Abstract
Over the past decades, the Hippo has been established as a crucial pathway involved in organ size control and cancer suppression. Dysregulation of Hippo signaling and hyperactivation of its downstream effector YAP are frequently associated with various human cancers. However, the underlying significance of such YAP activation in cancer development and therapy has not been fully characterized. In this study, we reported that the Hippo signaling deficiency can lead to a YAP-dependent oncogene addiction for cancer cells. Through a clinical compound library screen, we identified histone deacetylase (HDAC) inhibitors as putative inhibitors to suppress YAP expression. Importantly, HDAC inhibitors specifically targeted the viability and xenograft tumor growth for the cancer cells in which YAP is constitutively active. Taken together, our results not only establish an active YAP-induced oncogene addiction in cancer cells, but also lay the foundation to develop targeted therapies for the cancers with Hippo dysfunction and YAP activation.
Keywords: Hippo, YAP, oncogene addiction, cancer
Introduction
Precise manipulation of the genetic lesions that initiate and maintain cancer cell growth and survival is a key theme in cancer therapy[1]. Although multiple oncogenic abnormalities exist in cancer cells, a phenomenon termed “oncogene addiction” makes cancer cells majorly depend on individual oncogenes to sustain their malignancies, providing a rationale for targeted therapies[2–4]. Identification of such oncogene addiction in specific cancer types will provide opportunities to develop more effective and selective anti-cancer agents.
The Hippo pathway, which was initially identified and characterized in Drosophila, has been recognized as a key tumor suppressor pathway in organ size control and cancer inhibition[5–7]. To achieve this, the Hippo pathway suppresses cell proliferation and survival by restricting its downstream effector YAP. In mammals, a Ser/Thr kinase, MST functions together with its adaptor SAV1 to phosphorylate LATS, another Ser/Thr kinase, and its adaptor protein MOB1. In addition to MST, members in MAP4K and TAOK kinase families function redundantly to phosphorylate and activate LATS[8–12]. Activated LATS kinase in turn phosphorylates YAP and generates the binding sites for 14-3-3 proteins, by which YAP is sequestered in the cytoplasm and degraded in a proteasome-dependent manner. On the other hand, dysregulation of the Hippo pathway results in the dephosphorylation and nuclear translocation of YAP. As a transcriptional co-activator, YAP associates with TEAD transcription factors in the nucleus to promote the transcription of genes involved in various oncogenic activities, including cell proliferation, anti-apoptosis, cell mobility and metabolic alterations[6].
Importantly, both Hippo components and its upstream regulators are frequently mutated in different types of human cancers. For example, Hippo pathway component NF2 is one of the most frequently mutated genes in schwannomas, meningiomas and malignant mesothelioma[13, 14]. LATS1 and PTPN14 are found to be lost in skin basal cell carcinoma[15]. GNAQ/GNA11 in GPCR signaling are highly mutated in uveal melanoma and their constitutive activation results in a dramatic YAP activation[16, 17]. In KRAS mutated pancreatic cancer, YAP activation can drive KRAS-independent tumor relapse[18, 19]. Notably, in all these documented cancer types, YAP is highly activated and targeting YAP significantly suppresses the related tumorigenesis. These evidences suggest that YAP could be indispensable in the cancers with Hippo signaling dysfunction.
In this study, we demonstrated that Hippo signaling deficiency enables the development of an active YAP addiction in cancer cells. Hyperactivation of YAP is widely distributed in several tested human cancers, where cancer cells with the active YAP are sensitive to the loss of YAP. Through a clinical compound library screen, we identified histone deacetylase (HDAC) inhibitors as putative inhibitors for YAP by suppressing its gene transcription. Notably, HDAC inhibitors specifically targeted the viability and tumor growth in the cancer cells with the constitutively active YAP, suggesting that active YAP can serve as a biomarker to direct the application of HDAC inhibitors in cancer therapy. Taken together, our study not only established an active YAP addiction in cancer development, but also provided an opportunity to develop new targeted strategies for the cancers with deficient Hippo signaling.
Results
Inactivation of Hippo signaling addicts cells to YAP
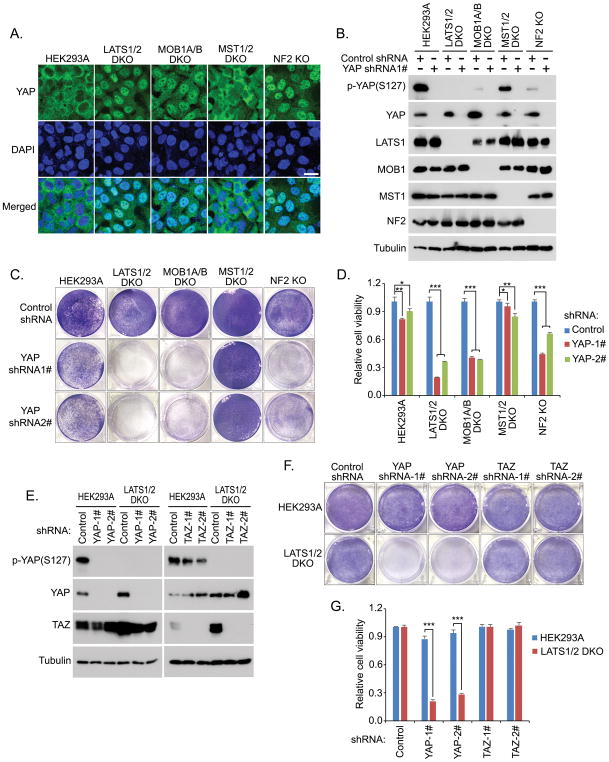
Given the well-established tumor suppressive role of the Hippo pathway, we generated a series of Hippo component knockout (KO) cells to mimic its dysfunction in cancer. In concordance with the previous studies[12], loss of Hippo pathway components, LATS1/2, MOB1A/B and NF2, dramatically induced a nuclear accumulation of YAP (Figure 1A) and significantly decreased the YAP phosphorylation (Figure 1B); deficiency of MST1/2 kinases only showed mild effect on YAP activity, because YAP is still mostly localized in the cytoplasm (Figure 1A) and had only a slight decrease of its phosphorylation (Figure 1B) in MST1/2 DKO cells. Next, we examined whether YAP is essential for these Hippo KO cells. Interestingly, cell proliferation assay revealed that loss of YAP resulted in a dramatic inhibitory effect on the growth of LATS1/2 DKO cells compared to that of wild-type cells (Figure S1). In addition, we used shRNA to downregulate YAP in each Hippo pathway component KO cells (Figure 1B) and found that loss of YAP specifically suppressed the viability of the LATS1/2 DKO, MOB1A/B DKO and NF2 KO cells, in which YAP is constitutively active, but only had a mild growth effect on both wild-type and MST1/2 DKO cells (Figures 1C and 1D). These results suggest that YAP is indispensable in the cells with the impaired Hippo signaling.
