting assay
LIMD1-54 RNA was 5′ end labeled with γ-[32P]-ATP-6000 Ci/mmol 150mCi/ml (PerkinElmer) using T4 PNK (New England BioLabs). RNA was then gel purified as stated previously. EMSA gel shifts were first used to determine optimal binding conditions (>90% binding, homogeneous complexes of RNA-protein). Binding buffer contained 0.01% Tween 20 (Sigma-Aldrich), 5 mM CaCl2, 5 mM KCl, 50 mM NaCl, 0.5 mM TCEP, 20 mM HEPES pH 8.0, 0.04 mg/ml yeast tRNA (Ambion), 0.2 mg/ml nuclease free Bovine Serum Albumin (BSA) (Ambion). A dilution series of SOX (8–0.5 μM) was incubated with 1 μl of radiolabeled LIMD1-54 in the presence of 0.1 unit of RNase T1 (Epicentre Illumina). Reactions were incubated at RT for a total of 10 min before being ethanol precipitated. RNA pellets were then resuspended in 5 μl of 95% formamide solution containing 10 mM EDTA and boiled for 5 min. Samples were then loaded onto a 10% analytical grade urea–PAGE gel and run at 22 W for 1.5 h. Gels were imaged and analyzed as stated above. In order to produce an RNase T1 ladder, 1 μL of LIMD1-54 was incubated with 0.1 units of RNase T1. Reactions were incubated at RT for 5 min before being quenched and prepared as stated previously. The LIMD1-54 hydrolysis ladder was generated as stated in the in-line probing methods.
Bio-layer interferometry (BLI) real-time binding kinetics
RNA probes (3′ end labeled with biotin) were synthesized from Dharmacon (GE healthcare) and HPLC and PAGE purified (See Supplementary Table S1). The Octet RED96e Bio-Layer Interferometry instrument and Streptavidin (SA) Biosensors were available from ForteBio (Menlo Park, CA, USA). All steps were performed in reaction buffer similar to EMSA binding conditions. Biosensors were incubated with 200 nM of the biotinylated RNA substrate for 150 sec and free RNA was washed away in EMSA buffer containing no RNA. SOX protein was incubated with the RNA conjugated biosensors for 200–400 s in order to reach saturation. Indicated protein concentrations for each bio sensor are located on corresponding binding curves. Complexes were dissociated for minimum of 15 min. Response curves for each biosensor were normalized against biosensors conjugated to RNA in the absence of SOX (buffer only control). Normalized response curves were processed using Octet Software version 7 by fitting the group of selected bio sensors to a nonlinear regression model (25). Dissociation constants (Kd) were determined from kon and kdis values derived from the fitted curves. A complete table of all values is provided in Supplementary Table S2.
RESULTS
KSHV SOX cleaves RNA substrates endonucleolytically as a monomer
In cells, the mRNA fragments resulting from the primary SOX endonucleolytic cleavage are predominantly cleared by the host 5′-3′ exonuclease XRN1, while in vitro, RNA fragments are rapidly degraded by 5′-3′ exonucleolytic activity intrinsic to purified SOX (9). Thus, it has been challenging to analyze the initial endonucleolytic cleavage event that is an essential component of mRNA target specificity in vivo. Here, we sought to develop a biochemical system to address these questions.
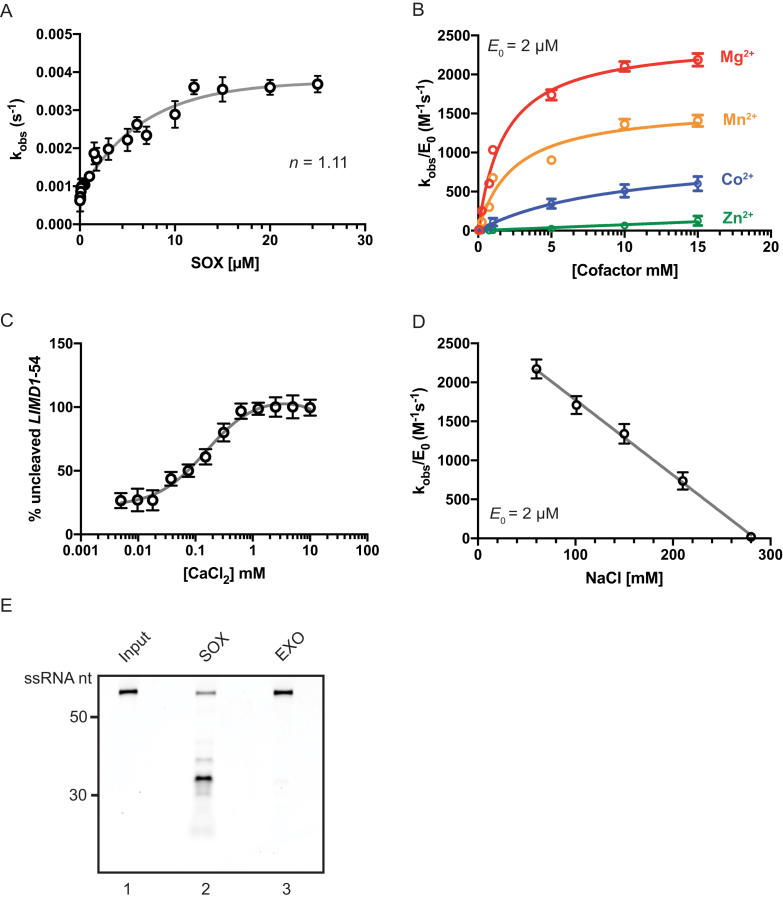
Our prior analysis of SOX targets in cells identified the human LIMD1 mRNA, which codes for a protein essential for P body formation and integrity, as being highly susceptible to cleavage by SOX (20). The minimum sequence required to directly cut the putative cleavage site in LIMD1 in cells was mapped to a 54-nucleotide segment (LIMD1-54), and we therefore chose this as our model substrate to study SOX targeting in vitro (20). We first expressed and purified KSHV SOX to greater then 95% purity from Sf9 insect cells (Supplementary Figure S1A). Using the LIMD1-54 substrate, we plotted the observed rate constant (kobs) as a function of SOX concentration, yielding a Hill coefficient of n = 1.11 (Figure 1A). Thus, in agreement with previous observations (9,10), SOX appears to function predominantly as