Abstract
Dead-end1 expression is restricted to the vertebrate germline where it is believed to activate translation of mRNAs required to protect and promote that unique lineage. nanos1 is one such germline mRNA whose translation is blocked by a secondary mRNA structure within the ORF. Dead-end1 contains a canonical mRNA Recognition Motif (RRM1) in its N-terminus but also contains a less conserved RRM2. Here we provide a mechanistic picture of the nanos1 mRNA-Dead-end1 interaction in the Xenopus germline. We show that RRM1, but not RRM2, is required for binding nanos1. Similar to the zebrafish homologue, Xenopus Dead-end1 possesses ATPase activity. Surprisingly, this activity appears to be within the RRM2, different from the C-terminal region where it is found in zebrafish. More importantly, we show that RRM2 is required for nanos1 translation and germline survival. Further, Dead-end1 functions as a homodimer and binds nanos1 mRNA just downstream of the secondary structure required for nanos1 repression. We propose a model in which the RRM1 is required to bind nanos1 mRNA while the RRM2 is required to promote translation through the action of ATPase. Dead-end1 appears to use RRMs to mimic the function of helicases.
Keywords: germline development, Dead-end, nanos1, translational regulation, Xenopus
Introduction
A challenge for all metazoans is how to preserve the full potential of their germline while surrounding somatic cells become increasingly more restricted as to their cell fates. Although the genes that play critical roles in this process are frequently conserved across phyla, the timing and means of their expression varies. In organisms that employ germ plasm to contain and move germline determinants into the presumptive primordial germ cells (PGCs) after fertilization, gene expression begins early during oogenesis when the germ plasm forms. At that time, specific maternal RNAs and proteins accumulate in the germ plasm, many of which are RNA binding proteins that function post-transcriptionally to either block somatic gene expression or to promote germline specific expression (Aguero, Kassmer, Alberio, Johnson, & King, 2017; Aguero et al., 2016).
In germlines, including Drosophila, Xenopus, zebrafish, mice, and humans, Nanos1 works with Pumilio to translationally repress specific mRNAs encoding proteins that promote somatic fates. Loss of Nanos1 activity in Xenopus results in misexpression of the endoderm determinant VegT, and ultimately the loss of the germline resulting in infertility (Lai, Singh, & King, 2012). Premature translation of nanos1 in the oocyte results in abnormal embryonic development consistent with a loss of VegT (Luo, Nerlick, An, & King, 2011; Zhang et al., 1998). Thus, the correct timing of nanos1 translation is critical to both somatic and germline fates. During oogenesis, nanos1 mRNA is sequestered within germinal granules and repressed through a secondary structure, located just 4 nucleotides (nts) downstream of the AUG site. This 73nt sequence, termed the Translational Control Element (TCE), prevents initiation by steric hindrance (Luo et al., 2011).
Most recently, we showed that the maternal RNA binding protein Dead-end1 (Dnd1) is required and sufficient to promote the translation of nanos1 mRNA after fertilization (Aguero, Jin, et al., 2017). Dnd1 activates nanos1 translation through a direct interaction with the eIF3 complex and nanos1 mRNA. Interestingly, Dnd1 also interacts with a subset of other germline RNAs, suggesting that Dnd1 may be a master regulator of translation initiation in the germline (Aguero, Jin, et al., 2017). In zebrafish, where Dnd1 was originally discovered, Dnd1-specific morpholino (MO) treatment results in PGC migration into ectopic sites, and eventually their loss either by differentiation into somatic cells or by apoptosis (Gross-Thebing et al., 2017; Weidinger et al., 2003). Similar phenotypes are also observed in Xenopus after dnd1 loss-of-function (Aguero, Jin, et al., 2017; Horvay, Claussen, Katzer, Landgrebe, & Pieler, 2006; Weidinger et al., 2003).
Dnd1 is germ plasm specific and a relatively recent addition to the evolution of the germline as it is exclusive to vertebrates (Weidinger et al., 2003). Dnd1 contains a canonical RNA Recognition Motif (RRM1) in its N-terminus and a less conserved RRM2, previously called the Dnd1 domain. Here we provide a mechanistic picture of the nanos1 mRNA-Dnd1 interaction. Similar to the zebrafish homologue, Xenopus Dnd1 possesses ATPase activity, Which is surprisingly mapped within the RRM2 site, very different from the C-terminal region where it is found in zebrafish (Liu & Collodi, 2010). Interestingly, an identical site to xDnd1 is found in mouse Dnd1. More importantly, we show that this site is required for nanos1 translation, but not RNA binding, whereas RRM1 is required for binding nanos1. Furthermore, Dnd1 functions as a homodimer and binds nanos1 mRNA just downstream of the TCE required for nanos1 repression. We propose a model where the RRM1 is required to bind nanos1 mRNA, while the RRM2 is required to promote translation through the action of the ATPase site.
RESULTS
Dnd1 protein directly binds nanos1 mRNA through RRM1
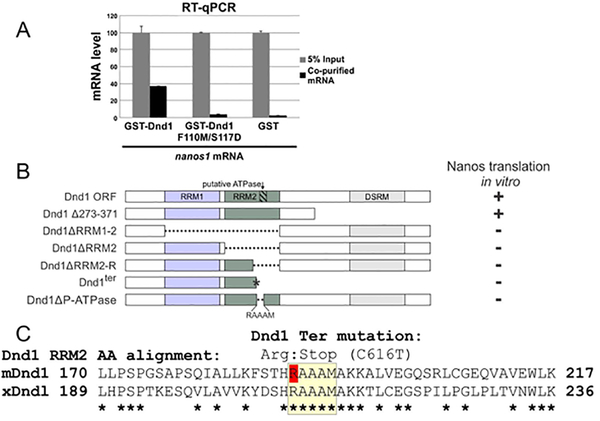
We have previously shown that the RNA-binding protein Dead-end1 (Dnd1) is required and sufficient to activate Xenopus nanos1 translation after fertilization (Aguero, Jin, et al., 2017). In that work, we showed that Dnd1 is capable of binding directly to nanos1 mRNA with a dissociation constant of ~140 nM (Aguero, Jin, et al., 2017). Dnd1 contains two RNA recognition motifs (RRM1 and RRM2) in its N-terminus (Marchler-Bauer et al., 2015; Weidinger et al., 2003). Point mutations within the RRM1 prevented RNA binding in zebrafish Dnd1 (Slanchev et al., 2009). This region is highly conserved from AA 76 to 153 (NCBI, #Q6DCB7). Therefore, to test whether the RRM1 was required to bind nanos1 mRNA, we generated Xenopus tropicalis Dnd1 recombinant protein containing two point mutations within this conserved region: Xt F110M/S117D, which corresponds to F106M/S113D in Xenopus laevis. GST tagged recombinant proteins Dnd1 and Dnd1-F110M/S117D were tested in RNA immunoprecipitation (RIP) assays and the amount of nanos1 RNA bound determined by RT-qPCR. GST alone served as a negative control. While wild-type Dnd1 bound nanos1 mRNA (40% of input), the double point mutation within the RRM1 was sufficient to prevent binding to nanos1 mRNA, confirming a role for RRM1 in nanos1 mRNA binding (Fig. 1A). As Dnd1 has two RRMs, these results also suggested that either RRM2 (also called Dnd1 motif) does not bind nanos1 mRNA at all or RRM2 alone is not sufficient for mRNA binding.
Figure 1. Dnd1 RRM1 and RRM2 are required for binding and translation of nanos1 RNA respectively.
