Abstract
RNA processing has emerged as a key mechanistic step in the regulation of the cellular response to environmental perturbation. Recent work has uncovered extensive remodeling of transcriptome composition upon environmental perturbation and linked the impacts of this molecular plasticity to health and disease outcomes. These isoform changes and their underlying mechanisms are varied - involving alternative sites of transcription initiation, alternative splicing, and alternative cleavage at the 3′ end of the mRNA. The mechanisms and consequences of differential RNA processing have been characterized across a range of common environmental insults, including chemical stimuli, immune stimuli, heat stress, and cancer pathogenesis. In each case, there are perturbation-specific contributions of local (cis) regulatory elements or global (trans) factors and downstream consequences. Overall, it is clear that choices in isoform usage involve a balance between the usage of specific genetic elements (ie. splice sites, polyadenlyation sites) and the timing at which certain decisions are made (ie. transcription elongation rate). Fine-tuned cellular responses to environmental perturbation are often dependent on the genetic makeup of the cell. Genetic analyses of inter-individual variation in splicing have identified genetic effects on splicing that contribute to variation in complex traits. Finally, the increase in the number of tissue types and environmental conditions analyzed for RNA processing is paralleled by the need to develop appropriate analytical tools. The combination of large datasets, novel methods and conditions explored promises to provide a much greater understanding of the role of RNA processing response in human phenotypic variation.
INTRODUCTION
The ability to interact with and respond to a range of environmental stimuli is an intrinsic property of any organism. On a cellular level, this response must be rapid, tunable, and dynamic to allow a cell to adapt quickly to a new environmental condition. While there is already a solid body of literature focusing on transcriptional gene regulatory responses, it is becoming increasingly clear that co- or post-transcriptional processing of mRNA molecules may also play large roles in regulating the composition and concentration of both the transcriptome and proteome upon a cellular response. Core RNA processing mechanisms and machinery are highly conserved (Gerstberger, Hafner, & Tuschl, 2014; Lee & Rio, 2015), with misregulation of constitutive patterns often having severe cellular consequences. For instance, mutations underlying mRNA splicing defects are implicated as causal effectors of many debilitating human diseases, such as muscular dystrophy, spinal muscular atrophy, cystic fibrosis, retinal degenerative disorders and many others (as reviewed in (Scotti & Swanson, 2016)). Analysis of differential exon usage across cellular contexts (175 individuals and 43 body sites) represented in the Genotype-Tissue Expression (GTEx) Consortium pilot dataset (Mele et al., 2015), identified gene expression as the main mechanism for cell specificity, with splicing variability likely playing a secondary or complementary role. Brain tissues are a notable exception, since they have very distinct splicing programs characterized by preferential exon inclusion. The high levels of splicing variability observed in the brain (specifically in the cerebellum) is supported by mechanisms such as the brain-specific expression of particular RNA-binding proteins. RNA processing decisions can be defining features of particular tissues and cell types and have the potential to determine cell fate (Hu et al., 2015; Xu, Wang, Luo, Zhao, & Zhou, 2017; Turner & D´ıaz-Muñoz, 2018). However, large variability in RNA processing across closely related species despite conservation of gene expression levels (Merkin, Russell, Chen, & Burge, 2012; Barbosa-Morais et al., 2012), suggests that RNA processing mechanisms are good substrates for regulatory plasticity to create fine-tuned transcriptome patterns. By extension, these pathways are likely prime candidates for coordinating the cascade of molecular events that occur when cells are faced with a change in their environment.
Broadly, mechanisms of RNA processing influence the final composition or function of an RNA molecule. This encompasses mechanisms such as 5′ capping, 3′ cleavage, 3′ polyadenylation, RNA modification RNA secondary structure, and mRNA splicing, all of which are likely to play a role in how cells respond to changing environments. In this review, we focus only on mechanisms influencing mRNA sequence or isoform composition, particularly alternative transcription initiation, alternative splicing, and alternative 3′ cleavage. These processes have been extensively studied, with well-characterized regulatory machinery, auxiliary factors, and sequence elements canonically associated with each mechanistic step (Figure 1). Alternative transcription initiation is governed by a decision to initiate transcription at an alternative start site. This process is likely determined by the local chromatin and transcription factor (TF) environment (Lai & Pugh, 2017), which is mediated by DNA sequence elements governing transcription factor binding. RNA Polymerase II (PolII) is recruited to initiate transcription, undergoing multiple phosphorylation checkpoints prior to entering productive transcript elongation (Harlen & Churchman, 2017). Processing at the 5′ end is completed with the addition of a 7-methylguanosine cap on the RNA molecule shortly after exit from the PolII exit channel (Bentley, 2014). During the pause between initiation and elongation, splicing regulatory factors (SRFs) are loaded onto the C-terminal domain of PolII and their presence is often necessary for productive elongation (Hsin & Manley, 2012). It is unclear whether the SRFs loaded onto PolII directly regulate downstream splice site choice, though the same factors are associated with the regulation of alternative splicing (Hsin & Manley, 2012). RNA splicing occurs through the recognition of 3 sites on an mRNA molecule (Lee & Rio, 2015): a 5′ splice site (9 nucleotide motif spanning the exon-intron boundary, represented by a mostly invariant GU defining the 5′ end of the intron), a branchpoint (an A nucleotide within a degenerate motif located approximately 20–50 nucleotides upstream of the 3′ end of an intron), and a 3′ splice site (approximately 15–20 nucleotide motif spanning the intron-exon boundary, starting with a polypyrimidine track and containing a mostly invariant AG defining the 3′ end of the intron). These sequences are initially bound by U1 and U2 small nuclear ribonucleoproteins (snRNPs; bound to the 5′ splice site and branchpoint, respectively), which then recruit the larger spliceosome complex that catalyzes the two sequential steps of splicing and excises an intron (Lee & Rio, 2015). Splice site choice can be regulated by the binding of SRFs to splicing regulatory elements (SREs) located in the intron or flanking exons, resulting in either enhanced usage or repression of given splice sites (Wang & Burge, 2008; Fu & Ares, 2014). Finally, cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) bind to a polyadenylation signal (PAS), usually found in the 3′ untranslated regions of mRNAs, to initiate cleavage of the mRNA approximately 30–40 nucleotides downstream of the PAS (Elkon, Ugalde, & Agami, 2013; Tian & Manley, 2013). An mRNA molecule is fully mature after the addition of the polyA tail to the 3′ end of a molecule.
Figure 1: Mechanistic details of RNA processing.
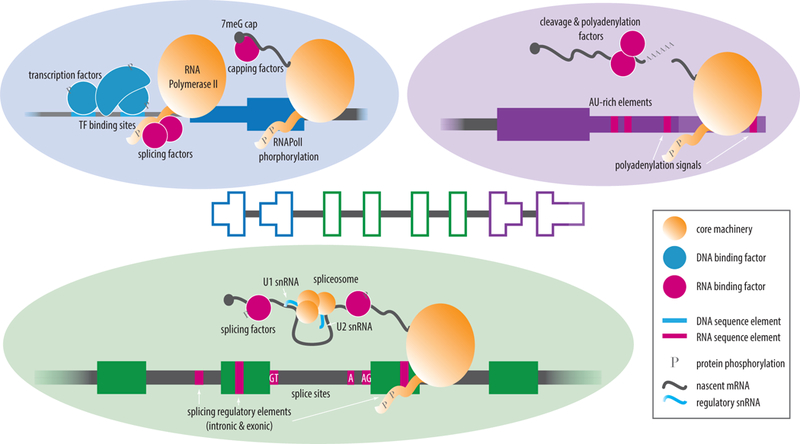
Isoform composition and complexity is regulated by core cellular machinery, DNA/RNA binding factors, and DNA/RNA regulatory sequences. A second layer of regulation is added by the dynamic mutational and modification events occurring at both RNA and protein levels. (A) Alternative transcription initiation is governed by transcriptional mechanisms regulating the usage of DNA promoters and transcription start sites. (B) Alternative splicing is governed by the spliceosome machinery and auxiliary splicing factors, which guide the recruitment of core snRNAs such as U1 and U2 to the nascent RNA molecule and direct 5′ or 3′ splice site selection. (C) Alternative cleavage and polyadenylation sites and factors direct decisions involved in 3′ end processing of nascent molecules.
Each of these stages in RNA processing are comprised of complex mechanisms involving multiple cis-acting nucleotide sequence elements and trans-acting regulatory factors, any of which can be targets in the process of regulating a cellular phenotype. Furthermore, while the processes of transcription initiation, mRNA splicing and RNA cleavage are commonly studied independently, their temporal relationships (Brugiolo, Herzel, & Neugebauer, 2013; Bentley, 2014), spatial proximity (Han, Xiong, Wang, & Fu, 2011; Bentley, 2014) and patterns of coordinated decisions (Anvar et al., 2018) suggest that they are co-regulated to influence final isoform levels. Here, we review the current literature on how RNA processing is affected upon changes in cellular environment (Figure 2), as well as the roles that various cis- or transacting regulators of RNA processing play in regulating the cellular response to environmental perturbations.
Figure 2: The diversity of RNA processing changes across environmental perturbations.