a monomer. Under conditions of half maximal activity (2 μM; Figure 1A), SOX displayed a strong preference for the ‘hard’ divalent metal Mg2+ and a weaker preference for the ‘softer’ and larger metals Mn2+, Co2+ and Zn2+ (Figure 1B). This is again consistent with other characterized members of the P/DExK family of enzymes (9,26). Notably, SOX activity in the presence of Mg2+ was inhibited in a dose-dependent manner upon competitive addition of Ca2+ (Figure 1C and Supplementary Figure S1D). This is likely the result of increased coordination partners engaged by Ca2+, which decreases the ability of catalytic residues to promote proper base hydrolysis (27–29). Finally, increasing the NaCl concentration above 100 mM led to substantially decreased SOX activity (Figure 1D), in accordance with the observation that high salt concentrations frequently inhibit nuclease activity by disrupting protein-protein or protein-substrate interactions (29). Given that recombinant SOX displays robust 5′-3′ exonuclease activity (9,10), we sought to confirm that LIMD1-54 was subject to endonucleolytic SOX cleavage, as this is the predominant event that directs mRNA turnover in SOX expressing cells (3,16). Both the 5′ and 3′ ends LIMD1-54 were blocked by capping the 5′ end with a Cy5 fluorophore and the 3′ end with an Iowa Black quencher (LIMD1-54 Flo). We confirmed this RNA was resistant to degradation by the 5′-phosphate dependent exonuclease terminator (Figure 1E, lane 3). However, in the presence of SOX, a cleavage product was observed that correlated with an endonucleolytic cut (Figure 1E, lane 2).
Figure 1.
Kinetic characterization of recombinant SOX. (A) The observed rate constant (kobs) was plotted as a function of SOX concentration using the 5′ 32P-labeled LIMD1-54 RNA substrate, showing a hill coefficient (n) of 1.11. (B) The catalytic efficiency of SOX (2 μM) in the presence of MgCl2, MnCl2, CoCl2 and ZnCl2 was plotted as a function of cofactor concentration. (C) The impact of adding increasing concentrations of CaCl2 on SOX-induced degradation of a 5′ 32P-labeled LIMD1-54 RNA probe. Reactions were carried out in the presence of 0.7 mM MgCl2. (D) SOX catalytic efficiency was determined under increasing concentrations of NaCl. (E) The 5′ and 3′ ends of LIMD-54 were blocked with Cy5 and Iowa Black, respectively, to prevent exonucleolytic degradation. Reactions were incubated for 30 min in the presence of SOX or, as a control, the Terminator 5′ exonuclease (EXO). Input refers to RNA in reaction buffer without enzyme.
To confirm this processing event was not a result of contamination, we purified a SOX mutant containing mutations within two key residues of the SOX active site (D221N/E244Q). Incubation of this mutant with LIMD1-54 over the course of 1.5 h yielded no RNA cleavage (Supplementary Figure S1E). Thus, recombinant SOX appears to target LIMD1-54 for endonucleolytic cleavage in vitro, as has been observed for this substrate in cells.
KSHV SOX shows RNA substrate selectivity in vitro
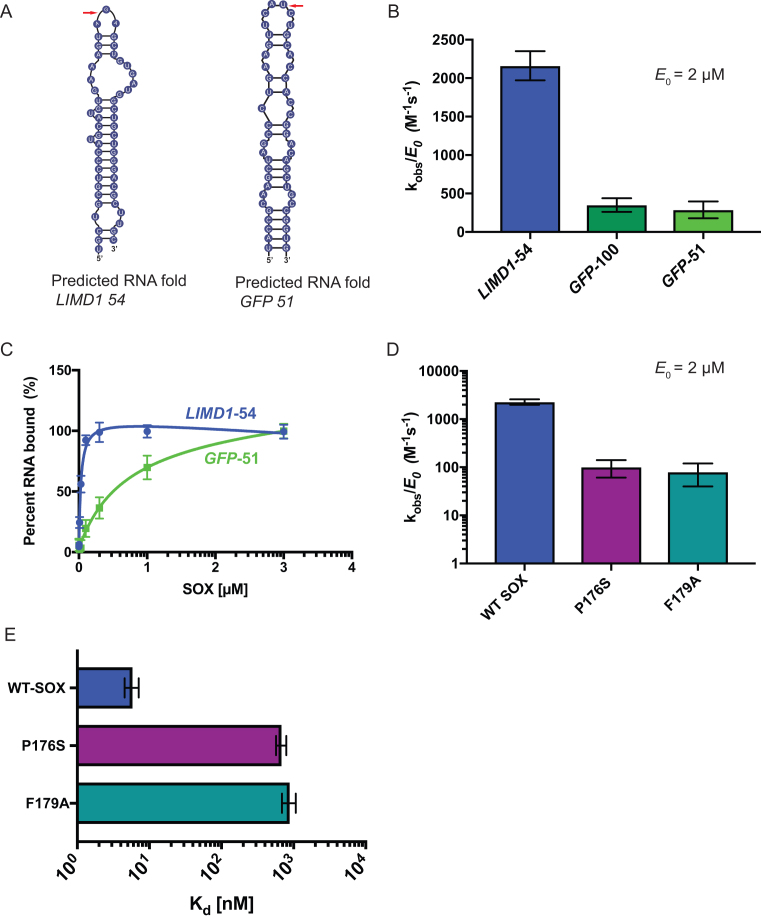
To analyze RNA substrate selectivity using our in vitro assay, we first compared SOX degradation of LIMD1-54 to a 51-nucleotide sequence of the mRNA encoding GFP (GFP-51). We have previously shown that GFP mRNA is cleaved by SOX in cells, and that GFP-51 is the minimal sequence required to elicit cleavage (15,16). The cleavage sites for LIMD1-54 and GFP-51 are predicted to occur in an open loop region (Figure 2A, red arrow). Upon direct comparison of these two RNAs, we observed a ∼6-fold increase in the catalytic efficiency of SOX for the LIMD1-54 substrate compared to GFP-51 (Figure 2B). This difference was not exclusively due to the fact that the GFP substrate was slightly shorter than LIMD1-54, as SOX also displayed a 5-fold reduction of catalytic efficiency on a longer, 100 nt GFP substrate (GFP-100; Figure 2B). Electrophoretic mobility shift assays (EMSA) further revealed a 10-fold increase in SOX binding to LIMD1-54 compared to GFP-51 (Figure 2C). Given that both substrates contain the requisite unpaired bulge at the predicted cleavage site (see Figure 2A and Supplementary Figure S2), these observations suggest that additional sequence or structural features impact SOX targeting efficiency on individual RNAs.