Figure 1. Hippo signaling deficiency induces the cell addiction to YAP. (See also Figure S1).
(A) YAP is activated and accumulated in the nucleus of the LATS1/2 DKO, MOB1A/B DKO and NF2 KO cells. Loss of MST1/2 does not affect YAP cellular localization. Nucleus was visualized by DAPI. Scale bar, 20 μm.
(B) Downregulation of YAP in the Hippo component KO cells. Western blotting was performed with indicated antibodies
(C and D) Loss of YAP specifically suppressed the viability of the LATS1/2 DKO, MOB1A/B DKO and NF2 KO cells. Cell viability was visualized by crystal violet staining (C) and quantified (mean ± s.d., n = 3 biological replicates) (D). * p < 0.05, ** p < 0.01, *** p < 0.001.
(E–G) Loss of YAP but not TAZ suppressed the LATS1/2 DKO cell viability. shRNA-mediated downregulation of YAP and TAZ was confirmed by Western blot in both wild-type HEK293A and LATS1/2 DKO cells (E). Cell viability was visualized by crystal violet staining (F) and quantified (mean ± s.d., n = 3 biological replicates) (G). *** p < 0.001.
In mammals, TAZ is an analog protein for YAP and is similarly regulated by the Hippo pathway. Although YAP and TAZ are both constitutively active in the LATS1/2 DKO cells[12], loss of YAP but not TAZ (Figure 1E) dramatically suppressed the LATS1/2 DKO cell viability (Figures 1F and 1G). Notably, a recent gene inactivation study comparing both YAP KO and TAZ KO cells further supports this finding, where loss of YAP showed greater effect on cell physiology than TAZ inactivation [20]. Together, at least under our experimental settings, these data indicate that Hippo signaling deficiency may addict the cells to YAP but not TAZ.
Cancer cells with the active YAP exhibit the YAP dependence
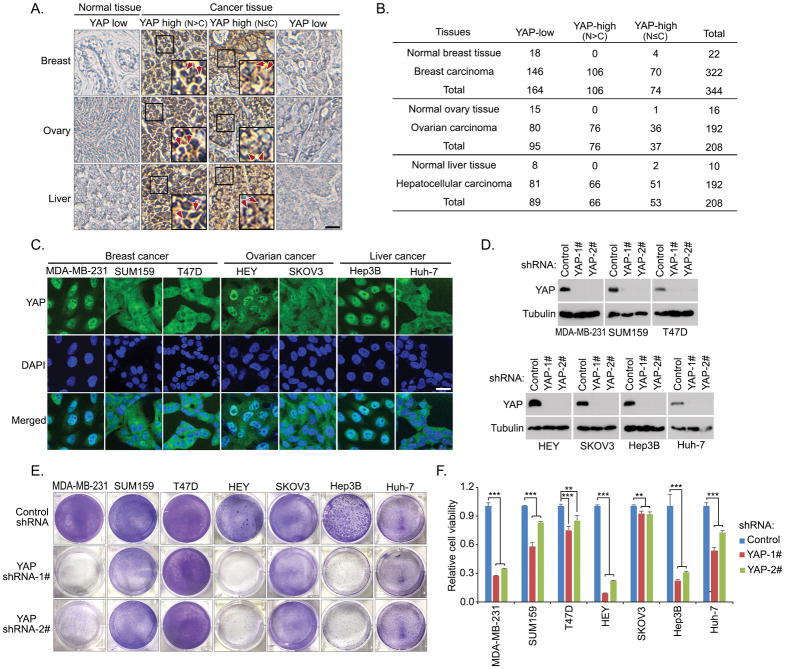
Next, we examined whether the active YAP addiction also exists in human cancers. Since dysregulation of the Hippo pathway results in a significant nuclear accumulation of YAP (Figure 1A), this nuclear enrichment of YAP can be taken as a readout for the YAP activity. First, we conducted immuohistochemical study to examine the YAP cellular localization in patient tissues from several major types of cancers. As shown in Figures 2A and 2B, YAP is highly expressed in the tested tumor tissues from breast (54.6%), ovarian (58.3%) and liver (57.8%) cancer patients. Among them, 32.9% of breast cancer samples, 39.6% of ovarian cancer samples and 34.4% of liver cancer samples show the nuclear enrichment of YAP (Figures 2A and 2B). To further determine the active YAP addiction in these cancers, a group of related cancer cells were used to examine the correlation between the YAP activity and their dependence on YAP. Immunofluorescence experiments showed that YAP is highly enriched in the nucleus of breast cancer cell line MDA-MB-231, ovarian cancer cell line HEY and liver cancer cell line Hep3B (Figure 2C), suggesting that YAP is activated in these cancer cell lines. As for the other tested cancer cells, YAP is either majorly localized in the cytoplasm (e.g. breast cancer cell lines SUM159 and T47D, liver cancer cell line Huh-7) or distributed evenly between the nucleus and cytoplasm (e.g. ovarian cancer cell line SKOV3) (Figure 2C). These findings suggest a heterogeneity of human cancer cells with a diverse Hippo/YAP activity.
