Abstract
In the present study, we included currently published evidence to comprehensively evaluate the influence of the rs5498 polymorphism within the ICAM1 (intercellular adhesion molecule 1) gene on the genetic risk of multiple sclerosis. STATA 12.0 software was utilized to carry out the heterogeneity assessment, association test, and Begg’s test as well as the Egger’s tests and sensitivity analyses. A total of 11 high-quality case–control studies were selected from the initially retrieved 2209 articles. The lack of high heterogeneity led to the use of a fixed-effect model in all genetic models. The results of the association test showed a reduced risk of multiple sclerosis in the allelic G vs A (Passociation = 0.036, OR = 0.91) and dominant AG+GG vs AA (Passociation = 0.042, OR = 0.85) but not in other genetic models (all Passociation > 0.05). In addition, the negative results were observed in further subgroup analyses based on ethnicity or Hardy-Weinberg equilibrium in all genetic models. Data from Begg’s and Egger’s tests further excluded the presence of remarkable publication bias, while sensitivity analysis data supported stable outcomes. Thus, we conclude that ICAM1 rs5498 may not be related to the risk of multiple sclerosis in Caucasian or Asian populations, which still merits further research.
Keywords: ICAM1, multiple sclerosis, rs5498, risk
Background
Multiple sclerosis is a type of chronic degenerative disease in the central nervous system (CNS) with the features of inflammatory demyelination-induced relapses and progressive neurological disability [1,2]. Multiple sclerosis (MS) mainly contains four phenotypic classifications, namely, relapsing-remitting MS (RRMS), primary progressive MS (PPMS), secondary progressive MS (SPMS), and clinically isolated syndrome (CIS) [3]. As a common autoimmune disease with neurodegeneration, immunological and genetic factors were involved in the initiation and development of multiple sclerosis [4–9]. For instance, as a primary risk allele, HLA DRB1*15 (HLA class II histocompatibility antigen, DRB1 beta chain *15) within the major histocompatibility complex (MHC) was associated with the susceptibility to multiple sclerosis [4]. A number of genetic variants, such as IL7R (interleukin 7 receptor) rs6897932, IL2RA (interleukin 2 receptor subunit alpha) rs2104286 and CD58 (cluster of differentiation 58) rs2300747 polymorphism, may be linked to the risk and progression of multiple sclerosis based on the data of large genome-wide association studies [5]. However, no specific molecular mechanism underlying the clinical course of multiple sclerosis has been uncovered. Herein, we aimed to comprehensively estimate the possible genetic impacts of the rs5498 polymorphism of the ICAM1 (intercellular adhesion molecule 1) gene on the susceptibility to multiple sclerosis.
The ICAM-1 protein, which is also referred to as CD54 (cluster of differentiation 54), participates in a group of biological processes regarding the immune responses [10]. As a member of the immunoglobulin superfamily, some membrane-bound and soluble ICAM-1 isoforms exist [10]. Abnormal cell surface ICAM-1 expression status and soluble ICAM-1 level are related to the pathogenesis of some clinical diseases (e.g. multiple sclerosis, asthma, rhinitis [11–13] etc).
Several polymorphic variants, such as rs5498 A/G (Lys469Glu), rs1799969 G/A (Gly241Arg) and rs5491 A/T (Lys56Met), were reported within the ICAM1 gene on chromosome 19 [14]. Several meta-analyses showed the different role of ICAM1 polymorphisms in some clinical diseases. For example, the ICAM1 rs5498 polymorphism was reported to be linked to a decreased risk of myocardial infarction (MI) [15]. The ICAM1 rs1799969 polymorphism may be associated with the occurrence of Bechet’s disease (BD), rheumatoid arthritis (RA) [16], and cancer [17]. As far as we know, two prior meta-analyses from 2000 [18] and 2003 [19] evaluated the association of ICAM1 rs5498 with the risk of multiple sclerosis; however, different conclusions were obtained. We therefore performed an updated meta-analysis for a comprehensive reassessment based on the available data.