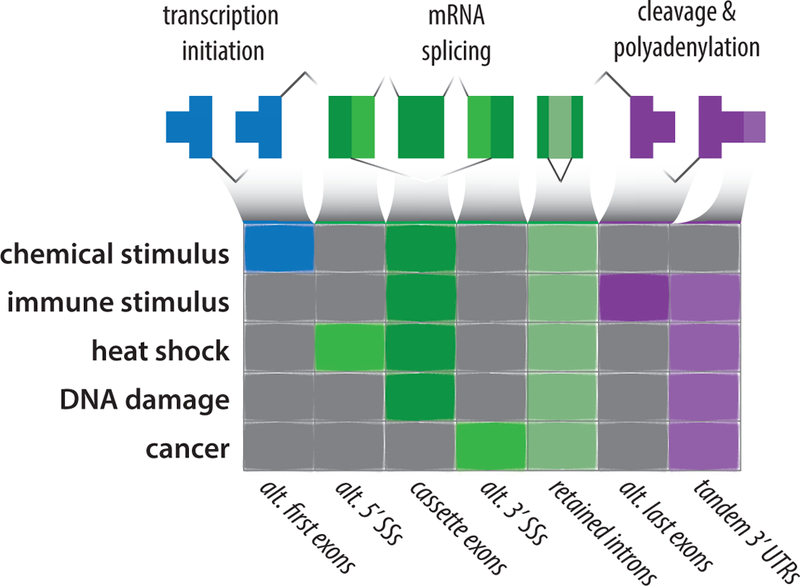
While each environmental context that has been studied so far shows substantial evidence for complex transcriptome changes across all RNA processing categories (grey boxes), each context also has characteristic global changes (dark colored boxes). These involve particular event types that are either (1) frequently observed to change, (2) change with a greater magnitude, or (3) change in a directional pattern after the environmental perturbation. Notably, retained introns are consistently enriched across all environmental perturbations, indicating that changes in environmental context affect splicing efficiency rather than only isoform choice. Global retention of annotated retained introns may serve as a sign that both regulated and canonically constitutive introns are being retained due to overall perturbation of the splicing process.
Shifts in RNA processing are treatment-specific
Environmental perturbations have large effects on both organismal and cellular traits, including gene expression and RNA processing. Analyses of RNA processing response have largely focused on genome-wide studies of response to individual chemical and biological agents or large scale screens of drug compound libraries with reporter minigenes.
Most of the individual agents investigated are relevant for cancer therapy, due to their ability to induce DNA damage (UV radiation treatment has also been investigated in this context) (Tresini, Marteijn, & Vermeulen, 2016). A genome-wide study of the splicing changes in human colon and breast cancer cell lines (HCT116 and MCF7) in response to the plant alkaloid camptothecin (CPT) used an exon microarray to study splicing (Sidebar 1) and identified alternative exon usage predominantly within splicing factors (Solier et al., 2010). The DNA topoisomerase Top1 is the primary target of CPT, and its clinical derivatives, topotecan and irinotecan are widely used as anticancer agents. Validation experiments of the genes with the largest splicing changes showed that the alternative exon usage induced by CPT is not observed with cisplatin or vinblastine, which are other two widely used chemotherapeutic agents. Genome-wide alternative splicing has also been observed in murine intestinal organoids in response to ER-stress (thapsigargin treatment) and nutrient starvation (Tsalikis et al., 2016). RNA-seq analysis showed that splicing changes shared between both treatments also occurred predominantly in splicing factors. A study of intron retention showed that certain introns that are stably detained in nuclear transcripts (without triggering nonsense-mediated decay even an hour after transcription) are more sensitive to drug-inhibition of Clk, a stress-responsive kinase (Boutz, Bhutkar, & Sharp, 2015). Similarly, these retained introns were often sensitive to changes after DNA damage, with consequences for effects on gene expression of the gene containing the retained intron. Overall the results of these studies suggest an auto-regulatory feedback loop regulating the splicing response that is common to different types of stress.
A large body of literature reports single gene, microarray and RNA-seq analyses of the transcriptional response to nuclear receptor ligands, including steroid hormones. The transcriptional response to nuclear receptor ligands such as progesterone, estrogen and vitamin D is predominantly the result of the nuclear receptor acting as a TF to directly regulate gene expression changes. However, several studies have demonstrated that NR ligands can control both transcription and mRNA splicing by recruiting receptor co-regulators (reviewed in (Zhou et al., 2015)).
Minigene reporter assays have typically been used to screen large panels of treatments for their effects on mRNA splicing (Sidebar 1). One study screened a library of 1,100 compounds (Prestwick) of Food and Drug Administration (FDA)-approved and other drugs and a smaller 340-compound library (BioMol) consisting of enzyme inhibitors and ion-channel antagonists. This study successfully identified many compounds that cause significant differential exon usage (Stoilov, Lin, Damoiseaux, Nikolic, & Black, 2008a) of the minigene construct in HEK293 cells. Several of these compounds are cardiotonic steroids, including digoxin, which is used to treat heart failure. A much larger library of compounds (> 23,000) was screened with a rapid-response splicing reporter assay in HeLa cells (Younis et al., 2010a), identifying compounds with splicing inhibitor function and validating these results with follow-up experiments.
Larger scale studies of RNA processing across different environmental contexts have been made possible by the widespread availability of high-throughput sequencing technology. In vitro studies of cellular response to environmental perturbations have the distinct advantage of allowing for tight control of the environmental conditions studied. Furthermore, the ability to perform different functional genomic assays in the same controlled context allows for a better understanding of the cellular mechanisms leading to variation in RNA processing. Recently, a high-throughput and cost-effective approach was developed to investigate transcriptional response to environmental perturbations in 250 cellular environments (Moyerbrailean et al., 2015, 2016). The high coverage RNA-seq data collected in 89 environmental conditions (including 5 cell types and 35 treatments that were chosen as the cell types and treatments with the largest transcriptional changes) provided the largest dataset to date to analyze RNA processing response (Richards et al., 2017). Using the probabilistic framework implemented in the software Mixture of Isoforms (MISO) (Katz, Wang, Airoldi, & Burge, 2010), Richards and co-workers identified 15,300 changes in RNA processing across conditions, representing a unique set of 8,489 events that significantly differ between at least one treatment and control conditions. This study cemented the idea that cellular response to environmental perturbations involves several different RNA processing mechanisms. Eight event types were considered: skipped exons (SE), retained introns (RI), alternative 3′ or 5′ splice sites (A3SS, A5SS), mutually exclusive exons (MXE), alternative first or last exons (AFE, ALE), and tandem untranslated regions (TandemUTR). The largest number of changes across all perturbations considered (after correcting for the number of events tested) occurred in AFEs, ALEs, and RIs, while A3SSs had the smallest number of changes. Overall, AFE usage changed the most across environments, though there was substantial variation in the extent to which each event type changed across treatments. For example, vitamin E, tunicamycin, and cadmium were enriched for different event types: vitamin E is enriched for A5SS while cadmium is depleted for RI.
The detailed analysis of a large number of environmental contexts in this work facilitated the evaluation of shared patterns of shifts across treatments and investigation of common regulatory mechanisms. AFEs, RIs and SEs consistently showed a directionality in their usage patterns, such that within an environmental context (cell type-treatment combination), there was often a preference for or against usage of the upstream transcription start site (TSS) leading to different AFEs, retention of an intron, or inclusion of a cassette exon. Eighteen environments were enriched for increased inclusion of retained introns, while only two contexts led to increased splicing of RIs. For AFEs, 34 environmental contexts led to increased usage of an upstream TSS, while only 7 contexts showed shifts towards usage of a downstream TSS. Richards et al. also analyzed the cis sequence elements and trans regulatory elements that may regulate these concerted RNA processing shifts. Changes in RIs correlated with differential gene expression of LARP7 and 13 other splicing factors in response to environmental perturbations, suggesting that increased expression of LARP7 could lead to more intron retention. Additionally, the presence of putative splicing factor binding sites near the RNA processing events is a predictive feature of SE and RI usage and provides a plausible cis-regulatory mechanism for these RNA-processing events. Analogously, this study found that differential expression and binding of transcription factors may regulate AFE usage in response to environmental perturbations. A total of 328 (out of 1,342) TF gene expression changes are correlated with shifts in AFE usage due to differential TSS preferences, and TF binding within 1Kb of the TSS is significantly predictive of TSS usage. Functional support for the importance of TF binding in determining AFE usage was provided by chromatin accessibility data collected in lymphoblastoid cell lines (LCLs) treated with selenium. Data from an Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) (Buenrostro, Giresi, Zaba, Chang, & Greenleaf, 2013), can be used to identify TF binding sites by inferring the footprint that they leave on chromatin (Pique-Regi et al., 2011). Significant differences in chromatin accessibility after selenium treatment were identified for 64 TF motifs located near alternative TSSs. Of these 64 motifs, 26 correspond to ETS transcription factor family members, including ELF2. These results provide support for a mechanism whereby environmental perturbations induce chromatin accessibility and differential TF binding at sites that regulate AFE usage.
Widespread RNA processing changes upon immune stimulation may be kinetically regulated
The immune response is perhaps the best known cellular program that an organism uses to mount responses to changes in environmental conditions. Infectious pathogenic agents or immune stimuli initiate a cascade of cell surface receptor interactions that trigger signaling responses to regulate a multitude of gene expression programs (Medzhitov & Janeway, 1998). Numerous studies have characterized the extensive transcriptional changes upon immune activation (Huang et al., 2001; Smale, 2010; Medzhitov & Horng, 2009), but less is known about the full extent to which changes in RNA processing impact cellular responses and how these transcriptome changes are regulated.
