Abstract
Tripartite motif (TRIM) proteins mediate antiviral host defenses by either directly targeting viral components or modulating innate immune responses. Here we identify a mechanism of antiviral restriction in which a TRIM E3 ligase controls viral replication by regulating the structure of host cell centrosomes and thereby nuclear lamina integrity. Through RNAi screen we identified several TRIM proteins, including TRIM43, that control the reactivation of Kaposi’s sarcoma-associated herpesvirus. TRIM43 was distinguished by its ability to restrict a broad range of herpesviruses and its profound upregulation during herpesvirus infection as part of a germline-specific transcriptional program mediated by the transcription factor DUX4. TRIM43 ubiquitinates the centrosomal protein Pericentrin thereby targeting it for proteasomal degradation, which subsequently leads to alterations of the nuclear lamina that repress active viral chromatin states. Our study identifies a role of the TRIM43-Pericentrin-lamin axis in intrinsic immunity, which may be targeted for therapeutic intervention against herpesviral infections.
Introduction
Infection by viruses of the family Herpesviridae is the cause of significant morbidity and mortality in humans worldwide, especially in immunocompromised individuals1. Furthermore, two of the eight human herpesviruses, Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV), are human carcinogens and cause a range of malignancies of lymphoproliferative and endothelial origin2, 3. Despite differences in pathogenesis and carcinogenicity, herpesviruses share common features in their replication strategies, most notably in the switch from a state of persistent latent infection with minimal viral gene expression to a state of lytic infection characterized by productive viral replication. While antiviral therapies targeting the lytic phase exist, a major obstacle to herpesvirus eradication is the persistence of these viruses in latently-infected cells. As such, molecular insight into the factors that modulate the latent-to-lytic transition of herpesviral replication is of fundamental importance. While major progress has been made toward identifying the viral factors that regulate the latent-to-lytic switch, our knowledge of host intrinsic factors that modulate the transition is still rudimentary.
Tripartite motif (TRIM) proteins have been increasingly appreciated as important antiviral factors that suppress the replication of a wide range of RNA and DNA viruses. TRIM proteins inhibit viral replication by either directly targeting viral components or modulating innate immune responses that result in antiviral gene expression4, 5. For example, TRIM19 (also known as promyelocytic leukemia protein (PML)) restricts multiple RNA viruses and DNA viruses, including herpesviruses, by organizing PML nuclear bodies6, 7. We postulated that hitherto unidentified TRIM proteins play critical roles in restricting herpesvirus lytic replication or reactivation.
Here, we identified several TRIM proteins that suppress KSHV reactivation. Among the TRIM proteins identified, TRIM43 was distinguished by its ability to restrict a broad range of herpesviruses. TRIM43 targets the centrosomal protein Pericentrin for degradation, which leads to the loss of nuclear envelope integrity and changes in viral chromatin that repress lytic replication.
Results
Identification of TRIM43 as a herpesvirus-specific antiviral protein
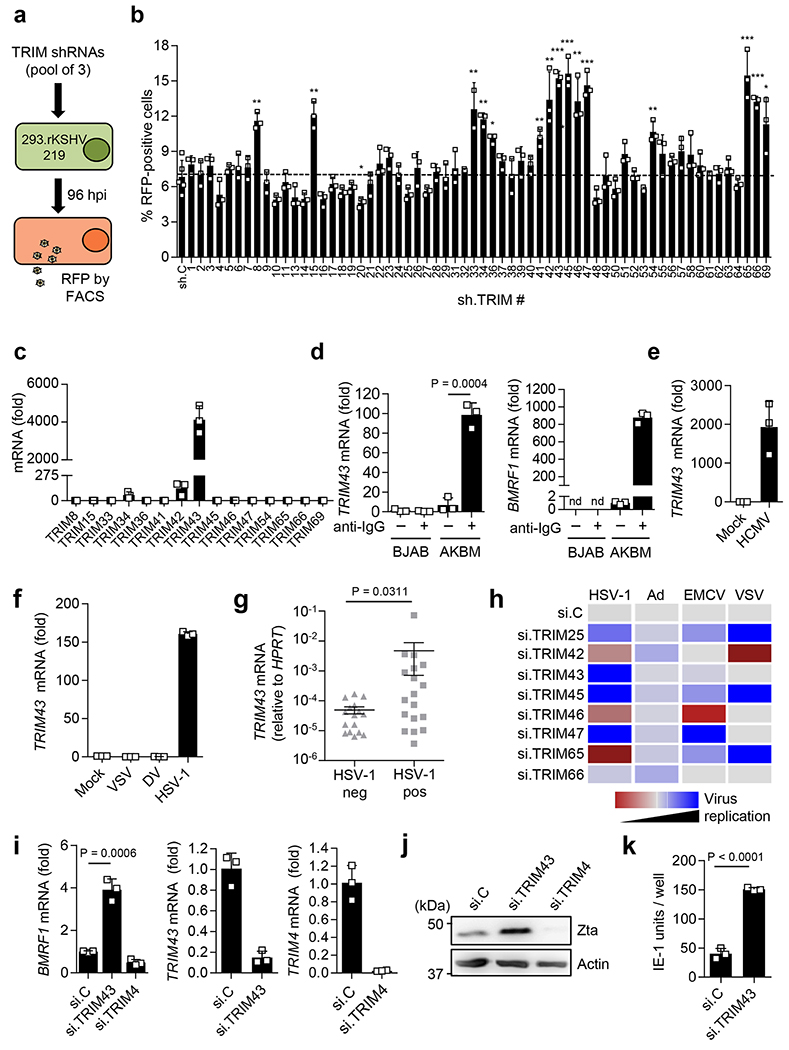
To identify TRIM proteins that modulate herpesvirus reactivation, we performed an RNAi screen targeting 62 human TRIM genes to test the effect of TRIM depletion on KSHV reactivation (Fig. 1a,b). Individual TRIM genes were silenced by lentiviral transduction of a pool of 3 different shRNAs per TRIM in HEK 293 cells latently infected with recombinant KSHV strain 219 (293-rKSHV.219), which expresses red fluorescent protein (RFP) under the control of a lytic viral promoter, allowing high-throughput detection of viral reactivation by monitoring RFP expression8. Depletion of 15 TRIM proteins showed a significant increase in RFP expression compared to cells transduced with control non-targeting shRNA (sh.C), suggesting that these TRIM proteins suppress KSHV reactivation and/or lytic replication (Fig. 1b). In contrast, the depletion of other TRIM proteins had minor or no effects on KSHV reactivation (Fig. 1b).