Materials and methods
Database search
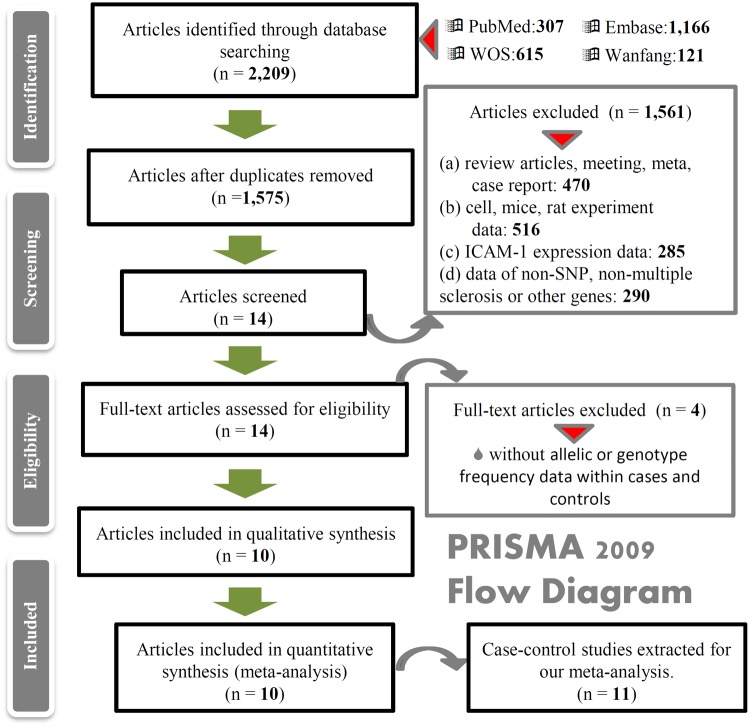
We designed and performed our meta-analysis prior to September 2018, following the preferred reporting items for systematic reviews and meta-analyses (PRISMA). The PRISMA-based analysis process was depicted in Figure 1. Four online databases, namely, PubMed, Embase, Web of Science (WOS) and Wanfang, were employed. The different terms ‘ICAM1’ and ‘multiple sclerosis’ were combined. The search terms in PubMed were as follows: (((((((Multiple Sclerosis) or Sclerosis, Multiple) or Sclerosis, Disseminated) or Disseminated Sclerosis) or MS (Multiple Sclerosis)) or Multiple Sclerosis, Acute Fulminating)) and (((((((Intercellular Adhesion Molecule 1) or Intercellular Adhesion Molecule 1) or ICAM1) or CD54 Antigens) or CD54 Antigen) or Antigen, CD54) or Antigens, CD54).
Figure 1. The selection process of eligible case–control studies.
Article screening
Adhering to the inclusion and exclusion criteria, we carefully screened the articles and finally evaluated the eligible case–control studies. Inclusion criteria: (a) case and control studies; (b) multiple sclerosis; (c) rs5498 polymorphism within the ICAM1 gene; and (d) allelic or genotype frequency data within cases and controls. Exclusion criteria: (a) review articles, meeting, meta, case report; (b) cell, mice, rat experiment data; (c) ICAM-1 expression data; and (d) data of non-SNP, non-multiple sclerosis or other genes.
Data collection
Of the eligible case–control studies, the first author, publication time, ethnicity, polymorphism, genotype frequency of control and case, control source, genotyping method, sample size, and P value of Hardy–Weinberg equilibrium were collected and summarized in the tables. The study quality was appraised using the system of NOS (Newcastle–Ottawa quality assessment Scale).
Statistical methods
Statistical analysis was conducted with STATA 12.0 (Stata Corporation, College Station, TX, U.S.A.). Briefly, we performed the I2-test and Q-statistical test to check the statistical heterogeneity of the studies. High heterogeneity (I2 > 50% or P < 0.05) led to the utilization of the DerSimonian and Laird method under the random-effect model, whereas the absence of remarkable heterogeneity (I2 < 50% or P > 0.05) led to the use of the Mantel–Haenszel method under a fixed-effect model.
Additionally, we computed the OR (odds ratio), 95% CI (95% confidence interval), and P value from the association tests under the allelic, homozygote, heterozygote, dominant, recessive, and carrier models, respectively. We performed the subgroup analyses based on the factors of ethnicity or Hardy–Weinberg equilibrium. We also used Begg’s test and Egger’s test to evaluate the potential publication bias. Sensitivity analysis with the sequential omission of each study was adopted as well to test the data stability.
Results
Eligible case–control studies
After the computerized retrieval of databases, a total of 2209 articles (PubMed with 307 articles, Embase with 1166 articles, WOS with 615 articles and Wanfang with 121 articles) were obtained. We then discarded 634 duplicates and 1561 articles depending on the exclusion criteria (shown in Figure 1). Another four articles without the allelic or genotype frequency data in case or control groups were ruled out. Consequently, we obtained a total of 11 case–control studies from ten articles [18–27] for the summarization of basic information. As listed in Table 1, all studies utilized population-based controls and showed high quality for analysis, in that the score of the NOS system in each study was greater than five.
