Abstract
Eastern equine encephalitis virus (EEEV) is a mosquito-transmitted alphavirus with a high case mortality rate in humans. EEEV is a biodefense concern because of its potential for aerosol spread and the lack of existing countermeasures. In this study, we identified a panel of 18 neutralizing murine monoclonal antibodies (mAbs) against the EEEV E2 protein, several of which had “elite” activity with 50% and 99% inhibitory concentrations (EC50 and EC99) of less than 10 and 100 ng/ml, respectively. Alanine-scanning mutagenesis and neutralization escape mapping analysis revealed epitopes for these mAbs in domains A or B of the E2 glycoprotein. A majority of the neutralizing mAbs blocked at a post-attachment stage, with several inhibiting viral membrane fusion. Administration of one dose of anti-EEEV mAbs protected mice from lethal subcutaneous or aerosol challenge. These experiments define the mechanistic basis for neutralization by protective anti-EEEV mAbs and suggest a path forward for treatment and vaccine design.
INTRODUCTION
Eastern equine encephalitis virus (EEEV) is a mosquito-transmitted New World alphavirus in the Togaviridae family and is closely related to the Western (WEEV) and Venezuelan (VEEV) equine encephalitis viruses. Although relatively few human infections are reported annually, EEEV is one of the most severe mosquito-transmitted diseases with a 50 to 70% mortality rate and significant brain damage in most survivors1–6. Florida is now considered one of the major sources of EEEV epidemics in the United States, as transmission occurs throughout the year7.
EEEV is an enveloped virus with a 11.5 kb single-stranded, positive-sense RNA genome that generates two RNA transcripts: a full-length genomic RNA and a subgenomic RNA encoding the structural genes, C-E3-E2–6K-E18. After translation, the structural polypeptide C-E3-E2–6K-E1 is cleaved at the endoplasmic reticulum (ER) into the capsid protein and E3-E2–6K-E1. Additional protein processing in the ER and the Golgi results in transport of E2-E1 heterodimers to the plasma membrane9 where encapsidation of the genomic viral RNA occurs. The mature virion surface displays 80 spikes of trimers of E2-E1 heterodimers10. Structural studies of related alphaviruses have established an architecture with T=4 icosahedral symmetry10–12. The E2 glycoprotein projects from the viral surface and is comprised of three domains: A, B, and C11,12. Binding of EEEV E2 to poorly characterized host receptors is believed to initiate entry and endocytosis13. The acidic environment of the endosome induces conformational changes in the alphavirus E1 and E2 glycoproteins, which allow for the exposure of the fusion loop, insertion into the host membrane11, and nucleocapsid escape into the cytoplasm.
Few anti-EEEV monoclonal antibodies (mAbs) have been described14–16 and only one has protective activity in mice17. These anti-EEEV mAbs have been mapped using peptides to three linear epitopes on E2: the N-terminus of domain A, the N- and C-terminal arches of domain B, and the C terminus of domain C14,15. In comparison, the epitopes of several murine and human mAbs against VEEV, WEEV, or the more distantly related arthritogenic alphaviruses (e.g., chikungunya virus [CHIKV]) with therapeutic efficacy in vivo have been mapped8,14,15,18,19. These neutralizing mAbs predominantly recognize epitopes in domains A (residues 58–80) or B (residues 180–215) of the E2 glycoprotein, and inhibit infection at multiple steps including viral attachment, entry, fusion, and egress18–23.
We isolated and purified a panel of murine mAbs against EEEV. Among these, 18 type-specific mAbs neutralized EEEV infection with 50% inhibitory concentration (EC50) values <100 ng/ml and did not bind to WEEV or VEEV. Ten of these mAbs potently inhibited infection with EC50 values <10 ng/ml. In cell culture, most inhibited EEEV predominantly by blocking viral infection at a post-attachment step. We localized the epitopes of the majority of potently neutralizing mAbs to two solvent-exposed regions in domains A and B of the E2 glycoprotein. In vivo studies demonstrated that many of the neutralizing mAbs could protect mice against lethal subcutaneous or aerosol challenges by EEEV. Our results define the molecular basis for neutralization by protective mAbs against EEEV and provide insight into the epitopes that could be targeted for immunotherapy and vaccine development against this highly lethal virus.
RESULTS
Generation of anti-EEEV mAbs.
We hypothesized that antibodies generated in the context of a live EEEV infection might have inhibitory activity. As EEEV is a BSL-3 select agent pathogen, performing B cell-myeloma cell fusions from infected animals has technical challenges. To circumvent these issues, we engineered a chimeric BSL-2 pathogen that incorporated the non-structural genes and RNA replication control elements of a Sindbis virus (SINV, strain TR339) with the structural genes (C-E3-E2–6K-E1) of an EEEV isolate (strain FL93–939) (Supplementary Fig 1a)24. SINV-EEEV replicated efficiently in cell culture but did not cause disease in outbred and Irf3−/− immunodeficient inbred mice (Supplementary Fig 1b-e, and see below).
To enhance the replication and immunogenicity of the attenuated SINV-EEEV in vivo, we inoculated Irf3−/− C57BL/6 mice25. After infection and homologous boosting 4 weeks later, serum from Irf3−/− mice had robust neutralizing activity against SINV-EEEV (endpoint titer > 1/10,000). Splenocytes were harvested from mice, fused to myeloma cells, and 76 hybridomas producing anti-EEEV antibodies were isolated (Fig 1a and Supplementary Table 1). Supernatant from 32 of the 76 hybridomas bound to EEEV virions purified from SINV-EEEV-infected cells and in a single endpoint dilution test, inhibited SINV-EEEV infection by 80% or more (Supplementary Table 1). These 32 mAbs were isotyped (all of the IgG2c or IgG3 subclass) and purified by protein A affinity chromatography for subsequent study.
Figure 1. Characterization of anti-EEEV mAbs.
(a) Supernatant from anti-EEEV hybridoma cells were screened for binding to a mixture of SINV-EEEV infected and uninfected BHK-21 cells by flow cytometry. Shown are antibody staining from representative negative (1A2) and positive (13B10, subcloned as EEEV-10) hybridomas. Data are representative of two independent experiments. (b) Recombinant EEEV E2 (residues 1–338) was refolded, and purified by size exclusion chromatography (left panel), and analyzed by SDS-PAGE under non-reducing and reducing conditions (right panel). Data are representative of two independent experiments. (c) Purified anti-EEEV mAbs were tested for binding to recombinant EEEV E2 protein by ELISA. Data are the mean and standard deviations (SD) of two independent experiments performed in duplicate. (d) HEK-293T cells were transfected with EEEV, VEEV, or WEEV pE2–6K-E1 structural genes and stained with EEEV mAbs, anti-VEEV mAb (3B4C-4), anti-WEEV mAb (WEEV-23), or an isotype control mAb (anti-HCV, H77.39). Data are from three independent experiments.
We evaluated the purified mAbs for their ability to recognize the EEEV E2 protein. To do this, we purified recombinant EEEV E2 protein after expression in bacteria and oxidative refolding (Fig 1b). Notably, 18 of 32 mAbs bound to the recombinant E2 protein in an ELISA (Fig 1c). We also tested a set of 24 mAbs for cross-reactivity with related VEEV or WEEV (55% and 56% amino acid identity in the structural proteins). None of these anti-EEEV mAbs cross-reacted with WEEV or VEEV structural proteins (Fig 1d).
Neutralizing activity of mAbs.
