Summary
The bacterial pathogen Legionella pneumophila modulates host immunity using effectors translocated by its Dot/Icm transporter to facilitate its intracellular replication. A number of these effectors employ diverse mechanisms to interfere with protein ubiquitination, a posttranslational modification essential for immunity. Here we found that L. pneumophila induces monoubiquitination of the E2 enzyme UBE2N by its Dot/Icm substrate MavC(Lpg2147). We demonstrate that MavC is a transglutaminase that catalyzes covalent linkage of ubiquitin to Lys92 and Lys94 of UBE2N via Gln40. Similar to canonical transglutaminases, MavC possess deamidase activity that targets ubiquitin at Gln40. We identified Cys74 as the catalytic residue for both ubiquitination and deamidation activities. Furthermore, ubiquitination of UBE2N by MavC abolishes its activity in the formation of K63-type polyubiquitin chains, which dampens NFκB signaling in the initial phase of bacterial infection. Our results reveal an unprecedented mechanism of modulating host immunity by modifying a key ubiquitination enzyme by ubiquitin transglutamination.
Keywords: Type IV secretion, ubiquitination, transglutamination, virulence, immunity, NFκB
Introduction
Legionella pneumophila extensively modulates host cellular processes by the hundreds of effectors injected by its Dot/Icm system, which results in the biogenesis of the Legionella-containing vacuole (LCV), a phagosome that supports bacterial replication 1. Infection by L. pneumophila activates the major immune regulator NFκB by at least two mechanisms: A transient Dot/Icm-independent activation most likely by immune agonists such as flagellin and LPS; this activation is more apparent when macrophages were challenged with bacteria at a high multiplicity of infection (MOI) 2,3. The second mechanism is mediated by the Dot/Icm transporter that persists throughout much of the intracellular life cycle of the bacterium 2,3. NFκB activated by the latter mechanism induces the expression of a large repertoire of genes, including those involved in cell survival, vesicle trafficking and immunity 2,4. The Dot/Icm effector LegK1 activates NFκB by directly phosphorylating IκBα and other members of this inhibitor family, including p100 5; LnaB also activates this transcriptional factor but its mechanism of action is unknown 6. Cell survival genes induced by NFκB activation is required for productive intracellular bacterial replication 2. The activation of immunity by ligands such as flagellin presumably is detrimental to the pathogen, how L. pneumophila counteracts such defense remains unknown.
Among other important functions, ubiquitination is essential in the regulation of immunity against infection 7. Classical ubiquitination is catalyzed by the action of E1, E2 and E3 enzymes that function coordinately to covalently attach ubiquitin to protein substrates 8. Interference of the host ubiquitin network is critical for the success of many microorganisms that have parasitic or symbiotic relationships with eukaryotic hosts and such interference often is achieved by virulence factors that function as deubiquitinases, E3 ubiquitin ligases 9, or as ubiquitin modification enzymes 10. More than 10 Dot/Icm effectors of L. pneumophila function to hijack host ubiquitin signaling11. Among these, members of the SidE effector family catalyze ubiquitination by a two-step process that completely differs from the canonical three-enzyme cascade. In the reaction catalyzed by SidEs, ubiquitin is first ADP-ribosylated at Arg42 to produce the reaction intermediate ADPR-Ub 12, which is utilized by a phosphodiesterase (PDE) activity also embedded in SidEs that transfers the phosphoribosylated ubiquitin to serine residues in their substrates 13,14.
In our efforts to identify eukaryotic proteins capable of catalyzing ubiquitination with mechanisms similar to those used by SidEs, we found that the E2 enzyme UBE2N can be modified by a ubiquitin mutant unable to be used by the canonical ubiquitination machinery. Further studies revealed that UBE2N was monoubiquitinated during L. pneumophila infection by the effector MavC (Lpg2147). We also demonstrate that MavC functions as a transglutaminase that catalyzes crosslinking between ubiquitin and UBE2N, leading to the inhibition of its activity in NFκB activation.
Results
Modification of UBE2N by a ubiquitin mutant that cannot be used by the canonical ubiquitination machinery
To identify potential eukaryotic enzymes capable of catalyzing ubiquitination by a mechanism similar to that of SidEs, we expressed 3xHA-Ub and 3xHA-Ub-AA (the last two glycine residues were replaced with alanine residues) in HEK293T cells, respectively. Although the number was drastically fewer than that modified by 3xHA-Ub, proteins potentially modified by 3xHA-Ub-AA were detected (Supplementary Fig. 1a). We thus performed immunoprecipitation from cells expressing 3xHA-Ub-AA with HA antibody and proteins in gels of relevant molecular weight (MW) were identified by mass spectrometric analysis. Among the proteins identified, the E2 enzyme UBE2N important for the formation of K63-type polyubiquitin chains15 was seen in multiple experiments (Supplementary Table 1). Subsequent experiments found that UBE2N was the only protein that can be detectably modified by 3xHA-Ub-AA in HEK293T cells (Supplementary Fig. 1b).
UBE2N is modified by the Dot/Icm effector MavC(Lpg2147) during L. pnuemophila infection
To identify the enzymes responsible for UBE2N ubiquitination with 3xHA-Ub-AA, we hypothesized that such enzymes may be induced under certain stress conditions. Clearly, the identification of such conditions would facilitate subsequent purification and characterization of the enzymes because of the potentially higher protein levels. Thus, we treated Raw264.7 cells with a variety of stresses and examined UBE2N ubiquitination. None of the tested physiochemical treatments led to an MW shift in UBE2N (Fig. 1a). In these experiments, we also included samples from cells infected with two L. pneumophila strains. Intriguingly, we observed an MW shift akin to monoubiquitination only in cells infected with the virulent strain (Fig. 1a). The requirement of the Dot/Icm transporter suggests that such modification is caused by one or more of its substrates. We chose first to pursue this potential effector-induced UBE2N modification because it is technically more straightforward than the identification of the mammalian proteins potentially involved in its modification.
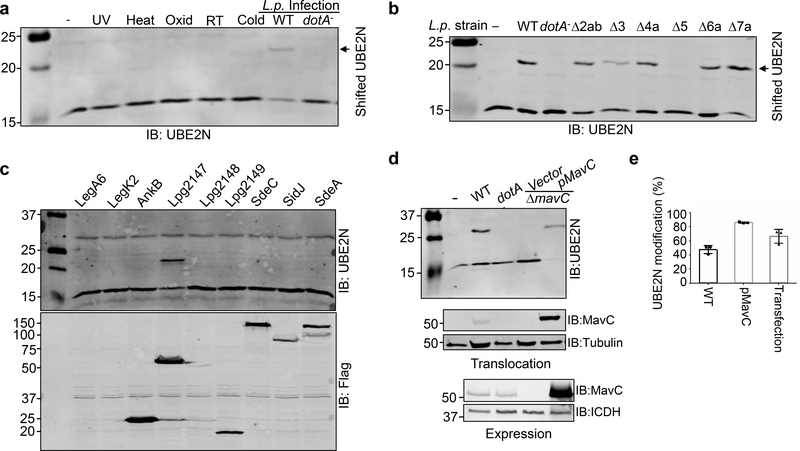
Fig. 1. L. pneumophila induces a molecular weight shift in UBE2N in a process that requires the Dot/Icm effector MavC.
a. Infection by virulent L. pneumophila but not several physiochemical stresses caused a molecular weight (MW) shift in UBE2N. Raw264.7 cells were subjected to the indicated treatments or were infected with wild type L. pneumophila or its dotA mutant defective in the Dot/Icm type IV transporter. Total proteins resolved by SDS-PAGE were probed with a UBE2N-specific antibody. Note the detection of a protein slightly above 20 KDa in samples infected with wild type bacteria. b. The gene responsible for L. pneumophila-induced UBE2N modification resides in the region removed from the deletion strain Δ5. Opsonized L. pneumophila were used to infect HEK293T cells transfected to express the FcγII receptor for 2 h at an MOI of 10. Total proteins separated by SDS-PAGE were probed with a UBE2N-specific antibody. Note that strain Δ5 cannot induce UBE2N modification. c. The Dot/Icm effector MavC (Lpg2147) caused UBE2N modification. A set of Dot/Icm substrates in the region missing in deletion strain Δ5 were individually expressed in HEK293T. The modification of UBE2N was detected by immunoblotting (upper level). The expression of the bacterial proteins was detected using a Flag-specific antibody (lower panel). Note that expression of MavC(Lpg2147) caused UBE2N modification. d. MavC is the only L. pneumophila protein responsible for UBE2N modification. HEK293 cells infected with the indicated bacterial strains, and proteins solubilized by saponin were probed for UBE2N (upper panel) or for translocated MavC (middle panel). The expression of MavC in bacteria was also analyzed (lower panel). e. Quantitation of modified UBE2N under different experimental conditions. Blots from at least three independent experiments performed similarly were quantified by Image J (mean ± s.e. from three replicates). Experiments in a-d were repeated at least three times and similar results were obtained.