Table 1. Basic information of the studies included in the meta-analysis.
| First author [reference] | Year | Region | Ethnicity | NOS | AA/AG/GG (control) | χ2 | PHWE | AA/AG/GG (case) | Methods |
|---|---|---|---|---|---|---|---|---|---|
| Killestein [18] | 2000 | Netherlands | Caucasian | 5 | 40/43/23 | 2.97 | 0.09 | 42/68/35 | NR |
| Li [20] | 2008 | China | Asian | 8 | 35/17/3 | 0.24 | 0.63 | 29/16/6 | PCR-LDR |
| Luomala [21] | 1999 | Finland | Caucasian | 7 | 27/59/25 | 0.45 | 0.50 | 34/45/25 | PCR-RFLP |
| Marrosu [22] | 2000 | Sardinia | Caucasian | 8 | 44/59/23 | 0.17 | 0.68 | 49/75/33 | PCR |
| Mousavi [23] | 2007 | Iran | Asian | 7 | 34/80/42 | 0.13 | 0.72 | 45/75/37 | PCR |
| Mycko [24] | 1998 | Poland | Caucasian | 6 | 23/20/25 | 11.50 | <0.05 | 42/23/14 | PCR |
| Nejentsev [19] | 2003 | Finland | Caucasian | 6 | 146/302/125 | 1.77 | 0.18 | 92/119/66 | PCR-RFLP |
| Spain | Caucasian | 6 | 33/47/33 | 3.19 | 0.07 | 49/55/36 | PCR-RFLP | ||
| Qiu [25] | 2013 | Australia | Caucasian | 7 | 318/678* | – | >0.05 | 240/460* | TaqMan |
| Sanadgol [26] | 2011 | Iran | Asian | 7 | 69/36/18 | 10.57 | <0.05 | 49/24/5 | PCR-SSP |
| Shawkatova [27] | 2017 | Slovakia | Caucasian | 8 | 68/101/39 | 0.02 | 0.89 | 75/133/40 | PCR-RFLP |
Abbreviations: HWE, Hardy–Weinberg equilibrium; LDR, ligase detection reaction; NOS, Newcastle–Ottawa Scale; NR, not reported; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; SSP, sequence-specific primers; *, the allelic frequency data of A/G; –, no data.
Meta-analysis result
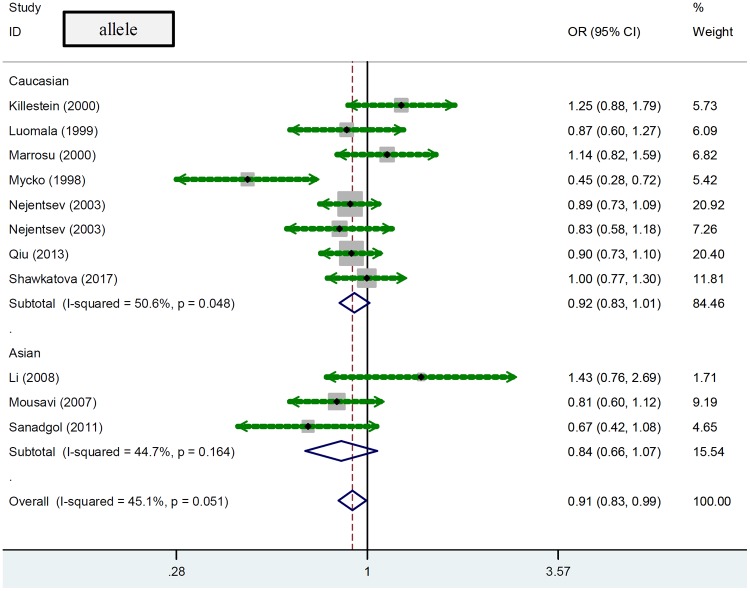
As shown in Table 2, 11 studies comprising 1786 cases and 2137 controls were included in the meta-analysis of allelic G vs A models, while ten case–control studies (1436/1639) were used for other genetic models. The lack of high inter-study heterogeneity (all Pheterogeneity > 0.05 and I2 < 50%) led us to use the fixed-effect model for all genetic models. After pooling the different studies together, we observed a reduced risk of multiple sclerosis in the allelic G vs A (Table 2, Passociation = 0.036, OR = 0.91) and dominant AG+GG vs AA (Passociation = 0.042, OR = 0.85) but not in other genetic models (all Passociation > 0.05), thereby suggesting that ICAM1 rs5498 is not a strong susceptibility locus for multiple sclerosis in the whole population.
