Abstract
Despite aptamers are very promising alternative to antibodies, very few of them are under clinical trials or are used as drugs. Among them, NU172 is currently in Phase II as anticoagulant in heart disease treatments. It inhibits thrombin activity much more effectively than TBA, the best-known thrombin binding aptamer. The crystal structure of thrombin-NU172 complex reveals a bimodular duplex/quadruplex architecture for the aptamer, which binds thrombin exosite I through a highly complementary surface involving all three loops of the G-quadruplex module. Although the duplex domain does not interact directly with thrombin, the features of the duplex/quadruplex junction and the solution data on two newly designed NU172 mutants indicate that the duplex moiety is important for the optimization of the protein-ligand interaction and for the inhibition of the enzyme activity. Our work discloses the structural features determining the inhibition of thrombin by NU172 and put the basis for the design of mutants with improved properties.
INTRODUCTION
Despite the widespread use of antithrombotic drugs for the prevention and treatment of arterial and venous thrombosis, thromboembolic diseases continue to be a major cause of death and disability worldwide (1,2). For this reason, further efforts must be made to combat these disorders. Oligonucleotide aptamer technology is extensively used to modulate the function of most of the factors involved in coagulation (3), the complex process by which blood forms clots upon the damage of a blood vessel wall. Coagulation pathway is the final step of haemostasis and is directed toward creating fibrin through a cascade of events, which are regulated by several coagulation factors (4). Anticoagulant nucleic acid aptamers are short, single-stranded DNA or RNA sequences that bind to coagulation factors with high affinity and specificity through their three-dimensional structures. They hamper the protein–protein recognition events that are the key processes of the coagulation cascade and have the unique ability to be therapeutically regulated, by either controlling their circulating half-life or reversing their function with an antidote (3,5–7). Some of them are evaluated in clinical studies (8–12). Thrombin is the central orchestrator of the coagulation cascade; it catalyses the conversion of fibrinogen to fibrin and activates procoagulant factors V, VIII, XI and XIII (13). Additionally, when bound to thrombomodulin, it activates protein C, an anticoagulant zymogen (14). The delicate balance between the two functions in the normal physiological state prevents clot formation in undamaged vessels and triggers the coagulation cascade in the damaged ones (15). Thrombin capability to perform such different functions relies on the ability to recognize a large variety of substrates, inhibitors and cofactors (16). This ability is finely regulated by two distinct regions on the protein surface, known as exosites I and II, which allosterically modulate thrombin function, thus providing specificity to the proteolytic activity of the protein (15). Therefore, thrombin represents an attractive target for the development of agents that effectively interfere with thrombogenesis. The most studied anti-thrombin aptamer is TBA (also known as HD1) (5′-GGTTGGTGTGGTTGG-3′), a DNA 15mer oligonucleotide that inhibits thrombin clotting activity at nanomolar concentration (17). This oligonucleotide adopts a unimolecular antiparallel G-quadruplex structure, composed of two TT loops and a TGT loop protruding from the opposite sides of a two G-quartet moiety (18,19). TBA is a strong anticoagulant in vitro, where it inhibits thrombin-catalysed activation of fibrinogen and thrombin induced platelet aggregation (17). Moreover, it is able to bind both free and clot-bound thrombin (20,21). For these reasons, TBA was considered as a promising anticoagulant drug and, with the name ARC183, reached Phase I clinical trials (Archemix Corp. and Nuvelo) as an anticoagulant for potential use in acute cardiovascular settings, such as coronary artery bypass graft (CABG) surgery. The finding that the amount of drug needed to achieve the desired anticoagulation for use in CABG surgery corresponds to a suboptimal dosing profile blocked the TBA development (3,22). Efforts have then been focused on a second-generation aptamer, designated NU172 (or ARC2172) (5′-CGCCTAGGTTGGGTAGGGTGGTGGCG-3′), which possesses a pharmacokinetic profile similar to that of TBA but with a significantly higher potency as an anticoagulant. NU172 has an IC50 value of 5–10 μg/ml in plasma in an ecarin clotting time assay obtained by using thromboelastography (23). Currently, NU172 is the only thrombin binding aptamer evaluated in Phase II clinical trials (ClinicalTrials.gov identifier NCT00808964) for anticoagulation in heart disease treatments by ARCA Biopharma, Inc. (3,11,24,25). It induces a short-acting anticoagulation effect and is well tolerated without serious adverse responses (12). Furthermore, due to its high affinity toward the target protein, which is about one order of magnitude greater than that of TBA (26), NU172 represents a promising biorecognition element for the development of high performance thrombin aptasensors (27).
Here, we present the crystallographic structure of the thrombin-NU172 complex that provides an accurate description of the mixed duplex/quadruplex fold of the aptamer and its mode of binding to thrombin exosite I. In particular, the details of the G-quadruplex loop organization on the protein surface suggests an interesting link to the specific antithrombotic properties of the aptamer. This has received further support by the folding and anticoagulation studies on NU172 and two variants. From these studies, it emerges the importance of the mixed duplex/quadruplex organization on modulating the NU172 properties.
MATERIALS AND METHODS
Sample preparation and crystallization
The human D-Phe-Pro-Arg-chloromethylketone (PPACK)-inhibited thrombin was purchased from Haematologic Technologies (USA). The oligonucleotides (Supplementary Figure S1) were purchased from Sigma-Aldrich (United Kingdom).