To identify the Dot/Icm substrate(s) responsible for UBE2N modification, we took advantage of the several cluster deletion mutants of L. pneumophila 16. With the exception of strain Δ5, infection by all of other mutants caused an MW shift in UBE2N (Fig. 1b). We thus identified the protein responsible for this modification by individually expressing effectors absent in strain Δ5 in HEK293T cells. These experiments revealed that Lpg2147 (MavC)17 was able to cause UBE2N modification (Fig. 1c). We thus constructed and tested strain Lp02ΔmavC and found that infection with wild type but not the mutant caused UBE2N modification (Fig. 1d). Furthermore, introduction of a plasmid expressing mavC restored the ability of the mutant to modify UBE2N (Fig. 1d). The level of UBE2N modification corresponds well to the expression level of MavC in bacterial strains. The relatively lower modification rates in transfected cells may be due to low transfection efficiency (Fig. 1e). Thus, mavC is the only gene responsible for UBE2N modification during L. pneumophila infection.
In macrophages, the ΔmavC strain grew at rates indistinguishable to that of the wild type strain (Supplementary Fig. 2a), indicating that similar to most Dot/Icm substrates, mavC is not required for proficient intracellular bacterial replication in commonly used tissue culture hosts. In broth grown bacteria, MavC was barely detectable in the lag and early exponential growth phases (OD600=0.05–2.2) but becoming highly expressed in exponential phase and continued to the post-exponential phase (OD600=2.6–3.6) (Supplementary Fig. 2b), suggesting that MavC functions in the initial phase of infection.
MavC induces UBE2N ubiquitination by a mechanism that does not require the host ubiquitination machinery
The modification of UBE2N induced by MavC led to an approximately 10 kDa increase in its MW, a size close to monoubiquitination (Fig. 1). To test whether it catalyzes ubiquitination, we incubated MavC, HA-Ub and 4xFlag-UBE2N with total lysates of HEK293T cells. A modified protein with a molecular weight similar to that of UBE2N-Ub was detected by HA-specific antibody (Fig. 2a), indicating that MavC utilizes HA-Ub to modify UBE2N. Importantly, we observed indistinguishable UBE2N modification by HA-Ub in reactions containing boiled total cell lysates (Fig. 2a, last lane), suggesting that the activity of MavC may not need the host ubiquitination machinery.
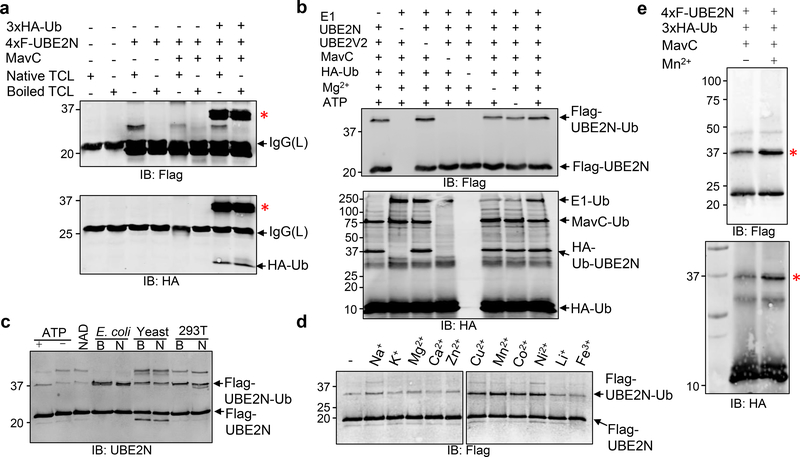
Fig. 2. MavC induces UBE2N ubiquitination independent of the canonical ubiquitination machinery.
a. MavC induces UBE2N ubiquitination. HA-Ub and Flag-UBE2N were incubated with native or boiled total cell lysates for 2 h at 37°C. Flag-UBE2N recovered by immunoprecipitation was probed by immunoblotting with antibody specific for Flag (upper panel) or HA (lower panel). Note that UBE2N was modified by HA-Ub even in reactions containing boiled total cell lysates (8th lane). *, modified UBE2N. b. MavC-induced ubiquitination of UBE2N occurred in the absence of E1, ATP or Mg2+. A series of reactions containing the indicated components were allowed to proceed for 2 h at 37°C. Samples resolved by SDS-PAGE were probed with the Flag (upper panel) and HA (lower panel) specific antibodies, respectively. Note that ubiquitinated UBE2N was detected in samples without E1 (1st lane), Mg2+ (6th lane) or ATP (7th lane). Self-ubiquitinated MavC was detected in all samples containing this protein and ubiquitin, regardless of other components required for canonical ubiquitination reaction (e.g. 1st, 2nd, 3rd, 6th and 7th lanes). c. A heat stable compound present in cells potentiates MavC-induced UBE2N ubiquitination. ATP, NAD, boiled or native total lysates from E. coli, yeast or HEK293T cells were added to reactions containing ubiquitin and Flag-UBE2N. Reactions were resolved by SDS-PAGE and probed with a Flag-specific antibody. d. The activity of MavC is induced by a few divalent metal ions. The indicated metal ions were added to reactions containing ubiquitin and Flag-UBE2N at a final concentration of 5 mM. Samples resolved by SDS-PAGE after 2 h incubation at 37°C were probed with a Flag antibody. Note that Cu2+, Mn2+, Co2+ or Ni2+ enhanced the activity of MavC. e. The metal ion Mn2+ potentiates the activity of MavC. Reactions containing the indicated components were established and the modification of UBE2N was probed by the formation of higher molecular weight conjugate (red *) detected with antibody specific for Flag (upper panel) or HA (lower panel). *, modified UBE2N. Experiments in a-e were repeated at least three times and similar results were obtained.
We next used biochemical assays to test the hypothesis that MavC is a ubiquitin ligase whose activity does not require components of the canonical ubiquitination reaction. To function as an E2 enzyme, UBE2N requires another protein such as UBE2V2 to form a heterodimer 18. In reactions that contained all the necessary components, modified UBE2N was produced (Fig. 2b, upper panel, last lane). Unexpectedly, ubiquitination of UBE2N also occurred in reactions that did not receive the E1 enzyme (Fig. 2b, upper panel, 1st lane). Such modification also occurred in reactions that did not receive Mg2+ or ATP (Fig. 2b, upper panel, 6th and 7th lanes). Importantly, self-ubiquitination occurred in MavC, again even in reactions that did not receive E1, Mg2+ or ATP (Fig. 2b, lower panel, 1st, 6th and 7th lanes). These results further suggest that MavC is capable of catalyzing ubiquitination by a mechanism that does not require components of the canonical ubiquitination machinery.
Infection with an L. pneumophila strain overexpressing MavC led to modification of about 80% of endogenous UBE2N (Fig. 1d). In contrast, the rates of modification were considerably lower in reactions containing purified proteins, suggesting that MavC requires one or more cellular factors for optimal activity. The addition of ATP or NAD did not increase the modification efficiency (Fig. 2c). Instead, lysates of prokaryotic or eukaryotic cells allowed the modification to proceed more completely and boiling treatment did not abolish such activity (Fig. 2c). Thus, one or more heat stable factors were required for full activity of MavC. We tested the effects of several metal ions and found that a few divalent ions including Cu2+, Mn2+, Co2+ and Ni2+ were able to enhance the activity of MavC (Fig. 2d-e). Together, these results indicate that MavC catalyzes ubiquitination by a mechanism that does not require components of the canonical ubiquitination machinery or even exogenous energy source. Furthermore, its activity can be potentiated by several divalent metal ions such as Mn2+ and Ni2+.