Table 2. Meta-analysis of ICAM1 rs5498 and multiple sclerosis risk.
| Genetic model | Sample size | Association analysis | Heterogeneity assessment | |||
|---|---|---|---|---|---|---|
| Study | Case/control | Passociation | OR (95% CI) | Pheterogeneity | I2 | |
| Allelic G vs A | 11 | 1786/2137 | 0.036 | 0.91 [0.83–0.99] | 0.051 | 45.1% |
| GG vs AA | 10 | 1436/1639 | 0.070 | 0.83 [0.68–1.02] | 0.104 | 38.1% |
| AG vs AA | 10 | 1436/1639 | 0.086 | 0.86 [0.73–1.02] | 0.147 | 32.6% |
| AG+GG vs AA | 10 | 1436/1639 | 0.042 | 0.85 [0.73–0.99] | 0.070 | 43.2% |
| GG vs AA+AG | 10 | 1436/1639 | 0.416 | 0.93 [0.78–1.11] | 0.173 | 29.5% |
| Carrier G vs A | 10 | 1436/1639 | 0.287 | 0.94 [0.83–1.06] | 0.660 | 0.0% |
Abbreviations: CI, 95% confidence interval; OR, odds ratio.
Subgroup analysis results
We also conducted a series of subgroup analyses based on the factors of ethnicity or Hardy–Weinberg equilibrium. As shown in Table 3, eight case–control studies (1500/1803) were included for the subgroup analysis of ‘Caucasian’ under the allelic model, while seven case–control studies (1150/1305) were enrolled for other genetic models. There was no statistically significant difference between cases and controls in Caucasian populations (Table 3, Passociation > 0.05). Similarly, no significant association between ICAM1 rs5498 and multiple sclerosis was observed in subgroup analyses of studies with Asian populations or studies with PHWE > 0.05 under any of the genetic models (Table 3, all Passociation > 0.05). Forest plots of subgroup meta-analysis by ethnicity are shown in Figure 2 (allele), Supplementary Figure S1 (homozygote), Supplementary Figure S2 (heterozygote), Supplementary Figure S3 (dominant), Supplementary Figure S4 (recessive), and Supplementary Figure S5 (carrier). As a consequence, ICAM1 rs5498 may not be associated with the risk of multiple sclerosis in Caucasian or Asian populations.
Table 3. Subgroup analysis of ICAM1 rs5498 and multiple sclerosis risk.
| Genetic model | Subgroup | Sample size | Association analysis | ||
|---|---|---|---|---|---|
| N | Case/control | Passociation | OR [95% CI] | ||
| Allelic G vs A | Caucasian | 8 | 1500/1803 | 0.094 | 0.92 [0.83–1.01] |
| Asian | 3 | 286/334 | 0.155 | 0.84 [0.66–0.99] | |
| PHWE > 0.05 | 9 | 1629/1946 | 0.253 | 0.95 [0.86–1.04] | |
| GG vs AA | Caucasian | 7 | 1150/1305 | 0.194 | 0.86 [0.69–1.08] |
| Asian | 3 | 286/334 | 0.131 | 0.68 [0.42–1.12] | |
| PHWE > 0.05 | 8 | 1279/1448 | 0.460 | 0.92 [0.74–1.14] | |
| AG vs AA | Caucasian | 7 | 1150/1305 | 0.132 | 0.87 [0.72–1.04] |
| Asian | 3 | 286/334 | 0.407 | 0.86 [0.59–1.24] | |
| PHWE > 0.05 | 8 | 1279/1448 | 0.131 | 0.87 [0.73–1.04] | |
| AG+GG vs AA | Caucasian | 7 | 1150/1305 | 0.093 | 0.86 [0.72–1.03] |
| Asian | 3 | 286/334 | 0.236 | 0.81 [0.58–1.15] | |
| PHWE > 0.05 | 8 | 1279/1448 | 0.190 | 0.89 [0.76–1.06] | |
| GG vs AA+AG | Caucasian | 7 | 1150/1305 | 0.682 | 0.96 [0.79–1.17] |
| Asian | 3 | 286/334 | 0.293 | 0.80 [0.52–1.22] | |
| PHWE > 0.05 | 8 | 1279/1448 | 0.857 | 1.02 [0.84–1.23] | |
| Carrier G vs A | Caucasian | 7 | 1150/1305 | 0.427 | 0.95 [0.83–1.08] |
| Asian | 3 | 286/334 | 0.422 | 0.89 [0.68–1.18] | |
| PHWE > 0.05 | 8 | 1279/1448 | 0.660 | 0.97 [0.86–1.10] | |
Abbreviations: CI, 95% confidence interval; HWE, Hardy–Weinberg equilibrium; N, number of studies; OR, odds ratio.
Figure 2. Forest plot of subgroup meta-analysis by ethnicity under the allelic model.
