ABSTRACT
Co-occurrence networks allow for the identification of potential associations among species, which may be important for understanding community assembly and ecosystem functions. We employed this strategy to examine prokaryotic co-occurrence patterns in the Amazon soils and the response of these patterns to land use change to pasture, with the hypothesis that altered microbial composition due to deforestation will mirror the co-occurrence patterns across prokaryotic taxa. In this study, we calculated Spearman correlations between operational taxonomic units (OTUs) as determined by 16S rRNA gene sequencing, and only robust correlations were considered for network construction (−0.80 ≥ P ≥ 0.80, adjusted P < 0.01). The constructed network represents distinct forest and pasture components, with altered compositional and topological features. A comparative analysis between two representative modules of these contrasting ecosystems revealed novel information regarding changes to metabolic pathways related to nitrogen cycling. Our results showed that soil physicochemical properties such as temperature, C/N and H++Al3+ had a significant impact on prokaryotic communities, with alterations to network topologies. Taken together, changes in co-occurrence patterns and physicochemical properties may contribute to ecosystem processes including nitrification and denitrification, two important biogeochemical processes occurring in tropical forest systems.
Keywords: forest, land use change; microorganisms; ecological network; pasture; amazon; physicochemical properties; nitrification; denitrification
Deforestation of Amazon rainforest alters soil microbial community and physicochemical properties, which subsequently alter microbially mediated biogeochemical processes, notably nitrogen cycle. The Amazon Rainforest is the largest terrestrial ecosystem on Earth and deforestation changes co-occurrence patterns of microbial communities with potential consequences to biogeochemical cycles
INTRODUCTION
The Amazon is the largest continuous rainforest ecosystem in the world and home to thousands of species (Dirzo and Raven 2003). Due to the growing demand for resources, mainly fuel and food, the Amazon rainforest has been facing continuous pressure over the last few decades, leaving its biological composition in jeopardy. One of the primary threats is forest-to-pasture conversion, with a considerable impact on communities of microorganisms, plants and animals (Bierregaard et al. 2001; Soares-Filho et al. 2009; Rodrigues et al. 2013; Ranjan et al. 2015; Navarrete et al. 2015a). Though ecologists have long been studying the roles of plant and animal communities to ecosystem functions in tropical forests (Eva et al. 2004; Feigl et al. 2006; Foley et al. 2007) and the effects of deforestation on these communities (Fearnside 1999; Feeley and Silman 2009), knowledge of the microbial ecology of deforestation is still rudimentary in the Amazon (Mirza and Rodrigues 2012; Rodrigues et al. 2013; Navarrete et al. 2011, 2015a and 2015b, Paula et al. 2014; Ranjan et al. 2015; Meyer et al. 2017; Khan 2016).
Soil is a heterogeneous environment that harbors enormously diverse microbial communities (Torsvik, Goksoyr and Daae 1990; Curtis, Sloan and Scannell 2002). Members of these communities interact in ways that affect their growth and metabolism. Such interactions can result in patterns of species abundance across space and time. Cooperative metabolic interactions can lead to increased growth of interacting bacteria and ultimately to positive co-occurrence patterns in abundance, while competition for the same resources may lead to an inverse pattern (Greenblum et al. 2013). Abundance patterns may also reflect the response of different species to a common environmental factor rather than their direct interactions (Zhou et al. 2011). A co-occurring microbial pair therefore indicates they are either interacting synergistically or they have similar responses to environmental factors. This inter-taxa relationship may help reveal the niche spaces shared by members of a prokaryotic community (Williams, Howe and Hofmockel 2014), which is particularly valuable in evaluating the impacts of environmental changes such as deforestation on microbial communities. To analyze co-occurrence patterns, organizing community data into networks can be helpful, where each node represents a species and the edges represent correlations in abundance (Zhou et al. 2011). Each network may be divided into sub-networks, termed modules, which contain a set of members e.g. microbial species that have a higher number of links among them than with members of other modules (Bascompte and Stouffer 2009). Groups belonging to a module may have similar ecological niches (Olesen et al. 2007). Although it is currently not possible to map out direct interactions in complex microbial communities, an empirical study reported that phylogenetic markers (e.g. 16S rRNA gene) properly predict the niche-defining properties (Fuhrman et al. 2006). Hence, microbial network studies provide a way forward to understand and test potential inter-taxa relationships in microbial communities (Zhou et al. 2011; Faust et al. 2012; Williams, Howe and Hofmockel 2014; Widder et al. 2014), and to enable posing questions about their niche spaces. Few recent studies have been conducted to explore the impact of deforestation on network structures, demonstrating the alteration of co-occurrence patterns of microbial communities in tropical forests (Navarrete et al. 2015b; Khan 2016; Wood et al. 2017; Tian et al. 2018). The identification of specific co-occurring bacterial taxa from the network study offers insights on the functional attributes and adaptation strategies of microbial communities in their ecosystems. Such information is essentially important for environments like Amazon soils, where the basic ecology and microbiology are mostly inconspicuous.
Characterization of soil physicochemical properties is important to explore the factors that affect microbial ecosystem and provide a better picture of how microbial communities and their diversity change (Guo and Gifford 2002; Don, Schumacher and Freibauer 2011; Navarrete et al. 2015b; Khan et al. 2018). Deforestation results in substantial alterations of the vegetation characteristics (e.g. plant biomass, species composition) and exerts substantial impacts on soil physicochemical properties (e.g. soil C, elemental stoichiometry, temperature, pH) (Guo and Gifford 2002; Don, Schumacher and Freibauer 2011). Consequently, microbial diversity and ecological processes are expected to be affected as well. For example, the slash-and-burn process of deforestation causes an initial loss of terrestrial nutrients, especially nitrate, through hydrologic leaching and emission of gases (Davidson et al. 2007), which most likely imposes a strong selective pressure on soil microbes to scavenge atmospheric nitrogen (Neill et al. 1995; Mirza and Rodrigues 2012; Paula et al. 2014). Therefore, it is important to incorporate these factors for better understanding of microbial ecology and any alterations in biogeochemical processes caused by anthropogenic activities such as deforestation.
By constructing correlation networks using 16S rRNA gene data, and estimating network defining topologies, we aimed: (i) to quantify the impact of prokaryotic diversity loss on the network structure and composition; (ii) to obtain information on the relative importance of individual prokaryotic species in the networks; (iii) to detect potential changes in important ecosystem processes that may result from forest-to-pasture conversion and (iv) to characterize differences in soil physicochemical properties, and evaluate the relationships with prokaryotic community structures and potentially, their ecosystem processes.
MATERIALS AND METHODS
Site description and sampling
The Amazon Rainforest Microbial Observatory (ARMO) study site is located at Fazenda Nova Vida in the State of Rondonia, Brazil (10°10′18.71″S, 62°47′15.67″W), representing the area with one of the highest rates of deforestation of the Brazilian Amazonia in the last two decades (INPE. 2011). Our study was conducted on a total of 10 soil samples collected from a primary forest (n = 5) and established pasture (n = 5; 38-year old) at the end of the rainy season, March 2010. Following the removal of litter, soil sampling was performed using a 5 cm-diameter corer to 10-cm depth with samples being transported on ice to the laboratory and stored at −80°C until soil DNA extractions. Soil physicochemical properties were analyzed at the Laboratorio de Fertilidade do Solo, Department of Soil Sciences, University of Sao Paulo. More detailed information about the sampling sites are provided in the supplementary materials and methods.
DNA extraction, amplification and sequencing
Ten grams of soil from each sample were used for extracting total genomic DNA using the PowerLyzer PowerSoil DNA isolation kit (MoBio Inc, Carlsbad, CA, USA). The concentration and purity of soil DNA were determined spectrophotometrically (NanoDrop Technologies Inc., Wilmington, DE). Prokaryotic primers 515F and 806R, which amplify the V4 hyper-variable region of 16S rRNA gene, were used for PCR amplification as described elsewhere (Caporaso et al. 2012). A unique 12-bp long barcode was incorporated to the forward primer for specific identification of each sample. The resulting bar-coded amplicons were pooled in an equimolar concentration for sequencing using the MiSeq platform to produce 300 bp length paired-end reads (Illumina, Inc., San Diego, California) at the DOE Joint Genome Institute (Walnut Creek, CA).