Initial studies focused on a few prominent immune genes and characterized how altered isoform usage after an immune response directly regulates cellular function. For instance, the CD45 antigen is alternatively spliced upon T-cell activation to exclude three alternative exons (Jacobsen et al., 2000, 2002). Increased skipping of these exons has been directly linked to reduction of CD45 phosphatase activity and attenuation of antigen receptor signaling in T-cells (Lynch & Weiss, 2000), with a broader role in preventing the development of auto-immune disease mediated by T-cell hyperactivation (Lynch & Weiss, 2000; Hermiston, Xu, Majeti, & Weiss, 2002). Similarly, the MAPK kinase MKK7 is also alternatively spliced upon T-cell activation to exclude its second exon. The resulting MKK7 isoform enhances JNK pathway activity by increasing c-Jun phosphorylation and ultimately leads to up-regulation of TNF-alpha (Martinez et al., 2015). Notably, this splicing event is part of a positive feedback loop promoting JNK-dependent signaling (Martinez et al., 2015), since the splicing event itself is dependent on JNK-induced expression of the splicing regulatory factor CELF2 (Martinez et al., 2015; Mallory et al., 2015). Approximately 25% of splicing changes after T-cell activation are likely regulated by this JNK-CELF2 autoregulatory axis during crucial stages of the immune response (Martinez et al., 2015; Mallory et al., 2015). These studies highlight how the regulation of RNA processing can attenuate the function of several individual regulators of the immune response (Martinez, Nicole M & Lynch, Kristen W, 2013).
On a genome-wide level, many studies have used either exon arrays or high-throughput sequencing (see Sidebar1 and Sidebar 2) to profile changes in RNA processing after various immune activations and stimulations. Upon T-cell activation, global profiling of alternative cassette exon usage identified 150–200 robust changes in exon usage - encompassing approximately 10% of known alternatively spliced exons expressed in T-cells (Ip et al., 2007; Martinez et al., 2012). These exons were enriched within genes related to both the immune response and cell-cycle regulation and where immediate responders upon activation (Ip et al., 2007). Strikingly, genes with activation-mediated alternative splicing were often distinct from genes with differential mRNA abundance despite sharing similar functional annotations, pointing to two independent regulatory programs affecting the transcriptome during immune activation (Ip et al., 2007). Similarly, a profile of human macrophages independently infected with either Listeria monocytogenes or Salmonella typhimurium also found that between 10–15% of alternatively processed exons expressed in macrophages change in usage upon infection, distributed across changes in all types of RNA processing events and enriched within genes with immune-related biological functions (Pai et al., 2016). Changes in 3′ UTRs and retained introns were the most prominent shifts, though the latter may be an indication of stress response similar to that observed during heat shock (described in detail below). Though genes with changes in isoform usage were also often differentially expressed, as many as 12–17% of genes expressed in macrophages change their isoform usage following infection without any detectable difference in gene expression levels (Pai et al., 2016).
Another RNA processing hallmark of both T-cell activation and bacterial infection is a shift towards usage of upstream polyadenylation signals, leading to shorter 3′ UTRs (Sandberg, Neilson, Sarma, Sharp, & Burge, 2008; Pai et al., 2016). Though this phenomenon is often associated with proliferative transitions (such as T-cell activation or cancer (Sandberg et al., 2008; Mayr & Bartel, 2009)), prominent 3′ UTR shortening after bacterial infection of macrophages - which are not proliferating - indicates that the immune response shares this regulatory strategy (Pai et al., 2016). In both T-cell activation and bacterial infection of macrophages, shorter 3′ UTRs were regulated to exclude portions of the 3′ UTR containing target sites for specific miRNAs (Sandberg et al., 2008; Pai et al., 2016). Thus, 3′ UTR shortening in the immune response might allow key genes to evade repression by immune-activated miRNAs, which otherwise could downregulate or translationally repress these genes.
Despite the numerous RNA processing changes that occur following immune activation or stimulation, the mechanisms regulating the immune-mediated changes in exon, splice site, or polyadenylation site usage are yet unclear. Though it is possible that each specific stimulus might trigger a different regulatory pathway, such as the up-regulation of CELF2 mediated by JNK-pathway signaling following T-cell activation, it is more likely that there are some shared regulatory responses across stimuli that affect the transcriptome. For instance, most immune responses lead to widespread changes in transcriptional activation and, thus, the timing of transcriptional events, both of which could influence the regulation of downstream RNA processing. Studies investigating nascent RNA production following pro-inflammatory response in macrophages induced by Lipid A stimulation found that there was an increased rate of transcript production, coupled with a large increase in chromatin-associated nascent transcripts (Bhatt et al., 2012; Pandya-Jones et al., 2013). Intriguingly, these chromatin-associated transcripts were enriched for full-length transcripts that had not yet been spliced - especially at time points shortly after stimulation - suggesting that the limiting step for mature mRNA production following immune stimulation was the timing or rate of mRNA splicing (Pandya-Jones et al., 2013). Similarly, nascent RNA profiling across a time course of T-cell activation found that splicing was initially lagging or inhibited, with increased efficiency at later stages (Davari et al., 2017). Productive splicing was associated with kinase-dependent phosphorylation of PolII that likely promoted co-transcriptional splicing (Davari et al., 2017). Thus, the kinetics of RNA processing might play a large role in fine-tuning the waves of gene expression and transcriptional activity characteristic of the immune response.
Differential protein phosphorylation and availability regulates RNA processing upon heat shock
Delayed or inhibited mRNA splicing is also a key feature of RNA processing regulation during the cellular response to heat shock. The profound influence that heat shock has on splicing regulation was first discovered in flies where un-spliced pre-mRNAs of two factors - Hsp83 (heat shock factor) and Adh (alcohol dehydrogenase enzyme) - accumulated following heat shock of Drosophila melanogaster cells (Yost & Lindquist, 1986). Broad inhibition of splicing after heat shock was later observed across many organisms and cell types, including yeast, trypanosomes, and HeLa cells (Bond, 1988; Yost, Petersen, & Lindquist, 1990; Yost & Lindquist, 1991). Severe heat shock in mammalian cells leads to widespread intron retention that causes nuclear retention of resulting transcripts (Shalgi, Hurt, Lindquist, & Burge, 2014). Introns in genes involved in protein folding or energy production are an exception and often escape this splicing inhibition (Shalgi et al., 2014), though heat shock also pauses global translational elongation (Shalgi et al., 2013). Introns that escape splicing inhibition tend to be co-transcriptionally spliced (Shalgi et al., 2014), suggesting heat-shock alters the timing or efficiency of splice site recognition and intron excision. The inhibition of splicing and widespread retention of transcripts may prevent production of aberrant proteins, reducing the burden on chaperone and proteasome machineries that are already highly burdened during heat shock. Instead, the stabilization of nuclear retained transcripts (as suggested by their detection at high levels) may serve as a reservoir of fully-transcribed mRNA molecules that could later be rapidly spliced, exported and translated to recover protein levels upon relief from stress conditions.
Though the details of the mechanisms underlying splicing inhibition during heat shock are unknown, there is evidence that Serine-rich (SR) splicing factors may play a crucial role. The loading of SR proteins onto the CTD of PolII during transcription is often necessary for proper transcription elongation and downstream splicing. They are known to bind splicing regulatory elements in exons and introns, acting as either splicing activators or repressors depending on their binding context. One SR protein, SRSF10, is customarily bound by 14–3-3 proteins (Shi & Manley, 2007), whose dissociation upon heat shock allows de-phosphorylation of SRSF10 by the phosphatase PP1 (Shi & Manley, 2007), which itself is activated by the dissociation of phosphatase inhibitors (Shi & Manley, 2007). SRSF10 de-phosphorylation upon heat shock is broadly correlated with splicing inhibition, since dephosphorylated SRSF10 interacts with the U1 snRNP and interferes with U1 snRNP recognition of 5′ splice sites (splice donors) (Shin, Feng, & Manley, 2004). Cells deficient for SRSF10 have increased temperature sensitivity and cannot recover after heat shock (Shin et al., 2004), which suggests a crucial role for SRSF10 in cell survival after heat-induced stress. As a group, SR proteins are broadly differentially phosphorylated after heat shock, due to heat-induced translocation of SR protein-specific kinases (ie. SRPK1) from the cytoplasm to the nucleus (Zhong, Ding, Adams, Ghosh, & Fu, 2009). Thus, it is possible that much of the differential splicing and widespread splicing inhibition seen upon heat shock is regulated by differential protein phosphorylation triggered by stress signaling pathways. Furthermore, some SR proteins, hnRNPs and other splicing factors have been shown to be sequestered into nuclear stress bodies during heat shock (Biamonti & Caceres, 2009; Biamonti & Vourc’h, 2010), while others have remained soluble in the nucleus. Since the state of co- vs. post-transcriptional splicing affects the fate of transcripts in heat shock, the ability of splicing factors to associated with chromatin under these conditions (and perhaps more broadly in cellular perturbation) may be critical. Thus, not only differential phosphorylation, but perhaps sequestration and/or availability of SRFs could regulate the specificity of heat-shock induced splicing inhibition or escape from it.
Similarly, 3′ cleavage and polyadenylation of mRNA molecules is also highly regulated in the context of stress responses. In heat shock, polyadenylation has been shown to be inhibited through the stress-induced post-translational modification of Poly(A) polymerase (PAP) leading to abrogation of PAP activity (Di Giammartino, Shi, & Manley, 2013). More recently, transcriptional read-through has been shown to occur in a very widespread, albeit differentially regulated manner in response to heat shock and several other stress conditions (Vilborg, Passarelli, Yario, Tycowski, & Steitz, 2015; Vilborg et al., 2017). These patterns represent a failure of polymerases to terminate properly, most likely due to inhibition of the cleavage and polyadenylation machinery. Read-through transcripts may actually function to maintain open chromatin upon stress, suggesting a beneficial role for polyadenylation inhibition (Vilborg et al., 2017). This regulated defect in 3′ end processing, which is strongly associated with stress response, is similar to observations of 3′ UTR shortening seen in proliferating cells, immune response, and cancer cells (Sandberg et al., 2008; Mayr & Bartel, 2009; Pai et al., 2016).