Figure 2. Cancer cells with the active YAP exhibit the YAP dependence.
(A and B) Immunohistochemical staining of YAP were performed in breast cancer, ovarian cancer and liver cancer tissue microarrays. Brown staining indicates positive immunoreactivity (A). Scale bar, 40 μm. The box region is twice enlarged. Arrows indicated nuclear staining of YAP. Correlation analysis of YAP expression/localization in the indicated human normal and tumor samples are shown as tables (B).
(C) YAP is activated and accumulated in the nuclei of a group of cancer cell lines. YAP localization in each cancer cell was examined by immunofluorescence. Nucleus was visualized by DAPI. Scale bar, 20 μm.
(D–F) Loss of YAP specifically suppressed the viability of the cancer cells with YAP dominantly localized in the nucleus. shRNA-mediated downregulation of YAP was confirmed by Western blot in the indicated cancer cells (D). Cell viability was visualized by crystal violet staining (E) and quantified (mean ± s.d., n = 3 biological replicates) (F). ** p < 0.01, *** p < 0.001.
To determine the active YAP addiction in human cancer cells, we used shRNA to downregulate YAP in all these tested cancer cell lines (Figure 2D) and examined their dependence on YAP. Interestingly, loss of YAP dramatically suppressed the viability for the cancer cells with YAP dominantly localized in the nucleus (e.g. MDA-MB-231, HEY, Hep3B), but only showed a certain extent of growth inhibitory effect on the cells with YAP mostly localized in the cytoplasm (e.g. SUM159, T47D, SKOV3, Huh-7) (Figures 2E and 2F). These results suggest that Hippo inactivation/YAP activation is associated with a YAP-dependent oncogene addiction in the tested cancer cells, which is consistent with our previous findings by using the Hippo KO cells (Figures 1C and 1D).
HDAC inhibitors suppress the YAP expression
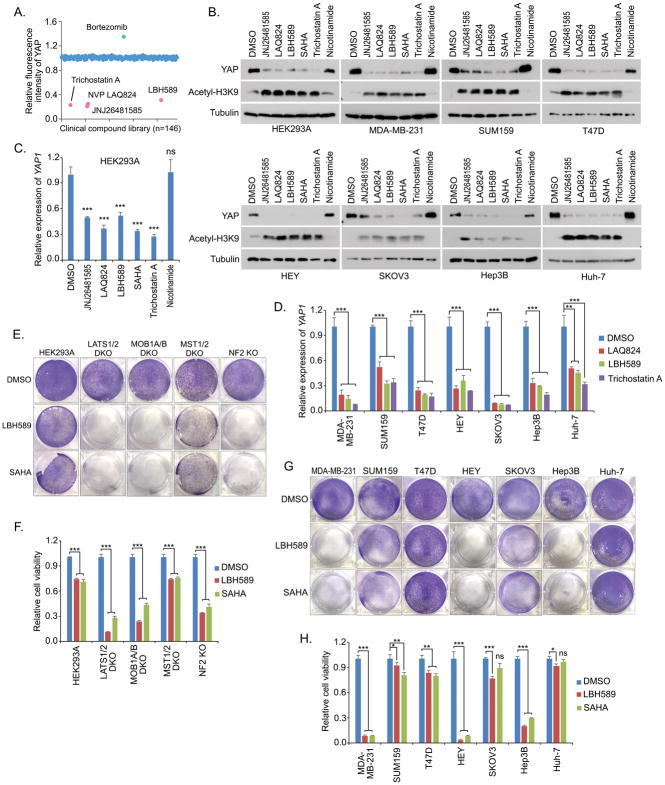
To develop a therapeutic method targeting the YAP-dependent cancers, we performed a clinical compound library screen using MDA-MB-231 cell line[21] (Figure 3A). Through it, we identified that several HDAC inhibitors, including LBH589 (Panobinostat), JNJ26481585 (Quisinostat), LAQ824 (Dacinostat) and Trichostatin A, can dramatically suppress the YAP expression in all the tested cancer cells (Figure 3B). Similar finding was also observed in the treatment of SAHA (Vorinostat) (Figure 3B), another HDAC inhibitor that is under clinical trial. As a control, treatment of nicotinamide, an inhibitor of the SIRT family deacetylase, did not affect the YAP expression (Figure 3B). Moreover, quantitative PCR assay revealed that these identified HDAC inhibitors dramatically inhibited the YAP gene transcription (Figures 3C and 3D) in a dose-dependent manner (Figure S2A), but did not affect that of TAZ (Figure S2B). These data demonstrate that HDAC inhibitors can inhibit YAP expression.
Figure 3. HDAC inhibitors suppress the YAP gene expression and the YAP-dependent cell viability. (See also Figure S2).
(A) A group of HDAC inhibitors were identified to suppress YAP expression. A clinical compound library (with 146 compounds) screen was performed in MDA-MB-231 cells. YAP immunofluorescent staining was performed and its relative fluorescent intensity was calculated. Several identified hits that affect YAP fluorescent intensity were indicated.
(B) HDAC inhibitors suppressed the YAP protein expression. Western blotting was performed with the indicated antibodies. Cells were treated for 24 hours with the indicated compounds (0.5 μM JNJ26481585, 0.5 μM LAQ824, 0.5 μM LBH589, 10 μM SAHA, 10 μM Trichostatin A, and 100 μM Nicotinamide).