Sequence processing and taxonomic assignment
All upstream and downstream analyses of raw Illumina sequences were carried out in the QIIME 1.8.0 environment as described elsewhere (Caporaso et al. 2010; Kuczynski et al. 2012). Raw sequences belonging to a specific sample were sorted based on the barcode sequences, followed by quality filtering to discard anonymous bases and removal of primer sequences. Sequences were assigned to Operational Taxonomic Units (OTUs) at a minimum of 97% sequence identity using de novo OTU-picking protocol with QIIME 1.8.0 (Caporaso et al. 2010) with the following controls before analysis: (i) singleton OTUs were removed from downstream analysis to reduce the possibility of sequencing error and to differentiate unique OTUs from potential data noise, (ii) resampling was performed 10 times and subsequent results were based on standard means of resampling. The algorithm Uclust (Edgar 2010) was used to cluster the quality-filtered reads against the GreenGenes database (DeSantis et al. 2006), which was followed by the assignment of taxonomy using the RDP classifier (Wang et al. 2007).
Taxonomic network construction
We constructed a network using the relative abundance data of the OTUs from 10 samples (five forest and five pasture samples), with forestation status was included as a binary variable. We considered only the OTUs that were observed in at least five samples. The relative abundance data for these OTUs were used to construct Spearman correlation coefficients between all possible pairs of OTUs across samples. The analysis was performed in R environment using the package ‘multtest’ package (Pollard, Sandrine and van der Laan 2005). A correlation coefficient was considered statistically robust if a numeric value was either ≥ 0.8 or ≤ −0.8 with significance of P ≤ 0.01. To reduce the chances of obtaining false positive results, the Benjamini and Hochberg (BH) false discovery rate correction was performed throughout the dataset (Benjamini and Hochberg 1995). When constructing networks, only significant pairwise relationships were used, with each node representing an OTU and each edge representing a significant pairwise association between them, a positive correlation between two OTUs denoted similar abundance patterns, while a negative correlation was characterized as opposite abundance patterns. Interacting nodes within networks represented co-occurrence across samples. The OTUs that were positively correlated with the forestation status were considered forest-associated and those that are negatively correlated were considered pasture-associated.
After the construction of networks, the following topological features were measured using Cytoscape 3.2.1 (Cline et al. 2007), and used to describe network properties: (i) connectivity, which is the number of links (i.e. edges) of a node to other nodes; (ii) clustering coefficient, a measure of interconnectivity in the neighborhood of a node; (iii) path length, which is the average number of edges on the shortest path connecting any two nodes and (iv) betweenness centrality, which reflects the number of times a node plays a role as a connector along the shortest path between two other nodes. While average values of the indexes are generally used to describe the overall features of the network, the relative betweenness centrality value of each node can indicate its relative importance in the network. Nodes with higher betweenness centrality values are likely to be situated in the core of the network and those with lower values are expected to have a more peripheral location (Greenblum et al. 2013), which may represent important ecological and biological insights.
Networks were visualized with Cytoscape 3.2.1 using the edge-weighted spring embedded layout to obtain an overall distribution pattern of the nodes, and group attributes layout, where nodes are separated into modules (Cline et al. 2007). Modules comprise groups of highly connected nodes and were detected using the Girvan–Newman algorithm in Gephi version 0.9.2 (Newman and Girvan 2004; Bastian, Heymann and Jacomy 2009). This algorithm estimates the edge betweenness of each edge in a network, which is defined as the number of the shortest paths between pairs of nodes. This algorithm divides the network into modules within which edges are most frequent between nodes (e.g. OTUs), and thus finds edges that are responsible for connecting many other nodes within a module. The modules are only loosely connected by a few inter-module edges with high edge betweenness as all the shortest paths from one module to another must run through them and removing these edges with lead to separate the modules from one another.
Predictive functional profiling
To determine the functional potential of the network members, we used the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) approach (Langille et al. 2013) and Functional Annotation of Prokaryotic Taxa (FAPROTAX; Louca, Parfrey and Doebeli 2016). Linear Discriminant Analysis (LDA) was performed using the LDA Effect Size (LEfSe) algorithm for comparing predictive functional categories between forest and pasture communities (Segata et al. 2011). More detailed information about the predictive functional profiling is provided in the supplementary materials and methods.
Quantitative PCR targeting bacterial taxa
Seven dominant bacterial taxa—Acidobacteria, Firmicutes, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria and Verrucomicrobia—were quantified by qPCR using phylum/class specific primer sets (Table S1, Supporting Information). We identified these taxa for quantitative analysis by qPCR as major variations were shown in their relative abundances between Amazon forest and pasture (Rodrigues et al. 2013; Ranjan et al. 2015). A total of 15 replicates (5 biological × 3 technical) were used for each land use and each taxon. Each 20 µl-reaction mixture contained 10 µl of SYBR Green Supermix (2X, Bio-Rad, Hercules, CA), 0.3 µM of each primer and 5 ng of DNA sample. Reactions were performed with the Applied Biosystems 7300 real-time PCR system and conditions were experimentally determined for each primer set (Table S1, Supporting Information). Negative controls were included with each reaction using PCR-grade water. Dissociation curve analysis of each post-reaction confirmed the specificity of the products. Standard curves (103 to 107 copies per reaction) were generated with the pCR2.1-TOPO vector containing a PCR-amplified fragment for each phylum or class (Invitrogen Corp., Carlsbad, CA). Reaction efficiency (E) was determined with the equation E = 10(−1/slope).
Statistics
Alpha diversity was calculated using species richness, and Bray–Curtis dissimilarity was used to calculate the pairwise dissimilarities (beta diversity) and perform principal coordinate analysis (PCoA) between OTUs dataset across samples. Analysis of similarity (ANOSIM) was performed to assess whether taxonomic compositions are significantly different across the terrestrial environments. To calculate the relationships of the soil physicochemical properties with the relative abundance of major taxa, alpha diversity (species richness) and species similarity (PCo1 loadings that explains highest variation within the dataset), we used Pearson correlation test as described elsewhere (Fierer and Jackson 2006). In addition, we calculated PCo1 loadings for each module, termed module eigengene (Langfelder and Horvath 2007), and correlated with the soil factors as previously described (Deng et al. 2012) to explore the relationships between individual modules and abiotic factors. To obtain the relationships between the prokaryotic communities and the soil physicochemical properties, a PCoA biplot was generated using the OTU-level abundance dataset, where soil factors were fitted as vectors using two R packages—‘ape’ (Paradis, Claude and Strimmer 2004) and ‘vegan’ (Oksanen et al. 2013).
Data accessibility
To determine whether differences in the relative abundances of major taxonomic categories between land uses are statistically significant, we used the Mann–Whitney test. Using the same statistical test, we compared the alpha diversities, and pairwise taxonomic distances between samples. Unless otherwise specified, QIIME 1.9.0 and GraphPad Prism 7.00 were used for performing statistical analyses and visualizing the results.
RESULTS
OTU filtering and data standardization
To reduce the noise caused by potential PCR or sequencing errors, singleton OTUs from each sample were filtered out prior to downstream analysis. Following filtering, the number of sequences obtained from different samples ranged from 27 014 to 34 659 with an average length of 250 bases. The abundance of OTUs is provided in Table S2 (Supporting Information). We calculated the relative abundance of the OTUs and then discarded the OTUs that were observed in less than five samples to keep the OTUs that are better representative of the two land uses. This resulted in a total number of 1660 OTUs, of which 1043 OTUs belong to forest and 1062 OTUs to pasture samples.