Splicing factors may also be moonlighting as regulators of the DNA damage response induced by UV radiation. There are many transcriptome changes upon DNA damage, with a spate of apoptotic genes expressing alternative isoforms that have functions antagonistic to the pre-damage isoform (Muñoz et al., 2009). One famous example is the alternative splicing of the BCL2L1 gene in humans upon DNA damage, where there is a splicing factor regulated shift away from the longer isoform that encodes an apoptotic inhibitor (BCL-xl) and increased expression of the shorter isoform that encodes an apoptotic activator (BCL-xs) (Shkreta, Toutant, Durand, Manley, & Chabot, 2016). In addition to their role in regulating RNA processing changes, splicing factors may also play a more direct, splicing-independent role in maintaining genome stability and repair, as reviewed in (Naro, Bielli, Pagliarini, & Sette, 2015). The dual role of splicing factors in regulating splicing and protection against DNA damage is likely facilitated by their co-localization and simultaneous interaction with both chromatin-wrapped DNA and nascent transcripts. This coordination may be enhanced by independent regulation of RNA PolII activity upon DNA damage. One study investigating transcriptional dynamics after UV irradiation found that UV induces hyper-phosphorylation of the CTD of RNA PolII, which inhibits both transcription elongation and mRNA 3′ end formation (Muñoz et al., 2009). Due to kinetic coupling between elongation rates and co-transcriptional splicing, UV-induced PolII modifications are likely to directly regulate a portion of the alternative RNA processing seen during a DNA damage response.
Mutations in the splicing machinery are a hallmark of cancer
Global transcriptional regulation has long been seen as a hallmark of cancer proliferation and tumor progression. Up-regulation of the transcription factor and proto-oncogene MYC during cancer progression results in an overall increase of the abundance of RNA molecules in a cell (Lin et al., 2012; Nie et al., 2012). The increase in pre mRNA-transcripts resulting from c-MYC up-regulation may cause an undue burden on the splicing machinery (Munding, Shiue, Katzman, Donohue, & Ares Jr, 2013), exemplified by the widespread intron retention commonly observed in tumors or cancerous cell lines. Abnormal splicing patterns in cancer include frequent retention of both alternative and constitutive introns (Dvinge & Bradley, 2015), with different sets of introns exhibiting alternative splicing in different cancer types. Notably, introns in genes encoding RNA splicing and export factors are frequently retained across many cancer conditions and often differentially expressed (Dvinge & Bradley, 2015), which could influence their trans-regulatory functions (ie. (Shapiro et al., 2011)). Genes within these pathways have been previously shown to be more likely to be differentially expressed and spliced during heat shock (Shalgi et al., 2014). Similarly, cancer cells may also be hijacking the use of widespread intron retention (Shalgi et al., 2014), which is an intrinsic aspect of the heat shock response (the pathway that governs the response to many proteotoxic stress conditions). Consistent with these observations, the increases in MYC activity have also been linked to the specific up-regulation of spliceosome components, core snRNP assembly genes, ribosomal proteins, and RNA binding protein genes (Koh et al., 2015), all of which are critical for the biogenesis of core snRNA and RNA-based regulatory machinery. In lymphomagenesis, MYC overexpression may increase the splicing fidelity for exons with weak 5′ splice sites (splice donors), resulting in splicing defects underlying proliferative and anti-apoptotic phenotypes commonly seen in lymphoma progression (Koh et al., 2015). Finally, cancer cells exhibit a shift towards increased usage of upstream polyadenylation sites, leading to shorter 3′ UTR regions (Mayr & Bartel, 2009). The shortening of 3′ UTRs, a hallmark of proliferative cellular phenotypes and immune activation (Sandberg et al., 2008; Pai et al., 2016), may lead to oncogene activation with consequences for cancer progression (Mayr & Bartel, 2009). Thus, cancer progression pathways may be co-opting stress or immune response regulatory mechanisms as a way of adapting to constant growth and proliferation during stressful environmental perturbations.
In addition to the widespread dysregulation of transcriptional activity and resulting consequences on the transcriptome in cancer, there is growing consensus that cancer progression is aided by the accumulation of somatic mutations within cancerous cells (Martincorena & Campbell, 2015; Dvinge, Kim, Abdel-Wahab, & Bradley, 2016; Tomasetti, Li, & Vogelstein, 2017). One of the most recurrently mutated genes in cancer is SF3B1 (Quesada et al., 2011; Darman et al., 2015), a key component of the spliceosome involved in branchpoint recognition (Gozani, Potashkin, & Reed, 1998). Mutated SF3B1 often promotes usage of alternative 3′ splice sites through the selection of aberrant branchpoints, resulting in tumor-specific RNA processing changes that often target transcripts for down-regulation by nonsense-mediated decay (Darman et al., 2015; Alsafadi et al., 2016). Mutations in another commonly mutated spliceosome component, U2AF1, similarly influence altered 3′ splice site selection in cancerous cells and tumors, contributing to altered transcriptome patterns but not widespread splicing failure (Ilagan et al., 2015). Somatic U2AF1 mutations have also been associated with tumor-specific alternative splicing of genes and regulatory RNAs involved in RNA processing (Shirai et al., 2015), including ribosomal RNAs, perhaps indirectly influencing the transcriptome patterns of hundreds of downstream targets. Interestingly, the most prevalent cancer-associated mutations in U2AF1 may also perturb the kinetic balance between the timing of splicing and 3′ cleavage decisions, causing splicing to occur entirely post-transcriptionally after cleavage and release of the RNA from the chromatin (Coulon et al., 2014). More broadly, the preponderance of somatic mutations that accumulate in RNA processing genes in cancerous cells, including many within tissue-specific splicing factors (Kim et al., 2015; Dvinge et al., 2016), point to a coupling between proliferation-driven mutagenesis and mis-regulation of RNA processing in order to drive cancer pathogenesis. This relationship underscores the need to better characterize the genetic regulation of RNA processing and the impact of both cis- and trans-acting polymorphisms on RNA processing regulation.
Genetic Regulation of RNA processing
Genetic variation in non-coding DNA can influence several molecular processes and phenotypes. Quantitative trait locus (QTL) mapping of molecular phenotypes is a popular approach to identify genetic variants associated with inter-individual variation in gene expression, chromatin accessibility and additional molecular traits - including RNA processing - collected through genomic and other high-throughput approaches. Although genetic studies of RNA processing are still limited, they are contributing important insights into two major areas: 1) understanding molecular mechanisms that are also used for RNA processing responses and 2) determining the genetic contribution to inter-individual variation in RNA-processing response, which has critical consequences in clinical phenotypes (e.g. response to drugs).
We can obtain an orthogonal assessment of the molecular mechanisms underlying RNA-processing plasticity in different environmental contexts by considering inter-individual variation of this molecular phenotype. Natural genetic variation can be considered nature’s perturbations and often mimic mild to extreme environmental perturbations. This is the framework that was used to interrogate the hypothesis that differential transcription factor binding can influence AFE usage (Richards et al., 2017) in response to treatment with selenium QTL mapping on AFE usage (measured as percent-spliced-in (PSI) values; AFE-QTL mapping) was performed across 200 individuals from the GEUVADIS study (Lappalainen et al., 2013) and demonstrated that AFE-QTLs in ELF2 motifs are enriched for single nucleotide polymorphisms (SNPs) computationally predicted to disrupt ELF2 binding.
Splicing QTLs (sQTLs) have been identified in humans and in model organisms (Li et al., 2016; Hasin-Brumshtein et al., 2016; Ongen & Dermitzakis, 2015; Gutierrez-Arcelus et al., 2015; Qu, Gurdziel, Pique-Regi, & Ruden, 2017; Saha et al., 2017). Analysis of alternative splicing in LCLs, fibroblasts and T-cells from 204 individuals showed that most splicing events are shared across cell types (up to 80%) and identified genetic variants correlated with inter-individual variation in splicing (Gutierrez-Arcelus et al., 2015). Furthermore, a significant correlation was found between splicing and promoter DNA methylation signals across individuals. Regions with DNA methylation and/or sQTLs were also enriched for CTCF binding, providing support for a mechanism where methylation-sensitive CTCF binding affects alternative splicing (Shukla et al., 2011). These results were replicated in a subsequent study by Li et al. that performed sQTL mapping (in addition to QTL mapping for other molecular phenotypes) in a panel of 70 LCLs (Li et al., 2016). Using a computational method that analyzes split-reads indicative of splice junctions in short sequencing reads (see Sidebar 2), this study identified almost 2,900 sQTLs, demonstrated that these QTLs are mostly independent from eQTLs, and provided strong evidence that genetic variation can affect splicing by altering chromatin-level traits. Finally, this study showed that sQTLs detected in LCLs are enriched for genetic variants associated with autoimmune diseases - similar to enrichments previously observed among eQTLs (expression quantitative trait loci) - further supporting the potential importance of splicing misregulation in complex traits. The prevalence of natural variation that influences splicing decisions also indicates that cells are able to tolerate some variance in splicing outcomes, though the magnitude of differential exon usage between two genetic alleles is often lower than that exhibited after a strong cellular perturbation or mis-regulation (Figure 3). All of these studies specifically focused only on mRNA splicing (alternative exon or splice site usage) and did not consider other types of RNA-processing events. Thus they were limited in their evaluation of the full potential for genetic variation to affect RNA processing and isoform composition.
Figure 3: The magnitude of RNA processing changes across different contexts.