Publication bias and sensitivity analysis
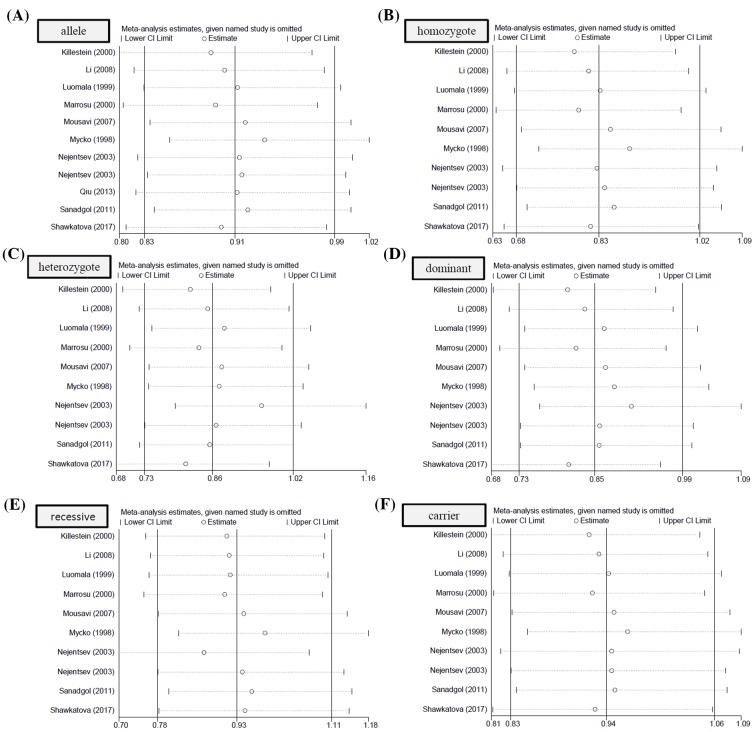
As shown in Table 4, no large publication bias was detected in all the above comparisons (all PBegg > 0.05, PEgger > 0.05). Publication bias plots were presented in Figure 3 (allele), Supplementary Figure S6 (homozygote), Supplementary Figure S7 (heterozygote), Supplementary Figure S8 (dominant), Supplementary Figure S9 (recessive), and Figure S10 (carrier). The relatively stable or credible outcomes were also observed in our sensitivity analyses under all genetic models (Figure 4A–F).
Table 4. Assessment of publication bias.
| Genetic model | Begg’s test* | Egger’s test | ||
|---|---|---|---|---|
| PBegg | z | PEgger | t | |
| Allelic G vs A | 0.755 | 0.31 | 0.758 | -0.32 |
| GG vs AA | 1.000 | 0.00 | 0.867 | -0.17 |
| AG vs AA | 0.858 | 0.18 | 0.577 | 0.58 |
| AG+GG vs AA | 0.474 | 0.72 | 0.825 | 0.23 |
| GG vs AA+AG | 1.000 | 0.00 | 0.442 | -−0.81 |
| Carrier G vs A | 0.858 | 0.18 | 0.661 | -0.46 |
*, continuity corrected.
Figure 3. Publication bias analysis under the allelic model.
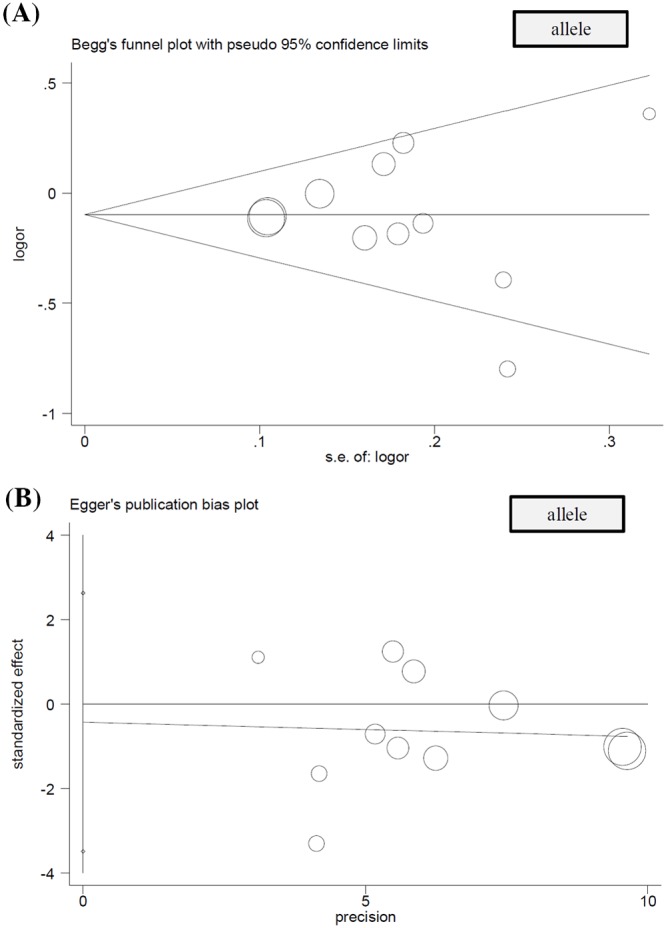
(A) Begg’s test and (B) Egger’s test.