Network and topological features
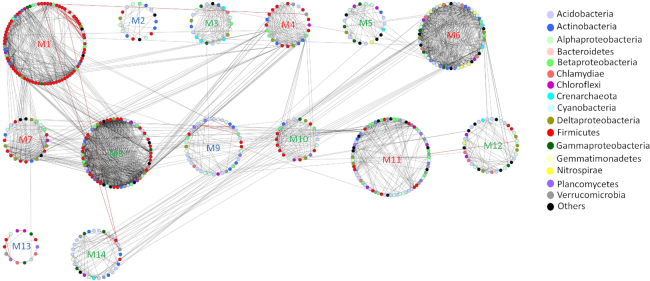
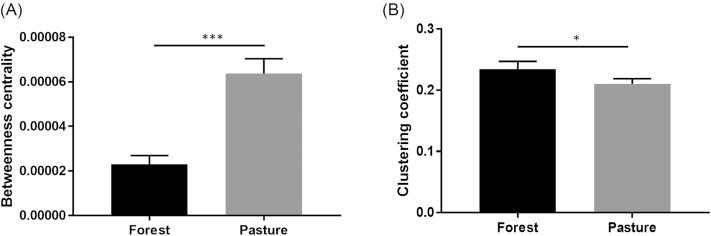
A correlation network, which included only significant OTU–OTU relationships based on their relative abundances, was constructed using both forest and pasture datasets. This network has a total of 1193 nodes and 2913 edges; where 2778 have positive and only 135 have negative correlations (Fig. S1, Supporting Information). In the network, we noticed a major connected part of 640 nodes and 2493 edges with distinct components of forest and pasture. We then visualized the giant connected part with modules, which provided a total of 14 modules (Fig. 1). Importantly, the calculated modules are mostly composed of either forest or pasture-associated nodes, and the rest of the network (disconnected part) mostly comprises the nodes that did not satisfy the correlation criteria with the forestation status (−0.80 ≥ ρ ≥ 0.80, BH-adjusted P < 0.01). The forest component has a lower number of nodes (255) and edges (1385) in comparison to pasture (848 nodes and 1645 edges). We then calculated the betweenness centrality and clustering coefficient of forest- and pasture-associated nodes, where forest nodes have significantly lower (P < 0.001) average betweenness centrality (Fig. 2A) and higher (P < 0.05) average clustering coefficient (Fig. 2B) scores compared to the pasture nodes.
Figure 1.
Network of co-occurring OTUs pairs based on a Spearman correlation (−0.80 ≥ Ρ ≥ 0.80) with significance of adjusted P ≤ 0.01. An interaction (edge) between nodes (OTUs) implies a significant correlation. Red edges represent negative correlations, while the grey edges represent positive correlations. The network is visualized using group attributes layout in Cytoscape 3.2.1, where every module is given an arbitrary number. Among 14 modules, 6 modules are forest associated (M3, M5, M8, M10, M12 and M14; green labeling), 5 modules are pasture associated (M1, M4, M6, M7 and M11; red labeling), and 3 modules comprise both types of nodes (M2, M9 and M13; blue labeling).
Figure 2.
Mean of topological features of the forest and pasture components of the network; (A), average betweenness centralities, (B), average clustering coefficients. Statistical significance was calculated using Mann–Whitney test. Error bars represent standard error. Symbols (*) and (***) indicate significance values of P < 0.05 and P < 0.001, respectively.
Network memberships
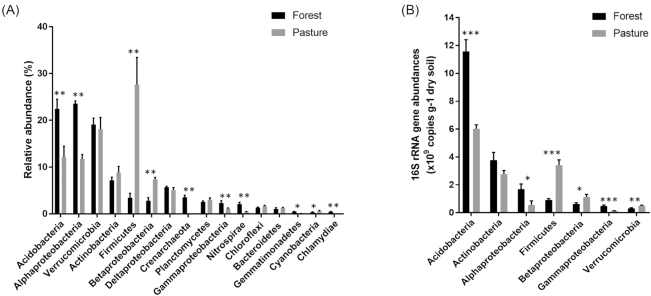
The distribution of major microbial phyla between forest and pasture components was not preserved. We determined that microbial communities comprising the networks were significantly different between forest and pasture. These differences were true at the phylum—[analysis of similarity (ANOSIM): R = 0.81, P < 0.01)], genus—[ANOSIM: R = 0.98, P < 0.02], and OTU-level [ANOSIM: R = 0.99, P < 0.02]. The largest variations in network phyla/class in response to forest-to-pasture conversion were observed for Alphaproteobacteria and Firmicutes (Fig. 3A). The former taxa decreased from the mean value of 23.56% [±1.01; α = 0.05 confidence intervals (CI)] to 11.69% (±2.5; α = 0.05 CI) and latter taxa increased from 3.44% (±2.2; α = 0.05 CI) to 27.60% (±10.7; α = 0.05 CI). Two other major groups, Acidobacteria (P < 0.05) and Nitrospirae (P < 0.01) showed significant decreases with forest-to-pasture conversion from 22.46% (±4.4; α = 0.05 CI) to 12.19% (±5.5; α = 0.05 CI) and 2.1% (±0.9; α = 0.05 CI) to 0.39% (±0.2; α = 0.05 CI), respectively, while the class Betaproteobacteria showed a significant increase (P < 0.01) from 2.9% (±1.6; α = 0.05 CI) to 7.3% (±0.8; α = 0.05 CI). Intriguingly, Crenarchaeota comprised 3.6% (±0.7; α = 0.05 CI) of the forest network, but none of the OTUs in the pasture network were assigned to this archaeal phylum.
Figure 3.
Abundances of major microbial taxa in forest and pasture soils. (A)Relative abundance of major microbial phyla and classes in forest (black) and pasture (grey) networks. These microbial taxa comprise more than 98% of 16S rRNA gene reads, (B) Copy numbers of seven major bacterial phyla and classes per gram of dry soil determined by quantitative PCR in two different land uses in the Amazon. Forest, black; pasture, grey. Error bars represent the standard error. Statistical significance was calculated using Mann–Whitney test. Symbols (*), (**) and (***) indicate significance values of P < 0.05, P < 0.01 and P < 0.001, respectively.
After binning OTUs into genera, 25.9% and 21.25% of all forest and pasture sequences, respectively, were assigned to the top 20 genera in each system (Figure S2, Supporting Information). Among these abundant genera, the largest percentile decrease for genera in response to the ecosystem conversion was observed with Rhodoplanes [14.05% (±0.5; α = 0.05 CI) in forest and 5.56% (±1.4; α = 0.05 CI) in pasture, P < 0.0001], which was counterbalanced by two endospore-forming genera, Sporosarcina [0.9% (±0.7; α = 0.05 CI) in forest and 7.8% (±3.3; α = 0.05 CI) in pasture, P < 0.01] and Bacillus [0.27% (±0.1; α = 0.05 CI) in forest and 2.17% (±1.08; α = 0.05 CI) in pasture, P < 0.01]. More noticeably, Candidatus Nitrososphaera (P < 0.0001) and Nitrospira (P < 0.001) contributed 3.1% (±0.9; α = 0.05 CI) and 1.05% (±0.2; α = 0.05 CI) in the forest network, respectively, whereas none of the OTUs in pasture network belong to these genera.
Quantitative PCR (qPCR)
We selected seven dominant bacterial taxa to be quantified by qPCR. Cycle threshold (Ct) values were obtained for each specific primer set using a representative 16S rRNA gene sequence per phylum or class. The Ct values were used to calculate the actual copy numbers of each microbial phylum or class standard curves (Table S3, Supporting Information). All seven qPCR assays showed R2 values above 0.97 with reaction efficiencies ranging between 1.79 and 2.35. The 16S rRNA gene copy numbers of Acidobacteria (P < 0.001), Alphaproteobacteria (P < 0.05) and Gammaproteobacteria (P < 0.001) were significantly higher for forest samples, while those numbers observed for Firmicutes (P < 0.001), Betaproteobacteria (P < 0.05) and Verrucomicrobia (P < 0.01) were higher for pasture samples. The 16S rRNA gene copy numbers for members of the phylum Actinobacteria were not significantly different between land uses (Fig. 3B).