(C and D) YAP gene transcription is suppressed by HDAC inhibitors. YAP gene transcription (mean ± s.d., n = 3 biological replicates) was examined in both HEK293A (C) and the indicated cancer cells (D) after 24 hour-treatment by the indicated compounds. ** p < 0.01, *** p < 0.001, ns, no significance.
(E and F) HDAC inhibitors dramatically targeted the viability of the LATS1/2 DKO, MOB1A/B and NF2 KO cells compared to that of the wild-type and MST1/2 DKO cells. Cell viability was visualized by crystal violet staining (E) and quantified (mean ± s.d., n = 3 biological replicates) (F). *** p < 0.001.
(G and H) HDAC inhibitors dramatically targeted the viability of the cancer cells with the active YAP addiction. Cell viability was visualized by crystal violet staining (G) and quantified (mean ± s.d., n = 3 biological replicates) (H). * p < 0.05, ** p < 0.01, *** p < 0.001, ns, no significance.
HDAC inhibitors specifically target the viability of cancer cells with the active YAP addiction
Since HDAC inhibitors suppress the YAP expression, next we tested their efficacy in targeting the cells with the active YAP addiction. As for the Hippo KO cells, HDAC inhibitors LBH589 and SAHA dramatically suppressed the viability of the LATS1/2 DKO, MOB1A/B DKO and NF2 KO cells, in which YAP is constitutively active; but only showed a certain extent of inhibitory effect on both wild-type and MST1/2 DKO cells, in which YAP is still normally controlled by the Hippo pathway (Figures 3E and 3F). As for the cancer cells, LBH589 and SAHA specifically suppressed the viability for MDA-MB-231, HEY and Hep3B cells (Figures 3G and 3H), which all showed the active YAP addiction (Figures 2E and 2F). As for the tested cancer cells with YAP mostly localized in the cytoplasm including SUM159, T47D, SKOV3 and Huh-7, cells are more resistant to the treatment of HDAC inhibitors (Figures 3G and 3H). Similarly, HDAC inhibitors induced cell apoptosis in MDA-MB-231 cells that are known deficient in NF2[22], but not in SUM159 cells, as indicated by the induced cleaved PARP (Figure S2C). To further confirm this finding, additional breast cancer cells were subjected to the treatment of HDAC inhibitors, where only the ones (MDA-MB-231 and HCC1806) with YAP dominantly localized in the nucleus were more sensitive to the treatment of HDAC inhibitors (Figure S2D). Together, these data indicate that the cancer cells with the active YAP addiction could be specifically targeted by HDAC inhibitors.
Hippo deficiency induces tumor vulnerability to HDAC inhibitors
Given the facts that Hippo deficiency can addict cancer cells to YAP and HDAC inhibitors preferably target the cancer cells with the active YAP addiction, one may expect that HDAC inhibitors could be applied to treat the cancers with the dysregulated Hippo signaling. To test this hypothesis, we took advantage of the Lats1/2 DKO 4T1 cells[23] to examine the efficacy of HDAC inhibitors in treatment of the Hippo-deficient tumor growth.
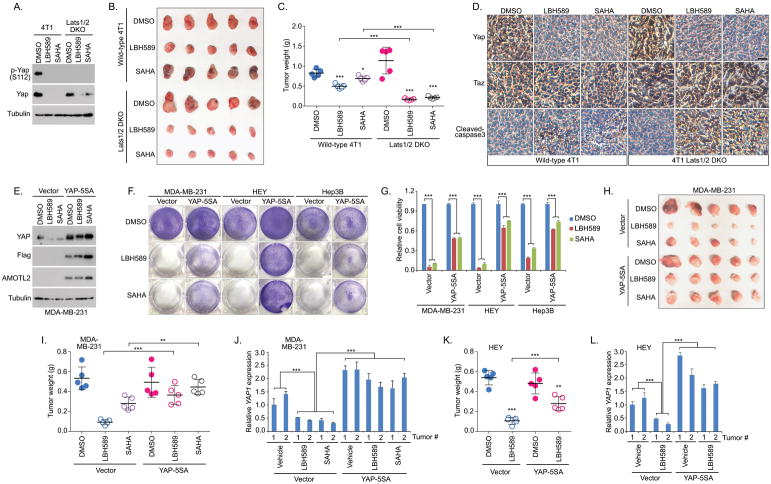
Consistently with the previous findings (Figure 3B), Western blot showed that Yap expression was dramatically suppressed in both wild-type 4T1 and the Lats1/2 DKO 4T1 cells upon the treatment of HDAC inhibitors LBH589 and SAHA (Figure 4A). Interestingly, although the wild-type 4T1-derived xenograft tumor growth was significantly suppressed by LBH589 and SAHA, loss of Lats1/2 further enhanced the tumor vulnerability in response to the treatment of LBH589 and SAHA (Figures 4B and 4C). Further immunohistologic analysis showed that Yap level was dramatically decreased in the LBH589 or SAHA-treated tumors that were derived from both wild-type 4T1 and Lats1/2 DKO 4T1 cells (Figure 4D). Although the expression of Taz was increased in the Lats1/2 DKO tumors, its level was not clearly changed upon the treatment of LBH589 or SAHA (Figure 4D). Moreover, LBH589 and SAHA treatment further enhanced the cell apoptosis (cleaved-caspase 3 staining) in the Lats1/2 DKO tumors compared to the wild-type 4T1 tumors under the same compound treated condition (Figure 4D). Taken together, these results suggest that Hippo signaling deficiency is able to sensitize tumors to HDAC inhibitors.
Figure 4. Hippo deficiency induces the tumor vulnerability to HDAC inhibitors.
(A) HDAC inhibitors suppressed Yap expression in both wild-type 4T1 and Lats1/2 DKO 4T1 cells. Western blotting was performed with the indicated antibodies.