Figure 2.
Substrate specificity, salt concentration and the bridge motif play important roles in SOX activity. (A) The predicted folding of the LIMD1-54 and GFP-51 RNAs was determined using mFold (35). Red arrows mark the predicted SOX cleavage site. (B) Catalytic efficiencies were determined for SOX (2 μM) in the presence of GFP-51, GFP-100 or LIMD1-54 substrates. Reactions were performed in triplicate. (C) Binding curves of SOX with GFP-51 and LIMD1-54 RNA. Percent binding of substrates was determined by EMSA, whereupon curves were fit to a single binding model from three independent measurements. (D) Catalytic efficiency of WT SOX or the host shutoff mutants P176S and F179A was determined at a constant enzyme concentration (2 μM) using a 5′ 32P-labeled LIMD1-54 RNA probe. Experiments were performed in triplicate. (E) EMSAs were used to determine percent binding of 5′ 32P-labeled LIMD1-54 RNA probe to WT SOX, P176S, and F179A. Curves were fit to a single binding model from three independent measurements.
Two SOX point mutants, P176S and F179A, located in an unstructured region of the protein that bridges domains I and II have been shown to be selectively required for its endonucleolytic processing of RNA substrates (Supplementary Figure S3A and S3B) (8,21). Structural data indicate that residue F179 forms a stacking interaction with an adenine base in the RNA, likely stabilizing the protein-RNA interaction, while P176 is hypothesized to contribute to structural rearrangements required for F179 engagement (21). We purified both mutants to evaluate their relative RNA processing and RNA binding activity against the optimal LIMD1-54 substrate. Both mutants displayed purity and elution profiles similar to wild type (WT) SOX (see Supplementary Figure S1A-C). However, the catalytic efficiency of each mutant was >10-fold less than WT SOX (Figure 2D). Furthermore, RNA binding was severely perturbed; the binding kinetics of WT SOX for LIMD1-54 are in the single digit nanomolar range (Kd = 7 nM), while P176S and F179A display >2 log defects (Kd = 702 nM and 831 nM, respectively) (Figure 2E and Supplementary Figure S4A–C). Thus, the large defect in RNA binding likely explains the decreased efficiency of RNA processing. Notably, while there was a dramatic decrease in the relative affinities of the two mutants for LIMD1-54, there was not a complete loss of binding or RNA processing. This could be a result of secondary nonspecific interactions and/or nonspecific exonucleolytic degradation by SOX from the 5′ monophosphorylated end of the probe.
Secondary structure determination of the LIMD1-54 substrate
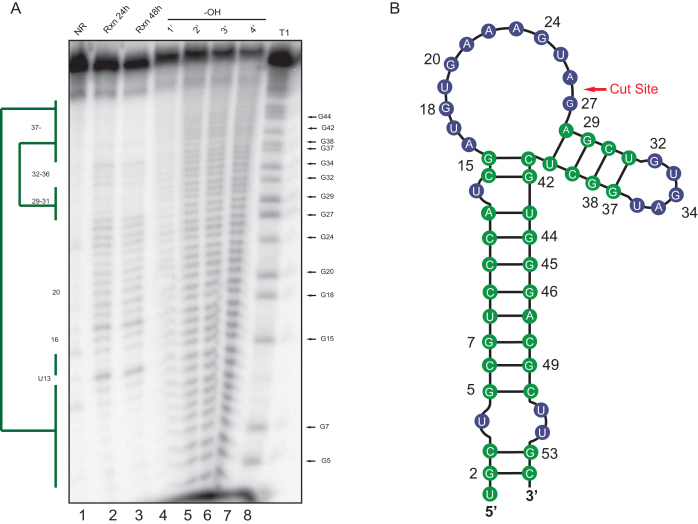
In silico RNA folding predictions of SOX targeting motifs, coupled with RNA mutagenesis experiments, have indicated that an RNA stem loop structure is an important determinant in SOX targeting both in vitro and in vivo (20,21). Given the importance of this predicted motif, and in particular the proposed requirement for unpaired sequence at the cut site, we sought to experimentally determine the structure of LIMD1-54 using chemical based in-line probing (Figure 3A). This showed that the LIMD1-54 structure contains a largely base paired stem region, followed by a loop at positions 15–27 that encompasses the predicted SOX cleavage site between nt 26 and 27, and a short hairpin structure at positions 29–40 (Figure 3B). Notably, some differences exist between the predicted and observed structures of LIMD1-54, including a larger loop region and the subsequent short stem-loop (compare Figure 3B to Figure 2A) However, in both cases the predicted cleavage site of SOX resides in a loop region.