Two stock solutions of NU172 were prepared at a concentration of about 0.1 mM, dissolving solid lyophilized oligonucleotides in 10 mM potassium phosphate buffer pH 7.4 and 100 mM KCl (K buffer) or 10 mM sodium phosphate buffer pH 7.4 and 100 mM NaCl (Na buffer). In order to induce folding, all aptamer samples were annealed by heating to 90°C for 5 minutes and then slow cooling down in 50–60 minutes and storing at 20°C overnight.
A standard protocol was followed for the preparation of the complexes between thrombin and NU172 in presence of potassium (thrombin–NU172–K) or sodium (thrombin–NU172–Na) ions (19,28–31). The thrombin-NU172 complexes were concentrated to about 10 mg/ml using 10 kDa-cutoff Centricon mini-concentrator (Vivaspin 500, Sartorius, Goettingen, Germany) and a refrigerated centrifuge (Z216MK, Hermle Labortechnik, Wehingen, Germany).
An initial extensive screening of sitting-drop crystallization experiments at 20°C in 96-well plates (Greiner Bio-One, Monroe, NC, USA) was carried out using an Automated Protein Crystallization Workstation (Hamilton Robotics) and precipitant solutions of a commercially available crystallization screen (Hampton Research Index). Optimization of starting conditions was performed by hanging drop vapour diffusion method mixing 0.5 μl complex solution with 0.5 μl reservoir solution at 20°C. Crystals suitable for X-ray diffraction data collection grew, for both complexes, in 50% (v/v) Tacsimate™ pH 7.0 (Supplementary Figure S2) (32).
Data collection, structure determination and refinement
Thrombin–NU172–K diffraction data were collected at the Institute of Biostructures and Bioimages (IBB), CNR, Naples, Italy, using a Saturn944 CCD detector. The X-ray radiation used was CuKα radiation (λ = 1.5418 Å) from a Rigaku Micromax 007 HF generator. Thrombin–NU172–Na diffraction data were collected at the ID30A-1 beamline of European Synchrotron Radiation Facility (ESRF), Grenoble, France, using λ = 0.9677 Å. In both cases, crystals were cryoprotected by addition of 20% (v/v) glycerol to the crystallization solution, flash-cooled at 100 K in supercooled N2 gas and maintained at this temperature during the data collection. Thrombin–NU172–K dataset was processed using HKL2000 (33), while that of thrombin–NU172–Na using iMOSFLM (34) and POINTLESS/AIMLESS (35,36).
Crystals of thrombin-NU172-K and thrombin-NU172-Na belong to the orthorhombic space group I222 and diffract X-rays up to 2.50 or 2.80 Å resolution, respectively. Matthews’ coefficient calculations suggested the presence of a 1:1 complex in the asymmetric unit and a solvent content of 74.7% in thrombin–NU172–K complex and 76.1% in thrombin–NU172–Na complex. Detailed statistics on data collection are reported in Supplementary Table S1.
The phase problem was solved by molecular replacement using Phaser (37). The coordinates of the native protein (PDB code: 1PPB) (38) were utilized as search model. The starting model was subjected to few cycles of rigid body refinement followed by several cycles of coordinate minimization and B-factor refinement using REFMAC5 program (39). Analysis of Fourier difference maps, calculated with (Fo – Fc) and (2Fo – Fc) coefficients, and manual model building performed with WinCoot program (40), allowed the rebuilding of the whole aptamer, of a sugar chain at the N-glycosylation site on thrombin, the positioning of ions and of several water molecules.
The final Rfactor/Rfree values were 16.9/20.5 and 15.9/20.3 for thrombin-NU172-K and thrombin-NU172-Na complexes, respectively. Detailed statistics of the refinement are reported in Supplementary Table S2.
Structural analysis
3DNA-dssr (41) was used to calculate local and overall geometric parameters of the aptamer. The Superpose program (42) from CCP4 package (43) was used to calculate root-mean-square deviations (RMSD). Features of the thrombin-NU172 interface and interactions between the two molecules were calculated by PISA (44), Contact (CCP4 package) (43) and CoCoMaps (45) programs. All the results were verified by visual inspection of the structure with WinCoot (40).
Molecular graphics figures were prepared with PyMOL (DeLano Scientific, Palo Alto, CA, USA). The coordinates of the structures were deposited in the Protein Data Bank (codes 6EVV and 6GN7 for thrombin-NU172-K and thrombin-NU172-Na, respectively).
Circular dichroism measurements
Oligonucleotides were suspended in K buffer at a concentration ranging between 1 and 40 μM and annealed, as reported above. CD spectra were recorded on a Jasco J-715 spectropolarimeter equipped with a Peltier temperature control, using a 1.0 or 0.1 cm path length cell. Spectra were registered at 10°C in the 210–320 nm range with 50 nm/min scanning speed, 2 s response time, 1 nm data pitch and 2.0 nm bandwidth. Each spectrum was obtained averaging three scans.