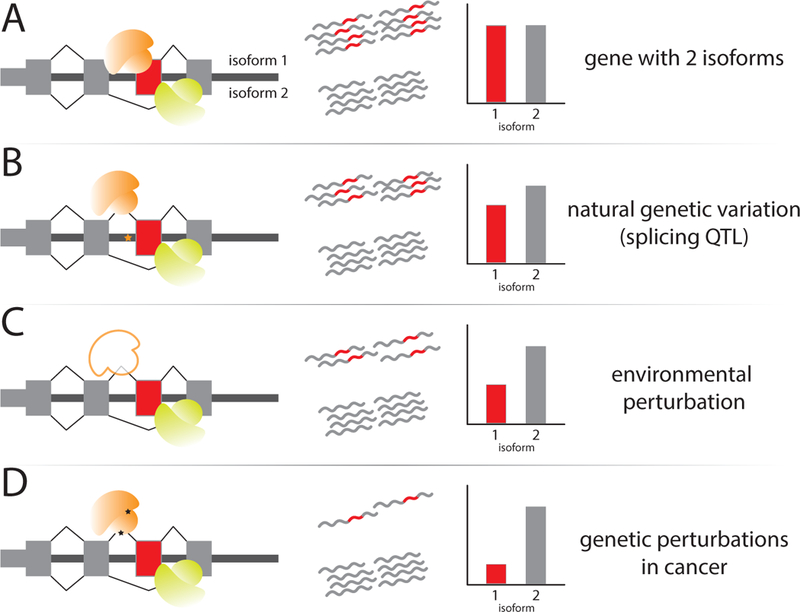
(A) Any two isoforms for a given gene are often both expressed at high levels, such that an isoform excluding a cassette exon (grey) may be expressed at an equal proportion to an isoform including the exon (red). (B) Natural genetic variation impacting isoform composition often has a small but consistent effect upon relative isoform levels. Most QTLs detected with current methods are common variants acting in cis, which are presumed to be drifting under nearly neutral selection rather than strong selective pressures. (C) RNA processing responses to common environmental perturbations activating a cellular defense system (such as stimuli or stresses) often have moderate effect sizes. These changes are characterized by global shifts in the usage of given sites or exons, putatively regulated by differential expression or post-translational modification of trans regulatory factors. (D) RNA processing changes in proliferative cancer cells are often quite severe and widespread, coupled with and likely regulated by deleterious somatic mutations in core spliceosome machinery or splicing factors.
Several studies have shown that genetic variants can alter molecular mechanisms that are important for response to environmental perturbations, thus contributing to inter-individual variation in response phenotypes. Pharmacogenetic studies have highlighted the clinical importance of such genotype-by-environment interactions that can modify an individual’s response to pharmacological treatment. The functional relevance of sQTLs has also been long recognized in the context of inter-individual differences in response to pharmaceutical drugs (for a review of sQTLs and pharmacogenetics see (Sadee et al., 2011)). Despite the increasing interest in splicing as an important mechanism for human health, the number of studies analyzing genetic and environmental effects causing differential splicing is still limited, especially relative to eQTL mapping. This is even more evident when considering studies of response sQTLs. Response eQTL mapping studies have demonstrated that genetic variation can modulate gene regulatory responses to environmental perturbations, such that the regulatory potential of a variant is unveiled only within a specific environmental context. Response QTL mapping studies not only uncovered context-specific functional genetic variants, but also uncovered the importance of personalized genomic characterizations in the study of environmental risk for human health (e.g. (Moyerbrailean et al., 2016; Mangravite et al., 2013; Knowles et al., 2018; Taylor et al., 2018; Knowles et al., 2017; Nedelec et al., 2016; Barreiro et al., 2011; Maranville et al., 2011)). Overall, our understanding of the role of genetic variation in modulating the RNA-processing response to environmental perturbations is still quite limited. There are a limited number of studies that have analyzed sQTLs in contexts relevant for human health. In two recent studies (Nedelec et al., 2016; Ye et al., 2017) over 1,000 sQTLs were identified in dendritic cells and macrophages treated with agents that stimulate the immune system (Salmonella, Listeria, Influenza, IFN-beta). The functional relevance of these sQTLs is strengthened by an enrichment for signatures of positive selection among these sites and high overlap with SNPs associated with autoimmune and neurodegenerative diseases as assessed by genome-wide association studies (GWAS). Another study identified over 100 cis-sQTLs in response to lead exposure in Drosophila melanogaster heads (Qu et al., 2017). The study also identified a potential trans-sQTL hotspot that regulated a cluster of transcripts in response to lead, indicating that trans-acting polymorphisms may have large impacts on isoform composition in different environmental contexts.
Sidebar 1: The challenges of measuring alternative RNA processing: experimental techniques
There are many experimental approaches that have been developed to assess RNA processing patterns. The usage and location of transcription start sites is typically measured using a cap-pulldown method, such as the Cap Analysis of Gene Expression (CAGE), either for a single gene or in a high-throughput fashion (Kodzius et al., 2006) (Figure 4A), or newer and more precise methods such as Cap-seq that isolate RNA based on pulldown of cap-binding complexes (Xie et al., 2013). Similarly, the position of the 3′ end of a transcript can be assayed using Rapid Analysis of cDNA Ends (RACE) methods (Frohman, Dush, & Martin, 1988) (Figure 4B), or newer methods like 3p-seq that provide more resolution for 3′ end identification (Jan, Friedman, Ruby, & Bartel, 2011). The majority of technology development has focused on measuring alternative splice site and exon usage. Classically, these experiments were performed with splicing reporter assays involving plasmids containing minigenes designed to resemble introns and exons from a real gene with known alternative splicing (ie. (Stoilov, Lin, Damoiseaux, Nikolic, & Black, 2008b) and (Somarelli et al., 2013)), or measure splicing in a synthetic gene construct (ie. inserting introns into Luciferase (Younis et al., 2010b) or a dual-signal reporter such as that encoding both green fluorescent protein (GFP) and red fluorescent protein (RFP) (Nasim & Eperon, 2006; Orengo, Bundman, & Cooper, 2006)) (Figure 4C). Minigenes contain all the sequence elements necessary to create a mature transcript (and often peptide fragment) and are quite useful to: (1) perform mutagenesis experiments to dissect the cis-elements responsible for splicing of the given intron or (2) screen the splicing response of the specific intron to treatment with many stimuli or conditions. The read-out for all perturbations (either mutagenesis or environmental conditions) can be quickly measured in a high-throughput fashion using either qRT-PCR, fluorescence assays, or cell sorting. However, these techniques are limited to assaying the response of a small number of genes and exons in a synthetic construct. More recently, microarrays - especially exon arrays - allowed for the measurement of hundreds of exons at a time. However, the limited sensitivity of exon arrays hindered the measurement of small changes in splicing and these arrays were still reliant on knowing all isoform possibilities.
Figure 4: Methods to assay isoform composition.

(A) Cap-analysis of gene expression (CAGE) protocols are commonly used to interrogate the location and usage of transcription initiation sites. The technique relies on capture of molecules with a 5′ 7-methylguanosine cap, commonly through selective biotinylation of the cap. Following selection, a 5′ adapter is added, containing a enzyme recognition site that allows cleavage of the cDNA at a given distance away from the adapter. The resulting product can be assayed through PCR and sequencing with either conventional or high-throughput methods. (B) Rapid analysis of cDNA ends (RACE) protocols allow the interrogation of the location and usage of mRNA cleavage and polyadenylation sites. Synthesis of cDNA is initiated by annealing of an oligo-dT primer that contains a known 3′ adapter sequence. Second strand synthesis is initiated using either a gene-specific or random primer and the resulting product can be assayed through PCR and sequencing with either conventional or high-throughput methods. (C) Splicing reporter assays are used to validate or interrogate sequences that affect splice site usage. A minigene construct containing either a real intron flanked by its corresponding exons or a synthetic construct encoding either luciferase, GFP or RFP is cloned into a reporter plasmid. The splicing of the introns in any of theses cases can be assayed using quantitative RT-PCR. For luciferase constructs, the increased splicing of intervening intron would result in increased luciferase production, which can be assayed using fluorescence based assays. Finally, the GFP/RFP construct often interrogates usage of individual splice sites, such that usage of an upstream splice site would encode GFP, while usage of the downstream splice site would encode RFP. The relative proportion of splice site usage can then be assayed by sorting cells based on markers for GFP and RFP. (D) Short reads from RNA-sequencing techniques can be used to identify and quantify RNA processing patterns across multiple event types. Quantification of whole isoforms or exons often rely on the quantification reads within all regions present in an isoform, while methods specifically interrogating alternative patterns rely on reads that are informative for usage of particular exons. The most informative reads are junction reads that are split between two exons and span the resulting junction. We note that reads for terminal exons could indicate either differential TSS or polyA site usage (for AFEs and ALEs, respectively) or differential splicing of terminal exons to internal exons.
Sidebar 2: The challenges of measuring alternative RNA processing: high-throughput advances
The advent of high-throughput sequencing of cDNA (RNA-seq) led to an explosion in splicing-related research, especially for assessing the extent to which there are global RNA processing changes in varying cellular conditions. RNA-seq enabled the genomewide assessment of changes in isoform usage, coupled with fine-scaled measurements of isoform levels and identification of novel isoform patterns. However, isoform analyses of RNA-seq data presented new computational and statistical challenges (Kakaradov, Xiong, Lee, Jojic, & Frey, 2012). This is epitomized by the multitude of algorithms that are still being designed to identify new isoforms, quantify their relative usage across multiple samples, and calculate statistical significance (Garber, Grabherr, Guttman, & Trapnell, 2011; Steijger et al., 2013; Liu, Loraine, & Dickerson, 2014; Hooper, 2014). These algorithms can be broadly categorized into three classes: (1) quantifying expression levels and differential expression of entire isoforms (Trapnell et al., 2010; Bray, Pimentel, Melsted, & Pachter, 2016; Patro, Duggal, Love, Irizarry, & Kingsford, 2017) , (2) quantifying exon usage and differential usage levels, based on locally informative reads (Katz et al., 2010; Vaquero-Garcia et al., 2016), and (3) quantifying sites of exon excision and differential splicing, based on split-junction reads spanning two non-contiguous transcript regions (Vaquero-Garcia et al., 2016; Li et al., 2018) (Figure 4D). Despite the ongoing development of computational tools in this area and lack of consensus about the ideal statistical models, all of these approaches have yielded reproducible results and generated many testable hypotheses. Combining the advantages of classical single-gene assays and more recent high-throughput approaches, studies are now using high-throughput sequencing to screen causal variants for RNA processing changes in reporter assays (ie. massively parallel reporter assays (MPRAs) (Rosenberg, Patwardhan, Shendure, & Seelig, 2015; Bogard, Linder, Rosenberg, & Seelig, 2018)) and assess the broader influences of in-vivo mutagenesis of splicing regulatory elements or factors (using techniques such as CRISPR (Kapahnke, Banning, & Tikkanen, 2016; Mou et al., 2017; Yue & Ogawa, 2018)).