To assess the inhibitory activity of the anti-EEEV mAbs more quantitatively, we performed focus reduction neutralization tests (FRNT) with Vero cells while maintaining mAbs in the medium both before and after virus inoculation (Pre/Post, Fig 2a and c). We determined the concentration of mAb that reduced infection by 50, 90, or 99% (EC50, EC90, or EC99 values, Table 1). Of the 33 mAbs tested, 18 inhibited SINV-EEEV with EC50 values less than 100 ng/ml, and 10 mAbs showed exceptional potency with EC50 values < 10 ng/ml and EC90 values < 100 ng/ml. Four of these mAbs (EEEV-18, EEEV-69, EEEV-82, EEEV-86) had “elite” neutralizing activity with EC99 values < 100 ng/ml.
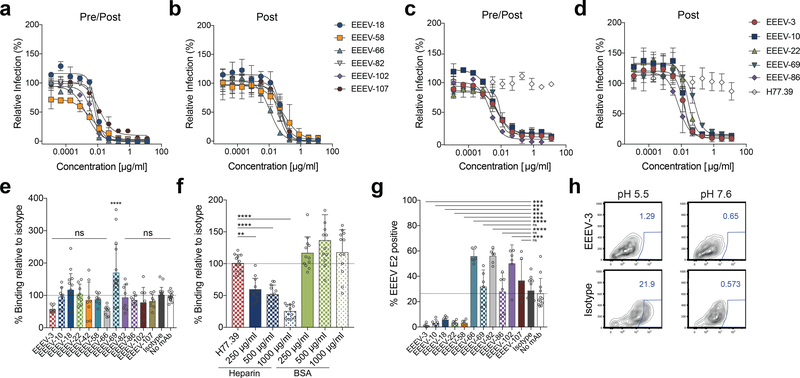
Figure 2. Neutralizing activity of anti-EEEV mAbs.
MAbs mapping to domain A and A/B (a, b) or domain B (c, d) were evaluated for neutralization. (a, c) Pre/post-attachment neutralization assay. Serial dilutions of anti-EEEV mAbs were incubated with SINV-EEEV and then added to a Vero cell monolayer. Infection was allowed to proceed for 18 h at which point infected foci were quantitated. Wells were normalized to infected cells containing no mAb. Data are the mean and SD of two independent experiments each performed in duplicate. (b, d) Post-attachment neutralization assay. SINV-EEEV was allowed to adsorb onto Vero cells at 4ºC. Unattached virus was removed by washing and diluted anti-EEEV mAbs were added. Infection and processing are as described in panels a and c. Data are the mean and SD of two independent experiments each performed in duplicate. (e-f) Attachment inhibition assay. SINV-EEEV was pre-incubated with anti-EEEV mAbs (1 μg/ml) (e), isotype control mAb (1 μg/ml) (e, f), heparin (f), or BSA (f). The virus-mAb complex was then added to Vero cells and incubated for at 4ºC. Cells were washed, and viral RNA was quantitated. Reduction in attachment by anti-EEEV mAbs or heparin was compared to an isotype control mAb (anti-HCV mAb H77.39). Experiments with EEEV-3, EEEV-10, EEEV-22, EEEV-58, EEEV-66, EEEV-82, EEEV-86, EEEV-102, and EEEV-107 are the mean and SD of four independent experiments performed in duplicate. Experiments with EEEV-18 and EEEV-69 data are the mean and SD of eight independent experiments performed in duplicate. The isotype and no antibody control data are the mean and SD of ten independent experiments performed in duplicate (one-way ANOVA with Dunnett’s post-test: *, P < 0.05; **, P < 0.01; *** P < 0.001; ****, P < 0.0001). (g-h) FFWO. SINV-EEEV was adsorbed to BHK-21 cells for 4ºC. Unbound virus was removed, and cells were incubated with anti-EEEV mAbs for at 4ºC. FFWO was induced by subjecting the cells to acidic pH (pH 5.5) and a 37ºC degree pulse. As a negative control, cells were subjected to a physiologically relevant pH (pH 7.6). Subsequently, cells were incubated in medium in the presence of NH4Cl to prevent subsequent endosomal acidification. Fusion inhibition (g) was determined from flow cytometry data (example with EEEV-3 in h) by staining for EEEV E2 positive cells (pH 5.5 condition) and the subtracting the background at pH 7.6 (average of 3.5%). Data with anti-EEEV mAbs are the mean and SD of three independent experiments performed in duplicate. The isotype and no antibody control are the mean and SD of six independent experiments performed in duplicate. Anti-EEEV mAbs were compared to isotype control (one-way ANOVA with Dunnett’s post-test: *, P < 0.05; **, P < 0.01; *** P < 0.001; ****, P < 0.0001).
Table 1.
Profiles of strongly neutralizing antibodies against EEEV
| Mutagenesis Mapping | Neutralization against SINV-EEEV | |||||
|---|---|---|---|---|---|---|
| Antibody | Isotypea | E2 Domainb | E2 Ala/Arg residues which reduced mAb binding | EC50 [ng/ml] | EC90 (ng/ml) | EC99 (ng/ml) |
| EEEV-3 | IgG2c | B | I180, H181 | 5.6 | 53.2 | 619.5 |
| EEEV-5 | IgG2c | A | K74 | 31.8 | 126.1 | 566.3 |
| EEEV-10 | IgG2c | B | I180, H181, S182 | 3.4 | 33.5 | 411.6 |
| EEEV-18 | IgG3 | A/B | M68, G192, A193, Q194, V195, K196, Y197 | 7.7 | 23.2 | 78.1 |
| EEEV-22 | IgG2c | B | I180, H181, S182 | 6.3 | 42.7 | 341.9 |
| EEEV-58 | IgG2c | A/B | K56, T57, D58, G59, D61, M68, K74, S75, L81, G192 | 4.3 | 66.3 | 1302 |
| EEEV-66 | IgG2c | A | D58, L81 | 1.9 | 19.6 | 244.8 |
| EEEV-69 | IgG3 | B | H213, T215 | 9.3 | 17.7 | 35.7 |
| EEEV-82 | IgG3 | A/B | M68, L81, G192 | 6.8 | 17.2 | 47.2 |
| EEEV-86 | IgG2c | B | I180, H181, S182 | 2.2 | 12.5 | 82.1 |
| EEEV-102 | IgG3 | A/B | M68, L81, G192, L227 | 4.3 | 20.3 | 110.7 |
| EEEV-107 | ND | A/B | T57, D58, M68, Q73, S75, L81, G192 | 11.4 | 96.2 | 985.3 |
Immunoglobulin isotype was determined by ELISA. ND = not done.
A indicates domain A; B indicates domain B; and A/B indicates domains A and B.
Antibody neutralization of alphaviruses can occur by inhibiting attachment, internalization, fusion, or by blocking assembly and budding18. To begin to define how the 11 most strongly neutralizing mAbs inhibited infection, we initially assessed whether they blocked virus attachment. Virus-mAb complexes were incubated with Vero cells at 4ºC and after extensive washing, viral RNA adsorbed to cells was detected by qRT-PCR18,26. Notably, the four anti-EEEV mAbs with “elite” neutralizing activity (EEEV-18, EEEV-69, EEEV-82, EEEV-86) did not reduce virus attachment (Fig 2e). A modest (43 to 48%) inhibition of attachment was observed for neutralizing mAbs EEEV-3 or EEEV-66, although statistical significance was not attained. As a positive control, pre-incubation of SINV-EEEV with soluble heparin, whose cell surface analog heparan sulfate, is an attachment factor for EEEV27, diminished virus binding to target cells in a dose-dependent manner (Fig 2f). Incubation with higher concentration of mAbs also failed to reduce virus attachment (Supplementary Fig 2). We next performed post-attachment neutralization assays in which mAbs were incubated with SINV-EEEV after absorption to cells (Post, Fig 2b and d). All of the potently neutralizing mAbs inhibited SINV-EEEV infection when added after virus was bound to cells, suggesting that at least part of their inhibitory activity was at a post-attachment step. We next tested whether our neutralizing mAbs could inhibit viral fusion using a plasma membrane fusion-from-without (FFWO) assay28. After allowing viral attachment to Vero cells at 4ºC, mAbs were added, and plasma membrane fusion was induced by a 37ºC pulse in an acidic (pH 5.5) medium. Subsequently, cells were propagated in medium supplemented with 20 mM NH4Cl to prevent de novo infection via the endocytic pathway and then stained for E2 antigen expression. Five of the mAbs tested (EEEV-3, EEEV-10, EEEV-18, EEEV-22, and EEEV-58) blocked virus plasma membrane fusion (Fig 2g-h). For reasons that remain unclear (see Discussion), EEEV-66, EEEV-82, EEEV-102, and EEEV-107 paradoxically enhanced plasma membrane fusion of the virus.