Figure 4. Sensitivity analysis data.
(A) allelic model, (B) homozygote model, (C) heterozygote model, (D) dominant model, (E) recessive model, and (F) carrier model.
Discussion
During the systematic database searches, we evaluated the inconclusive or controversial results on the genetic influence of the ICAM1 rs5498 polymorphism in the occurrence of multiple sclerosis. For example, the ‘G/G’ genotype of the ICAM1 rs5498 polymorphism was reportedly associated with a reduced risk of multiple sclerosis patients in Poland [24] rather than Finland [21]. Additionally, the ICAM1 rs5498 polymorphism was not related to the multiple sclerosis risk in Dutch [18] or Iranian [23,26] populations or in the Han nationality of Henan, China [20]. There is also no statistically significant difference in ICAM1 rs5498 frequencies between multiple sclerosis cases and negative control in the Slovak population [27]. However, the ‘G/G’ genotype may be associated with the development of multiple sclerosis at an earlier age [27]. These data deserve quantitative synthesis and a comprehensive assessment of the role of the ICAM1 rs5498 polymorphism in the susceptibility to multiple sclerosis.
In 2000, Killestein, J. enrolled three case–control studies [18,21,24] to conduct the first meta-analysis by the Mantel–Haenszel method and did not detect the genetic role of ICAM1 rs5498 polymorphism in the risk of multiple sclerosis [18]. In 2003, Nejentsev, S. et al. performed another meta-analysis, which involved five studies [18,19,21,22,24], and reported that the ‘A/A’ genotype of ICAM1 rs5498 may be associated with an enhanced susceptibility to multiple sclerosis [19]. In the present study, we obtained a total of 11 case–control studies from ten eligible articles and performed another updated meta-analysis and the following stratification analysis by ethnicity and Hardy–Weinberg equilibrium, under the models of allelic G vs A, GG vs AA (homozygote), AG vs AA (heterozygote), AG+GG vs AA (dominant), GG vs AA+AG (recessive), and carrier G vs A. Even though a weakly significant difference between multiple sclerosis cases and controls was observed in the overall meta-analysis under the allele and dominant models, no significant association between ICAM1 rs5498 and multiple sclerosis risk was detected in other inheritance models (homozygote, heterozygote, recessive, and carrier) of the overall meta-analysis and in neither of the inheritance models used in subgroup analyses. Hence, our data failed to support a strong connection between ICAM1 rs5498 polymorphism and the susceptibility to multiple sclerosis, which is in line with the conclusion of prior pooling analysis [18].
Several highlights exist in our study. First, population-based negative controls were utilized in all the eligible studies. Second, the results of Begg’s and Egger’s tests excluded the presence of large publication bias. Third, there is no evidence of high heterogeneity between studies in all meta-analyses. Fourth, stable results were detected in our sensitivity analyses. In addition, it is worth mentioning that, during the extraction of the genotype frequency data in the report by Luomala et al. in 1999 [21], we utilized the updated data in 2000 [18].
Nonetheless, we still need to note some disadvantages, which may affect our statistical power. First, as in other meta-analyses, small sample size was a factor in some comparisons. For instance, from 2209 articles, only eleven case–control studies were selected for statistical analysis, and only three case–control studies [20,23,26] were enrolled in the subgroup analysis of ‘Asian’. Less than ten case–control studies were enrolled in the subgroup analysis of ‘Caucasian’. Second, the genotype distribution of control groups in two studies [24,26] deviated from Hardy–Weinberg equilibrium. As a result, no significant association between ICAM1 rs5498 and multiple sclerosis risk was detected in the meta-analysis when only case–control studies with PHWE > 0.05 were enrolled. Third, of the eligible case–control studies, only allelic frequency data were extracted in one study [25]. Fourth, we only investigated the impact of the ICAM1 rs5498 polymorphism in the susceptibility to multiple sclerosis. Very limited data resulted in the failure of the relative meta-analysis concerning the combination of ICAM1 rs5498 and other potential functional variants, such as rs1799969 G/A polymorphism. Fifth, we should consider more adjusted factors (e.g. age, sex, exposure, clinical characteristic, or pharmacotherapy et al.) in the future, when more usable evidence is available.