(B and C) Loss of Lats1/2 sensitized the 4T1-derived xenograft tumors to HDAC inhibitors. Wild-type 4T1or Lats1/2 DKO 4T1cells were subjected to the xenograft tumor assay and treated with LBH589 and SAHA. Xenograft tumors are shown in (B) and the tumor weight was quantified (n = 5 mice, mean ± s.d.) (C). * p < 0.05, *** p < 0.001.
(D) Immunohistochemistry was performed with the indicated antibodies in the HDAC inhibitor-treated wild-type 4T1 and Lats1/2 DKO 4T1 xenograft tumors. Scale bar, 40 μm.
(E) HDAC inhibitors failed to suppress the expression of exogenously expressed YAP-5SA. Western blotting was performed with the indicated antibodies.
(F and G) Overexpression of the constitutively active YAP-5SA mutant rescued the viability of cells with the active YAP addiction under the treatment of HDAC inhibitors. Control or YAP-5SA mutant-transfected MDA-MB-231, HEY and Hep3B cells were treated with LBH589 and SAHA. Cell viability was visualized by crystal violet staining (F) and quantified (mean ± s.d., n = 3 biological replicates) (G).
(H–J) Overexpression of the constitutively active YAP-5SA mutant rescued MDA-MB-231 xenograft tumor growth under the treatment of HDAC inhibitors. Control or YAP-5SA mutant-transfected MDA-MB-231 cells were subjected to the xenograft tumor assay and treated with HDAC inhibitors LBH589 and SAHA. Xenograft tumors are shown in (H) and the tumor weight was quantified (n = 5 mice, mean ± s.d.) (I). YAP gene transcription was examined in two tumors (J) randomly selected from the indicated xenograft tumor groups (H). ** p < 0.01, *** p < 0.001.
(K and L) Overexpression of the constitutively active YAP-5SA mutant rescued HEY xenograft tumor growth under the treatment of HDAC inhibitor LBH589. Xenograft tumor weight was quantified (n = 5 mice, mean ± s.d.) (K). YAP gene transcription was examined in two randomly selected tumors (L). ** p < 0.01, *** p < 0.001.
HDAC inhibitors target YAP-addicted cancer cells through YAP suppression
Although HDAC inhibitors showed a promising efficacy in targeting the cancer cells with the active YAP addiction (Figures 3G, 3H, S2D and 4A–4D), it is still unclear whether this therapeutic effect was achieved by their ability to target YAP expression or not. To address this question, HDAC inhibitors were subjected to the treatment of MDA-MB-231 cells stably expressing the YAP-5SA active mutant[24]. Interestingly, Western blot revealed that HDAC inhibitors failed to suppress the expression of exogenously expressed YAP-5SA (Figure 4E), which provides us an ideal model to assess the role of YAP in the HDAC inhibitor-mediated targeted therapy.
Indeed, overexpression of YAP-5SA significantly rescued the viability of MDA-MB-231, HEY and Hep3B cells, which all exhibited an active YAP addiction (Figures 2E and 2F), under the treatment of LBH589 and SAHA (Figures 4F and 4G). Moreover, expression of YAP-5SA significantly rescued the MDA-MB-231 tumor growth compared to the vector control cells upon LBH589 and SAHA treatment (Figures 4H and 4I). Consistently, the transcription of YAP was largely decreased in the vector control tumors treated by LBH589 and SAHA, while YAP level was relatively stable in the YAP-5SA overexpressed tumors under the same treated conditions (Figure 4J). Similarly, expression of YAP-5SA significantly rescued the HEY-derived tumor growth upon LBH589 treatment (Figure 4K) by maintaining a relatively stable level of YAP (Figure 4L). Notably, expression of YAP-5SA did not completely rescue the cell viability (Figures 4F and 4G) and tumor growth (Figures 4H, 4I and 4K) upon HDAC inhibitor treatment, suggesting that YAP is at least a major target of HDAC inhibitors in treating cancers with the active YAP addiction.
Discussion
The Hippo has been established as a tumor suppressor pathway and its downstream effector YAP is known as an oncoprotein; however, compared to multiple genetic and epigenetic abnormalities during cancer development, role of Hippo signaling dysfunction/YAP activation in cancer has not been fully elucidated. In this study, we revealed an unexpected YAP addiction in the cancers driven by the Hippo inactivation. This finding was initiated in the study of the Hippo component KO cells that were generated within the same cell line, which largely decreased the genetic perturbation as caused by using different cell lines.
Notably, previous studies have implicated an essential role of YAP in the cancers with the Hippo deficiency. Genetic mouse studies demonstrated that inhibition of YAP expression or activity dramatically targeted the liver cancer formation as generated by the loss of Hippo component Nf2 or Mst1/2[25–27]. Inactivation of YAP dramatically inhibited the oncogenic activities of the NF2-deficient mesothelioma cells[28, 29]. Moreover, YAP is dominantly localized in the nucleus of the uveal melanoma cells with GNAQ/GNA11 mutations[16, 17]. Similarly, loss of YAP dramatically suppressed the cell viability and tumor formation for the uveal melanoma cells carrying GNAQ/GNA11 mutations, but did not affect the BRAF-mutated cells with the functional Hippo signaling[17]. These genetic and pathological evidences further support our current hypothesis that deficient Hippo signaling can result in a YAP addiction in cancers.
YAP activation is tightly controlled by the Hippo pathway (Figure 1A); however, genetic mutations of the Hippo pathway components in major human cancer types are rare[6], indicating that additional oncogenic alterations could exist to account for the YAP activation and subsequent tumorigenesis. These Hippo signaling perturbations deserve further investigation.