MavC is a transglutaminase that catalyzes crosslink between ubiquitin and UBE2N
We analyzed the biochemical mechanism underlying MavC-mediated ubiquitination by determining the sites of ubiquitination on UBE2N and the chemical bond that links these two molecules. The protein band corresponding to modified UBE2N was analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 3a), which led to the identification of two UBE2N peptides, -I86CLDILKDK94- and -D93KWSPALQIR102- that were cross-linked to the same ubiquitin fragment -E34GIPPDQQR40- (Fig. 3b-c). The fragmentation pattern and the fact that trypsin digestion did not occur at Lys92 and Lys94 in these UBE2N peptides suggest that the linkage is through a lysine residue. The tandem mass spectra also showed that ubiquitin is linked to the substrate through its Gln40 residue (Fig. 3b-c). By the mass difference, Gln40 of ubiquitin was linked to a lysine residue in UBE2N by eliminating an ammonia molecule, leading to the formation of an Nε-(γ-glutamyl)lysine isopeptide bond between Gln40 on ubiquitin and Lys92 or Lys94 of UBE2N (Fig. 3b-c).
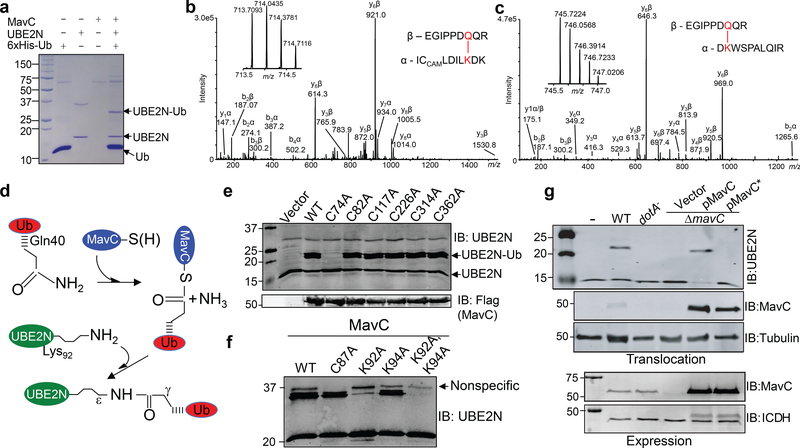
Fig. 3. MavC catalyzes ubiquitination of UBE2N by transglutamination.
a. MavC-induced UBE2N ubiquitination detected by Coomassie staining. Reactions containing the indicated components were allowed to proceed for 2 h and samples resolved by SDS-PAGE were detected by staining. b-c. MavC catalyzes the formation of an isopeptide bond between Gln40 of ubiquitin and Lys92 and Lys94 of UBE2N. Ubiquitinated UBE2N shown in panel a was subjected to mass spectrometric analysis. Tandem mass spectrum of the ubiquitin peptide -E34GIPPDQQR40- cross-linked with UBEN2 peptide –I86CLDILKDK94- (b) or -DK94WSPALQIR102-(right panel) (c). The inserts show the high-resolution measurements of parent ions, which correspond to mass errors of 0.8 (b) and 0.4 (c) parts per million compared to the calculated mass. Abbreviation: CAM, carbamidomethylation. d. A predicted reaction scheme of MavC-mediated protein crosslinking by transglutamination. A nucleophilic Cys residue on MavC attacks Gln40, a γ-glutaminyl residue in ubiquitin to form a thioester intermediate. The acylated MavC then reacts with the amine donor from the ε-lysine in UBE2N to form an intermolecular isopeptide bond. e. Cys74 is essential for the activity of MavC. Flag-tagged MavC or its mutants harboring mutations in each of its six Cys residues were expressed in HEK293T cells. The modification of UBE2N was probed by immunoblotting with a UBE2N-specific antibody (upper panel). The Cys74Ala mutation did not affect the expression and stability of MavC (lower panel). f. Lys92 is the major ubiquitination site in UBE2N. 4xFlag-UBE2N or its mutants were incubated with ubiquitin and MavC. The formation of Ub-UBE2N was detected by immunoblotting. Note that the UBE2NK92A mutant has largely lost the ability to be modified (3rd lane). g. Cys74 of MavC is essential for UBE2N ubiquitination induced by L. pneumophila. HEK293 cells were infected with the indicated bacterial strains for 2 h and the modification of UBE2N was probed by immunoblotting. The expression (lower panels) and translocation (middle panels) of MavC and its mutant were probed with MavC-specific antibodies. Experiments in a-c, e-g were repeated at least three times and similar results were obtained.
The chemical linkage that bridges ubiquitin and UBE2N is resembling those formed by transglutaminases (TGases), which often catalyze transamidation between the γ-carboxamide group of a glutamine residue in one protein and the ε-amino group of a lysine residue in another protein19. The reaction catalyzed by TGases requires a Cys-His-Asp catalytic triad in which the Cys residue is involved in the formation of a γ-glutamylthioester with the Gln-containing protein (ubiquitin here) (Fig. 3d)20. To identify the Cys residue critical for the activity of MavC, we constructed substitution mutants lacking each of its six Cys residues and examined their activity in UBE2N ubiquitination. Only mutation in Cys74 abolished the ability of MavC to induce UBE2N ubiquitination (Fig. 3e).
We confirmed the modification sites on UBE2N by testing mutants with substitutions in Lys92 or Lys94. Whereas incubation of the K94A mutant with MavC and ubiquitin still led to robust modification, ubiquitination of the K92A mutant was largely abolished and no modification was detected in reaction containing UBE2NK92AK94A (Fig. 3f). Mutation in C87, the residue important for UBE2N’s role as an E2 enzyme 18, did not affect MavC-mediated modification (Fig. 3f). Thus, the transglutamination reaction catalyzed by MavC links ubiquitin to Lys92 or Lys94 of UBE2N, of which Lys92 is the major modification site.
We next examined the role of the catalytic cysteine in ubiquitination of UBE2N during L. pneumophila infection. Unlike the ΔmavC mutant expressing MavC, expression of MavCC74A did not cause UBE2N modification despite the mutant protein was properly expressed and translocated into host cells (Fig. 3g).
We also determined the ability of MavC to modify several structurally similar E2s (harboring Cys87 and Lys92, not necessarily at the 87th and 92th residue, respectively) (Supplementary Fig. 3a). None of these E2s, including UBE2E2, 2K and 2S was detectably ubiquitinated by MavC in either biochemical reactions or in cells infected by L. pneumophila (Supplementary Fig. 3b-c). These results suggest that MavC specifically modifies UBE2N.
The involvement of Gln40 in the formation of the covalent bond suggests that residues important for ubiquitination catalyzed by the canonical mechanism are not required for MavC-induced UBE2N modification. Indeed, each of the lysine mutants (lacking one, more or all of the lysine residues) was active at levels comparable to wild type ubiquitin, so was the mutant in which the last two glycine residues were replaced with alanine (Supplementary Fig. 4).
MavC possesses deamidation activity against ubiquitin
In the absence of a suitable amine donor, the γ-glutamylthioester formed between the TGase and the substrate (ubiquitin here) can be hydrolyzed to a free glutamate residue, which corresponds to a net deamidation of the substrate21. We thus examined the product of the reaction containing MavC and ubiquitin. Incubation of ubiquitin with MavC altered the mobility of ubiquitin in a way similar to the product produced by bacterial ubiquitin deamidases10 (Supplementary Fig. 5a). Expectedly, deamidation by MavC required Cys74, a residue critical for the transglutamination activity (Supplementary Fig. 5c). Unlike Cif and CHBP10, MavC does not detectably attack NEDD8, indicative of substrate specificity (Supplementary Fig. 5a). In mass spectrometric analysis, deamidation can be easily mis-assigned by automated peptide identification algorithms with the 13C isotope of peptides because they differ from each other by only 0.019 Da. Yet, the high-resolution mass spectrometry analysis revealed a peak at m/z 520.7531, which is closer to the deamidated (delta=1.7 p.p.m) than the 13C isotope (delta=38.4 p.p.m) mass of the ubiquitin peptide -E34GIPPDQQR40- (Supplementary Fig. 5b). Further, only trace amounts of the unmodified peptide were detected at m/z 520.2608. In a series of reactions containing MavC and different amounts of ubiquitin. After 1 h incubation, modified ubiquitin became detectable in reactions in which the ratio between ubiquitin and MavC is 64 and the modification was almost complete when such ratio was 16 (Supplementary Fig. 5c). Thus, similar to other transglutaminases, MavC catalyzes a deamidation reaction on ubiquitin in the absence of UBE2N.