CONCLUSIONS
Eukaryotic transcriptomes are dynamic entities that react to the smallest changes in the cellular environment with rapid and consequential responses. It is clear that the interplay between transcriptional, RNA processing, and post-translational regulatory mechanisms is at the heart of the cellular response. While recent studies have started to scratch the surface of characterizing environment-specific transcriptomes and how they regulate cellular function in various contexts, there are still many challenges remaining. One question that remains unaddressed is whether RNA processing response to a certain environmental perturbation is cell type specific. The studies performed so far used different cell lines, therefore confounding comparisons across RNA processing responses in the few cases where the same treatment was used for more than one cell type. With the advent of new high-throughput sequencing technology, it is easier to profile hundreds of changes in RNA processing events across many cell types and environments. However, we are often unable to predict which changes have a direct phenotypic impact, their consequences, and the mechanisms by which these effects occur.
Mapping natural genetic variants that modulate RNA processing responses (response QTLs) represents a promising avenue to characterize mechanisms underlying isoform variation and gain a deeper understanding of inter-individual variation in the RNA-processing response to environmental perturbations. Examples of such context-specific genetic effects on splicing have been identified for over 50 genes in dendritic cells treated with Influenza and IFN-beta citeYe188961. However further studies with larger sample sizes and a wider range of environmental contexts are needed to paint a complete picture of these context-specific effects and their relevance for human health. As the cost of generating deep sequencing data continues to decrease, future efforts should focus on developing proper statistical and computational methods to robustly identify isoforms and quantify their abundance (Sidebar 2). Current methods have many technical gaps - for instance, they often lack the ability to account for experimental replicates, have limited power for differentiation of full length-transcripts, and results are often difficult to reproduce or interpret, especially across platforms or statistical methods. While many of these caveats are starting to be solved for differential expression quantification and analysis, computational approaches to study RNA processing are lagging behind in this regard. The development and availability of such tools will enable the study of how environmental and genetic perturbations coordinately affect all aspects of RNA-processing - beyond splicing and isoform expression - thus gaining a deeper understanding of cellular mechanisms relevant for health and disease in different environmental contexts.
Though it is clear that both cis and trans regulatory factors of RNA processing play a role in the coordinated response to environmental perturbations, we are still far from a complete characterization of the relative importance of such mechanisms. Results from sQTL mapping can help identify novel regulatory sequences and therefore cis-regulatory mechanisms in the RNA processing response. Additionally, these studies can inform predictive computational models for regulatory sequences, similar to those already available to predict non-coding variants that regulate gene expression. Ultimately such catalogs of regulatory variants influencing RNA processing response will contribute to the interpretation of association signals for complex traits in humans. These cis-regulatory sequences are the targets of trans factors that are often unknown. A full understanding of the mechanistic underpinnings of RNA-processing response will require that in parallel to cis-regulatory sequences, trans factors are characterized by means of experimental assays. High throughput assays to identify DNA and RNA binding sites and factors are already available and have enabled studies of steady-state binding patterns in many cell lines and tissues (ie. work of the ENCODE consortium and parallel efforts). Thus future studies should expand upon this work by focusing on performing these assays in a variety of cellular environments and contexts.
More broadly, it appears that variability in the gene regulatory response to environmental perturbations is reliant on a balance between cis and trans regulation. Differential usage of genetic loci influencing isoform composition (ie. transcription start sites, splice sites, polyadenylation sites) is canonically thought of as being regulated locally, such that the decision made at a particular locus is independent of genome-wide decisions. However, it is increasingly clear that many changes are happening in a concerted fashion across many loci. For instance, context-specific shifts towards usage of upstream sites across genes or increased inclusion of exons indicate a global regulatory change. Previous studies have found that these changes may be due to (1) changes in the stoichiometry of regulatory factors and their regulatory targets (Berg et al., 2012; Munding et al., 2013), (2) differential expression (or isoform usage) of RNA binding proteins that have downstream effects on RNA processing patterns (see above), and (3) signaling pathways that influence kinase activity to change protein phosphorylation of key RNA processing factors (see above). These observations underlie a need to better characterize how the transcriptional, RNA processing, and post-transcriptional or translational regulatory responses are all coordinated upon cellular perturbation. Furthermore, it is crucial to understand the timing at which regulatory decisions are occurring, since environmental stimuli may be directly acting on the speed or order at which RNA processing decisions are being made to regulate the final transcriptome. Finally, focusing on a genome-wide cellular milieu rather than individual events is crucial to delineate which variability has phenotypic consequences, rather than just being a consequence of cellular noise. Along these lines, emerging single-cell techniques for assaying gene expression and RNA processing (Shalek et al., 2013; Vu et al., 2018; Hayashi et al., 2018) will characterize these mechanisms at a higher resolution and may provide insights into the variability of cellular responses across a population of cells.
ACKNOWLEDGEMENTS
We thank Eleni Jaecklein, Allison Richards, Roger Pique-Regi and members of the Luca and Pai groups for helpful discussions. This work was partially supported by funds from the University of Massachusetts Medical School (AAP), NIH R01GM109215 (FL) and AHA 14SDG20450118 (FL).
References
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, … Stern M-H(2016, February). Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nature communications, 7 , 10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anvar SY, Allard G, Tseng E, Sheynkman GM, de Klerk E, Vermaat M, … ‘t Hoen PAC (2018, March). Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome biology , 19 (1), 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, … Blencowe BJ (2012, December). The Evolutionary Landscape of Alternative Splicing in Vertebrate Species. Science, 338 (6114), 1587–1593. [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, & Gilad Y (2011). Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. PNAS doi: 10.1073/pnas.1115761109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1115761109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014, March). Coupling mRNA processing with transcription in time and space. Nature Reviews Genetics, 15 (3), 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg MG, Singh LN, Younis I, Liu Q, Pinto AM, Kaida D, … Dreyfuss G (2012, July). U1 snRNP Determines mRNA Length and Regulates Isoform Expression. Cell , 150 (1), 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong A-J, Barozzi I, Lissner MM, Natoli G, … Smale ST (2012, July). Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell , 150 (2), 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G, & Caceres JF (2009, March). Cellular stress and RNA splicing. Trends in biochemical sciences, 34 (3), 146–153. [DOI] [PubMed] [Google Scholar]
- Biamonti G, & Vourc’h C (2010, June). Nuclear stress bodies. Cold Spring Harbor perspectives in biology , 2 (6), a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogard N, Linder J, Rosenberg AB, & Seelig G (2018). Predicting the impact of cis- regulatory variation on alternative polyadenylation. bioRxiv Retrieved from https://www.biorxiv.org/content/early/2018/04/12/300061 doi: 10.1101/300061 [Google Scholar]
- Bond U (1988, November). Heat shock but not other stress inducers leads to the disruption of a sub-set of snRNPs and inhibition of in vitro splicing in HeLa cells. The EMBO journal , 7 (11), 3509–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Bhutkar A, & Sharp PA (2015, January). Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes & development , 29 (1), 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, & Pachter L (2016, April). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology . [DOI] [PubMed] [Google Scholar]
- Brugiolo M, Herzel L, & Neugebauer KM (2013). Counting of co-transcriptional splicing. F1000Prime Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, & Greenleaf WJ (2013, December). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods, 10 (12), 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR, & Black DL (2014, October). Kinetic competition during the transcription cycle results in stochastic RNA processing. Elife, 3 , e03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, … Buonamici S (2015, November). Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3’ Splice Site Selection through Use of a Different Branch Point. Cell reports, 13 (5), 1033–1045. [DOI] [PubMed] [Google Scholar]
- Davari K, Lichti J, Gallus C, Greulich F, Uhlenhaut NH, Heinig M, … Glasmacher E (2017, April). Rapid Genome-wide Recruitment of RNA Polymerase II Drives Transcription, Splicing, and Translation Events during T Cell Responses. Cell reports, 19 (3), 643–654. [DOI] [PubMed] [Google Scholar]
- Di Giammartino DC, Shi Y, & Manley JL (2013, January). PARP1 represses PAP and inhibits polyadenylation during heat shock. Molecular cell , 49 (1), 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, & Bradley RK (2015). Widespread intron retention diversifies most cancer transcriptomes. Genome medicine, 7 (1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, & Bradley RK (2016, July). RNA splicing factors as oncoproteins and tumour suppressors. Nature reviews. Cancer , 16 (7), 413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, & Agami R (2013, July). Alternative cleavage and polyadenylation: extent, regulation and function. Nature reviews. Genetics, 14 (7), 496–506. [DOI] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, & Martin GR (1988, December). Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proceedings of the National Academy of Sciences of the United States of America, 85 (23), 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, & Ares M (2014, October). Context-dependent control of alternative splicing by RNA-binding proteins. Nature reviews. Genetics, 15 (10), 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Grabherr MG, Guttman M, & Trapnell C (2011, June). Computational methods for transcriptome annotation and quantification using RNA-seq. Nature methods, 8 (6), 469–477. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, & Tuschl T (2014, December). A census of human RNA-binding proteins. Nature reviews. Genetics, 15 (12), 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, & Reed R (1998, August). A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Molecular and cellular biology, 18 (8), 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Ongen H, Lappalainen T, Montgomery SB, Buil A, Yurovsky A, … Dermitzakis ET (2015, January). Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet, 11 (1), e1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, & Fu X-D (2011, June). Pre-mRNA splicing: where and when in the nucleus. Trends in cell biology , 21 (6), 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlen KM, & Churchman LS (2017, April). The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nature reviews. Molecular cell biology , 18 (4), 263–273. [DOI] [PubMed] [Google Scholar]