Gel electrophoresis (PAGE) experiment
A 20% polyacrylamide gel was prepared using 30% acrylamide/bis-acrylamide solution (24:1). The electrophoretic run was carried out in 90 mM Tris–borate buffer pH 8.0 and 20 mM EDTA (TBE buffer) on a 20 × 20 cm slab, at 4°C, by applying to the heads of the electrophoresis cell a 120 V voltage. For loading, 30 μl of 40 μM oligonucleotide sample, annealed in K buffer, were added to 20 μl of loading buffer (TBE buffer and 20% (v/v) glycerol). After a 4 h run time, bands were visualized by UV shadowing.
Anticoagulant activity experiments
Evaluation of the anticoagulant activity of TBA, NU172 and the mutants Des_NU172 and TBA_GT (Supplementary Figure S1) was performed by means of light scattering (LS) experiments monitoring the increase of the light scattered intensity upon conversion of fibrinogen to fibrin catalysed by thrombin (46). LS measurements were carried out with a home-made instrument. A 1.2 μM solution of fibrinogen in Phosphate Buffered Saline (PBS) was placed in a LS cuvette to which the oligonucleotide was added and left to equilibrate in the instrument for 20 min. Then, thrombin was added up to a final concentration of 5 nM and the light scattered intensity was registered every 20 s for at least 2 h. In the case of NU172 longer observation times were required. Each oligonucleotide, annealed in K buffer, was added to fibrinogen solutions before addition of thrombin at a concentration such that thrombin:oligonucleotide molar ratio was either 1:1, 1:2 or 1:5. All the experiments were performed in triplicate. Comparison of the anticoagulant activities was performed by plotting the normalized scattered intensity nI
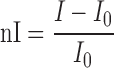 |
as a function of time and by linearly fitting the initial increase of nI upon the lag time. The slopes of the fitted lines give an estimation of the coagulation rate. The thrombin inhibiting properties of the different oligonucleotides were evaluated by calculating the ratio between the coagulation rate in the exclusive presence of thrombin and that in the presence of thrombin and oligonucleotides.
RESULTS
Crystal structure of the thrombin-NU172 complex
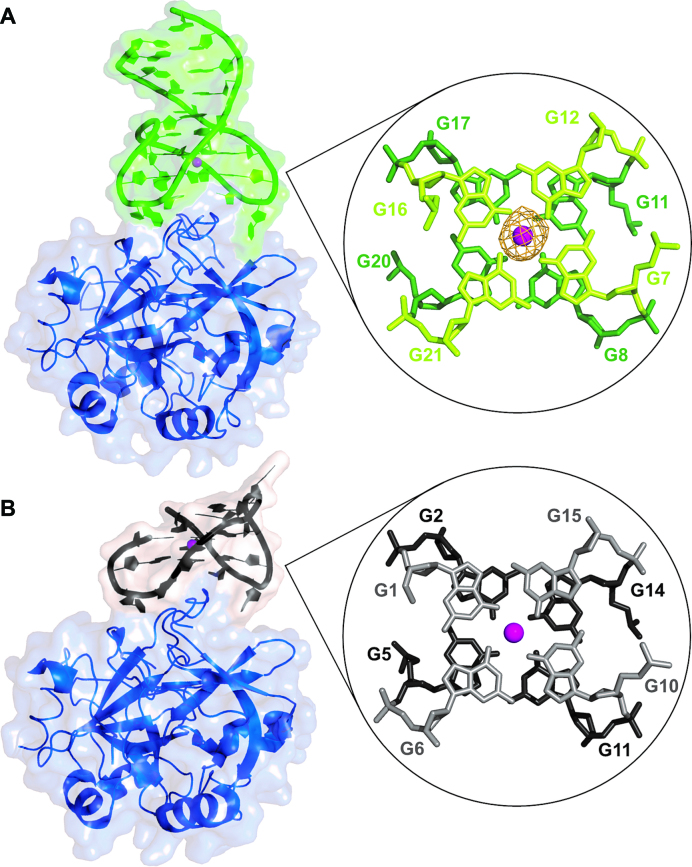
Orthorhombic I222 crystals with one 1:1 protein–aptamer complex (Figure 1A and Supplementary Figure S3) in the asymmetric unit were grown either in the presence of K+ (thrombin–NU172–K) or in the presence of Na+ (thrombin–NU172–Na). They are practically isomorphous and diffract X-rays up to 2.50 and 2.80 Å resolution, respectively. The two independently refined complexes, within the experimental errors, do not show significant differences and, therefore, only thrombin–NU172–K complex is analysed in detail.
Figure 1.
On the left, surface/cartoon representations of thrombin-NU172-K (A) and thrombin-TBA-K (PDB code: 4DII) (B) complexes. Thrombin is coloured in blue, NU172 is coloured in green and TBA is coloured in black. On the right, view of the two tetrads along the G-quadruplex axis of the two aptamers. The omit Fo – Fc electron density map (orange) of the potassium ion (magenta) in NU172 is contoured at 3.0 σ level.
As expected, the aptamer binding does not substantially modify the thrombin structure. After superposition of the Cα heavy chain and that of the PPACK inhibited enzyme (PDB code: 1PPB) (38) the value of RMSD is only 0.34 Å. The only structural novelty is the long N-linked sugar chain attached to Asn60G residue (Supplementary Figure S4) that is clearly visible in the electron density map of the complex.