Figure 3.
In-line structure probing of LIMD1-54. (A) An in-line reaction (Rxn) was performed at room temperature for 24 h (lane 2) or 48 h (lane 3) at pH 8.3 to identify structured regions of the LIMD1-54 RNA. Ladders were generated by subjecting the RNA to cleavage by RNase T1 (lane 8) or alkaline hydrolysis (–OH, lane 4–7). Products were separated by 8% urea PAGE, whereupon structured regions protected from cleavage were identified (green bars). No reaction (NR, lane1) refers to the input RNA. (B) Diagram showing the LIMD1-54 structure as deduced from the in-line probing gel. Green color corresponds to the structured regions denoted by the bars in (A), while blue refers to unstructured regions.
SOX binds to a region encompassing the unpaired stretch of adenosine repeats
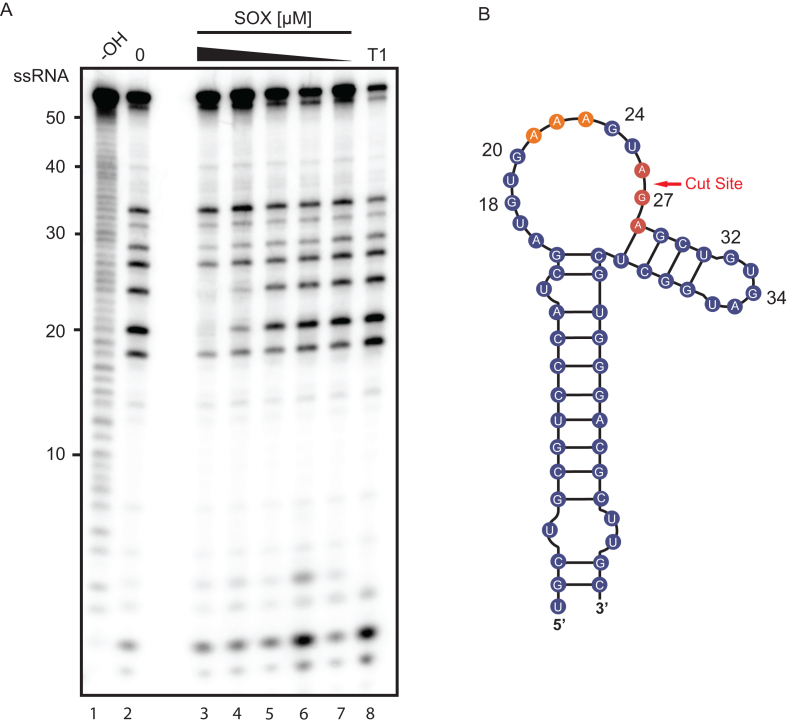
Recently, a high-resolution crystal structure was solved of SOX bound to a 31nt fragment of the KSHV pre-microRNA K12-2 (K2-31). In this structure, the only observed contacts between SOX and K2-31 occurred between the four active site residues of SOX (Y373, R248, C247, F179) and the UGAAG motif surrounding the cleavage site of the RNA (21). It was therefore hypothesized that no other residues beyond this unpaired UGAAG motif were involved in transcript recognition (21). However, the binding affinity we observed for LIMD1-54 was 200-fold stronger that what was previously reported for K2-31 (21), suggesting that a more extended interaction surface might distinguish optimal from sub-optimal RNA substrates. We therefore used RNA footprinting to map the SOX binding sites on LIMD1-54. Indeed, SOX protected a region of LIMD1-54 that included the three adenosine stretch (positions 20–24) from RNase T1 digestion in a dose dependent manner (Figure 4). Notably, this mapped binding region is the same region predicted from in vivo PARE-seq data to be important for SOX targeting, although the reason for its importance remained unknown (20). We also observed a modest protection of base 27 (G) located directly adjacent to the predicted cleavage site of SOX, which represents the region detected in the crystal structure of K2-31 bound to SOX. Collectively, these findings suggest that while SOX may interact with residues directly adjacent to the cut site, a more extensive interaction interface exists for its preferred in vivo targets.
Figure 4.
SOX binds to a stretch of adenosines upstream of the cleavage site. (A) An RNA footprinting assay was carried out by incubating 5′ 32P-labeled LIMD1-54 with RNase T1 in the presence (lanes 3–7) or absence (lane 2) of a dilution series of SOX (8–0.5 μM). Hydrolysis (–OH, lane 1) and RNase T1 (T1, lane 8) ladders of the RNA were also generated in order to map the location of protected sites. Lines on the right denote protected base pairs. (B) Diagram of LIMD1-54 indicating sites protected from RNase T1 cleavage by SOX. The upstream SOX binding site is colored orange while the protected residues surrounding the cut site are shown in red.