Epitope mapping by alanine-scanning mutagenesis.
We used alanine-scanning mutagenesis coupled with 293T cell-based expression and flow cytometry19,29 to identify residues in the E2 glycoprotein required for mAb binding (Fig 3a). Cells were transfected with plasmids encoding individual alanine (or serine for alanine residues) substitutions (360 residues) in the E2 gene in the context of a pE2–6K-E1 expression plasmid. We defined critical residues as those with <25% binding to a given individual mAb that retained >70% binding to an anti-EEEV oligoclonal antibody control (Table 1, Supplementary Fig 3, and Supplementary Table 2). We excluded from analysis mutations of cysteine residues and substitutions that globally altered E2 conformation, as defined by reduced binding of an oligoclonal antibody. A majority (13 of 16) of the neutralizing mAbs tested mapped to the “wing insertion” of domain A (residues 52–82) or the distal region of domain B (β-strands A, B, and E)11 of the E2 glycoprotein (Fig 3a-c). The key loss-of-binding residues were highly conserved between the four (I, II, III, and IV) EEEV subtypes (Fig 3a). Although the domain B residues (I180, H181, S182, H213, and T215) required for mAb binding showed clear loss-of-binding phenotypes (Fig 3d), some of the domain A residue changes (e.g., D58, G59, D61, and M68) resulted in only partial loss-of-binding phenotypes (Fig 3e). To extend these findings, we substituted selected residues in the A and B domains with bulkier and charged amino acids that might disrupt mAb interactions to a greater extent. We observed more profound loss-of-binding phenotypes when key domain B residues were substituted with arginine (Fig 3f). Similarly, when the residues in domain A (D58, G59, D61, M68, K74, and L81) were mutated to arginine or glutamic acid, more pronounced loss of mAb binding phenotypes were observed with EEEV-5, EEEV-58, EEEV-66, EEEV-82, EEEV-102, and EEEV-107 (Fig 3g, Supplementary Fig 4, and Supplementary Table 3). Mapping of the domain A and B residues onto the CHIKV E3-E2-E1 glycoprotein complex structure revealed continuous solvent exposed patches in each domain (Fig 3B and C).
Figure 3. Neutralizing mAbs map to domain A or B on the E2 glycoprotein.
(a) Epitope residues of anti-EEEV mAbs identified by alanine-scanning mutagenesis and viral escape are indicated on the EEEV subtype I (strain FL93–939, GenBank EF151502), subtype II (strain BR56-BeAn5122, GenBank AF159559), subtype III (strain PE-0.0155, GenBank DQ241304), and subtype IV (BR85–436087, GenBank AF159561) E2 protein sequences. Anti-EEEV mAbs mapped to domain A or A/B are depicted as circles and mAbs mapped to domain B are depicted as squares. Key domain B (b) and domain A (c) residues necessary for mAb engagement are highlighted in purple on the CHIKV p62-E1 monomer (PDB 3N41) and trimer (PDB 5ANY). The E1 glycoprotein is in gray, E2 glycoprotein is in cyan, and the E1 fusion loop is in orange. Binding data of key domain B (d) and domain A residues (e) identified from alanine-scanning mutagenesis are shown for potently neutralizing mAbs. Binding data of key domain B (f) and domain A residues (g) identified from arginine or glutamic acid mutagenesis are shown for potently neutralizing mAbs. Residues were identified as critical if <25% mAb binding was observed and >70% binding was retained by the oligoclonal EEEV mAb control. Data are the mean and SD from two independent experiments.
Epitope mapping by neutralization escape.
Alanine-scanning mutagenesis failed to map the epitopes of three inhibitory mAbs (EEEV-18, EEEV-82, and EEEV-102). As an alternative approach, we selected for neutralization escape mutants. We passaged SINV-EEEV in the presence of individual neutralizing mAbs until cytopathic effects were observed (3 to 4 passages), at which point the virus became resistant to neutralization. Remarkably, all three viral escape variants were reciprocally resistant to neutralization by the other mAbs in this group suggesting they bound to an overlapping or shared epitope (Fig 4a). To identify the escape mutations, we cloned and sequenced the viral RNA. Unexpectedly, all of the sequenced EEEV-18 escape variants (16 of 16 clones) contained a six-amino acid repeat insertion (192GAQVKY197) in domain B (Fig 4b-c and Supplementary Fig 5). All EEEV-82 escape variant clones (13 of 13 clones) contained a G192R mutation in E2, whereas the EEEV-102 escape variant contained mutations in both domain A (M68T; 3 of 4 clones) and domain B (L227R; 4 of 4 clones) (Fig 4b-c and Supplementary Fig 5). The M68R and G192R mutations were introduced individually into the pE2–6K-E1 plasmid to confirm the loss-of-function phenotype. Mutations in M68R or G192R of the E2 gene resulted in abolished binding of EEEV-18, EEEV-82, and EEEV-102 to cells transfected with the pE2–6K-E1 expression plasmid (Fig 4d). When the M68T, G192R, and L227R mutations were introduced into the SINV-EEEV infectious cDNA clone, the resultant viruses showed diminished neutralization by EEEV-18, EEEV-82, and EEEV-102 (Fig 4e). Finally, we tested whether the four neutralization escape variants were resistant to inhibition by the remaining potently neutralizing mAbs. Although all of the strongly neutralizing domain B mAbs (EEEV-3, EEEV-10, EEEV-22, EEEV-69, and EEEV-86) completely neutralized the escape variants with EC50 values similar to parental virus, domain A (EEEV-5 and EEEV-66) and domain A/B (EEEV-18, EEEV-58, and EEEV-107) mAbs failed to neutralize the escape variants as efficiently (Supplementary Fig 6).
Figure 4. Characterization of EEEV mAb escape mutants.
(a) Neutralization escape virus pools were tested for sensitivity to the mAbs used for selection. Serially diluted mAbs and 102 FFU of each passaged virus were incubated for 1 h and then added to Vero cell monolayers. Sixteen hours later, viral antigen containing foci were stained, and infection was normalized to infected wells containing no mAb. Data are the mean and SD of two independent experiments performed in duplicate. (b-c) Neutralization escape mutations were identified by Sanger sequencing. EEEV-18, EEEV-82, and EEEV-102 escape mutations are mapped onto the CHIKV p62-E1 (b) trimer (PDB 5ANY) and (c) monomer structure (PDB 3N41). The E1 glycoprotein is in gray, E2 glycoprotein is in cyan, and the E1 fusion loop is in orange. (d) Neutralization escape mutations were engineered into a structural gene (C-E3-E2–6K-E1) vector and expressed in HEK-293T cells. Cells were stained using the selection mAb and analyzed by flow cytometry. Data are the mean and SD from three independent experiments, with the exception of EEEV-18 (four experiments). (e) Escape mutations were engineered into the SINV-EEEV infectious cDNA clone. Mutant viruses were generated and tested for sensitivity to the mAbs used for selection (EEEV-18, EEEV-82, and EEEV-102) and a domain B mAb (EEEV-3). Data are the mean and SD of two independent experiments performed in duplicate.