Regarding the translational significance, it will be highly important to determine whether this newly identified active YAP addiction can be employed in targeted cancer therapy. We are more interested in identifying the existing compounds and/or clinically approved drugs in this setting. Interestingly, our clinical compound library screen revealed that HDAC inhibitors can dramatically suppress YAP expression (Figures 3A–3D) and target the YAP-dependent cancer cell viability (Figures 3E–3H) and tumor formation (Figure 4). As for the HDAC inhibitors, several key questions are remained to be elucidated. For example, since histone hyper-acetylation is believed to lead to transcriptional activation, it is hard to explain how the YAP gene transcription is inhibited by HDAC inhibitors. One possibility is that HDAC inhibitors may induce the expression of transcriptional factor(s) that can bind to YAP promoter and suppress the YAP gene transcription. Another possibility is that HDAC inhibitors may suppress the YAP gene transcription by upregulating miRNAs. Indeed, some miRNAs, which can target YAP transcription, are found to be regulated by HDAC inhibitors[30]. However, it is known that HDAC inhibitors can suppress the transcription of many other oncogenic proteins[31], but not specific to YAP. Based on this fact, we did not conduct an in-depth mechanistic study to further address this question; instead, we are more interested in determining the novel therapeutic roles of these compounds by taking YAP as a biomarker in cancer therapy.
Of note, as with any class of anticancer agents, HDAC inhibitors are also associated with toxicities. The most common adverse events with the use of HDAC inhibitors were thrombocytopenia, neutropenia, anemia, fatigue and diarrhea[32]. Nausea, vomiting, anorexia, constipation and dehydration were also seen in patients receiving HDAC inhibitors[32]. According to these facts, it will be critically important to identify the putative biomarkers to indicate the most effective response to HDAC inhibitors and direct the clinical use of this group of anticancer agents. If our hypothesis is correct, YAP could serve as a potential biomarker to direct the application of HDAC inhibitors for targeted cancer treatment and minimize the cytotoxic effects in patients.
Of course, the active YAP-induced oncogenic addiction could also be applicable to multiple human cancers, but not restricted to the tested breast cancer, ovarian cancer and liver cancer (Figure 2). In addition, HDAC inhibitors may not be the best compounds in targeting the active YAP addiction in clinical settings. Through our current study, we hope to not only conceptually establish the active YAP addiction as a promising biomarker in cancer diagnosis, but also outline a feasible translational strategy to utilize this newly identified oncogene addiction in cancer therapy.
Materials and Methods
Antibodies and chemicals
For Western blotting, anti-Flag (M2) (F3165-5MG, 1:5000 dilution) and anti-α-tubulin (T6199-200UL, 1:5000 dilution) monoclonal antibodies were obtained from Sigma-Aldrich. Anti-phospho-YAP (S127) (4911S, 1:1000 dilution), anti-LATS1(3477S, 1:1000 dilution), anti-MST1(3682S, 1:1000 dilution), anti-MOB1(3863S, 1:2000 dilution), anti-NF2 (12896S, 1:2000 dilution), anti-YAP/TAZ (8418S, 1:1000 dilution), anti-KIBRA (8774S, 1:1000 dilution), anti-PARP (S9542, 1:1000 dilution) and anti-acetyl-Histone H3 (Lys9) (9649S, 1:2000 dilution) antibodies were purchased from Cell Signaling Technology. Anti-LATS2 (A300-479A, 1:2000 dilution) polyclonal antibody was obtained from Bethyl. Anti-YAP antibody and anti-AMOTL2 antibody were raised by immunizing rabbits with bacterially expressed and purified GST-fused human full-length YAP protein and GST-fused human AMOTL2 protein (amino acids 1~675), respectively. The antisera were affinity-purified using the AminoLink Plus Immobilization and Purification Kit (Pierce). Both YAP and AMOTL2 antibodies were used at 1:1000 dilution for Western blotting. For immunostaining, an anti-YAP (sc-101199, 1:200 dilution) monoclonal antibody was purchased from Santa Cruz Biotechnology. For immunohistochemical staining, anti-YAP (14074S, 1:15 dilution) and anti-cleaved caspase-3 (S9661, 1:100 dilution) antibodies were purchased from Cell Signaling Technology. An anti-TAZ (HPA007415, 1:100 dilution) antibody was obtained from Sigma-Aldrich.
For HDAC inhibitors, JNJ26481585 (Quisinostat), LAQ824 (Dacinostat), LBH589 (Panobinostat) and Trichostatin A were purchased from SelleckChem. SAHA was obtained from Cayman Chemicals. Nicotinamide (N3376) was obtained from Sigma.
Constructs and viruses
YAP-5SA-SFB lentiviral expression vector was generated through a gateway-based LR reaction (Invitrogen). pLKO1-YAP shRNAs were purchased from Addgene (#27368 and #27369). pGIPZ-TAZ shRNAs were obtained from shRNA and ORFeome Core Facility at MD Anderson Cancer Center. The shRNA sequence is listed as follows:
Control shRNA: 5′-TCTCGCTTGGGCGAGAGTAAG-3′;
TAZ shRNA-1#: 5′-CAGACATGAGATCCATCACTA-3′;
TAZ shRNA-2#: 5′-TACTTCCTCAATCACATAGAA-3′.
The lentiviral supernatant was generated by transfecting HEK293T cells with pSPAX2 and pMD2G that were kindly provided by Dr. Zhou Songyang (Baylor College of Medicine) and collected 48 hours later. Supernatants were passed through a 0.45-μm filter and used to infect the cells with the addition of 8 μg/mL hexadimethrine bromide (Polybrene) (Sigma-Aldrich).