A. RRM1 of Dnd1 protein is required to bind to nanos1 mRNA. mRNAs were pulled down by recombinant GST-Dnd1 or GST-Dnd1-F110M/S117D proteins in an in vitro RIP assay and bound mRNA measured by qPCR. Ratio between the pulldown mRNAs and 5% of mRNA input is shown. GST protein served as a negative control. Data are shown as mean ± s.d. **p<0.01 of two independent experiments. B. Deletion mapping analysis of Dnd1 protein shows that the right half region of RRM2 (RRM2-R, Δ198–237) is the smallest region required to promote nanos1 translation. The putative ATPase site (RAAAM) lies within the RRM2-R region. Analysis was based on at least two independent in vitro translation experiments using wheat germ extracts. Findings were confirmed in vivo using oocytes from two different females. Raw data is shown in SFig.1. C. Alignment of the RRM2 right half region of mouse and Xenopus Dnd1. Putative ATPase region is highlighted in yellow. Note: the point mutation C616T that replaces an arginine creates a stop codon in the mouse Dnd1 ter mutation (red R). Asterisks indicate identity (Clustal Omega, EMBL-EBI, UK).
Dnd1 RRM2 is required for nanos1 translation
To determine what region(s) of Dnd1 protein was required to support nanos1 translation, we created a series of deletions in Dnd1 and sub-cloned them into the pCS2+ expression vector (Fig. 1B). All deletions were examined for Dnd1 expression by western blotting with anti-Dnd1 antibody and found to be expressed at comparable levels (Fig. 4B). We used two different assays, Xenopus oocytes and wheat germ (WG) extracts (Aguero, Jin, et al., 2017; Luo et al., 2011), to test each of the mutant Dnd1 proteins for their ability to promote nanos1 mRNA translation. Oocyte injections or in vitro translation (WG assay) yielded the same results (Supplemental Fig. 1) and these results are summarized in Fig. 1B. Zebrafish Dnd1 contains an ATPase site within the C-terminal region that is important for its function (Liu & Collodi, 2010). However, xDnd1 missing the C-terminal 100 amino acids (Δ273–371) still promoted nanos1 translation. Interestingly, deletion of the RRM2 (ΔRRM2) or just its C-proximal half (ΔRRM2-R; missing AA 198–237) resulted in the failure of Dnd1 to promote nanos1 translation (Fig. 1B and Supplemental Fig. 1).
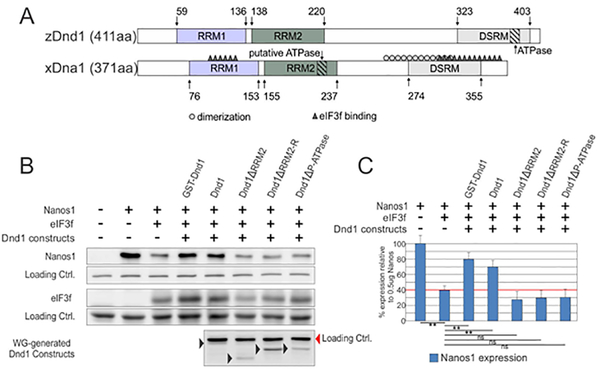
Figure 4. The putative xDnd1 ATPase site is required for nanos1 translation.
A. Sequence alignment of Dnd1 orthologues from zebrafish and Xenopus. Both single-strand RNA binding domains (RRM1 and 2) are depicted in colored rectangles. The double-stranded RNA-binding motif (DSRM) is localized at the C-terminal domain. Regions involved in dimerization (grey o) (see Fig. 5), binding to the translational repressor factor eIF3f (black Δ) (see (Aguero, Jin, et al., 2017)) and the putative ATPase domain are indicated. B. Wheat germ extract was incubated with 500ng of nanos1 mRNA with (+) or without (−) 1ug of the translational repressor eIF3f mRNA. Either purified (GST-Dnd) or wheat germ generated Dnd1 proteins, wild-type or mutant, were also included (+). Sample extracts were analyzed by western blotting with anti-Nanos1, anti-eIF3f, and anti-Dnd antibodies. Experiments were repeated a minimum of 3 times each. C. Quantification of Nanos1 expression shown in (B) for indicated samples. Nanos1 expression is relative to the nanos1 mRNA only sample which is defined as the 100% level of Nanos1 expression.
Interestingly, the Dnd1ter point mutation that results in tumors and infertility in mice and humans (Youngren et al., 2005; Zechel et al., 2013), generates a stop codon within the RRM2 domain. We created the identical mutant in Xenopus Dnd1 that results in a truncated protein by replacing the codon for arginine at position 209 with a stop codon (Fig. 1C). The xDnd1ter mutant did not promote nanos1 translation as tested in either assay, further supporting a role for the RRM2 in translation (Fig. 1B and Supplemental Fig. 1). These results suggested that the two Dnd1 RRM domains evolved to have different functions: one in RNA binding and the other in promoting translation.
Overexpression of xDnd1ter or Dnd1 ΔRRM2-R results in loss of PGCs.
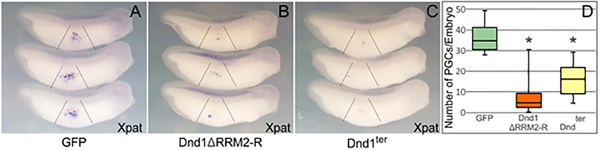
The failure of xDnd1ter and Dnd1 ΔRRM2-R to support nanos1 translation, prompted us to ask what the effects of these mutations might have on PGCs when overexpressed in the embryo. To assess the effect on PGCs, we injected one-cell stage embryos at the vegetal pole with either Dnd1 ΔRRM2-R or xDnd1ter RNA. GFP served as a negative control (Fig. 2A, B, C). The embryos were allowed to develop until stage 33 (late tailbud stage) when PGCs were assessed by whole mount in situ hybridization (WISH) with the PGC marker Xpat (Hudson & Woodland, 1998). Although PGC migration was not affected by either mutant when overexpressed, the number of PGCs were significantly reduced by both mutants within the context of an otherwise normal looking embryos (40% of embryos had less than 20 PGCs, and 50–60% less than 10; n=32) (Fig. 2D). These results show that the absence of AA 198–237 within the RRM2 (ΔRRM2-R) is sufficient to phenocopy the nanos1 loss-of-function mutant as well as the mDnd1ter mutation (Lai et al., 2012; Zechel et al., 2013). Taken together, our findings have narrowed the region required for nanos1 translation to 40 residues within the RRM2. We next asked what function might this region have.
Figure 2. Overexpression of xDnd1ter or Dnd1 ΔRRM2-R results in loss of PGCs.
A-C. One cell embryos were injected vegetally with 500pg of GFP (control) or the indicated dnd1 mRNA. Tailbud embryos (stage 32–35) were analyzed for Xpat expression by WISH. Representative images are shown. D. PGC number per embryo was quantified. GFP, n=16; Dnd1ΔRRM2-R, n=32; Dndter, n=16. *P<0.05, compared with GFP control. Analysis based on at least two independent experiments and shown as a box and whisker plot.
Dnd1 possesses ATPase activity
Zebrafish Dnd1 protein is known to possess ATPase activity. Furthermore, this activity is required for PGC development in zebrafish, although it remains unclear why this is the case (Liu & Collodi, 2010). We have previously shown that nanos1 translation is repressed by its secondary structure, TCE, and the loss of this structure is sufficient to allow translation (Luo et al., 2011). Since xDnd1 promotes nanos1 translation (Aguero, Jin, et al., 2017), we speculated that xDnd1 may also have ATPase activity, and that the ATPase activity may be required to melt the TCE to allow nanos1 translation. If this were true, it would explain the requirement of Dnd1 ATPase activity for normal PGC development as Nanos1 is required for PGCs survival (Lai et al., 2012).
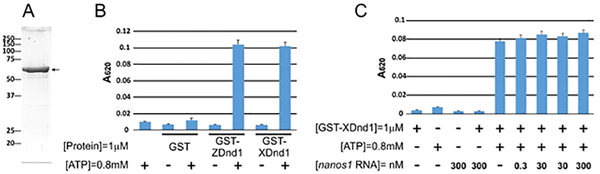
As a first step, we used purified recombinant GST-Dnd1 protein in a standard assay for ATPase activity (Fig. 3A, B). xDnd1 had the equivalent ATPase activity as that of recombinant zDnd1, used as a positive control. The amount of released phosphate from ATP by GST-Dnd1 increased in a time-dependent manner and required Mg2+ (Supplemental Fig. 2). The ATPase activity of the well-studied DEAD-box helicase family is also stimulated by its RNA substrate (Cordin, Banroques, Tanner, & Linder, 2006). To determine if Dnd1 shares similar properties to this family of helicases, we included nanos1 mRNA in the reaction and asked if it can synergistically stimulate the Dnd1 ATPase activity in the presence of Mg2+. As shown, ATPase activity was not further enhanced by the presence of nanos1 mRNA (Fig. 3C). Thus, although xDnd1 has ATPase activity and binds RNA, it does not function as a typical helicase, at least by this criterion.