The observation that Gln40 of ubiquitin provided the side chain required for ubiquitination predicted that a mutant with a substitution mutation in this residue can no longer be used in the reaction. Indeed, UBE2N cannot be modified by ubiquitinQ40E (Supplementary Fig. 5d). Thus, similar to canonical transglutaminases from eukaryotic cells, MavC deamidates the protein that provides the glutamine residue 22.
Transglutamination is the dominant activity of MavC in reactions containing UNE2N and ubiquitin.
We examined the catalytic activity of MavC by a series of reactions in which UBE2N and MavC were added at different molar ratios. Under our experimental conditions, the modification of UBE2N by MavC was still readily detectable within 1 h when the molar ratio between MavC and UBE2N was as low as 1:1900, and approximately 50% of UBE2N was modified when the ratio was 1:120 (Fig. 4a).
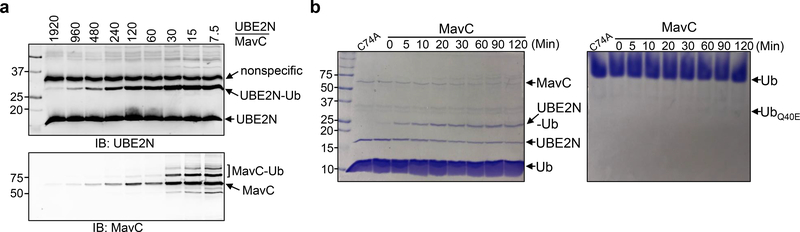
Fig. 4. Characteristics of MavC as transglutaminase and ubiquitin deamidase.
a. Dose-dependent modification of UBE2N by MavC. A series of reactions containing UBE2N and MavC at the indicated molar ratios were established and the reactions were allowed to proceed for 1 h before probing for the modified proteins (upper panel). MavC was also detected (lower panel). b. Transglutamination occurs faster than deamidation. A series of reactions with 1:60 molar ratio between MavC and UBE2N were allowed to proceed for the indicated time durations. The modification was detected by Coomassie staining. Note that Ub-UBE2N was detectable right after adding MavC (0 min left panel) and deamidated ubiquitin was barely detectable after 120 min incubation (right panel). Experiments in all panels were repeated at least three times and similar results were obtained.
To examine which activity is dominant, we established reactions in which the molar ratio of MavC to UBE2N and ubiquitin was 60 and 1024, respectively. A portion of the reaction was withdrawn at different time points after adding MavC to analyze the extent of UBE2N modification and ubiquitin deamidation. Ubiquitinated UBE2N became detectable right after (0 min) the addition of MavC and peaked at 20 min when approximately 50% of the protein was modified (Fig. 4b left panel). In contrast, deamidated ubiquitin was barely detectable even after the reaction had proceeded for 2 h (Fig. 4b right panel).
Deamidated ubiquitin can be detected in cells transfected to express MavC but not MavCC74A (Supplementary Fig. 6a-b); it is also detectable in cells infected with an L. pneumophila strain overexpressing MavC but not with wild type strain (Supplementary Fig. 6c-d), suggesting that the deamidase activity associated with MavC becomes detectable only when the protein level was artificially increased. Thus, MavC is highly active against UBE2N and that transglutamination is its dominant activity.
Ubiquitination of UBE2N by MavC abolished its E2 activity.
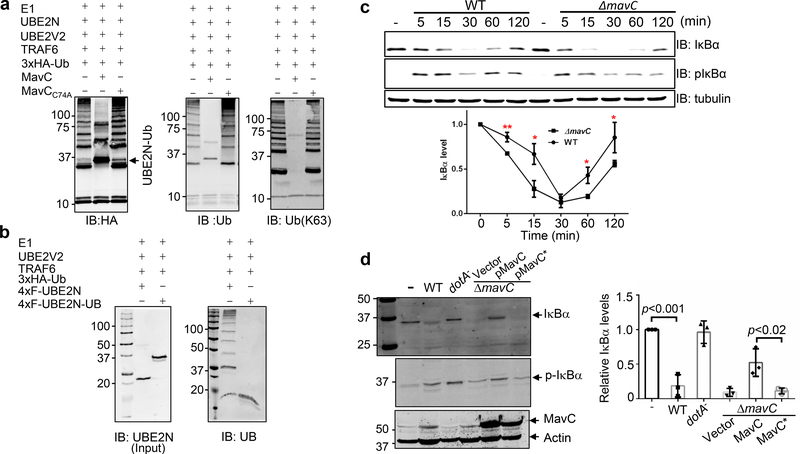
UBE2N forms heterodimers with proteins such as UBE2V1 and UBE2V2 to function as an E2 that catalyzes the elongation of K63-type ubiquitin chains18, which regulates multiple cellular processes 23. Inclusion of MavC, but not MavCC74A in reactions containing HA-Ub, the UBE2N-UBE2V2 complex and the E3 enzyme TRAF6 drastically reduced the production of polyubiquitin chains (Fig. 5a). The loss of E2 activity was equally apparent when purified UNE2N-Ub was used in reactions (Fig. 5b)
Fig. 5. The effects of MavC on UBE2N activity and on NFκB activation during L. pneumophila infection.
a. MavC blocks the formation of K63 polyubiquitin chains. Reactions containing the indicated components were allowed to proceed for 10 min at 37°C. UBE2N was preincubated with MavC or MavCC74A and ubiquitin for 4h at 37°C. Samples resolved by SDS-PAGE were probed with antibody specific for HA (left), ubiquitin (middle) or K63-type linkage (right). Note that few polyubiquitin chains were detected in reactions containing MavC. b. Ubiquitinated UBE2N is inactive. Purified Ub-UBE2N or its native form was incubated in reactions as described in A and the formation of polyubiquitin chains was detected with a ubiquitin-specific antibody (right panel). The amount of UBE2N and Ub-UBE2N used in the reactions was also probed (left panel). c. Kinetics of IκBα level in cells infected with wild type or the ΔmavC mutant of L. pneumophila. Raw264.7 cells infected with the indicated bacterial strains at an MOI of 5 were probed for IκBα or pIκBα. Tubulin was probed as a loading control. IκBα levels were quantitated from three independent experiments (lower panel) and the values were expressed as ratios of uninfected cells set at 1.0. *, p<0.05; **, p<0.01. Note the decrease of IκBα in the first 30 min of infection occurred faster in cells infected with the MavC-deficient mutant than wild type. d. Deletion of MavC led to reduction of IκBα in infected cells. Raw264.7 cells infected with the indicated bacterial strains for 1 h at an MOI of 10 were probed for IκBα by immunoblotting. Phosphorylated IκBα (pIκBα ) and translocated MavC were also probed. Relative levels of IκBα (lower panel) were the ratios between the intensity of the bands from uninfected cells and those of the indicated samples. Experiments in all panels were repeated at least three times and similar results were obtained. Quantitation shown was from three independent experiments (mean ± s.e. from three replicates). Statistical analysis used in c and d was Two sided, T-test, and no adjustment was made.
MavC dampens the NFκB pathway in the early phase of L. pneumophila infection.
The importance of K63-type polyubiquitin chains in NFκB activation prompted us to examine the levels of IκBα in Raw264.7 cells infected with wild type L. pneumophila and the ΔmavC mutant. Infections by both strains caused a rapid decrease in IκBα, which after reaching the lowest point at 30 min post-infection, began to recover. Importantly, such decrease occurred significantly faster in infections using the ΔmavC mutant (Fig. 5c). The recovery of IκBα in cells infected with the mutant was also slower (Fig. 5c). Furthermore, the decrease of cellular IκBα caused by mavC deletion can be restored by expressing MavC but not the C74A mutant (Fig. 5d). Notably, IκBα in cells infected with the complementation strain overexpressing MavC accumulated at significantly higher levels than those infected with wild type bacteria (Fig. 5d).