Base pairs surrounding the SOX binding and cleavage sites contribute to efficient substrate degradation
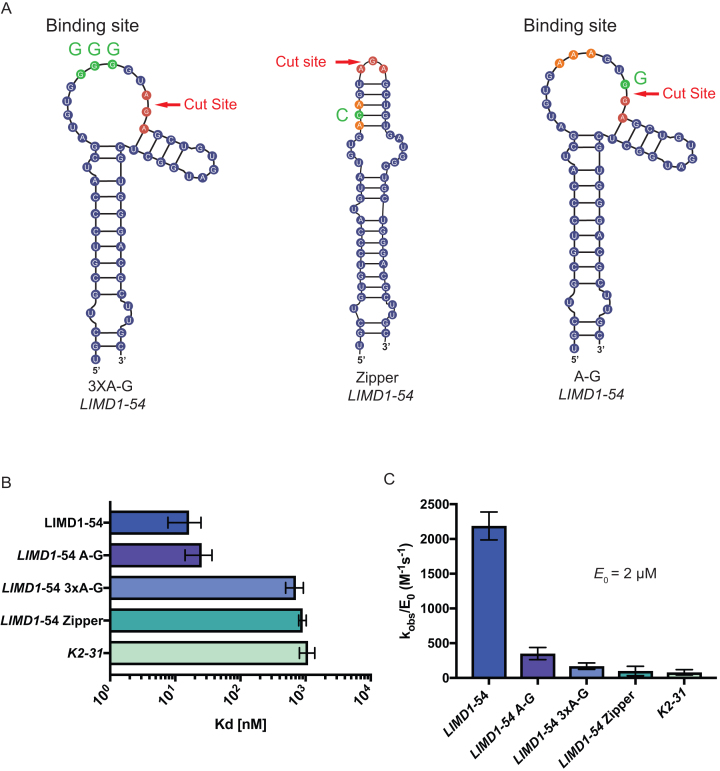
To explore the importance of the residues involved in SOX binding and cleavage, we engineered 3 mutants of the LIMD1-54 substrate (Figure 5A). First, we preserved the loop structure but replaced the three adenosines bound by SOX (residues 39–43) with guanosines (LIMD1-54 3xA-G). Second, we largely abolished the loop structure by providing complementary base pairing (LIMD1-54 Zipper). Third, we mutated the residue located at the predicted SOX cut site that was also protected in the footprinting assay (LIMD1-54 A-G). This mutant has been previously identified to block SOX cleavage in vivo (20). The predicted structures of the LIMD1-54 3XA-G and LIMD1-54 zipper mutants were verified by in-line probing (Figure 5, Supplementary Figure S6). Real-time binding kinetics for SOX with WT LIMD1-54 and each of the three mutant substrates were then measured using bio-layer interferometry (BLI). All RNA probes were 3′ biotinylated and immobilized to a streptavidin-coated BLI probe, whereupon the binding and dissociation of SOX was measured. To prevent degradation of the probe, excess calcium ion was used in place of magnesium (Supplementary Figure S1E). SOX retained similar binding affinity to the cut site mutant LIMD1 A-G (Kd = 25 nM) as to WT LIMD1-54 (Kd = 16.3 nM) (Figure 5B, Supplementary Figure S5A, B and Supplementary Table S2). In contrast, SOX exhibited dramatically reduced binding to both the predicted binding site mutant LIMD1-54 3xA-G (Kd = 710 nM) and to the LIMD1-54 zipper mutant lacking the loop region (Kd = 904 nM) (Figure 5B, Supplementary Figure S5C-D and Supplementary Table S2). To rule out the possibility that the effect on binding affinity to the LIMD1-54 zipper mutant was a result of altered residues within the binding site, we also engineered an additional zipper mutant (LIMD1-54 zipper 2) that did not disrupt the polyadenosine sequence. In agreement with the loop structure playing a critical role in target recognition, this LIMD1-54 zipper 2 mutant also displayed a substantial defect in binding (Kd = 2.09 μM; Supplementary Figure S7A, B, Supplementary Table S2). Finally, we measured SOX binding to the KSHV pre-miRNA sequence used to obtain the SOX-RNA co-crystal structure (K2-31) (21). Notably, the affinity of SOX for K2-31 was within the range of the LIMD1-54 structural mutants (Kd = 1.08 μM), suggesting that despite having an UGAAG motif upstream of a predicted bulge, this is unlikely to be a SOX target (Figure 5B, Supplementary Figure S5E and Supplementary Table S2).
Figure 5.
Mutations that disrupt LIMD1-54 structure reduce SOX binding affinity. (A) Diagram showing the location of mutations made within LIMD1-54, with the WT binding site and cut site residues labeled in orange and red, respectively. Binding site mutants and cut site mutants are labeled in green. (B) The binding affinity of SOX for each of the LIMD1-54 mutants was tested in parallel using Bio-Layer Interferometry (BLI). (C) SOX catalytic efficiency was tested at a constant concentration (2 μM) for all of the LIMD1-54 mutant substrates. All assays were performed in triplicate.
We next quantitatively measured the catalytic efficiency of SOX towards each of the above RNA substrates. Despite SOX having WT binding affinity for the predicted cleavage site mutant LIMD1-54 A-G, there was a 7-fold defect in its ability to degrade this mutant (Figure 5C). Even more marked defects in SOX catalytic efficiency were observed for the binding site mutant LIMD1-54 3XA-G, the loop mutants LIMD1-54 zipper and LIMD1-54 zipper 2, and the pre-miRNA K2-31 (Figure 5C and Supplementary Figure S7). Collectively, these data indicate that efficient RNA cleavage requires both an appropriate SOX binding site and a suitable cut site.