Figure 3. xDnd1 possesses ATPase activity.
A. GST-xDnd1 purified protein (arrow) visualized by Coomassie blue staining. B. Comparison of ATPase activity between recombinant GST-xDnd1 and GST-zDnd1 proteins in the presence and absence of ATP. The zebrafish Dnd1 protein served as a positive control. C. ATPase activity of GST-xDnd1 is not stimulated by nanos1 RNA. Analysis based on at least three independent experiments and shown as histograms.
xDnd1-mediated activation of nanos1 translation requires the putative ATPase site.
In a tour-de-force, Liu and Collodi (Liu & Collodi, 2010) mapped the precise location of the ATPase activity in zDnd1 to five residues, RAAAE (392–396), near the end of the non-conserved C-terminus (Fig. 4A). However, unexpectedly, deletion of 300nts (100 amino acid residues) from the xDnd1 C-terminus retained full ATPase activity (data not shown). To determine if the ATPase site was conserved, but found elsewhere in xDnd1, we blasted the Xenopus Dnd1 sequence for RAAAE. Only one related polypeptide was identified, RAAAM. RAAAM resides within the RRM2-R, a region required for nanos1 translation and conserved in mammals (Fig. 1B).
To determine if this putative ATPase (P-ATPase) site was required for nanos1 translation, synthetic transcripts encoding wild-type Dnd1 and the mutant missing the RAAAM site (Dnd1-ΔP-ATPase), were tested in WG extracts in the presence of nanos1 mRNA and its translational inhibitor, eIF3f (Aguero, Jin, et al., 2017). Nanos1 protein was detected by western blotting with anti-Nanos1 antibody. Dnd1 mutant proteins were translated at comparable levels to Dnd1-FL (Fig. 4B). In all cases, Dnd1 mutants, abrogating the putative ATPase site, failed to derepress nanos1 translation (Fig. 4B,C). These results strongly suggested that the putative ATPase site in Dnd1 was required for translational activation of nanos1 mRNA, and likely other germline RNAs as well(Aguero, Jin, et al., 2017).
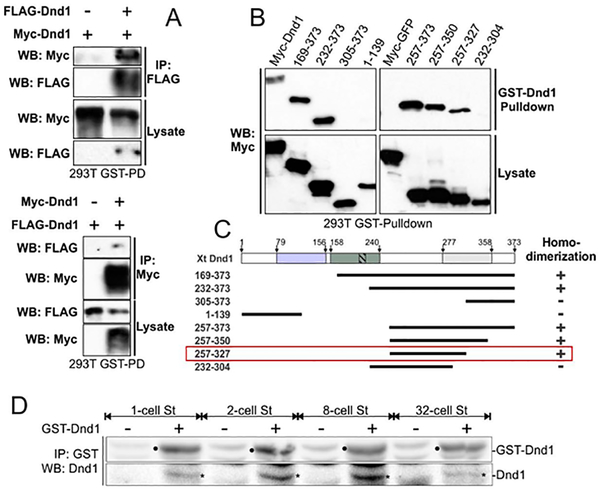
Dnd1 forms homodimers in vivo
Many RNA-binding proteins function as a dimer to bind RNA (Marchione, Leibovitch, & Lenormand, 2013; Wang et al., 1999). To determine if Dnd1 could form a homodimer, we generated plasmids containing Dnd1 tagged with either Flag or myc and transfected corresponding plasmids into HEK293T cells. In co-IP experiments Myc-Dnd1 could bring down Flag-Dnd1, and vice-versa, from cell lysates (Fig. 5A). Addition of RNase A into co-IP did not affect homodimerization of Dnd1 (data not shown). We extended the above studies by creating a series of Dnd1 deletions and mapping what regions in Dnd1 were required for its homodimerization. The smallest region able to form homodimers resides in the C-terminal region (Fig. 5B, summarized in C; Xt: 257–327AA). To determine if Dnd1 forms homodimers in embryos at the time of nanos1 translation, GST-Dnd1 purified protein was added to embryo extracts made from 1-, 2-, 8-, and 32-cell stage embryos. After an incubation period (1–2 hrs.), GST-Dnd1 was immunoprecipitated with anti-GST antibody and analyzed by western blotting for interaction with endogenous Dnd1 using an anti-Dnd1 antibody. The control lane without GST-Dnd1 added (−) revealed a non-specific protein that migrated similarly to Dnd1 (+). However, the non-specific protein was present at a much lower level, allowing us to conclude that GST-Dnd1 could bring down endogenous Dnd1 at all stages tested (Fig. 5D). Taken together, these results show that Dnd1 is capable of forming homodimers in embryos at the correct time to support nanos1 translation.
Figure 5. Dnd1 forms homodimers in vitro and in vivo.
A. Co-IP shows the interaction between myc-Dnd1 and FLAG-Dnd1 in transfected HEK293T cells. Anti-FLAG (top panel) or anti-Myc (bottom panel) antibodies were used to pulldown xDnd1 from cell lysates. B. Different Myc-tagged Dnd1 deletion constructs were transfected into HEK293T cells, lysed and incubated with GST-xDnd1. Co-purified Dnd1 was detected by western blot analysis using anti-Myc antibodies. C. Schematic summarizing experiments shown in (B). “+” for Dnd1 homodimerization; “-” lack of Dnd1 dimerization. Red rectangle highlights the smallest xDnd1 region required for homodimerization (see also Fig. 4A). Analysis based on at least two independent experiments. D. Dnd1 dimerizes in vivo. Extract from 50 1-, 2– 8- and 32-cell stage Xenopus embryos were made and incubated with GST-xDnd1 purified protein. Anti-GST antibody was used to pull down GST-xDnd1 and xDnd1 proteins were detected by western blotting using anti-xDnd antibody. Endogenous xDnd1 bands are indicated by asterisks, exogenous pulled-down GST-xDnd1 bands are indicated with black dots. Negative controls have a weak non-specific protein that migrates similarly to GST-xDnd1 protein. This experiment was repeated twice using embryos from four different females.
RRM1 and RRM2 have different structures
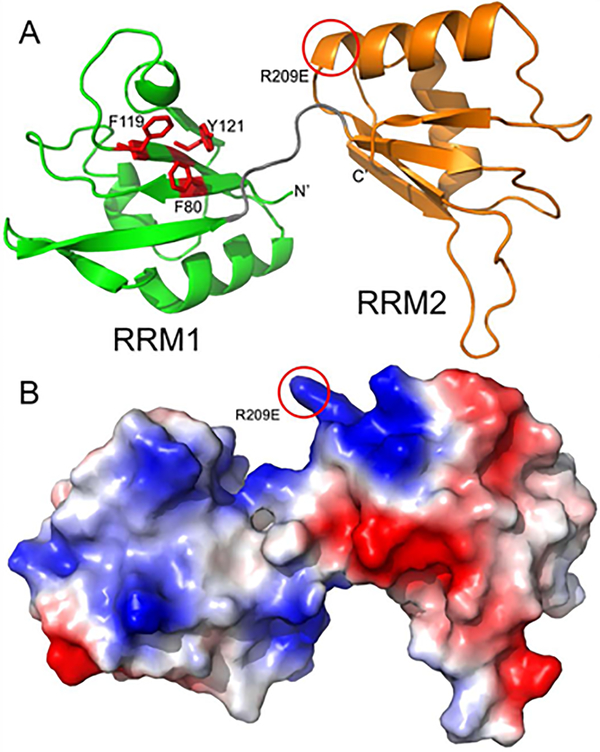
We created homology models of the two RRM domains to see if the basis for their functional differences was consistent with their predicted structure. Both domains adopt the classical RRM structure with a flat concave surface composed of four antiparallel beta strands supported by two helices on the back (Lunde, Moore, & Varani, 2007). RRM domains typically bind single-strand RNA along their concave beta sheet surface, stabilized by conserved aromatic residues that stack with the RNA bases. RRM1 (green) domain of Dnd1 has three aromatic residues (Fig. 6A; shown as red sticks) poised for stacking with RNA, while RRM2 (orange) has no aromatic side chains facing out from its beta sheet. The location of putative ATP binding on RRM2 is highlighted by a red circle. This model also shows the molecular surface and was colored to indicate the electrostatic potential (Fig. 5B; red is acidic, blue is basic). RRM1 presents a basic surface along its beta sheet which is conducive for RNA binding, while RRM2’s beta sheet has a very acidic patch that is unlikely to bind RNA. However, RRM domains are unusually versatile in their RNA binding modes (Maris, Dominguez, & Allain, 2005), and it is possible that the RRM2 binds RNA in a non-canonical manner via the basic patch near the helical regions. Taken together, the model suggests that only the N-terminal RRM1 domain is likely to bind RNA, consistent with our functional studies. RRM2 also has a basic non-standard P-loop/helix that is consistent with ATP binding sites, suggesting that an ATPase site resides in this domain.