In line with the above results, ectopic expression of mavC attenuated NFκB activation in response to multiple distinct stimuli. The activation of NFκB by both TNF-α and phorbol myristate acetate (PMA) was inhibited in cells expressing MavC but not MavCC74A (Supplementary Fig. 7a-b). Similarly, NFκB activation induced by ectopic expression of Nod1, Myd88, TRAF6 and Bcl10 24–26 was inhibited by MavC (Supplementary Fig. 7c-f).
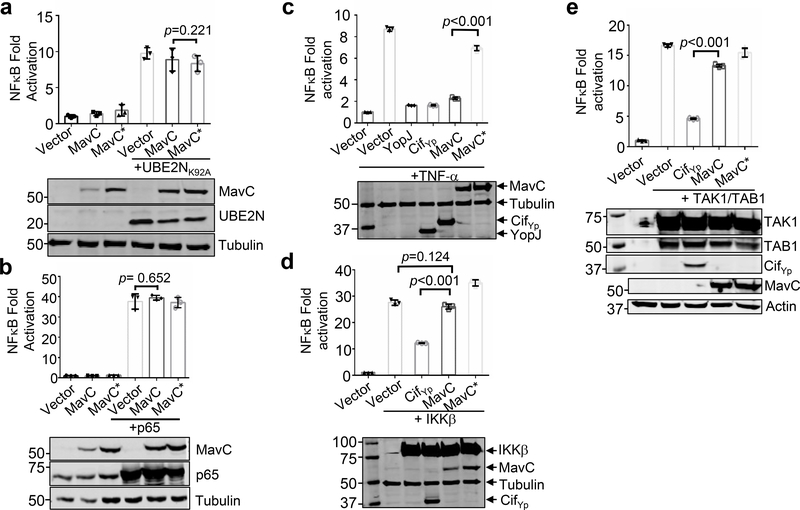
The observation that ubiquitination of UBE2N inhibited NFκB activation predicts that cells expressing UBE2NK92A will be resistant to MavC. Indeed, NFκB activation was not affected by MavC in cells expressing UBE2NK92A(Fig. 6a). Consistent with this observation, MavC cannot inhibit the activation induced by overexpressing p65 (Fig. 6b). We also compared MavC to YopJ and Cif, which inhibit NFκB activation by IKKβ acetylation and ubiquitin deamidation, respectively 10,27. These proteins exerted similar inhibitory effects (Fig. 6c). Yet, whereas IKKβ or TAK1/TAB1 suppressed the inhibitory effects caused by MavC, neither of them suppressed the inhibition imposed by Cif (Fig. 6d-e). Together, these results indicate that ubiquitination of UBE2N by MavC is responsible for the inhibition of NFκB activation.
Fig. 6. The effects of MavC on NFκB activation in cells overexpressing various relevant proteins.
a. UBE2NK92A suppresses the inhibition caused by MavC. HEK293T cells were transfected to express UBE2NK92A together with MavC or its mutant, NFκB activity was measured by luciferase activity. The expression of UBE2NK92A and MavC was probed by immunoblotting with specific antibodies. Tubulin was detected as a loading control. b. MavC cannot suppress the induction of NFκB by p65 overexpression. Cells were transfected to co-express UBE2NK92A and MavC or its mutant, and NFκB activity and protein expression was examined as described above. c. The inhibitory effect of MavC in NFκB activation is comparable to that of YopJ or Cif. Cells transfected to express the indicated proteins were treated with TNF-α and NFκB activity was measured by luciferase activity. d-e. MavC cannot inhibit NFκB activation induced by ectopic expression of IKKβ (d) or TAK1/TAB1 (e). For NFκB activity, in each case, similar results were obtained from at least three independent experiments done in triplicate (mean ± s.e. from three replicates); the immunoblots shown were one representative from at least three experiments. The expression of relevant proteins was probed by immunoblotting with specific antibodies against the protein of interest or an affinity tag and tubulin or actin was probed as a loading control. Statistical analysis was performed by Two sided, T-test, and no adjustment was made.
The inhibitory effects of MavC predict that its expression will lead to the sequestration of p65 in the cytosol upon stimulation. Indeed, in cells expressing MavC, TNF-α treatment did not cause p65 nucleus translocation (Supplementary Fig. 8). In contrast, in cells expressing the MavCC74A mutant, TNF-α stimulation led to p65 nucleus localization in almost every cell (Supplementary Fig. 8). Similarly, p65 nucleus translocation occurred in appropriately 75% of the cells infected with strain ΔmavC, which was significantly higher than those infected with wild type bacteria (Supplementary Fig. 9). Notably, infections with the ΔmavC strain overexpressing MavC reduced nucleus translocation of p65 to about 20% (Supplementary Fig. 9), which further supported its inhibitory role in NFκB activation. Taken together, these results indicate that MavC regulates host immunity by interfering with NFκB activation during L. pneumophila infection.
Discussion
Immune cells respond to damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) by various receptors, which dictates the outcome of the interaction between a pathogen and its hosts. Pathogens have evolved various strategies to counter host immune response to ensure successful infections. The bacterial pathogen L. pneumophila appears to activate the major immune regulator NFκB in different phases of infection by mechanisms that are either independent of or dependent upon its Dot/Icm transporter essential for virulence 2,3. The activation seen after the initial phase of infection likely attributes to Dot/Icm effectors such as LegK1 and LnaB 5,6. Whereas the activation at late phases of infection is of importance in multiple aspects, including the induction of genes involved in cell survival 2, virulence-independent activation of NFκB likely is detrimental to bacterial colonization. At least two lines of evidence suggest that the function of MavC is to counteract the effects of NFκB activation occurred in the early phase of infection. First, this gene is highly expressed in bacteria grown to post-exponential phase (Supplementary Fig. 2b). Second, MavC-induced reduction of IκΒα in infected cells reached the lowest level within the first 30 min after bacterial uptake (Fig. 5c).
E2 enzymes have emerged as important players in the formation of specific ubiquitin codes by directly controlling chain type synthesis, yet our understanding of their function and regulation still is limited28,29. Interestingly, although the enzymes involved remain unknown, UBE2N has been reported to be ISGylated at Lys92, which leads to inhibition of its E2 activity30,31. Similarly, UBE2T is automonoubiquitinated at Lys91, a modification that also inhibits its activity32. Other E2s such as UbcH7 have also been shown to be regulated by mechanisms such as ubiquitination, again at a lysine residue close to the active cysteine, leading to proteasome-mediated degradation33,34. Our results point to the importance of Lys92 in the activity and regulation of UBE2N, probably due to its close proximity to the catalytic Cys87 residue. The discovery of UBE2N monoubiquitination by MavC has added another layer of complexity to the regulation of E2 activity. The nature of the potential ubiquitination of UBE2N by Ub-AA observed in our initial experiments is unclear (Supplementary Fig. 1), nor is the enzyme involved in such modification. Ub-AA can be linked to UBE2N via Arg42, akin to that induced by members of the SidE family11, or by a Gln residue by enzymes similar to MavC or other yet uncharacterized mechanisms. Because TGases are widely present in eukaryotic cells19, it is possible that some of these enzymes also regulate cellular signaling by catalyzing ubiquitination.
Similar to established transglutaminases, MavC habors deamindase activity that attack ubiquitin (Supplementary Fig. 5), an activity also reported by a recent study 35. Several lines of evidence indicate that ubiquitination of UBE2N, rather than ubiquitin deamidation by MavC is responsible for its inhibition of the NFκB pathway. First, the inhibitory effects can be suppressed by co-expressing UBE2NK92A, a mutant that cannot be modified by MavC (Fig. 6a). Second, MavC cannot inhibit NFκB activation induced by TAK1/TAB1 or IKKβ, indicating that its point of action lies upstream of these kinases, which differs from deamindases that attack ubiquitin (Fig. 6d-e). Third, under identical reaction conditions, the transglutamination activity of MavC is much higher than the deamidation activity against ubiquitin (Fig. 4). Finally, whereas ubiquitination of UBE2N by MavC can be readily detected in cells infected with wild type L. pneumophila, deamidated ubiquitin was not detectable in such cells (Fig. 1 and Supplementary Fig. 6). Thus, the role of ubiquitin deamidation in the function of MavC, if any, should be minor.