MAb protection in mice.
We assessed whether the mAbs could confer protection against EEEV infection in vivo (Fig 5). We tested a subset of mAbs with differing neutralization potencies using a lethal challenge model in five week-old CD-1 mice with a highly pathogenic EEEV (strain FL93–939) engineered to express nanoluciferase (nLuc) with little effect upon virulence30. Mice received a single 100 μg (5 mg/kg) dose of EEEV mAbs via the intraperitoneal route either before (−24 h) or after (+ 24 h) subcutaneous (103 PFU of EEEV) or aerosol (50 to 100 x lethal dose 50%, LD50) inoculation of EEEV. Mice treated with neutralizing anti-EEEV mAbs (EEEV-3, EEEV-22, EEEV-43, EEEV-58, EEEV-73, EEEV-82, and EEEV-86; EC50 values of 2.2 to 761 ng/ml) prior to subcutaneous challenge had 80–100% survival rates, whereas administration of EEEV-26B, a poorly neutralizing mAb (EC50 >12,500 ng/ml) showed little protection (Fig 5a). When mice were subjected to a subcutaneous challenge and administered a single dose of mAb 24 h after infection (Fig 5b) most neutralizing mAbs (EEEV-3, EEEV-18, EEEV-43, EEEV-58, EEEV-73, and EEEV-82) exhibited moderate to high levels of protection (40–100% survival rates) whereas EEEV-22, EEEV-86, and EEEV-26B exhibited less protection. Unexpectedly, the modestly neutralizing EEEV-43 mAb (EC50 of 761 ng/ml) still conferred protection (70% survival rate) when administered as post-exposure therapy in this model. Additions of mAb combinations targeting domain A (EEEV-18) and domain B (EEEV-3) and subcutaneous challenge resulted in 100% protection as prophylaxis and 75% protection as post-exposure therapy (Fig 5a-b).
Figure 5. Anti-EEEV mAbs exhibit in vivo protection.
Five week-old female CD-1 mice were administered 100 μg of indicated mAbs via an intraperitoneal route either as prophylaxis (−24 h, left panels) or therapeutically (+24 h, right panels) and then challenged with EEEV FL93–939 via (a-b) subcutaneous (103 FFU) or (c-d) aerosol (50–100 LD50) route. Isotype control (a, n = 10; b, n = 20; c, n = 29; d, n = 22), EEEV-3 (a, n = 10; b, n = 10; c, n = 13; d, n = 13), EEEV-5 (c, n = 10; d, n = 10), EEEV-18 (b, n = 25; c, n = 40; d, n = 26), EEEV-22 (a, n = 10; b, n = 10; c, n = 5; d, n = 5), EEEV-26B (a, n = 10; b, n = 10; c, n = 10; d, n = 10), EEEV-43 (a, n = 10; b, n = 10; c, n = 5; d, n=5), EEEV-58 (a, n = 10; b, n = 10; c, n = 10; d, n = 10), EEEV-69 (c, n = 10; d, n = 10), EEEV-73 (a, n = 10; b, n = 10; d, n = 5), EEEV-82 (a, n = 10; b, n = 20; c, n = 15; d, n = 5), EEEV-86 (a, n = 10; b, n = 20; c, n = 15; d, n = 5), EEEV-3 + EEEV-18 (a, n = 16; b, n = 16; c, n = 16; d, n = 16); (one-sided log-rank test with Bonferroni multiple comparison correction: *, P < 0.05; **, P < 0.01; ****, P < 0.0001). (e) Four or five days post-infection, an in vivo imaging system (IVIS) was used to visualize EEEV-luciferase in mice that received prophylactic treatment and were challenged via an aerosol route. The total flux (photons/second) were quantified in the head region of each animal (Isotype, n = 5; EEEV-3, n = 7; EEEV-18, n = 20; EEEV-82, n = 4; EEEV-86, n = 4. (one-way ANOVA with Dunnett’s post-test: *, P < 0.05; **, P < 0.01; *** P < 0.001).
As EEEV is also highly pathogenic via an aerosol route, we examined the efficacy of the mAbs upon an aerosol challenge with 50–100 LD50 of EEEV FL93–939. Among the mAbs tested, a majority (EEEV-3, EEEV-5, EEEV-18, EEEV-58, and EEEV-82) protected against death (70–100% survival) when administered as prophylaxis (Fig 5c). Administration of mAb combination (EEEV-3 + EEEV-18) as prophylaxis resulted in a 94% survival rate (Fig 5c). In vivo imaging of mice treated with mAbs EEEV-3, EEEV-18, EEEV-82, and EEEV-86 but not the isotype control mAb, showed marked reductions in viral replication as judged by decrease in light signal 4 days post infection (Fig 5e). However, in the most stringent model of protection, post-exposure therapy at one day after aerosol challenge, lower survival rates (10–20%) were observed with individual neutralizing mAbs EEEV-3, EEEV-5, EEEV-18, EEEV-22, EEEV-58, EEEV-69, EEEV-82, and EEEV-86 or a combination of neutralizing mAbs (EEEV-3 + EEEV-18) (Fig 5d).
DISCUSSION
EEEV is a highly pathogenic, encephalitic alphavirus that lacks approved vaccines or therapies. We generated a panel of 76 mAbs that bound to EEEV-infected cells, including 18 strongly neutralizing mAbs. Ten of the 18 mAbs exhibited potent neutralizing activity with EC50 values of <10 ng/ml. Mapping studies show that these strongly neutralizing mAbs principally recognized epitopes in domains A and/or B of the E2 glycoprotein. Mechanism of action studies revealed that the most of the inhibitory mAbs blocked infection at a post-attachment stage, with a subset inhibiting viral fusion. Many of the neutralizing mAbs had protective activity against EEEV in vivo, as judged by outcome in lethal subcutaneous and aerosol challenge models in mice.
Although prior studies have generated mAbs against the EEEV proteins, these mAbs either lacked neutralization activity or were not characterized extensively due to biosafety limitations14–17. One cross-reactive, non-neutralizing anti-EEEV mAb that was evaluated had moderate protective efficacy (~50%) against VEEV challenge in mice17. Presumably, Fc effector functions contributed to the protection against VEEV by this mAb, as has been postulated for non-neutralizing antibodies against other arthritogenic alphaviruses, including Semliki Forest virus31 and CHIKV22. Whereas others have immunized mice with recombinant EEEV E2 protein or inactivated EEEV to obtain mAbs15,16, we speculate that we obtained a large number of neutralizing mAbs because mice were immunized with a replicating virus that displayed EEEV structural proteins in their native form. At present, it remains unclear why we obtained only type-specific neutralizing mAbs.
Neutralizing antibodies against alphaviruses inhibit infection at several stages in the viral replication cycle including attachment, entry, fusion, or egress. Our most inhibitory neutralizing mAbs to E2 domains A and/or B did not block viral attachment to cells, but instead inhibited infection at a post-attachment stage. Plasma membrane fusion assays showed these several of these mAbs block pH-dependent fusion with membranes. Among the mAbs tested that inhibited at a post-attachment step, generally, those recognizing epitopes in the domain B (EEEV-3, 10, 22, 69, and 86) showed less potency when antibody was added after the virus attached to cells. A previous study with domain B mAbs against CHIKV suggested that bivalent engagement of the virion was necessary for potent neutralization18. It is possible that the anti-EEEV mAbs also may require bivalent engagement for complete neutralization; this mode of recognition may be technically difficult to achieve once the virion has attached to cells because some epitopes are unavailable for binding. One of the neutralizing mAbs, EEEV-69, paradoxically increased virus attachment to Vero cells, and unexpectedly, increased plasma membrane fusion was observed with EEEV-66, EEEV-82, EEEV-102, and EEEV-107. These results are analogous to prior reports with anti-VEEV and anti-SINV mAbs, both of which increased attachment by stabilizing the interaction between the virus and cells32,33. The increase in fusion could be due to antibody-induced exposure of cryptic epitopes that facilitates virus binding to the plasma membrane, a mechanism previously reported with a flavivirus34. This phenomenon may not impact the neutralizing activity of these mAbs if (a) neutralization occurs at a stage in the entry pathway before fusion or (b) plasma membrane fusion is not equivalent to endosomal fusion.