Figure 6. Homology modeling of Xenopus laevis Dnd1 suggests that only the N-terminal RRM domain is likely to bind RNA.
A. Homology model of Dnd1 RRM domains is shown in a ribbon representation with RRM1 (residues 78–151) colored green and RRM2 (residues 158–233) colored orange. The linker between the two RRM domains is shown in grey, and the relative orientation between the two domains is likely variable and dependent on parts of Dnd1 not included in this model. RRM1 domain of Dnd1 has three aromatic residues (shown as red sticks) poised for stacking with RNA, while RRM2 has no aromatic side chains facing out from its beta sheet. Location of putative ATP binding on RRM2 is highlighted by a red circle. B. Homology model from panel A, shown as a molecular surface colored by electrostatic potential (red is acidic, blue is basic).
Dnd1 requires a region adjacent to the nanos1 TCE to promote nanos1 translation.
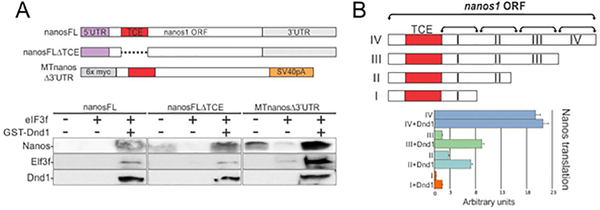
We next determined what region of nanos1 mRNA was required for Dnd1 to promote translation. Based on our previous findings, the simplest working model suggested that Dnd1 binds to the TCE, altering its structure and thus promoting translation (Luo et al., 2011). A series of deletions or substitution mutants were created within nanos1 mRNA, and these mutant mRNAs were tested, in WG extracts, to determine if Dnd1 protein could promote their translation. In these studies, we also sought to determine if the nanos1 TCE was required for Dnd1-mediated nanos1 translation. However, we could not simply investigate the ability of Dnd1 protein to promote translation of a nanos1Δ-TCE mutant because, unlike any other region, such a deletion dramatically promotes the translation of nanos1 (Luo et al., 2011). Furthermore, levels of nanos1 translation in recent commercial WG extracts were found to be variable (see Fig. 7A). To circumvent these problems, we blocked translation of nanos1 by including in the reaction the translational repressor, eIF3f, part of the eIF3 complex (Aguero, Jin, et al., 2017; Daubner, Clery, & Allain, 2013). As expected, in the presence of eIF3f, nanos1 translation was repressed, allowing us to test if Dnd1 could promote translation in the absence of the TCE (7; Fig. 7A). Addition of Dnd1 protein was able to enhance translation of the Δ-TCE mutant (Fig. 7A). Therefore, Dnd1 must interact with a region other than the TCE to promote translation.
Figure 7. Dnd1 requires a region adjacent to the nanos1 TCE to promote nanos1 translation.
A. Schematic of nanos1 deletions used in in vitro translation (WG) experiments shown below. WG extract was supplemented with 500 ng of nanos1 mRNA with or without purified Dnd1 protein and 1 ug of mRNA encoding the translational repressor eIF3f. Samples were analyzed for Nanos1, eIF3f and Dnd1 protein expression by western blotting. Experiments were repeated twice. Dnd1 protein promoted translation of each nanos1 mutant tested. B.Schematic of deletions within the nanos1 ORF downstream of the TCE (red box). Each transcript was tested for translation in the presence or absence of xDnd1 protein and eIF3f mRNA as shown in A. Results from blots are presented in the histogram. xDnd1 failed to promote nanos1 translation when region I (96 nt downstream of TCE) was deleted, suggesting that this region of nanos1 is required for Dnd1 function. Experiments were repeated twice using different wheat germ extracts.
Previously, we have shown that the nanos1 3’UTR is not required to promote translation (Aguero, Jin, et al., 2017). To rule out any possibility that the 3’UTR facilitates Dnd1 binding to nanos1 mRNA, we removed the 3’UTR and replaced it with the SV40pA to stabilize the transcript. Consistent with our previous findings, Dnd1 could promote nanos1 translation in the absence of the nanos1 3’UTR (Fig. 7A). To rule out any involvement of the 5’UTR, we replaced that region with six myc-tags. Dnd1 also greatly enhanced translation without the native 5’UTR. Therefore, Dnd1 required a region within the ORF downstream of the TCE to promote translation of nanos1.
Next, we did a more complete series of nanos1 deletions downstream of the TCE and within the ORF, dividing it into four regions (Fig. 7B; I-IV). A progressive series of deletions from the 3’end of the nanos1 ORF was tested in WG extracts for the ability of Dnd1 to significantly increase nanos1 translation (Fig. 7B; SFig. 3). Although all deletions were somewhat affected, the deletion of region I, 96-nt downstream of the TCE, had a profound negative effect on the ability of Dnd1 to promote nanos1 translation (Fig. 7B). One explanation for this result might be that, in the presence of the TCE, the shorter sequence has an altered structure that does not support optimal translation.
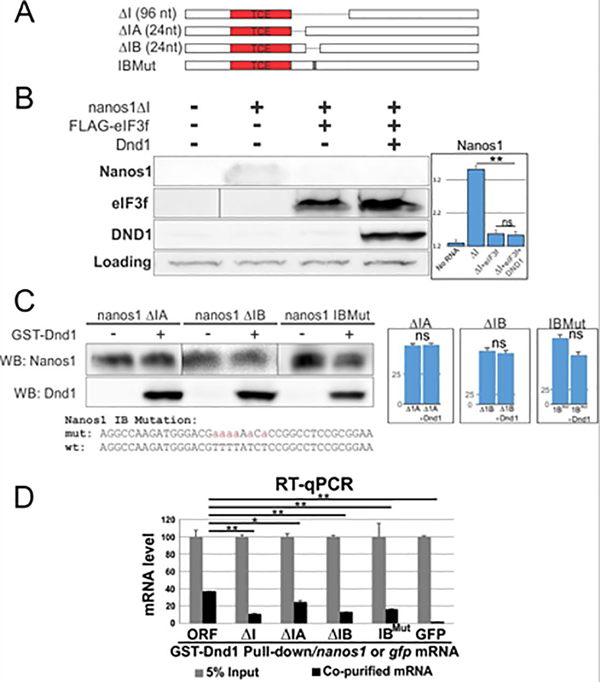
Therefore, to test region I alone as a putative site of Dnd1 interaction, we deleted it (Fig. 8A) while retaining all other regions (ΔI; Fig. 8A). We asked if region I was required for Dnd1 to promote nanos1 translation in the presence of eIF3f as detailed for Fig. 7. We found that Dnd1 protein could not rescue nanos1 translation when region I was deleted, and nanos1 translation remained at the fully repressed level (Fig. 8B). To determine even more precisely the site of interaction, we introduced two 24-nt deletions within region I, closest to the TCE (Fig. 8A; labeled A, B). Subregion B was of special interest as it contains a poly(U)- rich region, a putative Dnd1 binding site (Kedde et al., 2007; Ketting, 2007; Yamaji et al., 2017). To determine if the poly(U)- rich region was necessary for Dnd1 function, we also substituted uridines for adenines within subregion B (IB-Mut in Fig. 8A, C). Addition of Dnd1 protein to WG extracts had no effect on the translation of nanos1 mutants, as expected if region I was required for Dnd1 function promoting nanos1 translation (Fig. 8C).