We compared the abundance ratios of dominant bacterial taxa between forest and pasture as determined by the 16S rRNA gene sequencing and qPCR. While we notice variations in ratios of bacterial taxa, the alteration of abundances in these taxa following deforestation mostly followed similar trends as estimated in both approaches (Fig. S3A, Supporting Information), especially the taxa that showed major variations in abundances between forest and pasture. The abundance differences between qPCR and 16S rRNA gene sequencing on the bacterial taxa remained similar when 16S rRNA gene sequencing data are corrected for copy-number variation (Fig. S3B, Supporting Information).
Module comparisons
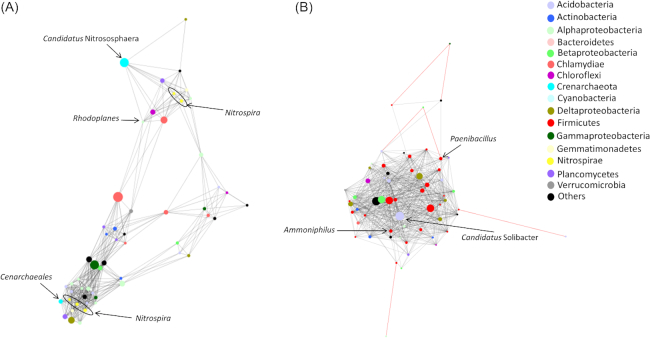
Among 14 modules, 6 modules comprised only forest associated (M3, M5, M8, M10, M12 and M14) and 5 modules comprised only pasture-associated (M1, M4, M6, M7 and M11) nodes, with 3 modules consisting of both types of nodes (M2, M9 and M13). Consistent with the overall network membership (Fig. 3A), pasture modules were dominated by Firmicutes nodes, whereas alphaproteobacterial, acidobacterial nodes mostly dominated in the forest modules (Fig. S4A, Supporting Information). For example, M6 is a pasture module with 61 nodes, where 39.3% of the nodes belonged to Firmicutes followed by Acidobacteria (9.8%) and Alphaproteobacteria (6.6%). On the other hand, M8 is a forest-associated module with 55 nodes, where Alphaproteobacteria and Acidobacteria comprised 18.2% and 10.9%, respectively. The M6 and M8 modules were most connected in their respective communities, with the average connectivity values of 10.6 and 7.9, respectively (Fig. S4B, Supporting Information). We further investigated these two modules by analyzing the betweenness centrality values of nodes to explore how land use change altered network co-ocurrence organization. The relative importance of individual nodes, as indicated by their betweenness centrality values, was compared between the forest (M8; Fig. 4A) and pasture (M6; Fig. 4B) modules. In both modules, most of the nodes were not classified to the genus level. Among the classified members with known functional attributes, Candidatus Nitrososphaera, Nitrospira, Cenarchaeales (an order of Thaumarchaeota) and Rhodoplanes, were distributed in the forest module (M8). Betweenness centrality values for these taxa were estimated to be 5.8 × 10−4, 7.27 × 10−5, 1.52 × 10−5 and zero, respectively (Fig. 4A). Similarly, we identified Candidatus Solibacter, Paenibacillus and Ammoniphilus in the pasture associated module (M6), with their centrality values of 1.26 × 10−3, 3.21 × 10−4 and 1.26 × 10−4, respectively (Fig. 4B).
Figure 4.
A representative module was obtained from (A), forest and (B), pasture components of the network for comparative analysis. The criteria used in comparing these modules were that they were most connected within their members and have high node size. Forest module (M8) is comprised of 55 nodes with average connectivity of 7.98, while the pasture module is comprised of 61 nodes with average connectivity of 10.62. The size of the nodes in each network is scaled according to their betweenness centrality values to indicate their relative importance in their networks.
Relationship between the community structures and soil physicochemical properties
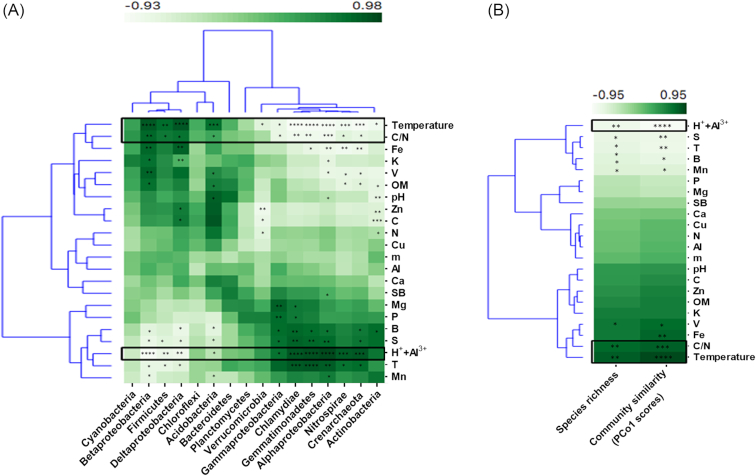
We have 22 soil variables incorporated in our analysis (Table S4, Supporting Information). Among them, B (P < 0.01), exchangeable acidity (H++Al+3; P < 0.01), Mn (P < 0.01), S (P < 0.01), cation exchange capacity (T; P < 0.01) were significantly higher and C (P < 0.01), C/N (P < 0.01), Fe (P < 0.05), Mg (P < 0.05), pH (P < 0.05), temperature (P < 0.01) and base saturation (V; P < 0.05) were significantly lower in forest soils compared to those in pasture. Among these statistically different variables, temperature, exchangeable acidity (H++Al+3) and C/N were calculated to have strongest correlations (as determined by Pearson correlation test) with most of the major taxonomy classes (Fig. 5A) and diversity metrics (species richness and community similarity by Bray–Curtis) (Fig. 5B). For community similarity, we used the PCo1 loadings from the principal coordinate analysis (PCoA), which explained 55.8% of the variation in the original dataset.
Figure 5.
Relationship between the microbial community structures and soil physicochemical parameters. (A), Pearson correlations between the relative abundances of major microbial taxa and soil variables. (B), Pearson correlations of the species richness (alpha diversity) and community similarity, the ordination score of the first axis of principal coordinate analysis (PCoA) (which explains 55.78% of the variance in the original data), with the soil variables. C, carbon; N, nitrogen; Fe, iron; V, base saturation; m, aluminum saturation; Ca, calcium; Mg, magnesium; Mn, manganese; B, boron; Cu, copper; T, cation exchange capacity; SB, sum of bases; H++Al3+, exchangeable acidity; OM, organic matter; P, phosphorus; S, sulfur. The color gradient represents the correlation strength of the relationships. Symbols (*), (**) and (***) indicate significance values of P < 0.05, P < 0.01 and P < 0.001, respectively.
Our statistical analyses showed that soil microbial composition and diversity were directly affected by soil physicochemical properties. A coordination plot revealed that pasture communities have positive associations with temperature (r2 = 0.97, P < 0.01), C/N (r2 = 0.95, P < 0.01) and Fe (r2 = 0.77, P < 0.05), while forest communities have positive associations with soil acidity (H++Al+3; r2 = 0.87, P < 0.01), S (r2 = 0.89, P < 0.01), base saturation (V; r2 = 0.91, P < 0.01) and cation exchange capacity (T; r2 = 0.77, P < 0.01) (Fig. S5, Supporting Information). We then asked what particular taxa are impacted by the soil factors and our analysis showed that soil acidity (H++Al3+), soil temperature and C/N ratio had the highest influences on the microbial composition and diversity in both land uses. Here, acidity (H++Al3+) was observed to have strong positive correlations with Crenarchaeota (r = 0.9, P < 0.001), Nitrospirae (r = 0.87, P < 0.0001), Alphaproteobacteria (r = 0.97, P < 0.0001), Gemmatimonadetes (r = 0.94, P < 0.0001), Chlamydiae (r = 0.91, P < 0.0001) and Gammaproteobacteria (r = 0.72, P < 0.05), Acidobacteria (r = 0.69, P < 0.05) and negative correlations with Deltaproteobacteria (r = −0.82, P < 0.01), Firmicutes (r = −0.78, P < 0.01), Betaproteobacteria (r = −0.92, P < 0.0001) (Fig. 5A). On the other hand, temperature and C/N had reverse relationships with the above microbial taxa. We also noticed that Cyanobacteria, Chloroflexi, Bacteroidetes and Planctomycetes were not correlated to any of the 22 soil variables measured in this study. Our results also demonstrated that the same three soil factors contributed to overall alterations in microbial diversity, with soil acidity being negatively correlated with species richness (r = −0.85, P < 0.001) and community similarity (r = −0.88, P < 0.001), while temperature (r = 0.82, P < 0.001 and r = 0.67, P < 0.001, respectively) and C/N ratio (r = 0.88, P < 0.001 and r = 0.74, P < 0.001) were positively correlated with the species richness and community similarity, respectively (Fig. 5B). Similarly, these soil variables, which in addition to sulfur, were estimated to have strongest correlations with individual modules (Fig. S6, Supporting Information).