Some reports have speculated that domains A and B on the E2 glycoprotein contain a site of receptor engagement for multiple alphaviruses11–13. A recent study mapped the binding site of Mxra8, a receptor for several arthritogenic alphaviruses, to residues within the A and B domains on CHIKV E2 protein24. Using a combination of alanine-scanning and targeted mutagenesis of E2 and neutralization escape selection, we mapped the epitopes for neutralizing anti-EEEV mAbs to residues within these domains. Regions in E2 domains A and B have been implicated as epitopes for neutralizing mAbs against other alphaviruses including VEEV, CHIKV, SINV, and Ross River virus18,22,35–37. Our most potently neutralizing mAbs (EEEV-5, EEEV-58, EEEV-66, EEEV-82, EEEV-102, and EEEV-107) recognize an epitope in the “wing region” (residues 51–81) on E2, a solvent exposed site at the distal tip of the A domain11. The neutralizing mAbs that mapped to domain B preferentially bound to two epitopes at residues 180–182 (EEEV-3, EEEV-10, EEEV-21, EEEV-22, and EEEV-86) or residues 213–215 (EEEV-4, EEEV-19, EEEV-21, EEEV-60, and EEEV-69). Cryo-EM studies with two neutralizing anti-VEEV mAbs (F5 and 3B4C-4) showed binding to sites proximal to and within the wing region of domain A (residues 73–120) or to residues (177–223) in domain B, respectively38. These mAbs are thought to neutralize VEEV infection by preventing the structural rearrangements required for fusion.
Through neutralization escape selection, we also mapped neutralizing mAbs (EEEV-18, EEEV-58, and EEEV-102) to residues spanning domains A and B (residues 68, 192–197, and 227). We note that the corresponding M68 residue on the CHIKV p62-E1 structure is located beneath the β-strand i6 (residues 74–79) and is not solvent exposed11. Residue M68 is tightly packed against residue L81, a key binding residue for mAbs EEEV-58, EEEV-66, EEEV-82, EEEV-102, and EEEV-107. We hypothesize that the mutation of either residue (M68 or L81) perturbs the conformational display of the domain A “wing region” epitope. Mutation of the solvent-exposed residue G192 markedly reduced binding and neutralization of mAbs EEEV-18, EEEV-58, and EEEV-102. In the CHIKV p62-E1 structure, the distance between residues M68 and G192 is ~28 Å11. This distance is sufficient for engagement by a Fab molecule, as the antigen binding site spans ~35 Å.
We also assessed whether the escape variants selected against domain A/B mAbs were susceptible to inhibition by the remaining potently neutralizing mAbs. The domain B mAbs (EEEV-3, EEEV-10, EEEV-22, EEEV-69, and EEEV-86) showed no loss in neutralization potency against the escape variants. However, four potently inhibitory mAbs, EEEV-5 (domain A), EEEV-58 (domain A/B), EEEV-66 (domain A), and EEEV-107 (domain A/B), showed reduced ability to neutralize the escape variants. The domain A-specific mAbs EEEV-5 and EEEV-66 did not neutralize the EEEV-18 and EEEV-102 escape variants, and the domain A/B-specific mAbs EEEV-58 and EEEV-107 failed to neutralize all three escape variants. Although we speculate that the binding site of EEEV-66 may be similar to or overlap that of mAbs EEEV-18, EEEV-82, and EEEV-102, higher resolution structural studies (e.g., x-ray crystallography or cryo-electron microscopy) will be required to determine the precise antibody footprints.
This composite A-B domain epitope, which bridges the two domains, is analogous to the site recognized by the neutralizing anti-CHIKV mAb (CHK-265), which binds and crosslinks these domains on adjacent spikes on the virion surface18. The cross-linking of two E2 subunits by CHK-265 restricts the domain B from undergoing conformational changes and prevents the exposure of the fusion loop located underneath in the E1 subunit. A similar mechanism may occur with the strongly neutralizing EEEV mAbs EEEV-18, EEEV-82, EEEV-102, and EEEV-107.
Several of our highly neutralizing mAbs showed substantial protective efficacy when mice were challenged with EEEV by a subcutaneous or aerosol route. In the lethal subcutaneous challenge models, mAb protection correlated most consistently with potent neutralization activity and binding to residues spanning domains A and B of the E2 protein (EEEV-18, EEEV-58, and EEEV-82). One strongly neutralizing domain B mAb (EEEV-3) also protected efficiently in these models. Most of these mAbs (EEEV-3, EEEV-18, and EEEV-58) neutralized infection at a post-attachment stage and efficiently blocked viral plasma membrane fusion. Unexpectedly, EEEV-43, a weakly neutralizing mAb (EC50 of 761 ng/ml), and EEEV-73 (EC50 of 49.7 ng/ml), a moderately neutralizing mAb, both protected when administered as prophylaxis or therapy. Analogously, a non-neutralizing anti-EEEV mAb protected against subcutaneous EEEV challenge in mice when administered one day before infection17. Although further studies are warranted, we speculate that Fc effector functions may contribute to the in vivo efficacy of weakly to moderately neutralizing protective mAbs. Alternatively, the neutralization assays with Vero cells may not fully reflect the inhibitory activity against cell targets in vivo.
The post-exposure mAb therapy trials in the context of aerosol challenge of mice showed limited efficacy. After aerosol challenge, encephalitic alphaviruses rapidly enter the brain from the olfactory neuroepithelium via olfactory neurons39,40. The treatment failure we observed in the context of aerosol challenge could be due to one of several reasons: (a) the virus spreads rapidly to the brain via olfactory neurons whereas antibody entry is limited by the blood-brain barrier41,42; (b) the combination of high levels of virus and limiting amounts of a single mAb in the brain may result in rapid neutralization escape. Indeed, the use of a single neutralizing anti-CHIKV mAb promoted escape variants in vivo22,43. However, as combination therapy with highly neutralizing domain A and domain B-reactive antibodies failed to improve clinical outcome after aerosol challenge, virus entry into the brain may represent a point after which mAb therapy has limited efficacy against EEEV in mice.
Currently, there are no approved vaccines against EEEV. Vaccine efforts against HIV, hepatitis C virus, and influenza virus focus on eliciting neutralizing antibodies to protective epitopes on viral envelope proteins through “reverse vaccinology”44–46. Our study identities specific epitopes on the E2 glycoprotein that can be engaged by potently neutralizing EEEV mAbs. Studies are planned to apply this information to the next generation of vaccine design against EEEV and other encephalitic alphaviruses.
METHODS
Animal ethics statement.
All animal procedures were carried out in accordance with AAALAC-approved institutional guidelines for animal care and use and approved by IACUC at the University of Pittsburgh and Washington University School of Medicine. Injections were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize suffering.
Cell lines and plasmids.
Vero, HEK-293T, and BHK-21 cells were obtained from the American Type Culture Collection and propagated in DMEM supplemented with 5% (Vero and BHK-21) or 10% (HEK-293T) FBS (Omega Scientific), 100 U/ml penicillin, 100 μg/ml streptomycin and 10 mM HEPES. All cell lines were tested and judged free of mycoplasma contamination using a commercial kit. The plasmids pKR780–2-EEEV, pKR780–2-VEEV, and pKR780–2-WEEV are comprised of the codon-optimized pE2–6K-E1 genes of EEEV FL93–939, VEEV TrD, and WEEV CB87, respectively, under the control of a chicken β-actin promoter, which have been cloned into the pCAGGS expression vector (Addgene). Replication-competent SINV chimeric viruses were constructed by replacing the SINV TR339 structural protein genes with EEEV FL93–939 structural protein genes under control of the SINV subgenomic promoter in the TR339 cDNA clone47. The cDNA clones of EEEV TrD FL93–939 WT and nanoluciferase-expressing challenge viruses have been described30,48.