Figure 8. Dnd1 binds to TCE adjacent U-rich region of nanos1 mRNA.
A. Diagram shows serial deletions of nanos1 mRNA used in in vitro translation and RIP assays. nanos1 ΔI region is missing 96 nt downstream of the TCE. Smaller deletions within region I were carried out (ΔIA and ΔIB, 24 nt each). B. WG extracts were supplemented with either 500 pg of nanos1 mRNA with or without 1ug of translation repressor eIF3f and 100 pg of xDnd1 protein and analyzed by western blot. A non-specific band served as a loading control. The presence of Dnd1 protein did not promote the translation of nanos1 mRNA missing region I (ΔI) after repression by eIF3f. Quantitation of blot shown on right. C. Wheat germ extracts with either 500 pg of nanos1ΔIA/B or nanos1 region IB with mutations in U-rich sequence. mRNAs were incubated with or without Dnd1 protein and analyzed by western blotting. Dnd1 does not improve nanos1 translation. Quantitation of blot shown on right. Experiments repeated twice. D. RIP analysis. Different nanos1 mRNAs with deletion or mutation were pulled down by GST-Dnd1 and measured by qPCR. Ratio between the pulldown and 5% of RNA input is shown. GFP served as a negative control. All experiments were repeated at least twice. Quantification of results shown in B, C are shown as mean ± s.d. Two-tailed t-tests were performed. ** P<0.01; * P<0.05; ns: no significant difference.
Lastly, to test region I and the poly(U)-rich site for Dnd1 protein binding, we carried out RIP analysis. We incubated purified recombinant GST-Dnd1 protein with nanos1 ΔI RNA. Dnd1 was precipitated and the levels of bound nanos1 mRNA was detected by RT-qPCR (Fig. 8D). RIP analysis showed that Dnd1 binding to nanos1 mRNA decreased only upon the deletion of region I (reduced to 25% of control). Deletion of IB and, importantly, the IB-Mut, reduced Dnd1 binding to similar levels as detected when the entire region I was deleted. Interestingly, deletion of IA caused a mild reduction in Dnd1 binding. We speculate that deletion of IA may influence the Dnd1 binding by altering the secondary of RNA around the poly(U)-rich site. Taken together, these results show that a small region downstream of the TCE is required for Dnd1 binding and promotion of nanos1 translation. Our results also suggest that Dnd1 binds the poly(U)- rich region within subregion B.
DISCUSSION
In this study, we provide evidence as to how nanos1 mRNA and Dnd1 interact to achieve proper translation of nanos1, an early event essential to the survival of the germline. We show that, similar to zebrafish, Xenopus Dnd1 possesses ATPase activity. However, surprisingly, the homologous ATPase site was mapped within the RRM2 or Dnd1 domain, and is very different from its C-terminus location in zebrafish (Liu & Collodi, 2010), but conserved in mouse. More importantly, we show that this site is required for nanos1 translation, but not RNA binding, whereas the canonical RRM1 is required for binding nanos1 mRNA. Overexpression of xDnd1 mutants lacking the putative RRM2-ATPase site caused a dominant negative effect, which resulted in the loss of the germline, further confirming its functional importance. Furthermore, xDnd1 forms as a homodimer in vivo and binds nanos1 mRNA just downstream of the TCE, a secondary structure required for nanos1 repression (Luo et al., 2011). We propose a model where the RRM1 is required to bind nanos1 mRNA while the RRM2 is required to promote translation through the action of ATPase. Thus, xDnd1 appears to use RRMs to achieve the same result as do DEAD box helicases.
Different roles for RRM1 in Dnd1?
In vertebrates, the RRM is the most abundant RNA-binding domain and its unusual versatility is well documented (Clery, Blatter, & Allain, 2008). As expected, the canonical RRM1 was found to bind nanos1 mRNA. Two conserved point mutations within the RRM1 rendered Dnd1 unable to bind nanos1 mRNA (Fig. 1A). Two additional functions suggested for the Dnd1 RRM1 appear to involve protein association. RRM1, a region required for RNA binding, is also required for eIF3f protein binding (Xt: 96–127 AA) (Aguero, Jin, et al., 2017). Furthermore, point mutations within the zebrafish RRM1 prevented Dnd1 in PGCs from shuttling between the nucleus and cytoplasm (Slanchev et al., 2009). As Dnd1 does not contain a nuclear localization signal, these results imply that the RRM1 may also interact with a co-factor for nuclear export after RNA binding.
xDnd1 has ATPase activity
zDnd1 has been shown to possess ATPase activity, although it does not share similarity to any known ATPase families. We showed that xDnd1 also exhibits ATPase activity that was commensurate with that of zebrafish. Consistent with other characterized ATPases, Mg2+ is required for the ATPase activity in both zebrafish (Liu & Collodi, 2010) and Xenopus (Fig. 3). Typically, RNA substrates can stimulate the ATPase activity of DEAD-box helicase proteins such as Vasa (Cordin et al., 2006; Liang, Diehl-Jones, & Lasko, 1994). Although Dnd1 interacted with nanos1 directly, the Dnd1 ATPase activity was not stimulated by nanos1 mRNA. Dnd1 may need additional factors in vivo to stimulate its activity in a manner similar to that of other RNA helicases. For example, the eIF4A ATPase activity increases 3-fold in the presence of accessory proteins eIF4B and eIF4H (Rogers, Richter, Lima, & Merrick, 2001). It will be interesting to determine if accessory factors, like DeadSouth, interact with Dnd1 in PGCs and enhance ATPase activity.
zDnd1 is a much larger protein than either mDnd1 or xDnd1 and contains large stretches in the C-terminus that are not well conserved ((Liu & Collodi, 2010) and Fig. 3A). It is in the non-conserved C-terminus where the zebrafish RAAAE ATPase site maps. Deletion of the C-terminal region from xDnd1 retained both ATPase activity (data not shown) and the ability to promote nanos1 translation (Fig. 1C). Interestingly, the homologous ATPase site to that found in zebrafish mapped to a second recently identified RRM2 or Dnd1 domain (Slanchev et al., 2009). RRM2 contains RAAAM, a site which is precisely conserved in chickens, mouse, and humans as to sequence and location with xDnd1 (Fig. 1A) (Aramaki, Kubota, Soh, Yamauchi, & Hattori, 2009; Aramaki et al., 2007; Kedde et al., 2007; Kito et al., 2010). The basic patch near the helical regions in RRM2 corresponds to the putative ATPase site, and potentially promotes interactions required for translation of nanos1. Consistent with that interpretation, deletions introduced into the RRM2 covering that site prevented Dnd1 from promoting nanos1 translation. Thus, our data support different functions for each of the two RRMs in Dnd1.