NU172 structural features
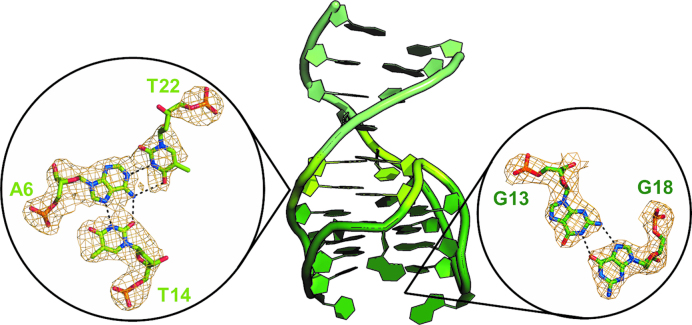
Fourier difference maps showed continuous electron density that allowed to build the whole oligonucleotide (Supplementary Figure S5). The refined model reveals the presence of the expected duplex and quadruplex domains (47), enchained to each other with interesting new features. In particular, the quadruplex domain (residues 7–21) is arranged in a chair-like anti-parallel fold, with guanines 8, 11, 17, 20 and 7, 12, 16, 21 forming G-tetrad I and II, respectively, and three edge-wise loops, TT (Thy9-Thy10), GT (Gua18-Thy19) and GTA (Gua13-Thy14-Ade15). A potassium ion is sandwiched between the two tetrads (Figure 1A). The quadruplex motif is very similar to that of TBA (Figure 1B) and its variants (19,29,31), which is formed by two G-tetrads (guanines 2, 5, 11, 14 for tetrad I and 1, 6, 10, 15 for tetrad II), surrounded by a TGT loop on one side and two TT loops on the opposite side. Differently from the analogous TGT loop of TBA-like aptamers that is highly mobile (19,29,31), the GTA loop is particularly well defined. In the latter, Gua13 is hydrogen bonded to Gua18 of the GT loop (Figure 2), a residue that replaces the thymine of the TBA-like aptamers, moreover Thy14 and Ade15 form reverse Hoogsteen base pairs with Ade6 and Thy5, respectively. Remarkably, Ade6 also participates to two non-linear Watson-Crick hydrogen bonds with Thy22, thus creating a TAT triad (Thy22/Ade6/Thy14). This triad and the base-pair Thy5/Ade15 mediate the transition from the quadruplex to the duplex domain, creating a continuous stacking of bases from one domain to the other. The duplex region of NU172 is composed by four Watson–Crick base pairs (Cyt1/Gua26, Gua2/Cyt25, Cyt3/Gua24 and Cyt4/Gua23), which define a regular B-type helix (Figure 2 and Supplementary Figure S6).
Figure 2.
Cartoon representation of NU172 architecture (duplex in light green, quadruplex in dark green and the junction in lemon-green). On the left, view of the TAT triad along the G-quadruplex axis. On the right, view of the interactions between guanine 13 (GTA loop) and guanine 18 (GT loop). The omit Fo – Fc electron density maps (orange) are contoured at 3.0 σ level. Dashed lines represent hydrogen bonds.
Thrombin-NU172 interface
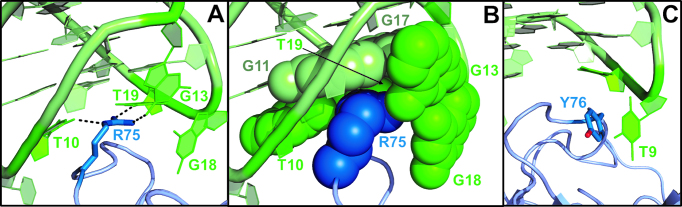
Resembling TBA and its variants (19,28,29,31,48,49), the two-residue loops of the G-quadruplex domain act as a pincer-like system that captures the protruding region of exosite I (Figure 1 and Supplementary Figure S7). However, recognition of exosite I by NU172 presents interesting new features. Indeed, in addition to the two-residue TT and GT loops, the three-residue GTA loop of the G-quadruplex is also involved in the binding. On the other hand, the duplex domain makes no contacts with the thrombin surface.
Focusing on exosite I, it can be described as formed by two sub-regions, referred to as A and B, respectively (48). In the former, a hydrophobic crevice on the protein surface forms a well-shaped joint that interlocks the Gua18–Thy19 loop. As mentioned above, Gua18 shares two hydrogen bonds with Gua13 of GTA loop. The strong interaction fixes Gua13 in a position most favourable to interact with Arg75, whose guanidinium group is fully surrounded by NU172 side chains and, in addition to Gua13, is hydrogen bonded to Thy19 and Thy10 (Figure 3A and B). This network of side chains contacts recruits the three-residue loop of the G-quadruplex for the binding to thrombin and represents a unique feature of NU172 that significantly enlarges the contact area between NU172 and the A-region, compared to the complexes between thrombin and other aptamers (Table 1 and Supplementary Table S3). As far as the TT loop is concerned, it interacts with B-region, which is less extended than the A-region (48), through a π-π stacking between Thy9 and Tyr76 (Figure 3C, Table 1 and Supplementary Table S3).
Figure 3.
Interface interactions between thrombin (in blue) and NU172 (in green). (A) Hydrogen bonds (dashed lines) connecting Arg75 to Gua13, Thy19 and Thy10. (B) Space-filling representation of Arg75 side chain and its environment. (C) π–π stacking between Thy9 and Tyr76.