Potential functional interpretation of taxonomic data
Procrustes analyses showed that two different OTU-picking algorithms, de novo vs closed-reference OTU lineages (M2 = 0.01, P < 0.01; Fig. S7A, Supporting Information), and PICRUSt-predicted KO gene profiles vs de novo OTU lineages (M2 = 0.29, P < 0.01; Fig. S7B, Supporting Information) produced similar clustering patterns. The coordinates in both cases explained most of the variances, estimated to be 78.9% and 87.07%, respectively. Following this analysis, we tested the impact of alterations in network membership between forest and pasture on specific metabolic pathways using PICRUSt (Fig. S8A, Supporting Information) and FAPROTAX (Fig. S8B, Supporting Information) based functional annotations. While we observed different functional categories being altered between forest and pasture soils, applying both approaches helped us gain information about pathways related to the nitrogen cycle. For example, PICRUSt showed a potential increase of genes associated with nitrogen metabolism in the forest soils, whereas FAPROTAX indicated a potential increase of nitrification and denitrification-related processes in the forest and nitrogen fixation in pastures.
DISCUSSION
Soil microbial communities are known to have diverse compositions and varied abundances, even when analyzed at local scales that matter to microorganisms (Bailey et al. 2013; Kuzyakov and Blagodatskaya 2015). These complex microbial patterns are not entirely random distributions, but the outcome of multiple ecological associations at different spatial and temporal scales (Lidicker 1979). In this study, we used a co-occurrence-based network analysis to represent association across multiple prokaryotic species, and identify alterations in potential relationships with forest-to-pasture conversion in the Amazon rainforest, the largest tropical ecosystem in the world. In addition, we employed soil physicochemical variables to explore the influences of specific edaphic factors affecting the prokaryotic composition and diversity.
Our results indicate that the network has distinct components of forest- and pasture-associated nodes, in which a major connected component contains 626 nodes and several small fragments of network, with nodes ranging 2 to 17 (Fig. S1, Supporting Information). Higher taxonomic and phylogenetic dissimilarity between forest and pasture communities probably explains why a major portion of the nodes remains disconnected (Rodrigues et al. 2013; Ranjan et al. 2015). These observations imply that a network is not a random combination of nodes, but rather organized as land use specific subnetworks (termed ‘module’) with unique functional significance in a complex web of associations between biotic and abiotic components.
We examined topological features of the forest- and pasture-associated nodes to inquire about the ecological traits of prokaryotic communities (Fig. 2A–B). We observed a decreased betweenness centrality score (P < 0.001) of forest component of network. This result implies that prokaryotic communities in forest tend to reside more in the interface between prokaryotic community and environment, where soil biotic and abiotic factors have a higher influence to prokaryotic community and vice versa. The oligotrophic nature and poor quality of complex organic substrates in the forest soils may require microbial syntrophy from multiple species, such as the uptake and utilization of these compounds would require more extracellular hydrolysis using microbial enzymes. This is further supported by higher tendency of forest nodes to tie together as indicated by higher clustering coefficient observed in forest in comparison to pasture. It is well established that certain microorganisms are functionally complementary to each other, such as microbial consortia for anaerobic methane oxidation, thermodynamically interdependent degradation and nutritional exchange, among others (Morris et al. 2013). In contrast, conversion of tropical forests to pastures containing only two grass species will reduce the variety of resources, even if the total amount of resources, specifically C-based compounds, increases in soil. There is evidence that this is occurring in our study system as we detected decreases in community dissimilarity at taxonomic, phylogenetic and functional gene levels (Mirza et al. 2014; Ranjan et al. 2015; Navarrete et al. 2015a; Hamaoui et al. 2016; Khan 2016) with increased values of total C in pastures (Cenciani et al. 2009).
Given the taxonomic and phylogenetic changes between forest and established pasture, we asked which groups were more susceptible to alterations. Our results indicate that the relative abundances of Alphaproteobacteria, Crenarchaeota, Acidobacteria and Nitrospirae were significantly decreased, while Firmicutes and Betaproteobacteria increased (Fig. 3A). We found direct support for these results with the use of qPCR (Fig. 3B). Our results contrast from previous work, in which no observed differences were found for these dominant taxa after deforestation. We attribute this difference to short-term impact of the deforestation (2–4 months, Navarrete et al. 2015b) in comparison to our sampling site where grasses have been established for 38 years. Next, we used 22 different soil physicochemical properties to identify potential reasons for our observed microbial community differences. Our results showed that soil temperature, C/N ratio and soil exchangeable acidity (H++Al3+) mostly predict the overall community composition and therefore module composition, and diversity of prokaryotic community (Fig. 5, Fig. S6, Supporting Inormation). The correlation between the prokaryotic composition/diversity and soil nutrient composition might actually be related to soil variables such as temperature and soil acidity (H++Al3+), which co-varies with soil C/N ratio. Similar correlation patterns of composition and diversity with soil acidity are evident in tropical soils (Navarrete et al. 2015b), while those correlational patterns associated with C/N ratio are frequently observed in the Arctic and temperate ecosystems (Ge et al. 2010; Chu et al. 2011) and in alkaline Tibetan permafrost soils (Zhang et al. 2014).