Dnd1 ATPase activity is required for PGC survival
Dnd1 ATPase activity is required for zebrafish PGC survival (Liu & Collodi, 2010), however, the mechanism for the involvement of such activity in germline development is not clear. We show that deletion of the homologous ATPase site in xDnd1 resulted in the loss of and the ability of Dnd1 to activate nanos1 translation and converted Dnd1 into a dominant negative mutant which interfered with PGC development (Figs. 1D). Our findings suggest that PGCs are lost after overexpression of Dnd1 ΔRRM2-R because ATPase activity is required to promote germline RNA translation essential for normal PGC development. We previously found that Dnd1 directly interacts with eIF3f, and through it, the whole eIF3 complex, which is required to initiate translation (Aguero, Jin, et al., 2017). Dnd1 may be recruited to nanos1 RNA and function as a chaperone that facilitates RNA structural remodeling at the cost of ATP (Jarmoskaite & Russell, 2011). In zebrafish PGCs, zDnd1 protects germ plasm-associated nanos1 mRNA from miRNA-mediated degradation that clears nanos1 from the soma (Kedde et al., 2007). However, a direct interaction between the zDnd1 and nanos1 mRNA has not been reported. Furthermore, whether the zDnd1 ATPase activity is involved in nanos1 protection is not clear. A recent study tethering zDnd1 to a reporter shows Dnd1 can act as a repressor of translation and this activity persists even if the ATPase site is deleted (Kobayashi, Tani-Matsuhana, Ohkawa, Sakamoto, & Inoue, 2017). xDnd1 interacts with eIF3f, a repressor of translation within the eIF3 complex at the earliest stages in development, and blocks its repressive activity, promoting nanos1 translation (Aguero, Jin, et al., 2017). Since the teleost genome is highly rearranged in comparison to other vertebrates such as mouse, chick, and Xenopus, it seems likely that the ATPase site moved to the end of the C’terminal region during teleost evolution and genome duplication. We would argue that the ATPase site within the RRM2 is the conserved site and functions to facilitate the role of Dnd1 in promoting translation.
xDnd1 forms homodimers
It is not uncommon for RRM-containing proteins to form homodimers (Marchione et al., 2013; Wang et al., 1999), and, in some cases, it is shown to be a requirement for RNA binding (van Gelder et al., 1993; Varani et al., 2000; Wang et al., 1999). We found that Dnd1 is capable of forming homodimers within the context of HEK293T cells as well as in embryos at the correct time for nanos1 translation. GST-Dnd1 co-immunoprecipitated with endogenous Dnd1 in embryos and did not require RNA, suggesting that dimerization occurs first. To complicate matters, Dnd1 is also capable of forming heterodimers with specific subunits of the eIF3 complex (Aguero, Jin, et al., 2017). xDnd1 directly binds eIF3f and in IP experiments, brings down eIF3m and eIF3h, which associate with eIF3f (Aguero, Jin, et al., 2017).
Although the RRM is required to form homodimers in some proteins (Crichlow et al., 2008; van Gelder et al., 1993), xDnd1 requires the C-terminus to form homodimers (Fig. 5) and to associate with eIF3f (Aguero, Jin, et al., 2017). In general, the C-terminus is less well conserved among different species, however the region that correlates with the ability to dimerize (304–350AA) contains two identical stretches of 10 residues in Xenopus, mouse, and humans that is not present in zebrafish. Finer mapping will be required to determine if these conserved regions are critical for dimerization. Deletion of the C-terminus in zDnd1 results in an 85% reduction in the number of PGCs, however, that phenotype is attributed to the loss of the ATPase site located in the last 91 AA. It will be important to determine if Dnd1 from other species, including zebrafish, functions as homodimers.
MODEL: nanos1 activation by xDnd1
We had anticipated that xDnd1 would bind the TCE in nanos1 to promote the melting of this repressive structure. However, our data identified a U-rich region immediately juxtaposed to the TCE required for Dnd1 binding. RIP analysis showed that substitution of these U’s greatly reduced the ability of xDnd1 to bind nanos1 mRNA (Fig. 8D). Previous studies proposed that Dnd1 binds to a stretch of U-rich sequences around the miR430-binding site in the 3’UTR and prevents access of miRNA machinery (Kedde et al., 2007). Similarly, in Xenopus, miR-18 is blocked by xDnd1 binding to U-rich sites within the 3’UTR that are embedded in mRNA localization signals (Koebernick, Loeber, Arthur, Tarbashevich, & Pieler, 2010). Here we propose another U-rich region within the ORF that binds xDnd1 and is required for nanos1 translation. Taken together, our results suggest a model whereby xDnd1 binds subunits of the eIF3 complex as well as RNA, promoting structural remodeling of the TCE at the cost of ATP. Disruption of the TCE allows the preinitiation complex to scan and translation to proceed. The temporal order in which these events occur and whether binding requires the dimerization of xDnd1 remains to be determined in future work.
Dnd1 as Master Regulator-Gatekeeper of the germline
In germ cells, Dnd1 appears to function at each one of three critical transition points for gene expression: in oocytes (Mei et al., 2013), just after fertilization (Aguero, Jin, et al., 2017), and during gastrulation as the maternal to zygotic transition peaks (Crichlow et al., 2008; Kedde et al., 2007; Mickoleit, Banisch, & Raz, 2011). In the fully-grown Xenopus oocyte, Dnd1 binds to trim36 RNA anchoring it within the vegetal cortex, a requirement for Trim36 E ligase protein to accumulate, microtubule arrays to form, and the dorsal/ventral axis to be established (Mei et al., 2013). Post-fertilization, Dnd1 is required to activate the translation of nanos1 mRNA through interaction with the eIF3 complex (this report and (Aguero, Jin, et al., 2017)). Nanos1 is required early on to translationally block maternally supplied somatic determinants and to preserve the germline (Lai et al., 2012). At the time of transition from maternal to zygotic gene expression, germline specific Dnd1 plays yet another role; protection of germ plasm associated nanos1 mRNA from miRNA-mediated degradation (Crichlow et al., 2008; Kedde et al., 2007).
Early development depends on the correct activation and regulation of maternal RNAs. Although we have focused on nanos1 mRNA, there is abundant evidence that Dnd1 binds many germline mRNAs ((Aguero, Jin, et al., 2017; Yamaji et al., 2017); and our unpublish data). Several germline mRNAs, including nanos1 mRNA, could be co-immunoprecipitated (RIP) with Dnd1 protein in early staged embryos. The storage of these messages involves formation of stable and detergent-resistant mRNP complexes. RNA helicases may not only play a role in RNA regulation, but also act as a remodeler to regulate RNA-protein complexes at the cost of ATP and promote translation. We propose that Dnd1, at the top of the regulatory pathway for germline mRNAs, may be the master regulator of gene expression in the germline.
EXPERIMENTAL PROCEDURES
Plasmids
The ORF of Xenopus dnd1 was cloned by RT-PCR (Invitrogen) from RNAs extracted from stage VI oocytes and sequenced. We identified an AUG start site upstream (MATS) of the start site originally reported by Horvay et al., 2006 (MELS) (Horvay et al., 2006). Our sequence adds 17 amino acids for a total number of 371 amino acids. Synthetic xdnd1 transcripts for oocyte injection or in vitro translation were generated by PCR and sub-cloned into a pCS2-MT vector at EcoRI/XhoI sites. dnd1 transcripts with Myc tags at the 5’ end were produced by SP6 transcription (mMESSAGE mMACHINE, Ambion). To obtain recombinant GST-xDnd1 protein, xdnd1 was sub-cloned into pGEX vector (GE Health) after the GST taget EcoRI/XhoI sites. Nanos1 full length, ΔTCE, and ORF deletions (I-IV) were generated by PCR and sub-cloned in pCS2+ vector. Nanos1 region I deletions (ΔI, ΔIA, ΔIB and B mut) were artificially synthesized and cloned into pUC57-Amp vector (Genewiz). Synthetic transcripts of mutants were generated in vitro by SP6 RNA polymerase (mMESSAGE mMACHINE, Ambion) using NotI-linearized plasmids as templates.
Protein expression and purification
Recombinant GST-xDnd1 protein was expressed in E. coli (BL21 stain, Invitrogen) at 37oC for 3 hours by IPTG induction (1 mM). The bacteria were harvested by centrifugation and subjected to cell disruption using a French Press. The protein extracts were loaded onto the GSTrap column (GE Health). The column was washed with phosphate buffer saline (PBS) before elution by addition of 10 mM reduced glutathione.