Table 1.
Interface area between protein and aptamer in thrombin–TBA–K (PDB code: 4DII) and thrombin–NU172–K complexes.
| TBA | NU172 | |
|---|---|---|
| Whole interface | 543 Å2 | 590 Å2 |
| Thrombin A-region | 282 Å2 (Thy3–Thy4)a | 324 Å2 (Gua13–Gua18–Thy19)a |
| Thrombin B-region | 183 Å2 (Thy12–Thy13)a | 215 Å2 (Thy9–Thy10)a |
aIn parentheses the aptamer nucleobases that make interactions with thrombin exosite I in A- and B-region.
It should be recalled that TBA can bind thrombin in two different ways that are practically isoenergetic. In one case, a TT loop interacts with the A-region and the second TT loop interacts with the B-region, in the second case these interactions are reversed through a rotation of 180° about the pseudo twofold axis of the aptamer (48,50). In the case of NU172, the replacement of a T for a G in one TT loop discriminates between the two binding modes, strongly favouring the one in which the GT loop interacts with the A-region of the protein.
Finally, it is worth to stress that protein-aptamer contacts do not change in the thrombin–NU172–Na structure (Supplementary Table S3), in contrast to what observed for thrombin-TBA complex (19), where the replacement of K+ with Na+ causes a small reorganization of the protein–aptamer interface that may explain the observed modification of the aptamer functionality.
Characterization of NU172 mutants
In order to gain a deeper knowledge of the structural features that are at the basis of the improved affinity of NU172 with respect to TBA-like aptamers, two mutants were constructed and characterized in solution. In particular, attention was focused on the role of the GT loop and that of the duplex module (Supplementary Figure S1), by studying the mutant TBA_GT of TBA with a T→G substitution in the appropriate loop (Thy12–Thy13), and the deletion mutant Des_NU172, in which only the G-quadruplex domain of NU172 is conserved. In particular, we tested their aggregation status, folding in solution and in vitro anticoagulant activity in comparison to TBA and NU172.
The polyacrylamide native gel electrophoretogram (Figure 4) exhibits a single band for TBA and NU172 (lanes 1 and 2, respectively), indicating the presence of a single structural species, with a relative mobility in fully agreement with their size. Vice versa, several bands are observed in lanes 3 and 4, suggesting that Des_NU172 and TBA_GT form non-homogeneous solution containing species with different molecularity.
Figure 4.

Gel electrophoresis analysis of (1) TBA, (2) NU172, (3) Des_NU172 and (4) TBA_GT. The 40 μM oligonucleotide samples were annealed the day before the run in 10 mM potassium phosphate buffer pH 7.4 and 100 mM KCl.
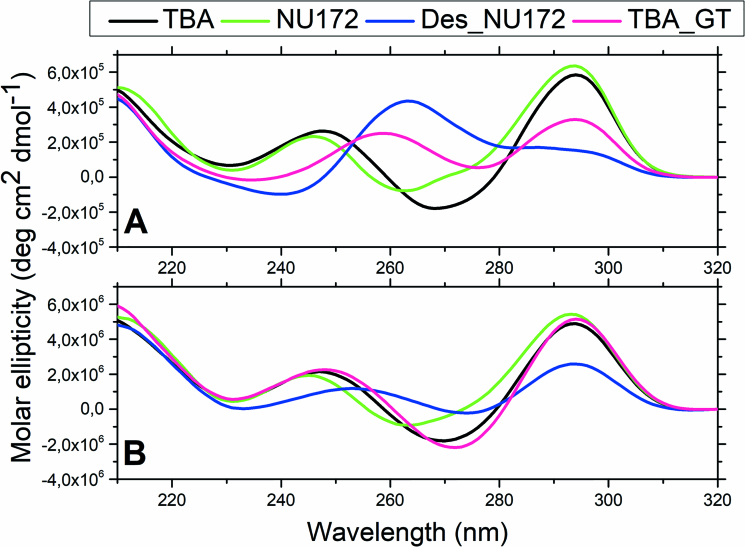
Similar conclusions can be drawn by the CD spectra recorded under the same conditions (Figure 5A). While the spectra of TBA and NU172 show features that indicate the presence of a canonical G-quadruplex structure, those of Des_NU172 and TBA_GT suggest conformational heterogeneity. Interestingly, at lower concentration (1 μM) the spectrum of TBA_GT becomes superimposable to that of TBA, whereas the spectrum of Des_NU172 still indicates the presence of more than one conformer (Figure 5B).
Figure 5.
Overlapping of CD spectra of TBA (black), NU172 (green), Des_NU172 (blue) and TBA_GT (magenta) in 10 mM potassium phosphate buffer pH 7.4 and 100 mM KCl. Spectra were recorded at 10°C on 40 μM (A) or 1 μM (B) annealed oligonucleotide samples.
These CD data, together with the electrophoretic results, indicate that the substitution of Thy12 with a guanine residue in the TT loop of TBA and, in particular, the deletion of the duplex region in NU172 induce a marked destabilization of the antiparallel unimolecular G-quadruplex conformation and an increase in its conformational flexibility.