In conclusion, we incorporated the current data for an updated pooling analysis, which indicated that the ICAM1 rs5498 polymorphism is not linked to the risk of multiple sclerosis in Caucasian and Asian populations. Given the limitations of our study, this negative conclusion still needs to be confirmed by more available evidence.
Supporting information
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
Figure S7.
Figure S8.
Figure S9.
Figure S10.
Abbreviations
- 95% CI
95% confidence interval
- BD
Bechet’s disease
- CD54
cluster of differentiation 54
- CD58
cluster of differentiation 58
- CIS
clinically isolated syndrome
- CNS
central nervous system
- HLA DRB1*15
HLA class II histocompatibility antigen, DRB1 β chain *15
- HWE
Hardy–Weinberg equilibrium
- ICAM1
intercellular adhesion molecule 1
- IL7R
interleukin 7 receptor
- IL2RA
interleukin 2 receptor subunit α
- MHC
major histocompatibility complex
- MI
myocardial infarction
- OR
odds ratio
- PPMS
primary progressive MS
- RA
rheumatoid arthritis
- RRMS
elapsing-remitting MS
- SPMS
secondary progressive MS
- WOS
Web of Science
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
C.J., C.X., and J.F. performed the database search and study screening. C.J., C.X., and M.H. extracted the data and performed the statistical analysis. C.X and C.J. wrote the manuscript. All the authors reviewed and approved the final manuscript.
References
- 1.Hemmer B., Kerschensteiner M. and Korn T. (2015) Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol. 14, 406–419 10.1016/S1474-4422(14)70305-9 [DOI] [PubMed] [Google Scholar]
- 2.Gholamzad M., Ebtekar M., Ardestani M.S., Azimi M., Mahmodi Z., Mousavi M.J.. et al. (2018) A comprehensive review on the treatment approaches of multiple sclerosis: currently and in the future. Inflamm. Res. 10.1007/s00011-018-1185-0 [DOI] [PubMed] [Google Scholar]
- 3.Lublin F.D., Reingold S.C., Cohen J.A., Cutter G.R., Sorensen P.S., Thompson A.J.. et al. (2014) Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 83, 278–286 10.1212/WNL.0000000000000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cree B.A. (2014) Multiple sclerosis genetics. Handb. Clin. Neurol. 122, 193–209 10.1016/B978-0-444-52001-2.00009-1 [DOI] [PubMed] [Google Scholar]
- 5.Didonna A. and Oksenberg J.R. (2015) Genetic determinants of risk and progression in multiple sclerosis. Clin. Chim. Acta 449, 16–22 10.1016/j.cca.2015.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jokubaitis V.G. and Butzkueven H. (2016) A genetic basis for multiple sclerosis severity: red herring or real? Mol. Cell. Probes 30, 357–365 10.1016/j.mcp.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 7.Baranzini S.E. and Oksenberg J.R. (2017) The genetics of multiple sclerosis: from 0 to 200 in 50 years. Trends Genet. 33, 960–970 10.1016/j.tig.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patsopoulos N.A. (2018) Genetics of multiple sclerosis: an overview and new directions. Cold Spring Harb. Perspect. Med. 8, 10.1101/cshperspect.a028951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotsapas C. and Mitrovic M. (2018) Genome-wide association studies of multiple sclerosis. Clin. Transl. Immunol. 7, e1018 10.1002/cti2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos T.N., Bullard D.C. and Barnum S.R. (2014) ICAM-1: isoforms and phenotypes. J. Immunol. 192, 4469–4474 10.4049/jimmunol.1400135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraus J., Oschmann P., Engelhardt B., Schiel C., Hornig C., Bauer R.. et al. (1998) Soluble and cell surface ICAM-1 as markers for disease activity in multiple sclerosis. Acta Neurol. Scand. 98, 102–109 10.1111/j.1600-0404.1998.tb01727.x [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay S., Malik P., Arora S.K. and Mukherjee T.K. (2014) Intercellular adhesion molecule-1 as a drug target in asthma and rhinitis. Respirology 19, 508–513 10.1111/resp.12285 [DOI] [PubMed] [Google Scholar]