Virus production.
All viruses were produced by plasmid linearization, in vitro transcription with Sp6 or T7 DNA-dependent RNA polymerase, and electroporation into BHK-21 cells. Virus mutants were generated using a QuikChange II XL Site-Directed Mutagenesis kit (Agilent) and verified by DNA sequencing. Virus supernatant (p0) was passaged in Vero cells and harvested 24 to 36 h after infection. Supernatant was overlaid onto a 20% sucrose gradient and concentrated at 30,000 rpm for 2 h using a SW32Ti rotor (Beckman Coulter). Viral pellets were resuspended in PBS and stored at −80ºC. Virus titer was determined by focus-forming or plaque assay.
MAb generation.
Six week-old Irf3−/− C57BL/6 female mice were infected and boosted with 105 FFU of SINV-EEEV and given a final intravenous boost with 106 FFU of SINV-EEEV three days prior to fusion with myeloma cells. Hybridomas that secreted antibodies reacting with SINV-EEEV-infected BHK-21 cells were identified by flow cytometry and cloned by limiting dilution. MAbs were isotyped by ELISA (Pierce) and hybridomas were sent for commercial preparation and purification by protein A affinity chromatography (Bio X Cell). All mAbs were screened initially with a single endpoint neutralization assay using neat hybridoma supernatant (∼10 μg/ml), which was incubated with 102 FFU of SINV-EEEV for 1 h at 37°C. MAb-virus complexes were added to Vero cell monolayers in 96-well plates. After 90 min, cells were overlaid with 1% (w/v) methylcellulose in Modified Eagle Media (MEM) supplemented with 2% FBS. Plates were harvested 18 h later and fixed with 1% PFA in PBS. The plates were incubated sequentially with murine mAb EEEV-10 and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG in PBS supplemented with 0.1% saponin and 0.1% BSA. SINV-EEEV-infected foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 Macroanalyzer (Cellular Technologies Ltd). Non-linear regression analysis was performed after comparison to wells infected with SINV-EEEV in the absence of mAb.
Protein expression and purification.
Residues 1–338 encoding the E2 gene of EEEV (strain FL93–939) were cloned into the pET-28a E. coli expression vector and transformed into BL21(DE3) chemically competent cells (Thermo Fisher). Cells were grown at 37ºC in LB to an OD600 of 0.9 and then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h. Bacteria were harvested, resuspended in 50 mM Tris-HCl, 1 mM EDTA, 0.01% NaN3, 1 mM DTT, 25% sucrose (TENDS), and lysed in 50 mM Tris-HCl, 1 mM EDTA, 0.01% NaN3, 1 mM DTT, 200 mM sodium chloride, 1% sodium deoxycholate, 1% Triton X-100. Inclusion bodies were obtained after centrifugation (6,000 x g for 30 min) and then washed in TENDS buffer supplemented with 100 mM NaCl and 0.5% Triton X-100. After a final wash in the same buffer without 0.5% Triton X-100, ~200 mg of inclusion bodies were denatured in 100 mM Tris-HCl, 6 M guanidinium hydrochloride and 20 mM β-mercaptoethanol for 1 h. Solubilized protein was refolded overnight at 4ºC into a buffer containing 400 mM L-arginine, 100 mM Tris-HCl, 5 mM reduced gluthathione, 0.5 mM oxidized gluthathione, 10 mM EDTA and 200 mM phenylmethylsulfonylfluoride. Refolded protein was concentrated using a 10 kDa molecular weight cut-off stirred cell concentrator (EMD Millipore) and purified by HiLoad 16/600 Superdex 75 size exclusion chromatography (GE Healthcare).
ELISA.
Recombinant E2 protein (5 μg/ml) was immobilized onto Maxisorp ELISA plates (Thermo Fisher) overnight in sodium bicarbonate buffer, pH 9.3. Plates were washed three times with PBS/0.05% Tween-20 and blocked with 5% BSA/PBS for 1 h at 37ºC. Anti-EEEV mAbs were diluted in 2% BSA in PBS and incubated for 1 h at room temperature. After serial washing, horseradish peroxide conjugated goat anti-mouse IgG (H+L) (1:2000 dilution), Jackson ImmunoResearch) was added and incubated for 1 h at room temperature. After washing, plates were developed with 3,3′−5,5′ tetramethylbenzidine substrate (Dako), the reaction was stopped with 2 N H2SO4 and absorbance was read at 450 nm with a TriStar Microplate Reader (Berthold). For virus capture ELISA, ultracentrifuged SINV-EEEV virions were immobilized directly onto Maxisorp ELISA plates for 1 h at room temperature. Virus ELISA were performed similarly as above but Tween-20 detergent was omitted from the wash buffer.
Expression of wild-type or mutant structural proteins.
Alanine-scanning mutagenesis was performed on EEEV E2 residues 1–360 with alanine residues mutated into serine. EEEV E2 alanine mutants that exhibited a partial loss-of-binding phenotype (residues 56–62, 64, 68, 73–79, 81, 192, 180–182, 212–213, and 215) were substituted with arginine residues. For residues with positive charges (K56 and K74), a glutamic acid substitution was made. Plasmids containing the codon-optimized EEEV, VEEV, or WEEV pE2–6K-E1 structural proteins or EEEV E2 alanine mutants were transfected in HEK-293T cells using Lipofectamine 3000 (Thermo Fisher). Sixteen hours post-transfection, cells were washed with PBS and fixed with Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher). Cells were washed twice with PBS followed by another wash with permeabilization buffer (Thermo Fisher). Cells were stained with anti-EEEV mAbs at 1 μg/ml in permeabilization buffer and incubated for 1 h at 4ºC. For cross-reactivity studies, anti-VEEV mAb 3B4C-420 and anti-WEEV mAb (WEEV-23; S.K.A. and M.S.D., unpublished results) were used as positive controls. After two washes with permeabilization buffer, antibodies were detected with Alexa Fluor 647 conjugated goat anti-mouse IgG (1:2000 dilution, Thermo Fisher). After two washes, cells were resuspended in 100 μl of permeabilization buffer and analyzed on a MACSQuant Analyzer (Miltenyi Biotec). Using previously published criteria, alanine mutants with <25% reactivity compared to wild-type that exhibited >70% reactivity to a polyclonal anti-EEEV mAb cocktail were deemed as key binding residues49.
Generation of virus escape mutants.
To generate neutralization escape mutants, SINV-EEEV (1.2 × 105 FFU) were incubated with 1 μg/ml of EEEV mAbs for 1 h at 37ºC. The virus-mAb complexes were added to Vero cells. One day post infection, half of the virus supernatant was incubated with 1 μg/ml of EEEV mAbs for 1 h at 37ºC and added to new Vero cells. The remaining half of the supernatant was frozen at −80ºC. This process was repeated for 9 days. Escape mutants were confirmed by focus forming neutralization assays. Viral RNA was isolated from bulk virus supernatant pools using a QIAamp Viral RNA Mini Kit (Qiagen) and cDNA was generated with an Oligo(dT)20 primer using the SuperScript III Reverse Transcriptase Kit (Thermo Fisher). Viral structural genes were amplified using the forward primer 5′-ATGTGCGTCCTGGCCAATATCACGTTTCC-3′ and the reverse primer 5′-GAACAAAACTAGGGCAACCACTGCTGTAGC-3′. The amplified structural genes were sequenced using four primer sets. Escape mutations were introduced into pKR780–2-EEEV containing the codon-optimized pE2–6K-E1 genes of EEEV FL93–939, expressed in HEK-293T cells, stained with anti-EEEV mAbs, and analyzed by flow cytometry as described above.