Anticoagulant activity of NU172 and its mutants
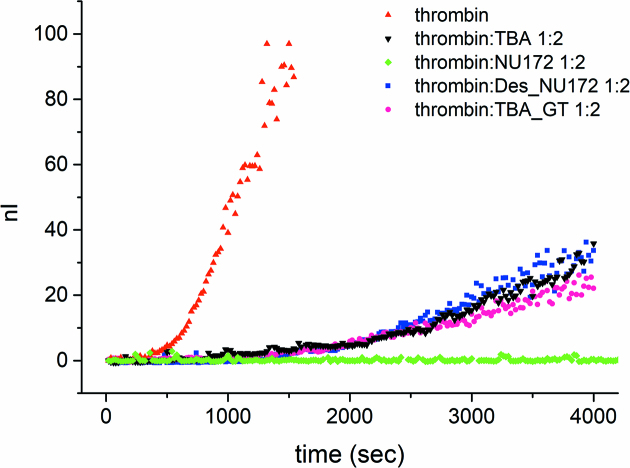
The anticoagulant activities were evaluated following the thrombin catalysed conversion of fibrinogen to fibrin by means of LS experiments (46). In Figure 6 the intensity of the scattered light, which measures the progression of fibrin formation, is reported as a function of time in the absence and in the presence of the oligonucleotides (1:2 thrombin:oligonucleotide molar ratio). The various curves clearly indicate the inhibition of thrombin coagulation activity exerted by all tested oligonucleotides. Remarkably, NU172 acts as a very potent thrombin inhibitor. Indeed, for a rather long time no change of the intensity is observed. In the case of the other oligonucleotides, the increase of the intensity can be noted much sooner and in few hours the samples gelifies, similarly to the samples in which only thrombin is present.
Figure 6.
Coagulation curves of fibrinogen in the presence of thrombin and different oligonucleotides (TBA; NU172; Des_NU172 and TBA_GT, see inset for details).
An estimate of the anticoagulant activity of each oligonucleotide was obtained by calculating the ratio between the initial slope of the normalized scattered intensities in the absence and in the presence of the oligonucleotide (Supplementary Table S4). The results show that NU172 is able to decrease the coagulation rate of about two orders of magnitude already at 1:1 thrombin:oligonucleotide molar ratio, while the two mutants are thrombin inhibitors as effective as TBA.
DISCUSSION
A fast and cost-efficient drug optimization program requires detailed structural information on the interaction between the pharmacologically active molecule and its target. In this respect, we have investigated the recognition between human alpha-thrombin and NU172, the only anti-thrombin aptamer currently in Phase II clinical trials for short-term anticoagulation (3,11,24,25). The crystal structure of the thrombin–NU172 complex reveals that the aptamer adopts a mixed duplex/quadruplex structure whose spine is a continuous stacking of bases from the duplex to the quadruplex region. Notably, a reverse Hoogsteen base pair and a base triad (TAT), involving residues of the quadruplex GTA loop, form the sharp transition between the two structural domains. It is worth to note that a TAT triad has been found as the junction motif between a parallel G-quadruplex and an antiparallel B-duplex structure in two duplex/quadruplex telomeric repeats, where it directly mediates the transition from one domain to the other (51). In our structure, TAT joins an antiparallel B-duplex with an antiparallel G-quadruplex, thus indicating that the triad is a versatile motif able to connect quadruplex and duplex domains with different topologies with no loss of stacking between adjacent bases.
The intramolecular compactness of the duplex/quadruplex architecture of NU172 generates a boundary surface particularly well suited to interact with thrombin exosite I in comparison to other thrombin-aptamer complexes (Supplementary Table S3) (19,29,31,48). The interface involves not only the two-residue loops, but also the GTA loop that in turn is directly involved in the stabilization of the duplex/quadruplex organization. Indeed, Gua13, the only GTA residue that does not participate to the duplex/quadruplex connection, forms a GG base pair with Gua18 of the GT loop, further locking the quadruplex structure and stiffening the assembly of the nucleotide bases around the guanidium group of Arg75 (Figure 3A and B). This double role qualifies GTA loop as a basic structural feature that confers to NU172 the improved functional properties in its interaction with thrombin. The compactness of the overall structure can also explain the observed relative insensitivity of the complex versus the K+/Na+ exchange, differently from what found in the case of the thrombin–TBA complex (19).
The observed strict interaction between duplex and quadruplex domains corroborates the results of a thorough investigation (47) on the conformational stability of bimodular anti-thrombin aptamers, which indicates a one-step unfolding mechanism for NU172, similar to that of RE31, an aptamer that presents a continuous stacking spine as well (31). Moreover, the characterization of Des_NU172 and TBA_GT mutants underlines the importance of the duplex domain, confirming the key role of this motif in the stabilization of the appropriate antiparallel G-quadruplex folding and in the organization of the inter-domains region that allows the participation of the GTA loop to the thrombin binding. Indeed, anticoagulant experiments in physiological mimicking conditions highlight the excellent inhibitory properties of NU172 that are about two orders of magnitude higher than those of TBA and both Des_NU172 and TBA_GT mutants. Moreover, they also prove that the presence of Gua13 and/or Gua18 in the absence of the duplex motif is not sufficient to reach the anticoagulant activity observed for NU172.