Co-occurrence patterns do not allow mapping of microbial interactions directly, but provide information on particular groups sharing habitats or performing similar ecological functions (Freilich et al. 2010). We then aimed at exploring the functional attributes of nodes based on the current knowledge of biology with their betweenness centrality scores. In particular, we focused our investigations on the two most connected modules, where one contains all forest-associated nodes and another contains all pasture-associated nodes—serving as two representative modules (or niches) of these contrasting ecosystems. While most of the nodes were not identified to the genus level, we noticed several nodes in the forest module are associated with nitrification including Candidatus Nitrososphaera, Cenarchaeales (an order of Thaumarchaeota), Nitrospira (Spang et al. 2002; Leininger et al. 2006; Stahl and de la Torre 2012; Daims et al. 2015; van Kessel et al. 2015) and denitrification including Rhodoplanes (Hiraishi and Imhoff 2005). In addition, Candidatus Nitrososphaera is estimated to have high betweenness centrality values suggesting its high importance in the forest ecosystem. (Fig. 4A). This observation is in agreement with our recently published findings that only thaumarchaeal sequences in the forest soils were retrieved for the gene amoA, encoding the α subunit of the ammonia monooxygenase enzyme, and consistent with our failed attempts to amplify amoA gene sequences associated with bacterial ammonia oxidizing microorganisms (Hamaoui et al. 2016). On the other hand, Rhodoplanes has betweenness centrality value of zero, which is categorized as a peripheral node implying its involvement in the initiation or termination processes according to Greenblum et al. (2013). Being denitrifier, the ability to produce atmospheric nitrogenous gases including nitrous oxide as byproducts justifies its role in the nitrogen cycle. Alternatively, the different set of microbial taxa in the pasture module may determine a change in ecosystem processes (Fig. 4B). A node with highest betweenness centrality value in the pasture module is related to Candidatus Solibacter, which has the genetic potential for the nitrate and nitrite reduction (Pearce et al. 2012). Two other nodes with low beweenness centrality values are associated with Paenibacillus and Ammoniphilus, of which the former is known to have at least 10 representative species for nitrogen fixation (Xie et al. 2012), and the latter is an ammonia dependent genus but does not have known representative for nitrification (Zaitsev et al. 1998). There are two important implications associated with our findings. At the compositional level, the network theory predicts that the nodes with high betweenness centrality values should be more vulnerable to disturbance (Solé and Montoya 2001), which might set the stage for other taxa to dominate in an altered microbial community. There is evidence for both the loss of Candidatus Nitrososphaera and Nitrospira (Fig. S2, Supporting Information), and the change of dominant prokaryotic taxa with forest-to-pasture conversion (Fig. S2, Supporting Information). At the functional level, ecological theory predicts that losses of important species, identified as hub nodes in the network theory, can have a large impact on ecosystem functioning (Chapin et al. 2000). There is evidence for the loss of nitrification potential in our study site as previous biogeochemical studies at Fazenda Nova Vida observed higher net nitrification rates in forest soils (1.32 to 3.51 μg N g−1 dry soil day−1) in comparison to pastures (0.02 to 0.77 μg N g−1 dry soil day−1) (Neill et al. 1995; Neill et al. 1997). These studies have hypothesized that a direct consequence of the change in net N mineralization is the decrease of NO and N2O emissions from pastures (Melillo et al. 2001), but the reasons for these alterations were not established. Therefore, higher nitrification and denitrification rates measured by others in our study site along with a lower nitrogen fixation in the forest soils compared to the pasture (Melillo et al. 2001) can be explained, at least partially, by the altered inter-taxa relationships represented by specific nodes and edges in co-occurrence networks. Predictive functional profiling using the PICRUSt and FAPROTAX further supports this observation (Fig. S8A–B, Supporting Information). Conversely, a recent study shows that land use change to agriculture (oil palm plantation) following deforestation-altered community composition but the central roles of Nitrospirae in the co-occurrence networks structure remain unaltered (Wood et al. 2017). This observation indicates that deforestation causes a substantial variation in soil physicochemical properties that depend on land use type, time since conversion, soil treatment and amendments and subsequent land use, which subsequently determine the functional predominance in the microbial communities. For example, newly deforested soils (2–4 months old) contained increased abundance of microbial functional genes that are related to the survival and adaptation in a changing environment (Navarrete et al. 2015b). On the other hand, previous reports and predictive functional profiling showed increased capacity for nitrogen fixation in established pastures (38-year old) (Mirza and Rodrigues 2012; Paula et al. 2014), implying that there is likely selective pressure on soil microbes to fix atmospheric nitrogen.
There is direct evidence that soil physicochemical properties not only influence the microbial structure, but also the rate of biogeochemical processes (Rastetter et al. 2005; Borken et al. 2006). Several studies reported that temperature is a key factor that regulates many terrestrial processes including soil respiration (Raich and Schlesinger 1992), N mineralization and nitrification (MacDonald, Zak and Pregitzer 1995), denitrification (Malhi, McGill and Nyborg 1990) and CH4 emission (Crill et al. 1988). In addition, C/N ratio has substantial impacts on nitrogen mineralization and nitrification rates (Strauss and Lamberti 2000; Knoepp and Swank 2002; Paul et al. 2003), and therefore on denitrification. Our report is in line with these observations. A study in stream sediments reported that heterotrophs outcompete nitrifiers for the available nitrogen under higher C/N and labile organic carbon conditions, and therefore nitrification rate declines (Strauss and Lamberti 2000). The higher C/N and availability of more easily degradable carbon sources in the pastureland are suggestive of a reduced nitrification rate (Neill et al. 1995; Neill et al. 1997). Strong negative correlations of nitrifiers Crenarchaeota and Nitrospirae with the C/N (r = −0.72, P < 0.01 and r = −0.65, P < 0.05, respectively) further suggest similar phenomenon in the pastureland compared to forest soils. On the other hand, a meta-analysis of experimental warming experiments at 32 sites reported that warming in the range 0.3–6.0°C resulted in an increase of soil respiration rates by 20% and net N mineralization rates by 46% (Rustad et al. 2001). Denitrification is also extremely sensitive to rising temperatures (Butterbach-Bahl et al. 2013). Here, temperature induced increased respiration would lead to a depletion of oxygen in the terrestrial ecosystem, which triggers soil anaerobiosis such as denitrification (Schaufler et al. 2010; Butterbach-Bahl et al. 2013). Particularly, the emission of greenhouse gas nitrous oxide responds more uniformly to temperature (Schindlbacher, Zechmeister‐Boltenstern and Butterbach‐Bahl 2004), which may be aided at low C/N. While temperature has a strong negative impact on the abovementioned nitrifiers (r = −0.9, P < 0.001 and r = −0.86, P < 0.01, respectively) (Fig. 5A), it has a strong positive impact on nitrogen mineralization (Rustad et al. 2001). The succession of temperature-sensitive microbial processes within the nitrogen cycle including nitrogen mineralization provide substrates for denitrification, suggesting multiplying effect of warming temperature on N2O fluxes from Amazon forest soil. This observation has an important implication. Our study in two adjacent land use types along the gradients of physicochemical factors provide an opportunity to explore the long-term impact of climate-driven variation in temperature on microbial structure and ecosystem processes in the Amazon. The variation in temperatures between our study sites (24.86°C in forest versus 27.14°C in pasture; Table S4, Supporting Information) is similar to what is predicted by 2050 in the Amazon (Nobre et al. 2016), with potential for increased emissions of greenhouse gases (nitrous oxide, nitric oxide) from the pristine forest. An implementation of these observations into global climate-change models may substantially alter predictions of greenhouse gas emissions and the severity of climate change.
Traditionally, biodiversity studies have relied on species richness and turnover, but ignore co-occurrence patterns, which are important for understanding how communities assemble and respond to changes. We identified prokaryotic groups serving as network hubs by their topological feature and predicted their functional profiles, coupling the role of prokaryotic taxa to the observed patterns of nitrogen dynamics, providing novel information regarding changes to biogeochemical processes following forest-to-pasture conversion. Importantly, the incorporation of the soil physicochemical properties in our study allowed us to identify the predominant factors (temperature, exchangeable acidity and C/N) affecting the microbial composition and diversities at the local scale, which differ from what was observed previously at continental scale. Despite the usefulness and strength of our current study, here we acknowledge few caveats. First, low sample size may hamper complete exploration of spatial variability in microbial profiles. However, previous studies at broader spatial scales in the pristine Amazon and deforested areas are consistent to our current reports, implying that higher sample sizes will further confirm the observed differences between forest and pasture. Second, samples were only collected at 0–10 cm depth, which may not capture the ‘true’ impact on microbial communities and therefore network structure by this study given that community patterns could be quite different with depth, depending on differences in rooting depth, architecture, leaching of dissolved organic carbon, etc. While the effect of sampling depth would be more pronounced in forest soils, this sampling depth contains majority of the root system of tropical grasses in the pastureland. For the purpose of comparison, we chose to collect samples with identical depth from both terrestrial ecosystems. Third, networks described here provide a snapshot of the co-occurring microbial communities at a given time, which may not explain some important phenomena, for example, responses to perturbation, succession, etc. Although the microbial communities are likely to change over time in their ecosystems, previous works reported that temporal variation is much lower than the spatial variability in modulating the composition of microbial community in terrestrial ecosystem (Krave et al. 2002; Fierer and Jackson 2006). Fourth, lateral gene transfer allows microbes to gain and lose genes rapidly, and it is common in microbial lineages that share similar ecologies (Smillie et al. 2011), therefore we have to be very cautious in coupling functional attributes and taxonomy. Nevertheless, the co-occurrence patterns described in our study provide guidance for isolation efforts for poorly characterized microbial species that share the same or complementary physiological traits with known species, and increase our limited understanding of associations involved in processes of community assembly, competition and habitat filtering.
Data Accessibility
The 16S rRNA gene sequence data has been deposited to Sequence Read Archive (SRA) under BioProject PRJNA490051. The sequence data (SRR7816680-SRR7816689) are available at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP160516.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the owners and staff of Agropecuaria Nova Vida for logistical support and permission to work on their property. We are grateful to Drs. Siu M. Tsai, Rebecca Mueller, Fabiana Paula and Babur Mirza, and Wagner Piccinini for assistance with fieldwork and Dr. James Grover for carefully reviewing this manuscript.