Mapping of mutations onto the CHIKV p62-E1 crystal structure.
Figures were prepared using the atomic coordinates of CHIKV p62-E1 monomer (PDB 3N41) and trimer (PDB 5ANY) using the program PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC).
Attachment inhibition assays.
Vero cells were seeded at 3 × 105 cells/well 24 h prior to assay. Anti-EEEV mAbs, heparin (Sigma, H3393), and BSA (Sigma) were diluted to specified concentrations and incubated for 1 h at 37ºC with SINV-EEEV at an MOI of 0.01. The mAb-virus complex was then chilled to 4ºC and added to pre-chilled Vero cells for 1 h at 4ºC. After six washes with chilled PBS, RNA was extracted using a RNeasy Mini Kit (Qiagen). EEEV RNA levels were determined using a Taqman RNA-to-Ct 1-Step Kit (Thermo Fisher) and a E2 specific primer/probe set26. EEEV RNA levels were normalized against GAPDH, and the relative fold change was compared to cells treated with an isotype control mAb.
Pre/post- and post-attachment neutralization assays.
Pre/post-attachment neutralization assays were performed by first incubating diluted anti-EEEV mAbs with 102 FFU of SINV-EEEV for 1 h at 37ºC. The mAb-virus complex was then added to Vero cells for 1.5 h at 37ºC. Cells were overlaid with 1% (w/v) methylcellulose in Modified Eagle Medium (MEM) supplemented with 2% FBS. Post-attachment neutralization assays were performed by first incubating Vero cells with 102 FFU of SINV-EEEV for 1 h at 4ºC. Cells were washed extensively with cold DMEM to remove unbound virus. Diluted anti-EEEV mAbs were added to virus adsorbed cells and incubated for 1 h at 4ºC. After a 15 min incubation at 37ºC to allow virus internalization, cells were overlaid with methylcellulose as previously described. Pre/post- and post-attachment neutralization assays were processed similarly to the single endpoint neutralization assay described above.
Fusion inhibition assays.
FFWO assays were performed by first allowing viral adsorption to BHK-21 cells (MOI 25) for 1 h at 4ºC. Unbound virus was removed by washing with chilled PBS. Diluted mAbs (50 μg/ml) were added to virus adsorbed cells for 30 min at 4ºC. Cells were washed with chilled PBS. FFWO was induced by pulsing with fusion medium (RPMI 1640, 10 mM HEPES, 0.2% BSA, and 30 mM succinic acid, pH 5.5) for 2 min at 37ºC. A non-fusion control was included using control media (RPMI 1640, 10 mM HEPES, 0.2% BSA, pH 7.6). After the 37ºC pulse, cells were washed twice with chilled PBS and incubated in DMEM supplemented with 5% FBS, 10 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin and 20 mM NH4Cl to prevent infection via endocytosis. Infection was allowed to proceed for 5 h and cells were detached and fixed with Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher). Cells were stained with human mAb EEEV-53 (L.E.W. and J.E.C, unpublished results) at 1 μg/ml in permeabilization buffer and incubated for 1 h at 4ºC. After two washes with permeabilization buffer, viral antigen was detected with Alexa Fluor 647 conjugated goat anti-human IgG (1:2000 dilution, Thermo Fisher). After two washes with permeabilization buffer, cells were resuspended in 100 μl and analyzed on a MACSQuant Analyzer (Miltenyi Biotec).
Mouse protection studies.
Five week-old female CD-1 mice (Charles River Laboratories) were administered 100 μg of anti-EEEV mAb or isotype control mAb via an intraperitoneal route 24 h pre- or post-challenge. For combined antibody testing, 100 μg of each antibody was given as described above. Mice were challenged with WT EEEV FL93–939 or a nanoluciferase-expressing version30 via a subcutaneous route (103 PFU) or an aerosol route (50–100 LD50). Aerosol exposures were performed as previously described50 using the AeroMP exposure system (Biaera Technologies) inside a class III biological safety cabinet. Infected mice were observed at 24 h intervals through 21 days post infection and at each time, mice were weighed and mortality was assessed. At 5 days post-challenge, some mice were injected with 10 μg Nano-Glo substrate (Promega) subcutaneously and imaged using the IVIS Spectrum CT instrument (Perkin Elmer) on the auto-exposure setting at 4 minutes post-substrate injection. The total flux (photons/second) in the head region, taken as a measure of brain replication, was calculated for animals in each treatment group based on the radiance (photons/second/cm2/steradian) and was quantified using Living Image Software (Perkin Elmer). The dynamic range of the IVIS imager signal from the heads of uninfected mice to highly infected mice was approximately one hundred-fold (~1–2 × 105 photons/second to ~1–2 × 107 photons/second, respectively). Samples sizes were estimated to determine a 50% reduction in lethality after mAb treatment. Blinding and randomization were not performed.
Statistical analysis.
Statistical significance was determined using GraphPad Prism version 7.0 (GraphPad). Attachment and fusion inhibition assays were analyzed using a one-way ANOVA test with Dunnett’s post-test. In vivo survival experiments were analyzed using a one-way log-rank test with a Bonferroni correction. Differences in IVIS signal were analyzed using a one-way ANOVA test with Dunnett’s post-test.
Data Availability.
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary information. The Supplemental Tables provide data on the newly-generated mAbs and mutagenesis (alanine and arginine) mapping of the mAb binding sites on EEEV E2 protein.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Defense Threat Reduction Agency (HDTRA1–15-1–0013 to M.S.D. and W.B.K. and HDTRA1–13-1–0034 to J.E.C) and NIH grant R01 AI095436 (W.B.K.).
Footnotes
COMPETING INTERESTS
M.S.D. is a consultant for Inbios and is on the Scientific Advisory Board of Moderna. J.E.C. has served as a consultant for Takeda Vaccines, Sanofi Pasteur, Pfizer, and Novavax, is on the Scientific Advisory Boards of CompuVax, GigaGen, Meissa Vaccines, PaxVax, and is Founder of IDBiologics.
REFERENCES
- 1.Garlick J et al. Locally Acquired Eastern Equine Encephalitis Virus Disease, Arkansas, USA. Emerging infectious diseases 22, 2216–2217, doi:10.3201/eid2212.160844 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukerji SS, Lam AD & Wilson MR Eastern Equine Encephalitis Treated With Intravenous Immunoglobulins. The Neurohospitalist 6, 29–31, doi:10.1177/1941874415578533 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong PM & Andreadis TG Eastern equine encephalitis virus--old enemy, new threat. The New England journal of medicine 368, 1670–1673, doi:10.1056/NEJMp1213696 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Eastern Equine Encephalitis, <https://www.cdc.gov/easternequineencephalitis/> (2018).