Cell culture and transfection
HEK293T cells were obtained from ATCC and maintained in Dulbecco’s modified essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in 5% CO2 (v/v). HEK293A cells were kindly provided by Dr. Jae-Il Park (MD Anderson Cancer Center). The MDA-MB-231, T47D, HCC1806, BT549 and HCC1937 cells were kindly provided by Dr. Mien-Chie Hung (MD Anderson Cancer Center). HEY and SKOV3 cells were kindly provided by Dr. Jingson Liu (MD Anderson Cancer Center). Hep3B and Huh-7 cells were kindly provided by Dr. Jian Chen (MD Anderson Cancer Center). SUM149 and SUM159 cells were kindly provided by Dr. Li Ma (MD Anderson Cancer Center) and cultured in DMEM/F12 medium supplemented with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone and 10 mM HEPES. MCF10A cells were purchased from ATCC and maintained in DMEM/F12 medium supplemented with 5% horse serum, 200 ng/ml epidermal growth factor, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxin and 10 μg/ml insulin. All the cells were passed the test of mycoplasma. Plasmid transfection was performed using a polyethylenimine reagent.
Generation of human Hippo pathway components knockout (KO) cell lines using CRISPR/Cas9
For each Hippo pathway component (MOB1A, MOB1B, LATS1 and LATS2), five distinct single-guide RNAs (sgRNA) were designed by CHOPCHOP website (https://chopchop.rc.fas.harvard.edu), cloned into lentiGuide-Puro vector (Addgene plasmid # 52963). As for NF2, sgRNA sequence was designed based on previous study[33].
sgRNAs were transfected into HEK293A cells with lentiCas9-Blast construct (Addgene plasmid # 52962). At the second day, cells were selected with puromycin (2 μg/ml) for two days and subcloned to form single colonies. Clones were screened by Western blotting to verify the loss of targeted proteins and subsequently confirmed by sequencing. MST1/2 DKO HEK293A cells and Lats1/2 DKO 4T1 cells were kindly provided by Dr. Kun-Liang Guan (UC San Diego).
The sequence information for sgRNAs is listed as follows:
NF2_sgRNA: AACCCAAGACGTTCACCGTG;
LATS1_sgRNA1: CGTGCAGCTCTCCGCTCTAA;
LATS1_sgRNA2: GTTGTGCAGGTGACCATCCA;
LATS1_sgRNA3: GTCTCCACATCGACAGCTTG;
LATS1_sgRNA4: CCTGTTCGTAGCAACACTTC;
LATS1_sgRNA5: ACAGACTGAAGCCATTAGAG
LATS2_sgRNA1: TACGCTGGCACCGTAGCCCT;
LATS2_sgRNA2: AGGTAGTCCACGTACGGCCG;
LATS2_sgRNA3: TTACGCCAGCCTGCCCACGA;
LATS2_sgRNA4: CGAAGCTTGGGCCCTCGTAG;
LATS2_sgRNA5: GTAGGACGCAAACGAATCGC;
MOB1A_sgRNA1: CTATTCTAAAGCGTCTGTTC;
MOB1A_sgRNA2: GCAGAAGCAACTCTAGGAAG;
MOB1A_sgRNA3: CAAGCTGTTATGTTGCCTGA;
MOB1A_sgRNA4: GTTTGAATGTTTTAGAAGAG;
MOB1A_sgRNA5: TGTTATGTTGCCTGAGGGAG;
MOB1B_sgRNA1: TGGCAGTGGCAACCTTCGGA;
MOB1B_sgRNA2: CACTTGGCAGTGGCAACCTT;
MOB1B_sgRNA3: TATACTCAAACGCCTCTTTA;
MOB1B_sgRNA4: TCATACTGGTGAGAACCCTC;
MOB1B_sgRNA5: TCATTGAGATCTTCCCCTTC.
Cell viability/proliferation assay
To examine the cell growth, same number of cells were seeded as triplicates in 12-well plates. As for the shRNA knockdown experiments, cells were cultured for 7 days; as for the drug treatment experiments, cells were cultured for 5 days. At the endpoint, cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet. After that, cells were washed three times and detained with acetic acid. The absorbance of the crystal violet solution was measured at OD 595nm and normalized to the control shRNA-transduced or vehicle-treated cells to indicate the relative cell viability/proliferation.
RNA extraction, reverse transcription and real-time PCR
Trizol reagent (Invitrogen) was used to prepare the RNA samples. We performed reverse transcription assay by using the Script Reverse Transcription Supermix Kit (Bio-Rad) according to the manufacturer’s instructions. Power SYBR Green PCR master mix (Applied Biosystems) was used for Real-time PCR. The 2−ΔΔCt method was used to quantify the gene expression. As for the normalization, GAPDH expression was examined along with the indicated genes.
The primer sequence information for real-time PCR analysis is listed as follows:
YAP-Forward: 5′-TTGGGAGATGGCAAAGACAT-3′;
YAP-Reverse: 5′-CTGTGACGTTCATCTGGGAC-3′;
TAZ-Forward: 5′-GGCTGGGAGATGACCTTCAC-3′;
TAZ-Reverse: 5′-ATTCATCGCCTTCCTAGGGT-3′;
GAPDH-Forward: 5′-ATGGGGAAGGTGAAGGTCG-3′;
GAPDH-Reverse: 5′-GGGGTCATTGATGGCAACAATA-3′.
Immunofluorescent staining
As described previously[34], cells were cultured on coverslips, fixed in 4% paraformaldehyde for 10 min at room temperature and extracted with 0.5% Triton X-100 solution for 5 minutes. After blocking with TBST containing 1% bovine serum albumin for 30 min, the cells were incubated with YAP antibody for 1 hour at room temperature. Cells were washed for three times with TBST and then incubated with fluorescein isothiocyanate-conjugated secondary antibody for 1 hour. After that, cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, 100 ng/mL) to visualize the nucleus. The cover slips were mounted onto glass slides with an anti-fade solution and subjected to the examination under a Nikon Eclipse Ti spinning-disk confocal microscope.