FUNDING
This work was supported by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02-05CH11231, the National Science Foundation—Dimensions of Biodiversity (grant numbers: DEB 14422214, NSF-FAPESP – 2014/50320-4), and by the Agriculture and Food Research Initiative Comfpetitive Grant 2009-35319-05186 from the US Department of Agriculture—National Institute of Food and Agriculture.
Conflicts of interest. None declared.
REFERENCES
- Bailey VL, McCue LA, Fansler SJ et al. . Micrometer-scale physical structure and microbial composition of soil macroaggregates. Soil Biol Biochem. 2013;65:60–68. [Google Scholar]
- Bascompte J, Stouffer DB. The assembly and disassembly of ecological networks. Philos Trans R Soc Lond B Biol Sci. 2009;364:1781–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. Int AAAI Conf Weblogs Soc Media. 2009;8:361–362. [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological). 1995;57:289–300. [Google Scholar]
- Bierregaard RO, Gascon C, Lovejoy TE et al. . Lessons from Amazonia: The Ecology and Conservation of a Fragmented Forest. New Haven: Yale University Press, 2001;1154–56. [Google Scholar]
- Borken W, Davidson EA, Savage K et al. . Effect of summer throughfall exclusion, summer drought, and winter snow cover on methane fluxes in a temperate forest soil. Soil Biol Biochem. 2006;38:1388–1395. [Google Scholar]
- Butterbach-Bahl K, Baggs EM, Dannenmann M et al. . Nitrous oxide emissions from soils: how well do we understand the processes and their controls?. Phil Trans R Soc B. 2013;368:20130122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA et al. . Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenciani K, Lambais MR, Cerri CC et al. . Bacteria diversity and microbial biomass in forest, pasture and fallow soils in the southwestern Amazon basin. Revista Brasileira de Ciência do Solo. 2009;33:907–916. [Google Scholar]
- Chapin FS, Zavaleta ES, Eviner VT et al. . Consequences of changing biodiversity. Nature. 2000;405:234–42. [DOI] [PubMed] [Google Scholar]
- Chu H, Neufeld JD, Walker VK et al. . The influence of vegetation type on the dominant soil bacteria, archaea, and fungi in a low Arctic tundra landscape. Soil Sci Soc Am J. 2011;75:1756–1765. [Google Scholar]
- Cline MS, Smoot M, Cerami E et al. . Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill PM, Bartlett KB, Harriss RC et al. . Methane flux from Minnesota peatlands. Global Biogeochem Cycles. 1988;2:371–84. [Google Scholar]
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99:10494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Lebedeva EV, Pjevac P et al. . Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EA, de Carvalho CJR, Figueira AM et al. . Recuperation of nitrogen cycling in Amazonian forests following agricultural abandonment. Nature. 2007;447:995–998. [DOI] [PubMed] [Google Scholar]
- Deng Y, Jiang YH, Yang Y et al. . Molecular ecological network analyses. BMC Bioinformatics. 2012;13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Philip H, Neils L et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirzo R, Raven PH. Global state of biodiversity and loss. Annu Rev Environ Resour. 2003;28:137–67. [Google Scholar]
- Don A, Schumacher J, Freibauer A. Impact of tropical land‐use change on soil organic carbon stocks–a meta‐analysis. Global Change Biol. 2011;17:1658–70. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–61. [DOI] [PubMed] [Google Scholar]
- Eva HD, Belward AS, De Miranda EE et al. . A land cover map of South America. Global Change Biol. 2004;10:731–744. [Google Scholar]
- Faust KJ, Fah S, Jacques I et al. . Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;7:e1002606–e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnside PM. Biodiversity as an environmental service in Brazil's Amazonian forests: risks, value and conservation. Environ Conserv. 1999;26:305–321. [Google Scholar]
- Feeley KJ, Silman MR. Extinction risks of Amazonian plant species. Proc Natl Acad Sci. 2009;106:12382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigl B, Cerri C, Piccolo M et al. . Biological survey of a low-productivity pasture in Rondônia state, Brazil. Outlook on Agriculture. 2006;35:199–208. [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci. 2006;103:626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JA, Asner GP, Costa MH et al. . Amazonia revealed: forest degradation and loss of ecosystem goods and services in the Amazon Basin. Front Ecol Environ. 2007;5:25–32. [Google Scholar]
- Freilich S, Kreimer A, Meilijson I et al. . The large scale organization of the bacterial interactions network. Nucleic Acid Research. 2010;38:3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS et al. . Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Nat Acad Sci USA. 2006;103:13104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Chen C, Xu Z et al. . Carbon/nitrogen ratio as a major factor for predicting the effects of organic wastes on soil bacterial communities assessed by DNA-based molecular techniques. Environ Sci Pollut Res. 2010;17:807–15. [DOI] [PubMed] [Google Scholar]
- Greenblum S, Chiu HC, Levy R et al. . Towards a predictive systems-level model of the human microbiome: progress, challenges, and opportunities. Curr Op Biotech. 2013;24:810–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LB, Gifford RM. Soil carbon stocks and land use change: a meta analysis. Global Change Biol. 2002;8:345–60. [Google Scholar]
- Hamaoui GS, Rodrigues JL, Bohannan BJ et al. . Land-use change drives abundance and community structure alterations of thaumarchaeal ammonia oxidizers in tropical rainforest soils in Rondônia, Brazil. Appl Soil Ecol. 2016;107:48–56. [Google Scholar]
- Hiraishi A, Imhoff JF, Genus Rhodoplanes. In Brenner DJ, Krieg NR, Staley JT, (eds.) Bergey's Manual of Systematic Bacteriology: Volume Two: the Proteobacteria, Part C. Springer Science & Business Media, New York, NY, 2005;545–49. [Google Scholar]
- INPE. Program for the Estimation of Amazon Deforestation (Projeto PRODES Digital). Brazilian National Institute for Space Research; 2011. [Google Scholar]
- Khan MAW. Response of microbial networks and microbiomes to the forest-to-pasture conversion in the Amazon soils. Ph.D. Dissertation. The University of Texas at Arlington library; 2016. [Google Scholar]
- Khan MAI, Biswas B, Smith E et al. . Microbial diversity changes with rhizosphere and hydrocarbons in contrasting soils. Ecotoxicol Environ Saf. 2018;156:434–442. [DOI] [PubMed] [Google Scholar]
- Knoepp JD, Swank WT. Using soil temperature and moisture to predict forest soil nitrogen mineralization. Biol Fertil Soils. 2002;36:177–182. [Google Scholar]
- Krave AS, Lin B, Braster M et al. . Stratification and seasonal stability of diverse bacterial communities in a Pinus merkusii (pine) forest soil in central Java, Indonesia. Environ Microbiol. 2002;4:361–373. [DOI] [PubMed] [Google Scholar]
- Kuczynski J, Stombaugh J, Walters WA et al. . Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Prot Microbiol. 2012;27(1):1E–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov Y, Blagodatskaya E. Microbial hotspots and hot moments in soil: concept and review. Soil Biol. Biochem. 2015;83:184–199. [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG et al. . Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech. 2013;31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M et al. . Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. [DOI] [PubMed] [Google Scholar]
- Lidicker WZ. A clarification of interactions in ecological systems. Bioscience. 1979;29:475–477. [Google Scholar]
- Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353:1272–1277. [DOI] [PubMed] [Google Scholar]
- MacDonald NW, Zak DR, Pregitzer KS. Temperature effects on kinetics of microbial respiration and net nitrogen and sulfur mineralization. Soil Sci Soc Am J. 1995;59:233–240. [Google Scholar]
- Malhi SS, McGill WB, Nyborg M. Nitrate losses in soils: effect of temperature, moisture and substrate concentration. Soil Biol Biochem. 1990;22:733–737. [Google Scholar]
- Melillo JM, Steudler PA, Feigl BJ et al. . Nitrous oxide emissions from forests and pastures of various ages in the Brazilian Amazon. J Geophys Res. 2001;106:34179–34188. [Google Scholar]