Site-specific endonucleolytic cleavage of target RNA occurs in vitro
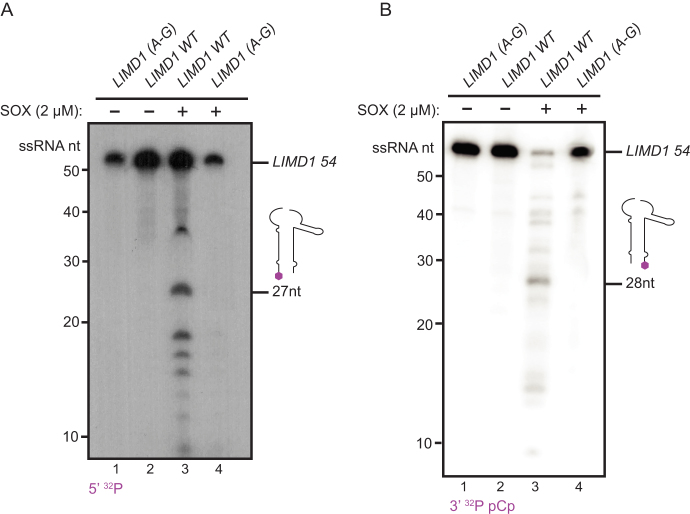
In cells, SOX cleaves its mRNA substrates site-specifically. Mutagenesis of residues in mapped cleavage sites generally abolishes SOX cleavage at that location (20). To determine if our in vitro assay faithfully recapitulated the site specificity of SOX endonucleolytic targeting observed in cells, we established reaction conditions that enabled trapping of the early cleavage events. By combining Ca2+ and Mg2+ in our reaction buffer, we were able to sufficiently slow SOX processing to visualize cleavage products derived from 5′ 32P labeled substrates. Indeed, we observed a predominant 27 nt band, which is the size of the product released upon LIMD1-54 cleavage at the predicted cut site (Figure 6A, lane 3). Additional bands also appeared, likely representing subsequent processing events. Importantly, when we incubate SOX with the cut site mutant LIMD1-54 A-G, there is a complete loss of this 27 nt product, as well as the additionally processed intermediates (Figure 6A, lane 4). Production of these cleavage intermediates required SOX, as no decay was observed in the RNA-only controls (Figure 6A, lanes 1–2).
Figure 6.
SOX endonucleolytically cleaves LIMD1-54 in a site-specific manner. (A) 5′ 32P-labeled LIMD1-54 and LIMD1 A-G were incubated for 10 min in 0.1 mM CaCl2 and 0.7 mM MgCl2 in the presence or absence of 2 μM SOX. The 5′ cleavage product with a size corresponding to cleavage at the mapped cut site (27 nt) is shown. (B) Cleavage assay was performed as in (A), except using 3′ 32P pCp-labeled LIMD1-54. The 3′ cleavage product with a size corresponding to cleavage at the mapped cut site (28 nt) is shown.
Finally, we sought to verify that the predominant 27 nt cleavage product we observed was a result of an endonucleolytic cleavage and not 5′ end processing. To this end, we generated a LIMD1-54 substrate containing a 3′ 32P pCp label and a free 5′ OH to block 5′ end processing. Again, in the presence of SOX, WT LIMD1-54 but not the A-G mutant produced a cleavage product whose size corresponded to cleavage at the predicted site (Figure 6B). Taken together, these data confirm that our in vitro assay faithfully recapitulates SOX cleavage site specificity on a true substrate.
DISCUSSION
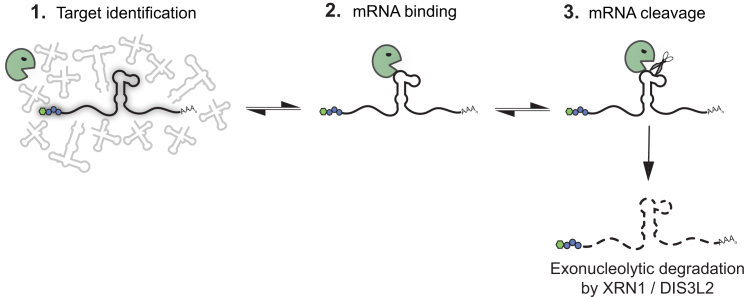
Endonuclease-directed mRNA degradation plays key roles in the lifecycle of gammaherpesviruses, yet the fundamental principles governing target specificity by SOX and other viral endonucleases are not well understood. Here, through the development of the first biochemical system to faithfully recapitulate the internal cleavage specificity observed for SOX in cells, we revealed how both RNA sequence and structure contribute to targeting. These findings resolve a central feature of the current model of SOX activity (Figure 7). Previous observations established that sequences flanking the cut site were required to direct cleavage by SOX (16,20). However, it was unresolved whether they played a strictly structural role in presenting an exposed loop for cleavage, served as a platform for SOX binding, or created a binding site for one or more cellular factors that then indirectly recruited SOX to its targets. Through a combination of mutational analyses, RNA structure probing, and RNA footprinting assays, we showed that efficient SOX targeting requires both an exposed loop structure and upstream sequences that serve as a SOX binding platform. This combination of sequence and structural features within the targeting motif helps explain why some mRNAs are efficiently cleaved by SOX, whereas others are weaker substrates.
Figure 7.