The finding that, despite the conformational heterogeneity of Des_NU172 at the oligonucleotide concentration used in the experiments, its anticoagulant activity is similar to that of TBA and TBA_GT could be ascribed to the thrombin ability to act as molecular chaperone (19,52), inducing the oligonucleotide to adopt a unimolecular antiparallel G-quadruplex conformation.
In conclusion, the structural characterization of thrombin-NU172 complex and some related mutants strongly indicates that the interesting properties of NU172 are associated to the concomitant presence of several motifs. Indeed, the coexistence of a duplex and a quadruplex domain interlocked through the TAT triad, the presence of guanines in two loops of the quadruplex structure and the GG inter-loop base pair are features that, taken together, synergistically cooperate in determining an overall structure capable to efficiently interact with the thrombin surface. We believe that these results represent an instructive example of the way in which several local features may concur to produce interesting new properties and should be taken in consideration particularly for the modification or the design of new aptamers, a class of substances that display an elevated structural flexibility.
DATA AVAILABILITY
Atomic coordinates and structure factors for thrombin–NU172–K and thrombin–NU172–Na complexes have been deposited with the Protein Data Bank under accession number 6EVV and 6GN7, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Dr Luigi Paduano (University of Naples ‘Federico II’, Naples, Italy) for helpful discussions and permission to use his home-made LS instrument. Giosuè Sorrentino, Maurizio Amendola (IBB, CNR, Naples, Italy) and members of ESRF (Grenoble, France) staff are gratefully acknowledged for technical assistance during data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: RFBR (Russian Foundation of Basic Research).
Conflict of interest statement. None declared.
REFERENCES
- 1. Wendelboe A.M., Raskob G.E.. Global burden of thrombosis: epidemiologic aspects. Circ. Res. 2016; 118:1340–1347. [DOI] [PubMed] [Google Scholar]
- 2. Collaborators, U.S.B.o.D. Mokdad A.H., Ballestros K., Echko M., Glenn S., Olsen H.E., Mullany E., Lee A., Khan A.R., Ahmadi A. et al. The state of US Health, 1990–2016: burden of diseases, injuries, and risk factors among US states. JAMA. 2018; 319:1444–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Woodruff R.S., Sullenger B.A.. Modulation of the coagulation cascade using aptamers. Arterioscler. Thromb. Vasc. Biol. 2015; 35:2083–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crawley J.T., Zanardelli S., Chion C.K., Lane D.A.. The central role of thrombin in hemostasis. J. Thromb. Haemostasis. 2007; 5(Suppl. 1):95–101. [DOI] [PubMed] [Google Scholar]
- 5. Dyke C.K., Steinhubl S.R., Kleiman N.S., Cannon R.O., Aberle L.G., Lin M., Myles S.K., Melloni C., Harrington R.A., Alexander J.H. et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006; 114:2490–2497. [DOI] [PubMed] [Google Scholar]
- 6. Chan M.Y., Rusconi C.P., Alexander J.H., Tonkens R.M., Harrington R.A., Becker R.C.. A randomized, repeat-dose, pharmacodynamic and safety study of an antidote-controlled factor IXa inhibitor. J. Thromb. Haemostasis. 2008; 6:789–796. [DOI] [PubMed] [Google Scholar]
- 7. Chan M.Y., Cohen M.G., Dyke C.K., Myles S.K., Aberle L.G., Lin M., Walder J., Steinhubl S.R., Gilchrist I.C., Kleiman N.S. et al. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008; 117:2865–2874. [DOI] [PubMed] [Google Scholar]
- 8. Cohen M.G., Purdy D.A., Rossi J.S., Grinfeld L.R., Myles S.K., Aberle L.H., Greenbaum A.B., Fry E., Chan M.Y., Tonkens R.M. et al. First clinical application of an actively reversible direct factor IXa inhibitor as an anticoagulation strategy in patients undergoing percutaneous coronary intervention. Circulation. 2010; 122:614–622. [DOI] [PubMed] [Google Scholar]
- 9. Povsic T.J., Vavalle J.P., Aberle L.H., Kasprzak J.D., Cohen M.G., Mehran R., Bode C., Buller C.E., Montalescot G., Cornel J.H. et al. A Phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur. Heart J. 2013; 34:2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nimjee S.M., White R.R., Becker R.C., Sullenger B.A.. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017; 57:61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ospina-Villa J.D., Zamorano-Carrillo A., Castanon-Sanchez C.A., Ramirez-Moreno E., Marchat L.A.. Aptamers as a promising approach for the control of parasitic diseases. Braz. J. Infect. Dis. 2016; 20:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou J., Rossi J.. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017; 16:181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Cera E. Thrombin. Mol. Aspects Med. 2008; 29:203–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esmon C.T. The protein C pathway. Chest. 2003; 124:26S–32S. [DOI] [PubMed] [Google Scholar]
- 15. Di Cera E. Thrombin as procoagulant and anticoagulant. J. Thromb. Haemostasis. 2007; 5(Suppl. 1):196–202. [DOI] [PubMed] [Google Scholar]
- 16. Bock P.E., Panizzi P., Verhamme I.M.. Exosites in the substrate specificity of blood coagulation reactions. J. Thromb. Haemostasis. 2007; 5(Suppl. 1):81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J.. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992; 355:564–566. [DOI] [PubMed] [Google Scholar]