- Hasin-Brumshtein Y, Khan AH, Hormozdiari F, Pan C, Parks BW, Petyuk VA, … Smith DJ (2016, September). Hypothalamic transcriptomes of 99 mouse strains reveal trans eQTL hotspots, splicing QTLs and novel non-coding genes. Elife, 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, & Nikaido I (2018, February). Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nature communications, 9 (1), 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, & Weiss A (2002, January). Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. The Journal of clinical investigation, 109 (1), 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JE (2014, January). A survey of software for genome-wide discovery of differential splicing in RNA-Seq data. Human genomics, 8 (1), 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J-P, & Manley JL (2012, October). The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & development , 26 (19), 2119–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Warri A, Jin L, Zwart A, Riggins RB, Fang H-B, & Clarke R (2015, January). NF-κB signaling is required for XBP1 (unspliced and spliced)-mediated effects on antiestrogen responsiveness and cell fate decisions in breast cancer. Molecular and cellular biology , 35 (2), 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, … Hacohen N (2001, October). The plasticity of dendritic cell responses to pathogens and their components. Science (New York, N.Y.), 294 (5543), 870–875. [DOI] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, & Bradley RK (2015, January). U2AF1 mutations alter splice site recognition in hematological malignancies. Genome research, 25 (1), 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, & Lynch KW (2007, April). Global analysis of alternative splicing during T-cell activation. RNA (New York, N.Y.), 13 (4), 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M, Hoffmann S, Cepok S, Stei S, Ziegler A, Sommer N, & Hemmer B (2002, June). A novel mutation in PTPRC interferes with splicing and alters the structure of the human CD45 molecule. Immunogenetics, 54 (3), 158–163. [DOI] [PubMed] [Google Scholar]
- Jacobsen M, Schweer D, Ziegler A, Gaber R, Schock S, Schwinzer R, … Hemmer B (2000, December). A point mutation in PTPRC is associated with the development of multiple sclerosis. Nature genetics, 26 (4), 495–499. [DOI] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, & Bartel DP (2011, January). Formation, regulation and evolution of Caenorhabditis elegans 3’UTRs. Nature, 469 (7328), 97– 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakaradov B, Xiong HY, Lee LJ, Jojic N, & Frey BJ (2012, April). Challenges in estimating percent inclusion of alternatively spliced junctions from RNA-seq data. BMC bioinformatics, 13 Suppl 6 , S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahnke M, Banning A, & Tikkanen R (2016, December). Random Splicing of Several Exons Caused by a Single Base Change in the Target Exon of CRISPR/Cas9 Mediated Gene Knockout. Cells, 5 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, & Burge CB (2010, December). Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods, 7 (12), 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC-W, Ramakrishnan A, … Abdel-Wahab O (2015, May). SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer cell , 27 (5), 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DA, Burrows CK, Blischak JD, Patterson KM, Serie DJ, Norton N, … Gilad Y (2018, May). Determining the genetic basis of anthracycline-cardiotoxicity by response QTL mapping in induced cardiomyocytes. Elife, 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles DA, Davis JR, Edgington H, Raj A, Fave MJ, Zhu X, … Battle A (2017, July). Allele-specific expression reveals interactions between genetic variation and environment. Nat. Methods, 14 (7), 699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, … Carninci P (2006, March). CAGE: cap analysis of gene expression. Nature methods, 3 (3), 211–222. [DOI] [PubMed] [Google Scholar]
- Koh CM, Bezzi M, Low DHP, Ang WX, Teo SX, Gay FPH, … Guccione E (2015, July). MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature, 523 (7558), 96–100. [DOI] [PubMed] [Google Scholar]
- Lai WKM, & Pugh BF (2017, September). Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nature reviews. Molecular cell biology , 18 (9), 548–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, ‘t Hoen PA., Monlong J, Rivas MA, … Hamosh A (2013, September). Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501 (7468), 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, & Rio DC (2015). Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annual review of biochemistry , 84 , 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, & Pritchard JK (2018, January). Annotation-free quantification of RNA splicing using LeafCutter. Nature genetics, 50 (1), 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, … Pritchard JK (2016, April). RNA splicing is a primary link between genetic variation and disease. Science, 352 (6285), 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lov´en J, Rahl PB, Paranal RM, Burge CB, Bradner JE, … Young RA (2012, September). Transcriptional amplification in tumor cells with elevated c-Myc. Cell , 1 5′ (1), 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Loraine AE, & Dickerson JA (2014, December). Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC bioinformatics, 15 , 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW, & Weiss A (2000, January). A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Molecular and cellular biology , 20 (1), 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory MJ, Allon SJ, Qiu J, Gazzara MR, Tapescu I, Martinez NM, … Lynch KW (2015, April). Induced transcription and stability of CELF2 mRNA drives widespread alternative splicing during T-cell signaling. Proceedings of the National Academy of Sciences of the United States of America, 112 (17), E2139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangravite LM, Engelhardt BE, Medina MW, Smith JD, Brown CD, Chasman DI, … Krauss RM (2013, October). A statin-dependent QTL for GATM expression is associated with statin-induced myopathy. Nature, 502 (7471), 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranville JC, Luca F, Richards AL, Wen X, Witonsky DB, Baxter S, … Di Rienzo A (2011). Interactions between glucocorticoid treatment and cis-regulatory polymorphisms contribute to cellular response phenotypes. PLoS genetics, 7 , e1002162. doi: 10.1371/journal.pgen.1002162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, & Campbell PJ (2015, September). Somatic mutation in cancer and normal cells. Science (New York, N.Y.), 349 (6255), 1483–1489. [DOI] [PubMed] [Google Scholar]
- Martinez NM, Agosto L, Qiu J, Mallory MJ, Gazzara MR, Barash Y, … Lynch KW (2015, October). Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation. Genes & development , 29 (19), 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA, Heyd F, … Lynch KW (2012, May). Alternative splicing networks regulated by signaling in human T cells. RNA (New York, N.Y.), 18 (5), 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Nicole M, & Lynch Kristen W. (2013). Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunological reviews, 253 (1), 216–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, & Bartel DP (2009, August). Widespread Shortening of 3UTRs by Alternative Cleavage and Polyadenylation Activates Oncogenes in Cancer Cells. Cell , 138 (4), 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, & Horng T (2009, October). Transcriptional control of the inflammatory response. Nature Reviews Immunology , 9 (10), 692–703. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, & Janeway CA (1998, October). Innate immune recognition and control of adaptive immune responses. Seminars in immunology , 10 (5), 351–353. [DOI] [PubMed] [Google Scholar]
- Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, … Guigo R (2015, May). Human genomics. The human transcriptome across tissues and individuals. Science, 348 (6235), 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, & Burge CB (2012, December). Evolutionary Dynamics of Gene and Isoform Regulation in Mammalian Tissues. Science, 338 (6114), 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Smith JL, Peng L, Yin H, Moore J, Zhang X-O, … Xue W (2017, June). CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome biology , 18 (1), 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean GA, Davis GO, Harvey CT, Watza D, Wen X, Pique-Regi R, & Luca F (2015, October). A high-throughput RNA-seq approach to profile transcriptional responses. Sci Rep, 5 , 14976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyerbrailean GA, Richards AL, Kurtz D, Kalita CA, Davis GO, Harvey CT, … Luca F (2016, December). High-throughput allele-specific expression across 250 environmental conditions. Genome Res, 26 (12), 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munding EM, Shiue L, Katzman S, Donohue JP, & Ares M Jr (2013, August). Competition between Pre-mRNAs for the Splicing MachineryDrives Global Regulation of Splicing. Molecular Cell , 5′ (3), 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz MJ, P´erez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, … Kornblihtt AR (2009, May). DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell , 137 (4), 708–720. [DOI] [PubMed] [Google Scholar]
- Naro C, Bielli P, Pagliarini V, & Sette C (2015). The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability. Frontiers in genetics, 6 , 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim MT, & Eperon IC (2006). A double-reporter splicing assay for determining splicing efficiency in mammalian cells. Nature protocols, 1 (2), 1022–1028. [DOI] [PubMed] [Google Scholar]