- Meyer KM, Klein AM, Rodrigues JL et al. . Conversion of Amazon rainforest to agriculture alters community traits of methane‐cycling organisms. Mol Ecol. 2017;26:1547–1556. [DOI] [PubMed] [Google Scholar]
- Mirza BS, Rodrigues JL. Development of a direct isolation procedure for free-living diazotrophs under controlled hypoxic conditions. Appl Environ Microbiol. 2012;AEM–00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza BS, Potisap C, Nüsslein K et al. . Response of free-living nitrogen-fixing microorganisms to land use change in the Amazon rainforest. Appl Environ Microbiol. 2014;80:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BE, Henneberger R, Huber H et al. . Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. [DOI] [PubMed] [Google Scholar]
- Navarrete AA, Taketani RG, Mendes LW et al. . Land-use systems affect archaeal community structure and functional diversity in western Amazon soils. Revista Brasileira de Ciência do Solo. 2011;35:1527–1540. [Google Scholar]
- Navarrete AA, Venturini AM, Meyer KM et al. . Differential response of Acidobacteria subgroups to forest-to-pasture conversion and their biogeographic patterns in the western Brazilian Amazon. Front Microbiol. 2015a;6:1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete AA, Tsai SM, Mendes LW et al. . Soil microbiome responses to the short‐term effects of Amazonian deforestation. Mol Ecol. 2015b;24:2433–2448. [DOI] [PubMed] [Google Scholar]
- Neill C, Piccolo MC, Steudler PA et al. . Nitrogen dynamics in soils of forests and active pastures in the western Brazilian Amazon Basin. Soil Biol Biochem. 1995;27:1167–1175. [Google Scholar]
- Neill C, Melillo JM, Steudler PA et al. . Soil carbon and nitrogen stocks following forest clearing for pasture in the southwestern Brazilian Amazon. Ecol Appl. 1997;7:1216–1225. [Google Scholar]
- Newman ME, Girvan M. Finding and evaluating community structure in networks. Physical review E. 2004;69:026113. [DOI] [PubMed] [Google Scholar]
- Nobre CA, Sampaio G, Borma LS et al. . Land-use and climate change risks in the Amazon and the need of a novel sustainable development paradigm. Proc Nat Acad Sci USA. 2016;113:10759–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al. . Package ‘vegan’. Community ecology package, version,2013;2. [Google Scholar]
- Olesen JM, Bascompte J, Dupont YL et al. . The modularity of pollination networks. Proc Nat Acad Sci USA. 2007;104:19891–19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. [DOI] [PubMed] [Google Scholar]
- Paul KI, Polglase PJ, O'connell AM et al. . Defining the relation between soil water content and net nitrogen mineralization. Eur J Soil Sci. 2003;54:39–48. [Google Scholar]
- Paula FS, Rodrigues JLM, Zhou J et al. . Land use change alters functional gene diversity, composition and abundance in Amazon forest soil microbial communities. Mol Ecol. 2014;23:2988–2999. [DOI] [PubMed] [Google Scholar]
- Pearce DA, Newsham K, Thorne M et al. . Metagenomic analysis of a southern maritime Antarctic soil. Front Microbiol. 2012;3:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Sandrine D, van der Laan MJ. Multiple testing procedures: the multtest package and applications to genomics. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer, 2005;249–271. [Google Scholar]
- Raich JW, Schlesinger WH. The global carbon dioxide flux in soil respiration and its relationship to vegetation and climate. Tellus B. 1992;44:81–99. [Google Scholar]
- Ranjan K, Paula FS, Mueller RC et al. . Forest-to-Pasture conversion increases the diversity of the phylum Verrucomicrobia in Amazon rainforest soils. Front Microbiol. 2015;6:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastetter EB, Perakis SS, Shaver GR et al. . Terrestrial C sequestration at elevated CO2 and temperature: the role of dissolved organic N loss. Ecol Appl. 2005;15:71–86. [Google Scholar]
- Rodrigues JLM, Pellizari VH, Mueller R et al. . Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Nat Acad Sci USA. 2013;110:988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustad LEJL, Campbell J, Marion G et al. . A meta-analysis of the response of soil respiration, net nitrogen mineralization, and aboveground plant growth to experimental ecosystem warming. Oecologia. 2001;126:543–562. [DOI] [PubMed] [Google Scholar]
- Schindlbacher A, Zechmeister‐Boltenstern S, Butterbach‐Bahl K. Effects of soil moisture and temperature on NO, NO2, and N2O emissions from European forest soils. J Geophysi Res: Atmospheres. 2004;109. [Google Scholar]
- Schaufler G, Kitzler B, Schindlbacher A et al. . Greenhouse gas emissions from European soils under different land use: effects of soil moisture and temperature. Eur J Soil Sci. 2010;61:683–696. [Google Scholar]
- Segata N, Izard J, Waldron L et al. . Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie CS, Smith MB, Friedman J et al. . Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241. [DOI] [PubMed] [Google Scholar]
- Soares-Filho BS, Nepstad DC, Curran LM et al. . Modeling conservation in the Amazon basin. Nature. 2009;440:520–523. [DOI] [PubMed] [Google Scholar]
- Solé RV, Montoya JM. Complexity and fragility in ecological networks. Proc Biol Sci. 2001;268:2039–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Poehlein A, Offre P et al. . The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2002;14:3122–45. [DOI] [PubMed] [Google Scholar]
- Stahl DA, de la Torre JR. Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol. 2012;66:83–101. [DOI] [PubMed] [Google Scholar]
- Strauss EA, Lamberti GA. Regulation of nitrification in aquatic sediments by organic carbon. Limnol Oceanogr. 2000;45:1854–1859. [Google Scholar]
- Tian J, He N, Kong W et al. . Deforestation decreases spatial turnover and alters the network interactions in soil bacterial communities. Soil Biol Biochem. 2018;123:80–86. [Google Scholar]
- Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kessel MA, Speth DR, Albertsen M et al. . Complete nitrification by a single microorganism. Nature. 2015;528:555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al. . Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widder S, Besemer K, Singer GA et al. . Fluvial network organization imprints on microbial co-occurrence networks. Proc Nat Acad Sci USA. 2014;111:12799–12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RJ, Howe A, Hofmockel KS. Demonstrating microbial co-occurrence pattern analyses within and between ecosystems. Front Microbiol. 2014;5:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SA, Gilbert JA, Leff JW et al. . Consequences of tropical forest conversion to oil palm on soil bacterial community and network structure. Soil Biol Biochem. 2017;112:258–268. [Google Scholar]
- Xie JB, Bai LQ, Wang LY et al. . Phylogeny of 16S rRNA and nifH genes and regulation of nitrogenase activity by oxygen and ammonium in the genus Paenibacillus. Microbiology. 2012;81:702–709. [PubMed] [Google Scholar]
- Zaitsev GM, Tsitko IV, Rainey FA et al. . New aerobic ammonium-dependent obligately oxalotrophic bacteria: description of Ammoniphilus oxalaticus gen. nov., sp. nov. and Ammoniphilus oxalivorans gen. nov., sp. nov. Int J Syst Evol Microbiol. 1998;48:151–163. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xu S, Li C et al. . The soil carbon/nitrogen ratio and moisture affect microbial community structures in alkaline permafrost-affected soils with different vegetation types on the Tibetan plateau. Res Microbiol. 2014;165:128–139. [DOI] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Luo F et al. . Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio. 2011;2:e00122–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
To determine whether differences in the relative abundances of major taxonomic categories between land uses are statistically significant, we used the Mann–Whitney test. Using the same statistical test, we compared the alpha diversities, and pairwise taxonomic distances between samples. Unless otherwise specified, QIIME 1.9.0 and GraphPad Prism 7.00 were used for performing statistical analyses and visualizing the results.
The 16S rRNA gene sequence data has been deposited to Sequence Read Archive (SRA) under BioProject PRJNA490051. The sequence data (SRR7816680-SRR7816689) are available at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP160516.