Model of mRNA targeting by SOX. SOX is able to distinguish mRNA from other types of RNA in cells by an as yet unknown mechanism. Subsequently, it endonucleolytically cleaves its targets at specific sites, whereupon the fragments are degraded by host exonucleases such as XRN1 and DIS3L2. Here, we revealed that in addition to the requirement for an unpaired loop at the cleavage site, additional upstream RNA sequences increase the affinity of SOX for individual targets, thereby controlling cleavage efficiency.
A key open question related to SOX function is how it can target the majority of mRNAs in cells, yet with significant site specificity. Our observations suggest that there must be specific mRNA features that influence targeting. Indeed, PARE-seq analyses of cleavage intermediates in SOX expressing cells revealed that cleavage sites were associated with a degenerate sequence motif (20). Sequences proximal to the cleavage site were predicted to be un-base paired and frequently contained a polyadenosine stretch followed by a purine (20). The requirement for these sequence features for SOX targeting was validated for the LIMD1 transcript in cells (20). Because LIMD1 has been established as a particularly robust SOX target in cells (20), we reasoned that it must contain features optimal for SOX processing and therefore would be an ideal substrate to dissect biochemically why these features are important. Indeed, SOX binding to LIMD1-54 was 10-fold better than to the commonly used reporter substrate GFP, and ∼100-fold better than to the K2-31 pre-miRNA, which has not been demonstrated to be processed by SOX in cells. Importantly, these binding differences correlated with the efficiency of SOX cleavage in vitro, arguing that the ability to bind the targeting motif is a key step in target recognition. Through RNA footprinting assays, we were able to show that SOX binds to a bulge structure proximal to the cleavage site containing the polyadenosine stretch previously predicted to be important for mRNA cleavage by SOX in cells (20). Mutating either just the bulge structure (LIMD1-54 zipper 2) or maintaining the bulge but mutating the polyadenosine stretch (LIMD1-54 3xA-G) resulted in a ∼100-fold reduction in binding affinity, correlating with a dramatic decrease in cleavage efficiency. Collectively, these data demonstrate that variability in the efficiency of SOX targeting observed in cells is likely due to differences in RNA sequences that mediate SOX binding.
A recent crystal structure of SOX bound to the K2-31 pre-miRNA captured the importance of the exposed loop region for SOX cleavage (21). However, the structure did not reveal additional interactions between SOX and the RNA beyond the three residues surrounding the cut site. Our data suggest that this is likely because the K2-31 RNA lacks the additional residues necessary for SOX binding site found in both LIMD1 and GFP. While the K2-31 RNA does contain adenosines upstream of the cleavage site, structural predictions indicate these residues are within a stem region (21), rather than in an exposed loop as is the case for LIMD1 and GFP. Together, these observations indicate that while upstream adenosines are important for binding, they must be present in an unpaired state to promote SOX binding. It is notable that prior studies reported much weaker interactions between SOX and RNA (Kd = 75 μM) compared to its DNA substrates (Kd = 1 μm) (9,10,21). However, in these cases binding assays were conducted with scrambled RNA sequences. We found that SOX binding affinities to RNA substrates vary over several orders of magnitude, in a manner that correlates with cleavage efficiency. Interestingly, the crystal structure of SOX bound to DNA showed more dynamic interactions along the length of the protein (∼480 Å2 interaction surface), when compared to the K2-31 RNA bound structure (∼240 Å2 interaction surface). It is therefore possible that more interaction along the length of SOX protein might occur with optimal substrates such as LIMD1 that are more tightly bound.
The fact that purified SOX endonucleolytically cleaved LIMD1-54 at the precise site observed in SOX-expressing cells demonstrates that cleavage site selection on an mRNA is not mediated by a cellular cofactor. Instead, targeting at particular RNA motifs is strongly influenced by the strength of SOX binding. Our observation that the P176S and F179A SOX mutants display significant RNA binding defects indicates that their failure to cleave mRNAs in cells is due to an inability to efficiently bind the targeting motif.
The mechanism by which SOX initially distinguishes RNA polymerase II transcribed mRNAs from other types of RNA in cells remains an important open question, as this feature of SOX selectivity is not preserved in vitro. We hypothesize that cellular co-factors, perhaps though interactions with SOX, enable this distinction. More broadly, endonucleases are instrumental in RNA processing and degradation. Nuclease processing defects lead to several human pathologies ranging from cancer to neurodegeneration (30–34), and our study provides a framework for better understanding the mechanistic features governing endonuclease targeting.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Glaunsinger lab for their suggestions and critical reading of the manuscript. We would like to thank the University of California, Berkeley tissue culture facility for Sf9 cell maintenance and the University of California San Francisco Quantitative Biosciences Institute, Antibiome Center for use of their Octet Red96e for binding kinetics measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health( NIH) [R01CA136367 to B.A.G.]; DAAD P.R.I.M.E. program [57222872 to C.V.]. B.A.G. is an investigator of the Howard Hughes Medical Institute. Funding for open access charge: NIH [R01 CA136367].
Conflict of interest statement. None declared.