- Nedelec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, Dumaine A, … Barreiro B (2016, October). Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell , 167 (3), 657–669. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, … Levens D (2012, September). c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell , 1 5′ (1), 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongen H, & Dermitzakis ET (2015, October). Alternative Splicing QTLs in European and African Populations. Am. J. Hum. Genet, 97 (4), 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo JP, Bundman D, & Cooper TA (2006). A bichromatic fluorescent reporter for cell-based screens of alternative splicing. Nucleic acids research, 34 (22), e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Baharian G, Pag´e Sabourin A, Brinkworth JF, N´ed´elec Y, Foley JW, … Barreiro LB (2016, September). Widespread Shortening of 3’ Untranslated Regions and Increased Exon Inclusion Are Evolutionarily Conserved Features of Innate Immune Responses to Infection. PLoS genetics, 12 (9), e1006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya-Jones A, Bhatt DM, Lin C-H, Tong A-J, Smale ST, & Black DL (2013, June). Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA (New York, N.Y.), 19 (6), 811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, & Kingsford C (2017, April). Salmon provides fast and bias-aware quantification of transcript expression. Nature methods, 14 (4), 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, & Pritchard JK (2011, March). Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res, 21 (3), 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu W, Gurdziel K, Pique-Regi R, & Ruden DM (2017). Identification of Splicing Quantitative Trait Loci (sQTL) in Drosophila melanogaster with Developmental Lead (Pb2+) Exposure. Front Genet , 8 , 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordo´ñez GR, Jares P, Bassaganyas L, … Lo´pez-Ot´ın C (2011, December). Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics, 44 (1), 47–52. [DOI] [PubMed] [Google Scholar]
- Richards AL, Watza D, Findley A, Alazizi A, Wen X, Pai AA, … Luca F (2017, October). Environmental perturbations lead to extensive directional shifts in RNA processing. PLoS Genet, 13 (10), e1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg AB, Patwardhan RP, Shendure J, & Seelig G (2015, October). Learning the sequence determinants of alternative splicing from millions of random sequences. Cell , 163 (3), 698–711. [DOI] [PubMed] [Google Scholar]
- Sadee W, Wang D, Papp AC, Pinsonneault JK, Smith RM, Moyer RA, & Johnson AD (2011, March). Pharmacogenomics of the RNA world: structural RNA polymorphisms in drug therapy. Clin. Pharmacol. Ther, 89 (3), 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Kim Y, Gewirtz ADH, Jo B, Gao C, McDowell IC, … Zhu J (2017, November). Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res, 27 (11), 1843–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, & Burge CB (2008, June). Proliferating Cells Express mRNAs with Shortened 3’ Untranslated Regions and Fewer MicroRNA Target Sites. Science, 320 (5883), 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, & Swanson MS (2016, January). RNA mis-splicing in disease. Nature Reviews Genetics, 17 (1), 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, … Regev A (2013, June). Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature, 498 (7453), 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, & Burge CB (2013, February). Widespread regulation of translation by elongation pausing in heat shock. Molecular Cell , 49 (3), 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Lindquist S, & Burge CB (2014, June). Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell reports, 7 (5), 1362–1370. [DOI] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay H, … Gertler FB (2011, August). An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS genetics, 7 (8), e1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, & Manley JL (2007, October). A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Molecular cell , 28 (1), 79–90. [DOI] [PubMed] [Google Scholar]
- Shin C, Feng Y, & Manley JL (2004, February). Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature, 427 (6974), 553–558. [DOI] [PubMed] [Google Scholar]
- Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, … Walter MJ (2015, May). Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer cell , 27 (5), 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkreta L, Toutant J, Durand M, Manley JL, & Chabot B (2016, November). SRSF10 Connects DNA Damage to the Alternative Splicing of Transcripts Encoding Apoptosis, Cell-Cycle Control, and DNA Repair Factors. Cell reports, 17 (8), 1990– 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, … Oberdoerffer S (2011, November). CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature, 479 (7371), 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST (2010, March). Selective transcription in response to an inflammatory stimulus. Cell , 140 (6), 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solier S, Barb J, Zeeberg BR, Varma S, Ryan MC, Kohn KW, … Pommier Y (2010, October). Genome-wide analysis of novel splice variants induced by topoisomerase I poisoning shows preferential occurrence in genes encoding splicing factors. Cancer Res, 70 (20), 8055–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somarelli JA, Schaeff D, Bosma R, Bonano VI, Sohn JW, Kemeny G, … Garcia-Blanco MA (2013, January). Fluorescence-based alternative splicing reporters for the study of epithelial plasticity in vivo. RNA (New York, N.Y.), 19 (1), 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engstro¨m PG, Kokocinski F, RGASP Consortium, Hubbard TJ, … Bertone P (2013, December). Assessment of transcript reconstruction methods for RNA-seq. Nature methods, 10 (12), 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, & Black DL (2008a, August). A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl. Acad. Sci. U.S.A, 105 (32), 11218–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Lin C-H, Damoiseaux R, Nikolic J, & Black DL (2008b, August). A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proceedings of the National Academy of Sciences of the United States of America, 105 (32), 11218–11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DL, Knowles DA, Scott LJ, Ramirez AH, Casale FP, Wolford BN, … Collins FS (2018). Interactions between genetic variation and cellular environment in skeletal muscle gene expression. PLoS ONE , 13 (4), e0195788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, & Manley JL (2013, June). Alternative cleavage and polyadenylation: the long and short of it. Trends in biochemical sciences, 38 (6), 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Li L, & Vogelstein B (2017, March). Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science (New York, N.Y.), 355 (6331), 1330– 1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, … Pachter L (2010, May). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology , 28 (5), 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M, Marteijn JA, & Vermeulen W (2016). Bidirectional coupling of splicing and ATM signaling in response to transcription-blocking DNA damage. RNA Biol , 13 (3), 272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalikis J, Pan Q, Tattoli I, Maisonneuve C, Blencowe BJ, Philpott DJ, & Girardin SE (2016, August). The transcriptional and splicing landscape of intestinal organoids undergoing nutrient starvation or endoplasmic reticulum stress. BMC Genomics, 17 , 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M, & D´ıaz-Muñoz MD. (2018, February). RNA-binding proteins control gene expression and cell fate in the immune system. Nature immunology , 19 (2), 120–129. [DOI] [PubMed] [Google Scholar]
- Vaquero-Garcia J, Barrera A, Gazzara MR, Gonza´lez-Vallinas J, Lahens NF, Hogenesch JB, … Barash Y (2016, February). A new view of transcriptome complexity and regulation through the lens of local splicing variations. eLife, 5 , e11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, & Steitz JA (2015, August). Widespread Inducible Transcription Downstream of Human Genes. Molecular cell , 59 (3), 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Sabath N, Wiesel Y, Nathans J, Levy-Adam F, Yario TA, … Shalgi R (2017, October). Comparative analysis reveals genomic features of stress-induced transcriptional readthrough. Proceedings of the National Academy of Sciences of the United States of America, 114 (40), E8362–E8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TN, Wills QF, Kalari KR, Niu N, Wang L, Pawitan Y, & Rantalainen M (2018, February). Isoform-level gene expression patterns in single-cell RNA-sequencing data. Bioinformatics (Oxford, England: ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, & Burge CB (2008, May). Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA (New York, N.Y.), 14 (5), 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Li M, Vilborg A, Lee N, Shu M-D, Yartseva V, … Steitz JA (2013, December). Mammalian 5’-capped microRNA precursors that generate a single microRNA. Cell , 155 (7), 1568–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wang Y, Luo J, Zhao W, & Zhou X (2017, December). Deep learning of the splicing (epi)genetic code reveals a novel candidate mechanism linking histone modifications to ESC fate decision. Nucleic acids research, 45 (21), 12100–12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CJ, Chen J, Villani A-C, Subramaniam M, Gate RE, Bhangale T, … Hacohen N (2017). Genetic analysis of isoform usage in the human anti-viral response reveals influenza-specific regulation of erap2 transcripts under balancing selection. bioRxiv Retrieved from https://www.biorxiv.org/content/early/2017/10/31/188961 doi: 10.1101/188961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost HJ, & Lindquist S (1986, April). RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell , 45 (2), 185–193. [DOI] [PubMed] [Google Scholar]
- Yost HJ, & Lindquist S (1991, February). Heat shock proteins affect RNA processing during the heat shock response of Saccharomyces cerevisiae. Molecular and cellular biology , 11 (2), 1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost HJ, Petersen RB, & Lindquist S (1990, July). RNA metabolism: strategies for regulation in the heat shock response. Trends in genetics : TIG , 6 (7), 223–227. [DOI] [PubMed] [Google Scholar]
- Younis I, Berg M, Kaida D, Dittmar K, Wang C, & Dreyfuss G (2010a, April). Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol. Cell. Biol, 30 (7), 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Berg M, Kaida D, Dittmar K, Wang C, & Dreyfuss G (2010b, April). Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Molecular and cellular biology , 30 (7), 1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M, & Ogawa Y (2018, March). CRISPR/Cas9-mediated modulation of splicing efficiency reveals short splicing isoform of Xist RNA is sufficient to induce X-chromosome inactivation. Nucleic acids research, 46 (5), e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X-Y, Ding J-H, Adams JA, Ghosh G, & Fu X-D (2009, February). Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes & development , 23 (4), 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chun RF, Lisse TS, Garcia AJ, Xu J, Adams JS, & Hewison M (2015, April). Vitamin D and alternative splicing of RNA. J. Steroid Biochem. Mol. Biol, 148 , 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
FURTHER READING
- Pereria MA, Imada EL, Guedes RLM. (2017, September) RNA-seq: Applications and Best Practices. In Marchi (Ed.), Applications of RNA-Seq and Omics Strategies: From Microorganisms to Human Health IntechOpen. Available from: https://www.intechopen.com/books/applications-of-rna-seq-and-omics-strategies-from-microorganisms-to-human-health/rna-seq-applications-and-best-practices. This chapter works through applications of RNA-seq technology and experimental considerations while designing studies to measure RNA processing.
- Li WV, Li JJ. (2018, May) Modeling and analysis of RNA-seq data: a review from a statistical perspective. arXiv. Available from https://arxiv.org/pdf/1804.06050.pdf. This review details statistical considerations, limitations, and potential advancements for the analysis of RNA-seq data, across both gene expression, isoform, and localized RNA processing differential analyses. [DOI] [PMC free article] [PubMed]