Immunohistochemical analysis
The patient tissue arrays were obtained from US Biomax, deparaffinized and rehydrated. Unmask Solution (Vector Laboratories) were used to retrieve the antigens by steaming for 40 min. After that, the sections were treated with 3% hydrogen peroxide for 30 min to block endogenous peroxidase activity. The tissue samples were pre-incubated by 10% goat serum to prevent non-specific staining and then incubated with an antibody at 4°C overnight. The tissue samples were further incubated with SignalStain Boost detection reagent (Cell Signaling Technology) for 30 min at room temperature. To visualize the staining, samples were treated with SignalStain 3,3′-diaminobenzidine chromogen-diluted solution (Cell Signaling Technology) and counterstained with Mayer hematoxylin.
To quantify the results, a total score of protein expression was calculated from both the percentage of immunopositive cells and immunostaining intensity. High and low protein expressions were defined using the mean score of all samples as a cutoff point. By using this criteria, the staining for some tissues could be interpreted as “low”, even the immunohistochemical signal was hardly detected.
Xenograft Assays
All the xenograft tumor experiments were strictly followed institutional guidelines that are approved by the UC Irvine Institutional Animal Care and Use Committee (IACUC). Four-week-old female nude mice were obtained from Jackson Laboratory and kept in a pathogen-free environment. 4T1 (wild-type and Lats1/2 DKO) or MDA-MB-231 (vector and YAP-5SA) cells (2×106) were injected bilaterally into the inguinal mammary fat pad of nude mice. HEY (vector and YAP-5SA) cells (2×106) were injected subcutaneously into the nude mice. When tumors were approximately 50mm3 in size, 10 mice for each cell line will be randomly assigned into two groups (5 mice per group) and subjected to the indicated compound treatment (vehicle control or 10 mg/kg/day LBH589 or 50 mg/kg/day SAHA) through intraperitoneal administration every other day. After 15 days for adaptation, mice were euthanized for tumor collection and analyses.
Statistical analysis
We repeated each experiment twice or more, unless otherwise noted. There were no samples or animals excluded for the analyses in this study. As for the mouse experiments, there was no statistical method used to predetermine sample size. We assigned the samples or animals randomly to different groups. A laboratory technician was blinded to the group allocation and tumor collections during the animal experiments as well as the data analyses. The Student t-test was used to analyze the differences between groups. A P value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Steven H. Lin and Junjie Chen (MD Anderson Cancer Center) for the clinical compound library. W.W. is a recipient of an American Association for Cancer Research Career Development Award for Translational Breast Cancer Research supported by the Breast Cancer Research Foundation (16-20-26-WANG). This work was supported by University of California, Irvine Chao Family Comprehensive Cancer Center (Pilot Project 2018) and in part by a NIH grant (R01 GM126048) and a Research Scholar Grant (RSG-18-009-01-CCG) from the American Cancer Society to W.W.
Footnotes
Author contributions
W.W. conceived and supervised study. H.H. and W.W. designed the experiments and analyzed the data. H.H. and B.Y. performed all the experiments with the assistance from H.J.N., J.Y., Y.Z., K.L.N., A.T.B., and T.K.M.. W.W. wrote the manuscript.
Conflict of interest
The authors declare no competing conflict of interests.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. discussion 3080. [DOI] [PubMed] [Google Scholar]
- 3.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 4.Pagliarini R, Shao W, Sellers WR. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Rep. 2015;16:280–296. doi: 10.15252/embr.201439949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y, Wang W, Liu B, Deng H, Uster E, Pan D. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev Cell. 2015;34:642–655. doi: 10.1016/j.devcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, et al. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Cho YS, Yue T, Ip YT, Jiang J. Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discov. 2015;1:15038. doi: 10.1038/celldisc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plouffe SW, Meng Z, Lin KC, Lin B, Hong AW, Chun JV, et al. Characterization of Hippo Pathway Components by Gene Inactivation. Mol Cell. 2016;64:993–1008. doi: 10.1016/j.molcel.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrilli AM, Fernandez-Valle C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene. 2016;35:537–548. doi: 10.1038/onc.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 16.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plouffe SW, Lin KC, Moore JL, Tan FE, Ma S, Ye Z, et al. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. Journal of Biological Chemistry. 2018 doi: 10.1074/jbc.RA118.002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Li N, Li X, Tran MK, Han X, Chen J. Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins. Cell Rep. 2015;13:524–532. doi: 10.1016/j.celrep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 23.Moroishi T, Hayashi T, Pan WW, Fujita Y, Holt MV, Qin J, et al. The Hippo Pathway Kinases LATS1/2 Suppress Cancer Immunity. Cell. 2016;167:1525. doi: 10.1016/j.cell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang WQ, Dai YY, Hsu PC, Wang H, Cheng L, Yang YL, et al. Targeting YAP in malignant pleural mesothelioma. J Cell Mol Med. 2017;21:2663–2676. doi: 10.1111/jcmm.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuno T, Murakami H, Fujii M, Ishiguro F, Tanaka I, Kondo Y, et al. YAP induces malignant mesothelioma cell proliferation by upregulating transcription of cell cycle-promoting genes. Oncogene. 2012;31:5117–5122. doi: 10.1038/onc.2012.5. [DOI] [PubMed] [Google Scholar]
- 30.Jung DE, Park SB, Kim K, Kim C, Song SY. CG200745, an HDAC inhibitor, induces anti-tumour effects in cholangiocarcinoma cell lines via miRNAs targeting the Hippo pathway. Sci Rep. 2017;7:10921. doi: 10.1038/s41598-017-11094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Seto E. HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med. 2016:6. doi: 10.1101/cshperspect.a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mottamal M, Zheng SL, Huang TL, Wang GD. Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents. Molecules. 2015;20:3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Chen L, Ding Y, Jin J, Liao K. Centrosome separation driven by actin-microfilaments during mitosis is mediated by centrosome-associated tyrosine-phosphorylated cortactin. J Cell Sci. 2008;121:1334–1343. doi: 10.1242/jcs.018176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.