UBE2N is the main E2 enzyme that specifies the formation of K63-type ubiquitin chains 15,28,29, which plays an important role in the activation of NFκB, a process that clearly is required during L. pneumophila infection 2,4. Thus, the activity of MavC mandates the requirement of enzymes that function to reverse the modification on UBE2N. Such enzymes may be of host or bacterial origin, or both, and should serve to impose temporal regulation of UNE2N activity together with MavC. Alternatively, other yet unidentified E2 enzymes not attacked by MavC may be responsible for NFκB activation seen in infected cells after the initial phase of infection 2. The identification of MavC as an inhibitor for NFκB activation reveals that L. pneumophila modulates the activity of this immune regulator by effectors with converse biological consequences, which is akin to the reversible regulation of other cellular events such as vesicle trafficking by effectors of opposite biochemical activities 11,36. Interestingly. the Shigella effector OspI deamidates UBE2N to interfere with its activity 26. Differing from the effects of MavC, which inhibits all most signaling branches that activate NFκB (Supplementary Fig. 10), deamidation of UBE2N at Gln100 by OspI only blocks its function in response to PMA via the diacylglycerol (DAG)-CBM (CARD–BCL10–MALT1) axis 26. The targeting of UBE2N by multiple pathogens highlights its importance in immunity. Further study is required to analyze the regulation of the effects imposed by MavC and the structural basis of the mechanism of the inhibition by this unique ubiquitination.
Methods
Media, bacteria strains, plasmid constructions and cell lines
L. pneumophila strains used in this study were derivatives of the Philadelphia 1 strain Lp0237 and were grown and maintained on CYE (charcoal-yeast extract) plates or in N-(2-Acetamido)-2-aminoethane (ACES) buffered yeast extract (AYE) broth as previously described37. The mavC in-frame deletion strain was constructed as described 38. mavC and mavCC74A genes were cloned into pZL507 39 for complementation. All the L. pneumophila cluster deletion strains were gifts from Dr. Ralph Isberg (Tufts University) 16. The E. coli strains XL1-Blue and BL21(DE3) were used for expression and purification of all the recombinant proteins used in this study. E. coli strains were grown in Luria Broth (LB). Genes for protein purifications were cloned into pQE30 (QIAGEN) or pETSUMO (Invitrogen) for expression. For ectopic expression of proteins in mammalian cells, genes were inserted into the 4xFlag CMV vector 12 or the 3xHA pCDNA3.1 vector 40. HEK293, 293T cells were cultured in Dulbecco’s modified minimal Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS). Raw264.7 cells were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium supplemented with 10% FBS. All cell lines are purchased from ATCC and authenticated. All cell lines were regularly checked for potential mycoplasma contamination by the universal mycoplasma detection kit from ATCC (cat# 30–1012K).
Protein purification
Ten mL overnight E. coli cultures were transferred to 400 mL LB medium supplemented with 100 μg/mL of ampicillin or 30 μg/mL of kanamycin, and the cultures were grown to OD600 of 0.5~0.7 prior to the induction with 0.2 mM isopropyl thio-D-galactopyranoside (IPTG). Cultures were further incubated at 18ºC for 16~18 h. Bacteria were collected by centrifugation at 4,000g for 10 min, and were lysed by sonication in 35 mL phosphate-buffered saline (PBS). Bacteria lysates were centrifuged twice at 18,000g at 4ºC for 20 min to remove insoluble fractions and unbroken cells. Supernatant containing recombinant proteins were incubated with 1 mL Ni2+-NTA beads (Qiagen) at 4 ºC for 2 h with agitation. Ni2+-NTA beads with bound proteins were washed with PBS buffer containing 20 mM imidazole for 3 times, 30x of the column volume each time. Proteins were eluted with PBS containing 300 mM imidazole. Proteins were dialyzed in buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl and 1 mM dithiothreitol (DTT) for 16–18 h.
Transfection, infection, immunoprecipitation
Lipofectamine 3000 (Thermo Fisher Scientific) was used to transfect HEK293 or HEK293T cells grown to about 70% confluence. Different plasmids were transfected into 293T cells respectively. Transfected cells were collected and lysed with the Radioimmunoprecipitation assay buffer (RIPA buffer, Thermo Fisher Scientific) at 16–18 h post transfection. Cell lysates were resolved by SDS-PAGE gels, transferred to nitrocellulose membranes for immunoblotting analysis with the specific antibodies. When needed, immunoprecipitation was performed with lysates of transfected cells with Flag- or HA-specific antibody coated agarose beads (Sigma, cat#F1804 and cat#A2095, respectively) at 4°C for 4 h. Beads were washed with pre-cold RIPA buffer or respective reaction buffers for 3 times and resolved by SDS-PAGE gels and followed by immunoblotting analysis with the specific antibodies or silver staining following the manufacturer’s protocols (Sigma, cat# PROTSIL1).
For all L. pneumophila infection experiments, L. pneumophila strains were grown to the post-exponential phase (OD600=3.2–3.8) in AYE broth. Complementation strains were induced with 0.2 mM IPTG for 4 h at 37 ºC before infection. Raw 264.7 cells were infected with L. pneumophila strains at an MOI of 10 for 2 h. HEK293 cells transfected to express the FcγII receptor were infected with opsonized L. pneumophila strains at an MOI of 10 for 2 h. L. pneumophila strains were opsonized by incubating bacteria with the L. pneumophila specific anti-sera at a dilution of 1:500. Cells were collected and lysed with 0.2% Saponin on ice for 30 min. Cell lysates were resolved by SDS-PAGE and followed by immunoblotting analysis with the specific antibodies respectively. L. pneumophila bacteria lysates were resolved by SDS-PAGE followed by immunoblotting with the MavC specific antibody to examine the expression of MavC, and isocitrate dehydrogenase (ICDH) was probed as loading control with antibodies previously described 39. For intracellular growth, infection was performed at an MOI of 0.05 and the total bacterial counts were determined at 24-h intervals as described38. For immunostaining, Raw264.7 cells were challenged by L. pneumophila at an MOI of 2 for 2 h. To detect deamidated ubiquitin in cells, HEK293 cells were transfected for 14 h and U937 cells were infected with relevant L. pneumophila strains for 2 h at an MOI of 10.
In vitro ubiquitination assays
For canonical ubiquitination dropout assays, 5 μg His6-3xHA-ubiquitin, 0.2 μg E1, 1 μg UBE2N, 1 μg UBE2V2, 1 μg MavC, 2 mM ATP and 5 mM Mg2+ were added into a 25 μL reaction containing 50 mM Tris-HCl (pH 7.5) and 1 mM DTT. When needed, a specific component was withdrawn from the reaction. The reactions were performed at 37°C for 2 h.
For non-canonical MavC-mediated ubiquitination reaction, 5 μg His6-3xHA-ubiquitin or His6-ubiquitin, 0.5 μg UBE2N, 0.05 μg MavC and 5 mM Mn2+ were included in a 25 μL reaction system containing 50 mM Tris-HCl (pH 7.5) and 1 mM DTT. Reaction was allowed to proceed for 1 h at 37°C. When needed, 50 μg native or boiled lysates of E. coli, yeast or 293T cells were added into reactions. For obtaining native lysates, E. coli, yeast cells were lysed by sonication, HEK293T cells were lysed by RIPA buffer without EDTA. Boiled lysates were prepared by treating cells at 100°C for 5 min. Concentrations of lysates were measured by the Bradford assay.
For ubiquitin derivatives ubiquitination assays, 5 μg His-tagged wild type or ubiquitin mutants was incubated with 0.5 μg UBE2N, 0.05 μg MavC or MavCC74A in 25 μL reactions, each containing 50 mM Tris-HCl (pH 7.5), 5 mM Mn2+ and 1 mM DTT at 37 °C for 1 h. All ubiquitin derivatives were obtained from Boston Biochem.