- 13.Mitosek-Szewczyk K., Stelmasiak Z., Bartosik-Psujek H. and Belniak E. (2010) Impact of cladribine on soluble adhesion molecules in multiple sclerosis. Acta Neurol. Scand. 122, 409–413 10.1111/j.1600-0404.2010.01330.x [DOI] [PubMed] [Google Scholar]
- 14.Anbarasan C., Bavanilatha M., Latchumanadhas K. and Ajit Mullasari S. (2015) ICAM-1 molecular mechanism and genome wide SNP’s association studies. Indian Heart J. 67, 282–287 10.1016/j.ihj.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P., Dai T., Yu W., Luo Y. and Huang S. (2017) Intercellular adhesion molecule 1 rs5498 polymorphism is associated with the risk of myocardial infarction. Oncotarget 8, 52594–52603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y.H. and Bae S.C. (2016) Intercellular adhesion molecule-1 polymorphisms, K469E and G261R and susceptibility to vasculitis and rheumatoid arthritis: a meta-analysis. Cell. Mol. Biol. 62, 84–90 [DOI] [PubMed] [Google Scholar]
- 17.Cheng D. and Liang B. (2015) Intercellular adhesion molecule-1 (ICAM-1) polymorphisms and cancer risk: a meta-analysis. Iran J. Public Health 44, 615–624 [PMC free article] [PubMed] [Google Scholar]
- 18.Killestein J., Schrijver H.M., Crusius J.B., Perez C., Uitdehaag B.M., Pena A.S.. et al. (2000) Intracellular adhesion molecule-1 polymorphisms and genetic susceptibility to multiple sclerosis: additional data and meta-analysis. Ann. Neurol. 47, 277–279 10.1002/1531-8249(200002)47:2%3c277::AID-ANA29%3e3.0.CO;2-Y [DOI] [PubMed] [Google Scholar]
- 19.Nejentsev S., Laaksonen M., Tienari P.J., Fernandez O., Cordell H., Ruutiainen J.. et al. (2003) Intercellular adhesion molecule-1 K469E polymorphism: study of association with multiple sclerosis. Hum. Immunol. 64, 345–349 10.1016/S0198-8859(02)00825-X [DOI] [PubMed] [Google Scholar]
- 20.Li C.B., H. L., Y. Z.S. and L. Z.W. (2008) The research on the association of the ICAM-1 K469E gene polymorphism with multiple sclerosis in Han nationality of Henan. Shandong Med. J. 48, 12–13 [Google Scholar]
- 21.Luomala M., Elovaara I., Koivula T. and Lehtimaki T. (1999) Intercellular adhesion molecule-1 K/E 469 polymorphism and multiple sclerosis. Ann. Neurol. 45, 546–547 10.1002/1531-8249(199904)45:4%3c546::AID-ANA22%3e3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- 22.Marrosu M.G., Schirru L., Fadda E., Mancosu C., Lai M., Cocco E.. et al. (2000) ICAM-1 gene is not associated with multiple sclerosis in sardinian patients. J. Neurol. 247, 677–680 10.1007/s004150070109 [DOI] [PubMed] [Google Scholar]
- 23.Mousavi S.A., Nikseresht A.R., Arandi N., Borhani Haghighi A. and Ghaderi A. (2007) Intercellular adhesion molecule-1 gene polymorphism in Iranian patients with multiple sclerosis. Eur. J. Neurol. 14, 1397–1399 10.1111/j.1468-1331.2007.01956.x [DOI] [PubMed] [Google Scholar]
- 24.Mycko M.P., Kwinkowski M., Tronczynska E., Szymanska B. and Selmaj K.W. (1998) Multiple sclerosis: the increased frequency of the ICAM-1 exon 6 gene point mutation genetic type K469. Ann. Neurol. 44, 70–75 10.1002/ana.410440113 [DOI] [PubMed] [Google Scholar]
- 25.Qiu W., Pham K., James I., Nolan D., Castley A., Christiansen F.T.. et al. (2013) The influence of non-HLA gene polymorphisms and interactions on disease risk in a Western Australian multiple sclerosis cohort. J. Neuroimmunol. 261, 92–97 10.1016/j.jneuroim.2013.04.022 [DOI] [PubMed] [Google Scholar]
- 26.Sanadgol N., Nikravesh A., Motalleb G., Roshanzamir F., Balazade T., Ramroodi N.. et al. (2011) Evaluation of the association between ICAM-1 gene polymorphisms and sICAM-1 serum levels in multiple sclerosis (MS) patients in Southeast Iran. Int. J. Genet. Mol. Biol. 3, 81–86 [Google Scholar]
- 27.Shawkatova I., Javor J., Parnicka Z., Bucova M., Copikova-Cudrakova D., Michalik J.. et al. (2017) Analysis of ICAM1 gene polymorphism in Slovak multiple sclerosis patients. Folia Microbiol. 62, 287–293 10.1007/s12223-017-0499-6 [DOI] [PubMed] [Google Scholar]