REFERENCES
- 1. Abernathy E., Glaunsinger B.. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology. 2015; 479–480:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glaunsinger B.A. Modulation of the Translational Landscape During Herpesvirus Infection. Annu. Rev. Virol. 2015; 2:311–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaglia M.M., Covarrubias S., Wong W., Glaunsinger B.A.. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012; 86:9527–9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S.. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009; 16:1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayashi T., MacDonald L.A., Takimoto T.. Influenza A Virus Protein PA-X Contributes to Viral Growth and Suppression of the Host Antiviral and Immune Responses. J. Virol. 2015; 89:6442–6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rabkin C.S., Biggar R.J., Horm J.W.. Increasing incidence of cancers associated with the human immunodeficiency virus epidemic. Int. J. Cancer. 1991; 47:692–696. [DOI] [PubMed] [Google Scholar]
- 7. Kaplan L.D. Human herpesvirus-8: Kaposi sarcoma, multicentric Castleman disease, and primary effusion lymphoma. Hematol. Am Soc Hematol. Educ. Program. 2013; 2013:103–108. [DOI] [PubMed] [Google Scholar]
- 8. Glaunsinger B., Chavez L., Ganem D.. The exonuclease and host shutoff functions of the SOX protein of Kaposi's sarcoma-associated herpesvirus are genetically separable. J. Virol. 2005; 79:7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dahlroth S.L., Gurmu D., Schmitzberger F., Engman H., Haas J., Erlandsen H., Nordlund P.. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 2009; 276:6636–6645. [DOI] [PubMed] [Google Scholar]
- 10. Bagneris C., Briggs L.C., Savva R., Ebrahimi B., Barrett T.E.. Crystal structure of a KSHV-SOX-DNA complex: insights into the molecular mechanisms underlying DNase activity and host shutoff. Nucleic Acids Res. 2011; 39:5744–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richner J.M., Clyde K., Pezda A.C., Cheng B.Y., Wang T., Kumar G.R., Covarrubias S., Coscoy L., Glaunsinger B.. Global mRNA degradation during lytic gammaherpesvirus infection contributes to establishment of viral latency. PLoS Pathog. 2011; 7:e1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abernathy E., Clyde K., Yeasmin R., Krug L.T., Burlingame A., Coscoy L., Glaunsinger B.. Gammaherpesviral gene expression and virion composition are broadly controlled by accelerated mRNA degradation. PLoS Pathog. 2014; 10:e1003882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rowe M., Glaunsinger B., van Leeuwen D., Zuo J., Sweetman D., Ganem D., Middeldorp J., Wiertz E.J., Ressing M.E.. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee Y.J., Glaunsinger B.A.. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol. 2009; 7:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glaunsinger B., Ganem D.. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell. 2004; 13:713–723. [DOI] [PubMed] [Google Scholar]
- 16. Covarrubias S., Gaglia M.M., Kumar G.R., Wong W., Jackson A.O., Glaunsinger B.A.. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog. 2011; 7:e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clyde K., Glaunsinger B.A.. Deep sequencing reveals direct targets of gammaherpesvirus-induced mRNA decay and suggests that multiple mechanisms govern cellular transcript escape. PLoS One. 2011; 6:e19655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hutin S., Lee Y., Glaunsinger B.A.. An RNA element in human interleukin 6 confers escape from degradation by the gammaherpesvirus SOX protein. J. Virol. 2013; 87:4672–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muller M., Glaunsinger B.A.. Nuclease escape elements protect messenger RNA against cleavage by multiple viral endonucleases. PLoS Pathog. 2017; 13:e1006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaglia M.M., Rycroft C.H., Glaunsinger B.A.. Transcriptome-wide cleavage site mapping on cellular mRNAs reveals features underlying sequence-specific cleavage by the viral ribonuclease SOX. PLoS Pathog. 2015; 11:e1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee H., Patschull A.O.M., Bagneris C., Ryan H., Sanderson C.M., Ebrahimi B., Nobeli I., Barrett T.E.. KSHV SOX mediated host shutoff: the molecular mechanism underlying mRNA transcript processing. Nucleic Acids Res. 2017; 45:4756–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hui E., Cheung J., Zhu J., Su X., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I. et al. . T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017; 355:1428–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., Stroud R.M., Walter P.. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009; 457:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Regulski E.E., Breaker R.R.. In-line probing analysis of riboswitches. Methods Mol. Biol. 2008; 419:53–67. [DOI] [PubMed] [Google Scholar]
- 25. Cingolani G., Duncan T.M.. Structure of the ATP synthase catalytic complex (F(1)) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 2011; 18:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laganeckas M., Margelevicius M., Venclovas C.. Identification of new homologs of PD-(D/E)XK nucleases by support vector machines trained on data derived from profile-profile alignments. Nucleic Acids Res. 2011; 39:1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., McCabe K.A., Bell C.E.. Crystal structures of lambda exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mordasini T., Curioni A., Andreoni W.. Why do divalent metal ions either promote or inhibit enzymatic reactions? The case of BamHI restriction endonuclease from combined quantum-classical simulations. J. Biol. Chem. 2003; 278:4381–4384. [DOI] [PubMed] [Google Scholar]
- 29. Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Stroud R.M., Zhang C., Shokat K.M., Walter P.. Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1. BMC Biol. 2011; 9:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morton D.J., Kuiper E.G., Jones S.K., Leung S.W., Corbett A.H., Fasken M.B.. The RNA exosome and RNA exosome-linked disease. RNA. 2018; 24:127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gillespie A., Gabunilas J., Jen J.C., Chanfreau G.F.. Mutations of EXOSC3/Rrp40p associated with neurological diseases impact ribosomal RNA processing functions of the exosome in S. cerevisiae. RNA. 2017; 23:466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reis F.P., Pobre V., Silva I.J., Malecki M., Arraiano C.M.. The RNase II/RNB family of exoribonucleases: putting the ‘Dis’ in disease. Wiley Interdiscip. Rev. RNA. 2013; 4:607–615. [DOI] [PubMed] [Google Scholar]
- 33. Popp M.W., Maquat L.E.. Nonsense-mediated mRNA decay and cancer. Curr. Opin. Genet. Dev. 2018; 48:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuzmiak H.A., Maquat L.E.. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 2006; 12:306–316. [DOI] [PubMed] [Google Scholar]
- 35. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.