K63 poly-ubiquitin chain synthesis assay
Ten μg His6-3xHA-ubiquitin, 0.5 μg UBE2N were pretreated with 0.05 μg MavC or MavCC74A in 25 μL reaction system containing 50 mM Tris-HCl (pH 7.5), and 1 mM DTT at 37 °C for 4 h. After pretreatment, reactions were supplemented with 0.5 μg E1, 0.5 μg UBE2V2, 2.5 μg TRAF6, 2 mM ATP and 5 mM Mg2+, and incubated at 37 °C for additional 10 min.
Antibodies and Immunoblotting
Purified His6-MavC was used to raise rabbit specific antibodies using a standard protocol (Pocono Rabbit Farm & Laboratory). The antibodies were affinity purified as describe38. For immunoblotting, samples resolved by SDS-PAGE were transferred onto 0.2 μm nitrocellulose membranes (Pall Life Sciences cat# 66485). Membranes were blocked with 5% non-fat milk, incubated with the appropriate primary antibodies: anti-UBE2N (Cell signaling, cat# 6999S, Thermo Fisher Scientific, cat# 37–1100), 1:1,000; anti-Flag (Sigma, Cat# F1804), 1: 2000; anti-HA (Santa Cruz, cat# sc-7392 1:1,000, Roche, cat# 11867423001 1:5,000), anti-ICDH39, 1:10,000, anti-actin (Sigma, cat# A2103), 1:5,000, anti-tubulin (DSHB, E7) 1:10,000, anti-Ub K63 (Millipore, cat# 05–1308), 1:1,000, anti-IκBα (Cell signaling, cat# 9242S), 1:1,000, anti-Ub (Santa Cruz cat#sc-8017), 1:1,000, anti-p-IκBα (Cell Signaling, cat# 9246S), 1:1000, anti-UBE2K (Cell Signaling, cat# 8226S), 1:1000, anti-UBE2S (Cell Signaling, cat# 11878S), 1:1000, anti-UBE2E2 (Abcam, cat# Ab177485), 1:1000, anti-His (Sigma, cat# H1029), 1:10,000, anti-p65 (Cell signaling, cat# 8242S), 1:500. Membranes were then incubated with an appropriate IRDye infrared secondary antibody and scanned using an Odyssey infrared imaging system (Li-Cor’s Biosciences).
Liquid chromatography-tandem mass spectrometry analysis
Protein bands corresponding to MavC treated ubiquitin were excised from SDS-PAGE and digested with trypsin as described elsewhere41. Mass spectrometry experiments were performed at the Pacific Northwest National Laboratory (PNNL). The digested peptides were analyzed in C18 reversed phase column connected to a UPLC (ACQUITY, Waters) by separating with a gradient acetonitrile/0.1% formic acid (solvent B) vs. 0.1% formic acid in water (solvent A): 1% B to 8% B over 5 min and 8% B to 40% B over 95 min at 300 nL/min. Eluting peptides were directly analyzed in an Orbitrap mass spectrometer (Q-Exactive Plus, Thermo Fisher Scientific), by targeting the top 12 most intense ions to HCD fragmentation with a normalized collision energy of 30.
Samples for protein identification obtained by immunoprecipitation and crosslinking products between ubiquitin and UBE2N were analyzed at the Biological Mass Spectrometry of the Chemistry Department at Indiana University, Bloomington. Protein mixture was reduced and alkylated, followed by addition of trypsin at a 1:100 (w/w) ratio. After digestion overnight at 37°C, the resulting peptides were desalted by Zip-tip and injected into an Easy-nLC 100 HPLC system coupled to an Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific, Bremen, Germany). Peptide samples were loaded onto a house-made C18 trap column (75 μm × 20 mm, 3 μm, 100 Å) in 0.1% formic acid. The peptides were separated using an Acclaim PepMapTM RSLC C18 analytical column (75 μm × 150 mm, 2 μm, 100 Å) using an acetonitrile-based gradient (Solvent A: 0% acetonitrile, 0.1% formic acid; Solvent B: 80% acetonitrile, 0.1% formic acid) at a flow rate of 300 nL/min. A 30 min gradient was as follows: 0–0.5 min, 2–6% B; 0.5–24 min, 6–40% B; 24–26 min, 40–100% B; 26–30 min, 100% B, followed by re-equilibration to 2% B. The electrospray ionization was carried out with a nanoESI source at a 260°C capillary temperature and 1.8 kV spray voltage. The mass spectrometer was operated in data-dependent acquisition mode with mass range 400 to 1600 m/z. The precursor ions were selected for tandem mass (MS/MS) analysis in Orbitrap with 3 sec cycle time using HCD at 28% collision energy. Intensity threshold was set at 4e5. The dynamic exclusion was set with a repeat count of 1 and exclusion duration of 30 s. The resulting data was searched in Protein Prospector (http://prospector.ucsf.edu/prospector/mshome.htm) against the 6xHis-3HA-UB-Q40E and 6xHis-UBE2N sequences. Carbamidomethylation of cysteine residues was set as a fixed modification. Protein N-terminal acetylation, oxidation of methionine, protein N-terminal methionine loss, and pyroglutamine formation were set as variable modifications. A total of three variable modifications were allowed. Trypsin digestion specificity with one missed cleavage was allowed. The mass tolerance for precursor and fragment ions was set to 10 ppm for both. The crosslinked peptide search option was selected in Protein Prospector, with the crosslinking reagent designated as “EDC” (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride). EDC-style cross linking involves the formation of a zero-length isopeptide bond between an amine group and a carboxylic acid, as would be expected for ubiquitin conjugation Peptide and protein identification cut-off scores were set to 15 and 22, respectively. All putative cross-linked spectra were manually inspected. In both case, data were analyzed manually by de novo sequencing or StavroX v.3.6.0.142 using default parameters.
To detect deamidated ubiquitin from cells, cells were lysed in 50 μL of a buffer containing 50 mM NH4HCO3, 8 M urea and 5 mM dithiothreitol and incubated for 30 min at 37°C. After this incubation period samples were diluted 8-fold with 50 mM NH4HCO3 and CaCl2 was added to a final concentration of 1 mM from a 1 M stock solution. The digestion was carried out overnight at 37°C with 2 μg of sequencing-grade trypsin (Promega). The samples were desalted using 1-mL C18 cartridges (50 mg, Stracta, Phenomenex) under the manufacturer recommendations. The samples were dried in a vacuum centrifuge, resuspended in water and quantified by the bicinchoninic acid assay (Thermo Fisher Scientific). A total of 500 ng of digested peptides were loaded in a trap C18 column and the separation was carried out in a C18 column (70 cm x 360 μm ODx75 μm ID silica tubing, Polymicro, packed with 3-μm Jupiter C18 stationary phase, Phenomenex). Peptides were eluted at a flow rate of 300 nL/min with a gradient of acetonitrile (ACN) in water, both containing 0.1% formic acid: 1–8% ACN solvent in 2 min, 8–12% ACN in 18 min, 12–30% ACN in 55 min, 30–45% ACN in 22 min, 45–95% ACN in 3 min, hold for 5 min in 95% ACN and 99–1% ACN in 10 min. Eluting peptides were analyzed in an Orbitrap mass spectrometer (Velos, Thermo Fisher Scientific) by targeting specific of peptides to fragmentation (PRM – parallel-reaction monitoring). The normalized collision energy was set to 30 in the HCD cell and spectra were collected with a m/z range of 50–2000 and resolution of 15,000 at 400 m/z. Collected spectra were analyzed using Xcalibur v2.2 (Thermo Fisher Scientific). Extracted-ion chromatograms were plotted using a tolerance of ±10 mDa and smoothed with 11 points using the Gaussian function. Quantification was done by comparing peak areas and significantly differences in ubiquitin deamidation profiles were determined by T-test assuming two tails and equal variance between samples.
In vitro deamidation of ubiquitin and Nedd8, and Native PAGE
Ten μg ubiquitin or Nedd8 was incubated with 1 μg MavC or MavCC74A in 50 μL reaction in a buffer containing 50 mM Tris-HCl (pH 8.8) at 37 °C for 2 h. 20 μL reaction mixtures were mixed with 5 μL 5x native gel loading buffer (pH 8.8), and loaded onto 10% native gels, gels were run in a Tris-glycine buffer (pH 8.8). Protein bands were visualized by Coomassie brilliant blue staining.