- 5.Lindsey NP, Staples JE & Fischer M Eastern Equine Encephalitis Virus in the United States, 2003–2016. The American journal of tropical medicine and hygiene 98, 1472–1477, doi:10.4269/ajtmh.17-0927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyler KL Acute Viral Encephalitis. The New England journal of medicine 379, 557–566, doi:10.1056/NEJMra1708714 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Tan Y et al. Large scale complete genome sequencing and phylodynamic analysis of eastern equine encephalitis virus reveal source-sink transmission dynamics in the United States. Journal of virology, doi:10.1128/jvi.00074-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt AR, Frederickson S, Maruyama T, Roehrig JT & Blair CD The first human epitope map of the alphaviral E1 and E2 proteins reveals a new E2 epitope with significant virus neutralizing activity. PLoS neglected tropical diseases 4, e739, doi:10.1371/journal.pntd.0000739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman MB & Weaver SC Structure of the recombinant alphavirus Western equine encephalitis virus revealed by cryoelectron microscopy. Journal of virology 84, 9775–9782, doi:10.1128/jvi.00876-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R et al. 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. The EMBO journal 30, 3854–3863, doi:10.1038/emboj.2011.261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voss JE et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468, 709–712, doi:10.1038/nature09555 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Li L, Jose J, Xiang Y, Kuhn RJ & Rossmann MG Structural changes of envelope proteins during alphavirus fusion. Nature 468, 705–708, doi:10.1038/nature09546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith TJ et al. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proceedings of the National Academy of Sciences of the United States of America 92, 10648–10652 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J et al. Phage display identifies an Eastern equine encephalitis virus glycoprotein E2-specific B cell epitope. Veterinary immunology and immunopathology 148, 364–368, doi:10.1016/j.vetimm.2012.06.021 (2012). [DOI] [PubMed] [Google Scholar]
- 15.EnCheng S et al. Analysis of murine B-cell epitopes on Eastern equine encephalitis virus glycoprotein E2. Applied microbiology and biotechnology 97, 6359–6372, doi:10.1007/s00253-013-4819-8 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Roehrig JT et al. Identification of monoclonal antibodies capable of differentiating antigenic varieties of eastern equine encephalitis viruses. The American journal of tropical medicine and hygiene 42, 394–398 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Pereboev AV, Razumov IA, Svyatchenko VA & Loktev VB Glycoproteins E2 of the Venezuelan and eastern equine encephalomyelitis viruses contain multiple cross-reactive epitopes. Archives of virology 141, 2191–2205 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Fox JM et al. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 163, 1095–1107, doi:10.1016/j.cell.2015.10.050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SA et al. Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell host & microbe 18, 86–95, doi:10.1016/j.chom.2015.06.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt AR, Frederickson S, Hinkel C, Bowdish KS & Roehrig JT A humanized murine monoclonal antibody protects mice either before or after challenge with virulent Venezuelan equine encephalomyelitis virus. The Journal of general virology 87, 2467–2476, doi:10.1099/vir.0.81925-0 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Hulseweh B et al. Human-like antibodies neutralizing Western equine encephalitis virus. mAbs 6, 718–727, doi:10.4161/mabs.28170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pal P et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS pathogens 9, e1003312, doi:10.1371/journal.ppat.1003312 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin J et al. Neutralizing Monoclonal Antibodies Block Chikungunya Virus Entry and Release by Targeting an Epitope Critical to Viral Pathogenesis. Cell reports 13, 2553–2564, doi:10.1016/j.celrep.2015.11.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang R et al. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574, doi:10.1038/s41586-018-0121-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schilte C et al. Cutting edge: independent roles for IRF-3 and IRF-7 in hematopoietic and nonhematopoietic cells during host response to Chikungunya infection. Journal of immunology (Baltimore, Md. : 1950) 188, 2967–2971, doi:10.4049/jimmunol.1103185 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Armstrong PM, Prince N & Andreadis TG Development of a multi-target TaqMan assay to detect eastern equine encephalitis virus variants in mosquitoes. Vector borne and zoonotic diseases (Larchmont, N.Y.) 12, 872–876, doi:10.1089/vbz.2012.1008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner CL, Ebel GD, Ryman KD & Klimstra WB Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proceedings of the National Academy of Sciences of the United States of America 108, 16026–16031, doi:10.1073/pnas.1110617108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edwards J & Brown DT Sindbis virus-mediated cell fusion from without is a two-step event. The Journal of general virology 67 ( Pt 2), 377–380, doi:10.1099/0022-1317-67-2-377 (1986). [DOI] [PubMed] [Google Scholar]
- 29.Fong RH et al. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. Journal of virology 88, 14364–14379, doi:10.1128/jvi.01943-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun C, Gardner CL, Watson AM, Ryman KD & Klimstra WB Stable, high-level expression of reporter proteins from improved alphavirus expression vectors to track replication and dissemination during encephalitic and arthritogenic disease. Journal of virology 88, 2035–2046, doi:10.1128/jvi.02990-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wust CJ, Crombie R & Brown A Passive protection across subgroups of alphaviruses by hyperimmune non-cross-neutralizing anti-Sindbis serum Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine; (New York, N.Y.) 184, 56–63 (1987). [DOI] [PubMed] [Google Scholar]
- 32.Roehrig JT, Hunt AR, Kinney RM & Mathews JH In vitro mechanisms of monoclonal antibody neutralization of alphaviruses. Virology 165, 66–73 (1988). [DOI] [PubMed] [Google Scholar]
- 33.Flynn DC, Olmsted RA, Mackenzie JM Jr. & Johnston RE Antibody-mediated activation of Sindbis virus. Virology 166, 82–90 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Haslwanter D, Blaas D, Heinz FX & Stiasny K A novel mechanism of antibody-mediated enhancement of flavivirus infection. PLoS pathogens 13, e1006643, doi:10.1371/journal.ppat.1006643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agapov EV et al. Localization of four antigenic sites involved in Venezuelan equine encephalomyelitis virus protection. Archives of virology 139, 173–181 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Pence DF, Davis NL & Johnston RE Antigenic and genetic characterization of Sindbis virus monoclonal antibody escape mutants which define a pathogenesis domain on glycoprotein E2. Virology 175, 41–49 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Vrati S, Fernon CA, Dalgarno L & Weir RC Location of a major antigenic site involved in Ross River virus neutralization. Virology 162, 346–353 (1988). [DOI] [PubMed] [Google Scholar]
- 38.Porta J et al. Locking and blocking the viral landscape of an alphavirus with neutralizing antibodies. Journal of virology 88, 9616–9623, doi:10.1128/jvi.01286-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson AM et al. Ribbon scanning confocal for high-speed high-resolution volume imaging of brain. PloS one 12, e0180486, doi:10.1371/journal.pone.0180486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charles PC, Walters E, Margolis F & Johnston RE Mechanism of neuroinvasion of Venezuelan equine encephalitis virus in the mouse. Virology 208, 662–671, doi:10.1006/viro.1995.1197 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Honnold SP et al. Eastern equine encephalitis virus in mice I: clinical course and outcome are dependent on route of exposure. Virology journal 12, 152, doi:10.1186/s12985-015-0386-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honnold SP et al. Eastern equine encephalitis virus in mice II: pathogenesis is dependent on route of exposure. Virology journal 12, 154, doi:10.1186/s12985-015-0385-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal P et al. Chikungunya viruses that escape monoclonal antibody therapy are clinically attenuated, stable, and not purified in mosquitoes. Journal of virology 88, 8213–8226, doi:10.1128/jvi.01032-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou T et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science (New York, N.Y.) 329, 811–817, doi:10.1126/science.1192819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law M et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nature medicine 14, 25–27, doi:10.1038/nm1698 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Julien JP, Lee PS & Wilson IA Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunological reviews 250, 180–198, doi:10.1111/imr.12005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klimstra WB, Ryman KD & Johnston RE Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. Journal of virology 72, 7357–7366 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aguilar PV et al. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. Journal of virology 82, 4920–4930, doi:10.1128/jvi.02514-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith Scott A. et al. Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell Host & Microbe 18, 86–95, doi:10.1016/j.chom.2015.06.009 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardner CL et al. Antibody Preparations from Human Transchromosomic Cows Exhibit Prophylactic and Therapeutic Efficacy against Venezuelan Equine Encephalitis Virus. Journal of virology 91, doi:10.1128/jvi.00226-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary information. The Supplemental Tables provide data on the newly-generated mAbs and mutagenesis (alanine and arginine) mapping of the mAb binding sites on EEEV E2 protein.