In vitro RNA pull-down assay (RIP)
GST-xDnd1-bound glutathione beads were incubated with 10 μg yeast tRNA in 1ml RIP buffer (50 mM Tris pH7.6, 125 mMNaCI, 1 mM EDTA, 0.25% NP-40, 0.2% glycerol, 0.1 mM DTT, and 100 U/ml RNasin) at 4°C for 1 hour for pre-absorption. 100 ng synthesized mRNAs were added to 1ml RIP buffer containing pre-absorbed beads and incubated for an additional 4 hours at 4°C. The mixture was then centrifuged and 50 μl supernatants were set aside as “5% of mRNA input”. Beads were washed five times with RIP buffer and once with RIP buffer without NP-40 and DTT (50 mMTris pH7.6, 125 mMNaCl, 1 mM EDTA, 0.2% glycerol, and 100 U/ml RNasin). mRNAs were extracted from beads using Trizol reagent, and subjected to cDNA synthesis. Subsequently, qPCR was performed using Power SYBR Green Master Mix (Life Technologies) on an Applied Biosystems 7500 real-time PCR system. The ratio between pulled down mRNA and 5% of mRNA input was used to determine binding of mRNAs by GST-Dnd1. GST alone served as the negative control. Primers for nanos1 were 5’-gggaggcgctgtctctatac-3’ and 5’-ctctggggatctctgaggag-3’. Primers for GFP were 5’-ctgaagttcatctgcaccac-3’ and 5’-gtccttgaagaagatggtgc-3’.
Oocytes, embryos and micro-injection
Xenopus oocytes and embryos were obtained and microinjected as described (Aguero, Newman, & King, 2018; Newman, Aguero, & King, 2018; Sive, Grainger, & Harland, 2000). For injection, mRNAs were synthesized using mMESSAGE mMACHINE Kit (Ambion) and the plasmid templates described above.The protocol for Xenopus studies has been approved by the University of Illinois Institutional Animal Care and Use Committee (#14249) and by the University of Miami IACC (#12–276).
In vitro translation
In vitro translation was carried out using wheat germ extracts according to the manufacturer’s instructions (Promega). Either 0.5 μg (nanos1) or 1.0 μg (eIF3f and xdnd1) capped-mRNA and GST-Dnd1 protein were used per reaction.
Phosphate release assay
The assay was performed as outlined in the manufacturer’s instructionsusing BioMol Green Reagent (BIOMOL). Recombinant Dnd1 proteins (1 μM) and ATP (0.8 mM) were added to reaction buffer (10 mM Tris-HCl 7.5, 75 mMKCl, 0.2 mM EDTA with indicated cations) and incubated at room temperature for 20 min. 200 ul BioMol Green Reagent was subsequently added to allow the color reaction. After 10 min, the absorbance was measured at 620 nm.
Homology modeling
Homology models of Dnd1 (Uniprot entry Q6DCB7, residues 76 to 235) were constructed using the SWISS-MODEL web server (Biasini et al., 2014). Figure was generated using Pymol (http://www.pymol.org).
Homodimer analysis in vitro and in vivo
Homodimer in vitro analysis was performed by GST-pull down as describled previously (Jin et al., 2015). For in vivo analysis, total embryo extracts from 1, 2, 8, and 32-cell stage embryos were incubated with purified GST-Dnd1 protein on ice for 1 hour (Aguero, Jin, et al., 2017). For IP, embryo extracts were incubated with anti-GST antibody for 3 hrs, followed by an overnight incubation at 4°C with protein G-magnetic nanobeads (Nvigen). Co-precipitated endogenous Dnd1 proteins were detected by western blot using anti-Dnd1 antibody 1:1000 (Aguero, Jin, et al., 2017).
Cell culture, transfection, antibodies, co-IP and western blot
HEK293T cells, which were authenticated and tested for contamination, were cultured and transfected as previously described (Jin et al., 2009). Protocols for CoIP and western blot were as previously described (Jin et al., 2009). Antibodies were: anti-myc (#5546, Sigma-Aldrich, 1:1,000), anti-FLAG (#F1804, Sigma-Aldrich, 1:1,000), anti-α-tubulin (#T5293, Sigma-Aldrich, 1:2,500), anti-xDnd1 ((Mei et al., 2013), 1:1000), anti-Nanos1 ((Luo et al., 2011), 1:1000), anti-mouse eIF3f (#390413, Santa Cruz, 1:500), anti-GST (ab19256, abcam). All blots consistently showed a high molecular weight non-specific band, which was used as a loading control.
Supplementary Material
Supplemental Figure 1. Dnd1 RRM2 is required for nanos1 RNA translation. Overexpressed wild-type or mutant Dnd1 proteins were tested for their ability to promote nanos1 translation in (A) oocytes and (B) wheat germ extracts. Nanos1 and Dnd1 proteins were detected by western blot analysis with either Nanos1 or Dnd1 antibody. These results are summarized in Fig. 1B.
Supplemental Figure 2. Dnd1 possesses ATPase activity that is stimulated by Mg2+. (A) Time course for ATPase activity for recombinant GST-Dnd1 protein. (B) ATPase activity of GST-zDnd1 and GST-xDnd1 is stimulated by Mg2+.
Supplemental Figure 3. A region adjacent to theTCE is required for Dnd1 to promote nanos1 translation. Each nanos1 transcript (I-IV) shown in Fig. 7B, was tested for translation in WG extracts in the presence of the translational inhibitor eIF3f and a bacterial-produced GST-Dnd1 protein. These images support the summary diagram presented in Figure 7B.
AKNOWLEDGMENTS
We greatly appreciate the insights provided by Dr. Luo at Department of Ophthalmology, Shanghai First People’s Hospital, Shanghai Jiao Tong University School of Medicine and Dr. Jain for providing guidance and the control for the ATPase assay. We thank Dr. Amanda Butler for her insightful comments on our work. Our work was supported by grants R01GM102397 to MLK, and R01GM111816 to JY.
Grants: This work is supported by NIH grants R01GM102397 to MLK, and R01GM111816 to JY
Funding sources: Our work was supported by grants from the National Institutes of Health (RO1GM102397 to M.L.K. and R01GM111816 to J.Y.)