NF-κB luciferase reporter assay
HEK293T were grown to 70% confluence in 24 well plate, and transfected with 75 ng NF-κB reporter plasmids43 and 150 ng 4xFlag CMV empty vector, 4xFlag-MavC or 4xFlag-MavCC74A vectors. For NF-κB epistasis analysis, 150 ng corresponding plasmids were used. 75 ng plasmids that directs the expression of Renila luciferase in pRL-SV40 (Promega) was co-transfected as internal controls. At 16–18 h post transfection, cells with treated with 20 ng/mL TNF-α or 50 nM PMA for 4 h. Then cells were collected and lysed for NF-κB luciferase reporter assay following the manufacturer’s protocols (Promega, cat#E1910).
Immunostaining
HEK293 cells or Raw264.7 cells were seeded at 2×105 per well. HEK293 cells were transfected with corresponding plasmids and Raw264.7 cells were infected with corresponding strains at an MOI of 2. 16–18 h post HEK293 cells transfection and 2 h post Raw264.7 cells infection. Cells were washed with PBS and fixed by 4% formaldehyde solution for 15 min at room temperature, and premeablization by 0.2% triton solution for 5 min at RT, and blocked with 4% goat serum for 30 min at 37 °C. Bacteria were stained with the anti-rat L. pneumophila serum at a dilution of 1:10,000 for 1 h, p65 was stained with the p65 specific antibody(Cell signaling, cat# 8242S) at a dilution of 1:500, Flag peptide was stained with the Flag peptide specific antibody(Sigma, Cat# F1804) at a dilution of 1:400 for 4 h, nucleus was stained with DAPI. Then stained with Alexa Flour 594 or Alexa Flour 488 correspondingly (Thermo Fisher Scientific). At least 300 cells were scored.
Data quantitation and statistical analyses
Student’s t-test was used to compare the mean levels between two groups each with at least three independent samples.
Supplementary Material
Acknowledgements
We thank Dr. Ralph Isberg (Tufts School of Medicine, Boston, MA USA) for bacterial strains. Dr. Feng Shao (National Institute for Biological Sciences, Beijing) for plasmids. Ron Moore and Anil Shukla at the Pacific Northwest National Laboratory (PNNL) for technical support. This work was supported by National Institutes of Health grants R01AI127465(ZQL), R21AI117205 (PJH and ZQL) and GM103493. Part of this work was performed in W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a Department of Energy (DOE) office of Biological and Environmental Research (BER) national user facility located at the PNNL.
Footnotes
The authors declare no conflict of interest.
Reporting Summary. Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability. The data that support the conclusions of this study are included in this published article along with its Supplementary Information files, and are also available from the corresponding author upon request.
References
- 1.Isberg RR, O’Connor TJ & Heidtman M The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7, 13–24, doi:10.1038/nrmicro1967 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losick VP & Isberg RR NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med 203, 2177–2189 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartfeld S et al. Temporal resolution of two-tracked NF-kappaB activation by Legionella pneumophila. Cell Microbiol 11, 1638–1651, doi:10.1111/j.1462-5822.2009.01354.x (2009). [DOI] [PubMed] [Google Scholar]
- 4.Abu-Zant A et al. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol 9, 246–264 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Ge J et al. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci U S A 106, 13725–13730, doi:10.1073/pnas.0907200106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losick VP, Haenssler E, Moy MY & Isberg RR LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol 12, 1083–1097, doi:10.1111/j.1462-5822.2010.01452.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X & Chen ZJ The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol 12, 35–48, doi:10.1038/nri3111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershko A & Ciechanover A The ubiquitin system. Annu Rev Biochem 67, 425–479, doi:10.1146/annurev.biochem.67.1.425 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Maculins T, Fiskin E, Bhogaraju S & Dikic I Bacteria-host relationship: ubiquitin ligases as weapons of invasion. Cell Res 26, 499–510, doi:10.1038/cr.2016.30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J et al. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329, 1215–1218, doi:10.1126/science.1193844 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu J & Luo ZQ Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15, 591–605, doi:10.1038/nrmicro.2017.67 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Qiu J et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124, doi:10.1038/nature17657 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhogaraju S et al. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 167, 1636–1649 e1613, doi:10.1016/j.cell.2016.11.019 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kotewicz KM et al. A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host Microbe 21, 169–181, doi:10.1016/j.chom.2016.12.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodge CD, Spyracopoulos L & Glover JN Ubc13: the Lys63 ubiquitin chain building machine. Oncotarget 7, 64471–64504, doi:10.18632/oncotarget.10948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor TJ, Adepoju Y, Boyd D & Isberg RR Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A 108, 14733–14740, doi:10.1073/pnas.1111678108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L et al. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13, 227–245, doi:10.1111/j.1462-5822.2010.01531.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddins MJ, Carlile CM, Gomez KM, Pickart CM & Wolberger C Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol 13, 915–920, doi:10.1038/nsmb1148 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Lorand L & Graham RM Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol 4, 140–156, doi:10.1038/nrm1014 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Keillor JW, Clouthier CM, Apperley KY, Akbar A & Mulani A Acyl transfer mechanisms of tissue transglutaminase. Bioorg Chem 57, 186–197, doi:10.1016/j.bioorg.2014.06.003 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Klock C & Khosla C Regulation of the activities of the mammalian transglutaminase family of enzymes. Protein Sci 21, 1781–1791, doi:10.1002/pro.2162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckert RL et al. Transglutaminase regulation of cell function. Physiol Rev 94, 383–417, doi:10.1152/physrev.00019.2013 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen ZJ Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7, 758–765, doi:10.1038/ncb0805-758 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H et al. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 427, 167–171, doi:10.1038/nature02273 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Fitzgerald K, Kurt-Jones E, Finberg R & Knipe DM Herpesvirus tegument protein activates NF-kappaB signaling through the TRAF6 adaptor protein. Proc Natl Acad Sci U S A 105, 11335–11339, doi:10.1073/pnas.0801617105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanada T et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626, doi:10.1038/nature10894 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee S et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214, doi:10.1126/science.1126867 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Stewart MD, Ritterhoff T, Klevit RE & Brzovic PS E2 enzymes: more than just middle men. Cell Res 26, 423–440, doi:10.1038/cr.2016.35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Y & Rape M Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10, 755–764, doi:10.1038/nrm2780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou W et al. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun 336, 61–68, doi:10.1016/j.bbrc.2005.08.038 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi T & Yokosawa H ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun 336, 9–13, doi:10.1016/j.bbrc.2005.08.034 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Machida YJ et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell 23, 589–596, doi:10.1016/j.molcel.2006.06.024 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Rape M & Kirschner MW Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 432, 588–595, doi:10.1038/nature03023 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Ravid T & Hochstrasser M Autoregulation of an E2 enzyme by ubiquitin-chain assembly on its catalytic residue. Nat Cell Biol 9, 422–427, doi:10.1038/ncb1558 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Valleau D et al. Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila. Cell Rep 23, 568–583, doi:10.1016/j.celrep.2018.03.060 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Urbanus ML et al. Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila. Mol Syst Biol 12, 893, doi:10.15252/msb.20167381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger KH & Isberg RR Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7, 7–19 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Luo ZQ & Isberg RR Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 101, 841–846 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L et al. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog 6, e1000822, doi:10.1371/journal.ppat.1000822 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheedlo MJ et al. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc Natl Acad Sci U S A, doi:10.1073/pnas.1514568112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shevchenko A, Tomas H, Havlis J, Olsen JV & Mann M In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860, doi:10.1038/nprot.2006.468 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Gotze M et al. StavroX--a software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom 23, 76–87, doi:10.1007/s13361-011-0261-2 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Li X et al. Negative Regulation of Hepatic Inflammation by the Soluble Resistance-Related Calcium-Binding Protein via Signal Transducer and Activator of Transcription 3. Front Immunol 8, 709, doi:10.3389/fimmu.2017.00709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.