- 18. Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J.. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F.. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012; 40:8119–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Griffin L.C., Tidmarsh G.F., Bock L.C., Toole J.J., Leung L.L.. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993; 81:3271–3276. [PubMed] [Google Scholar]
- 21. Li W.X., Kaplan A.V., Grant G.W., Toole J.J., Leung L.L.. A novel nucleotide-based thrombin inhibitor inhibits clot-bound thrombin and reduces arterial platelet thrombus formation. Blood. 1994; 83:677–682. [PubMed] [Google Scholar]
- 22. Mayer G., Rohrbach F., Potzsch B., Muller J.. Aptamer-based modulation of blood coagulation. Hamostaseologie. 2011; 31:258–263. [DOI] [PubMed] [Google Scholar]
- 23. Keefe A.D., Pai S., Ellington A.. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010; 9:537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Becker R.C., Povsic T., Cohen M.G., Rusconi C.P., Sullenger B.. Nucleic acid aptamers as antithrombotic agents: opportunities in extracellular therapeutics. Thromb. Haemostasis. 2010; 103:586–595. [DOI] [PubMed] [Google Scholar]
- 25. Ni X., Castanares M., Mukherjee A., Lupold S.E.. Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem. 2011; 18:4206–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zavyalova E., Ustinov N., Golovin A., Pavlova G., Kopylov A.. G-quadruplex aptamers to human thrombin versus other direct thrombin inhibitors: the focus on mechanism of action and drug efficiency as anticoagulants. Curr. Med. Chem. 2016; 23:2230–2244. [DOI] [PubMed] [Google Scholar]
- 27. Rangnekar A., Nash J.A., Goodfred B., Yingling Y.G., LaBean T.H.. Design of potent and controllable anticoagulants using DNA aptamers and nanostructures. Molecules. 2016; 21:e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Russo Krauss I., Merlino A., Randazzo A., Mazzarella L., Sica F.. Crystallization and preliminary X-ray analysis of the complex of human alpha-thrombin with a modified thrombin-binding aptamer. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2010; 66:961–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Russo Krauss I., Merlino A., Giancola C., Randazzo A., Mazzarella L., Sica F.. Thrombin-aptamer recognition: a revealed ambiguity. Nucleic Acids Res. 2011; 39:7858–7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russo Krauss I., Pica A., Merlino A., Mazzarella L., Sica F.. Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013; 69:2403–2411. [DOI] [PubMed] [Google Scholar]
- 31. Russo Krauss I., Spiridonova V., Pica A., Napolitano V., Sica F.. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016; 44:983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McPherson A., Cudney B.. Searching for silver bullets: an alternative strategy for crystallizing macromolecules. J. Struct. Biol. 2006; 156:387–406. [DOI] [PubMed] [Google Scholar]
- 33. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 34. Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011; 67:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bode W., Turk D., Karshikov A.. The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992; 1:426–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu X.J., Olson W.K.. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008; 3:1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 43. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mihel J., Sikic M., Tomic S., Jeren B., Vlahovicek K.. PSAIA - protein structure and interaction analyzer. BMC Struct. Biol. 2008; 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vangone A., Spinelli R., Scarano V., Cavallo L., Oliva R.. COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics. 2011; 27:2915–2916. [DOI] [PubMed] [Google Scholar]
- 46. Riccardi C., Russo Krauss I., Musumeci D., Morvan F., Meyer A., Vasseur J.J., Paduano L., Montesarchio D.. Fluorescent thrombin binding aptamer-tagged nanoparticles for an efficient and reversible control of thrombin activity. ACS Appl. Mater. Interfaces. 2017; 9:35574–35587. [DOI] [PubMed] [Google Scholar]
- 47. Russo Krauss I., Napolitano V., Petraccone L., Troisi R., Spiridonova V., Mattia C.A., Sica F.. Duplex/quadruplex oligonucleotides: role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2018; 107:1697–1705. [DOI] [PubMed] [Google Scholar]
- 48. Pica A., Russo Krauss I., Merlino A., Nagatoishi S., Sugimoto N., Sica F.. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013; 280:6581–6588. [DOI] [PubMed] [Google Scholar]
- 49. Dolot R., Lam C.H., Sierant M., Zhao Q., Liu F.W., Nawrot B., Egli M., Yang X.. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018; 46:4819–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pica A., Russo Krauss I., Parente V., Tateishi-Karimata H., Nagatoishi S., Tsumoto K., Sugimoto N., Sica F.. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017; 45:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russo Krauss I., Ramaswamy S., Neidle S., Haider S., Parkinson G.N.. Structural insights into the quadruplex-duplex 3′ interface formed from a telomeric repeat: a potential molecular target. J. Am. Chem. Soc. 2016; 138:1226–1233. [DOI] [PubMed] [Google Scholar]
- 52. Baldrich E., O'Sullivan C.K.. Ability of thrombin to act as molecular chaperone, inducing formation of quadruplex structure of thrombin-binding aptamer. Anal. Biochem. 2005; 341:194–197. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for thrombin–NU172–K and thrombin–NU172–Na complexes have been deposited with the Protein Data Bank under accession number 6EVV and 6GN7, respectively.