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Aguero T, Jin Z, Chorghade S, Kalsotra A, King ML, & Yang J (2017). Maternal Dead-end 1 promotes translation of nanos1 by binding the eIF3 complex. Development, 144(20), 3755–3765. doi:10.1242/dev.152611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguero T, Kassmer S, Alberio R, Johnson A, & King ML (2017). Mechanisms of Vertebrate Germ Cell Determination. Adv Exp Med Biol, 953, 383–440. doi:10.1007/978-3-319-46095-6_8 [DOI] [PubMed] [Google Scholar]
- Aguero T, Newman K, & King ML (2018). Microinjection of Xenopus Oocytes. Cold Spring Harb Protoc, 2018(2), pdb prot096974. doi:10.1101/pdb.prot096974 [DOI] [PubMed] [Google Scholar]
- Aguero T, Zhou Y, Kloc M, Chang P, Houliston E, & King ML (2016). Hermes (Rbpms) is a Critical Component of RNP Complexes that Sequester Germline RNAs during Oogenesis. J Dev Biol, 4(1). doi:10.3390/jdb4010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramaki S, Kubota K, Soh T, Yamauchi N, & Hattori MA (2009). Chicken dead end homologue protein is a nucleoprotein of germ cells including primordial germ cells. J Reprod Dev, 55(2), 214–218. [DOI] [PubMed] [Google Scholar]
- Aramaki S, Sato F, Kato T, Soh T, Kato Y, & Hattori MA (2007). Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res, 330(1), 45–52. doi:10.1007/s00441-007-0435-1 [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, . . . Schwede T (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res, 42(Web Server issue), W252–258. doi:10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clery A, Blatter M, & Allain FH (2008). RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol, 18(3), 290–298. doi:10.1016/j.sbi.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, & Linder P (2006). The DEAD-box protein family of RNA helicases. Gene, 367, 17–37. doi:10.1016/j.gene.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Crichlow GV, Zhou H, Hsiao HH, Frederick KB, Debrosse M, Yang Y, . . . Braddock D (2008). Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. Embo J, 27(1), 277–289. doi:10.1038/sj.emboj.7601936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Clery A, & Allain FH (2013). RRM-RNA recognition: NMR or crystallography...and new findings. Curr Opin Struct Biol, 23(1), 100–108. doi:10.1016/j.sbi.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Gross-Thebing T, Yigit S, Pfeiffer J, Reichman-Fried M, Bandemer J, Ruckert C, . . . Raz E, (2017). The Vertebrate Protein Dead End Maintains Primordial Germ Cell Fate by Inhibiting Somatic Differentiation. Dev Cell, 43(6), 704–715 e705. doi:10.1016/j.devcel.2017.11.019 [DOI] [PubMed] [Google Scholar]
- Horvay K, Claussen M, Katzer M, Landgrebe J, & Pieler T (2006). Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol, 291(1), 1–11. doi:10.1016/j.ydbio.2005.06.013 [DOI] [PubMed] [Google Scholar]
- Hudson C, & Woodland HR (1998). Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mech Dev, 73(2), 159–168. [DOI] [PubMed] [Google Scholar]
- Jarmoskaite I, & Russell R (2011). DEAD-box proteins as RNA helicases and chaperones. Wiley Interdiscip Rev RNA, 2(1), 135–152. doi:10.1002/wrna.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Chung JW, Mei W, Strack S, He C, Lau GW, & Yang J (2015). Regulation of nuclear-cytoplasmic shuttling and function of Family with sequence similarity 13, member A (Fam13a) by B56-containing PP2As and Akt. Mol Biol Cell. doi:10.1091/mbc.E14-08-1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Shi J, Saraf A, Mei W, Zhu GZ, Strack S, & Yang J (2009). The 48-kDa alternative translation isoform of PP2A:B56epsilon is required for Wnt signaling during midbrain-hindbrain boundary formation. J Biol Chem, 284(11), 7190–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, . . . Agami R (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell, 131(7), 1273–1286. doi:10.1016/j.cell.2007.11.034 [DOI] [PubMed] [Google Scholar]
- Ketting RF (2007). A dead end for microRNAs. Cell, 131(7), 1226–1227. doi:10.1016/j.cell.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Kito G, Aramaki S, Tanaka K, Soh T, Yamauchi N, & Hattori MA (2010). Temporal and spatial differential expression of chicken germline-specific proteins cDAZL, CDH and CVH during gametogenesis. J Reprod Dev, 56(3), 341–346. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Tani-Matsuhana S, Ohkawa Y, Sakamoto H, & Inoue K (2017). DND protein functions as a translation repressor during zebrafish embryogenesis. Biochem Biophys Res Commun, 484(2), 235–240. doi:10.1016/j.bbrc.2017.01.080 [DOI] [PubMed] [Google Scholar]
- Koebernick K, Loeber J, Arthur PK, Tarbashevich K, & Pieler T (2010). Elr-type proteins protect Xenopus Dead end mRNA from miR-18-mediated clearance in the soma. Proc Natl Acad Sci U S A, 107(37), 16148–16153. doi:10.1073/pnas.1004401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Singh A, & King ML (2012). Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development, 139(8), 1476–1486. doi:10.1242/dev.079608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, & Lasko P (1994). Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development, 120(5), 1201–1211. [DOI] [PubMed] [Google Scholar]
- Liu W, & Collodi P (2010). Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development. FASEB J, 24(8), 2641–2650. doi:10.1096/fj.09-148403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, & Varani G (2007). RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol, 8(6), 479–490. doi:10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Nerlick S, An W, & King ML (2011). Xenopus germline nanos1 is translationally repressed by a novel structure-based mechanism. Development, 138(3), 589–598. doi:10.1242/dev.056705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchione R, Leibovitch SA, & Lenormand JL (2013). The translational factor eIF3f: the ambivalent eIF3 subunit. Cellular and Molecular Life Sciences, 70(19), 3603–3616. doi:10.1007/s00018-013-1263-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, . . . Bryant SH (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res, 43(Database issue), D222–226. doi:10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, & Allain FH (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J, 272(9), 2118–2131. doi:10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- Mei W, Jin Z, Lai F, Schwend T, Houston DW, King ML, & Yang J (2013). Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development, 140(11), 2334–2344. doi:10.1242/dev.094748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickoleit M, Banisch TU, & Raz E (2011). Regulation of hub mRNA stability and translation by miR430 and the dead end protein promotes preferential expression in zebrafish primordial germ cells. Dev Dyn, 240(3), 695–703. doi:10.1002/dvdy.22571 [DOI] [PubMed] [Google Scholar]
- Newman K, Aguero T, & King ML (2018). Isolation of Xenopus Oocytes. Cold Spring Harb Protoc, 2018(2), pdb prot095851. doi:10.1101/pdb.prot095851 [DOI] [PubMed] [Google Scholar]
- Rogers GW Jr., Richter NJ, Lima WF, & Merrick WC (2001). Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem, 276(33), 30914–30922. doi:10.1074/jbc.M100157200 [DOI] [PubMed] [Google Scholar]
- Sive H, Grainger R, & Harland R (2000). Early Development of Xenopus laevis; A Laboratory Manual (1st ed.). Cold Spring Harbor: Cold Spring Harbor Press. [Google Scholar]
- Slanchev K, Stebler J, Goudarzi M, Cojocaru V, Weidinger G, & Raz E (2009). Control of Dead end localization and activity--implications for the function of the protein in antagonizing miRNA function. Mech Dev, 126(3–4), 270–277. doi:10.1016/j.mod.2008.10.006 [DOI] [PubMed] [Google Scholar]
- van Gelder CW, Gunderson SI, Jansen EJ, Boelens WC, Polycarpou-Schwarz M, Mattaj IW, & van Venrooij WJ (1993). A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. Embo J, 12(13), 5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani L, Gunderson SI, Mattaj IW, Kay LE, Neuhaus D, & Varani G (2000). The NMR structure of the 38 kDa U1A protein - PIE RNA complex reveals the basis of cooperativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol, 7(4), 329–335. doi:10.1038/74101 [DOI] [PubMed] [Google Scholar]
- Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, & Krug RM (1999). RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. Rna, 5(2), 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, . . . Raz E (2003). dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol, 13(16), 1429–1434. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Jishage M, Meyer C, Suryawanshi H, Der E, Yamaji M, . . . Tuschl T (2017). DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature, 543(7646), 568–572. doi:10.1038/nature21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, . . . Matin A (2005). The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature, 435(7040), 360–364. doi:10.1038/nature03595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechel JL, Doerner SK, Lager A, Tesar PJ, Heaney JD, & Nadeau JH (2013). Contrasting effects of Deadend1 (Dnd1) gain and loss of function mutations on allelic inheritance, testicular cancer, and intestinal polyposis. BMC Genet, 14, 54. doi:10.1186/1471-2156-14-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Houston DW, King ML, Payne C, Wylie C, & Heasman J (1998). The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell, 94(4), 515–524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Dnd1 RRM2 is required for nanos1 RNA translation. Overexpressed wild-type or mutant Dnd1 proteins were tested for their ability to promote nanos1 translation in (A) oocytes and (B) wheat germ extracts. Nanos1 and Dnd1 proteins were detected by western blot analysis with either Nanos1 or Dnd1 antibody. These results are summarized in Fig. 1B.
Supplemental Figure 2. Dnd1 possesses ATPase activity that is stimulated by Mg2+. (A) Time course for ATPase activity for recombinant GST-Dnd1 protein. (B) ATPase activity of GST-zDnd1 and GST-xDnd1 is stimulated by Mg2+.
Supplemental Figure 3. A region adjacent to theTCE is required for Dnd1 to promote nanos1 translation. Each nanos1 transcript (I-IV) shown in Fig. 7B, was tested for translation in WG extracts in the presence of the translational inhibitor eIF3f and a bacterial-produced GST-Dnd1 protein. These images support the summary diagram presented in Figure 7B.