Fig. 1. TRIM43 is a herpesvirus-specific antiviral factor.
a, Schematic representation of the TRIM shRNA screen in 293.rKSHV.219 cells. b, rKSHV.219 reactivation from a following lentiviral transduction of non-targeting control shRNA (sh.C) or TRIM-specific shRNAs (x axis) by analyzing RFP-positive cells using fluorescence-activated cell sorting (FACS). c, TRIM transcripts in iSLK.219 cells treated with doxycycline (1 μ g ml-1) for 3 days to induce rKSHV.219 reactivation, determined by qRT-PCR. d, Left, TRIM43 transcripts in EBV-infected AKBM or EBV-negative BJAB cells that were mock-treated or treated with 100 μ g ml-1 anti-human immunoglobulin G (IgG) for 24 h to induce EBV reactivation, determined by qRT-PCR. Right, EBV reactivation determined by analyzing BMRF1 transcripts by qRT-PCR. e, TRIM43 transcripts in human foreskin fibroblast (HFF) cells infected with HCMV (MOI of 3) for 24 h, determined by qRT-PCR. f, TRIM43 transcripts in Huh7 cells infected with VSV (MOI 0.01), DV (MOI 1) or HSV 1 (MOI 1) for 18 h, assessed by qRT-PCR. g, TRIM43 mRNA expression in BAL samples from patients with acute pulmonary infection, assessed by qRT-PCR. h, Heatmap summarizing the results from the siRNA mini-screen to test the effect of TRIM knockdown on the replication of HSV-1, Ad, EMCV and VSV (Supplementary Fig. 2d–g). i, Left, EBV BMRF1 transcripts in AGS-EBV cells transfected with non-targeting control siRNA (si.C), si.TRIM43 or si.TRIM4 (negative control), determined by qRT-PCR at 96 h post-transfection. Middle and right, knockdown of TRIM43 and TRIM4, determined by qRT-PCR at 48 h after siRNA transfection. j, EBV Zta-protein abundance of AGS-EBV cells from i, determined by immunoblot (IB) using anti-Zta at 120 h after siRNA transfection. k, HCMV IE-1 protein-positive HFF cells following transfection with si.TRIM43 or si.C and subsequent infection with HCMV (50 IE-1 units) for 24 h, determined by immunofluorescence (IF). Data represent mean and s.d. of n = 3 (biological replicates) (b,c–f,i,k), or mean and s.d. of 18 HSV-1-positive or 16 HSV-1-negative BAL samples (g). Statistical significance was calculated by unpaired two-tailed t-test (b,d,i,k), or two-tailed Mann-Whitney U test (g). *P < 0.05, **P < 0.01, ***P < 0.001. Exact P values for b are provided in Supplementary Table 1. Results are representative of one screen (b) or three independent experiments (c–f,h–k). nd, not detectable.
A hallmark of many antiviral factors is inducible gene expression in response to the virus(es) they restrict9. Therefore, we next examined whether the expression of the 15 candidate TRIM proteins identified in our screen was inducible by KSHV reactivation (Fig. 1c). This showed that the transcript levels of TRIM43 were profoundly (> 10,000-fold) upregulated upon KSHV reactivation; in comparison, the mRNA levels of the other candidate TRIM proteins were much less, or not, induced by KSHV reactivation (Fig. 1c). TRIM43 mRNA expression was also strongly induced upon reactivation of EBV, a gamma-herpesvirus closely related to KSHV, or upon de novo infection by human cytomegalovirus (HCMV) (of the Betaherpesvirinae subfamily) or herpes simplex virus type 1 (HSV-1) (of the Alphaherpesvirinae subfamily) (Fig. 1d–f and Supplementary Fig. 1a). In contrast, infection with adenovirus (Ad), a dsDNA virus unrelated to herpesviruses, or with the RNA viruses vesicular stomatitis virus (VSV) or dengue virus (DV), did not induce TRIM43 mRNA expression (Fig. 1f and Supplementary Fig. 1b).
We next asked whether TRIM43 is also upregulated in herpesvirus-associated diseases in humans. TRIM43 mRNA was significantly upregulated in bronchoalveolar lavage (BAL) human patient samples that were positive for HSV-1 as compared to HSV-1-negative BAL samples (Fig. 1g). Investigation of TRIM43 protein expression in Kaposi’s sarcoma (KS) tissues derived from AIDS patients showed that TRIM43 was moderately or highly expressed in 10 of 17 cases analyzed, whereas TRIM43 expression was minimal in all healthy skin tissues (Supplementary Fig. 1c). Taken together, these results show that TRIM43 gene expression is robustly induced upon herpesviral infection, which prompted us to investigate whether TRIM43 has anti-herpesviral activity.
To test this, we first asked whether TRIM43 knockdown induces productive KSHV replication in cells that are usually refractory to efficient KSHV lytic replication, such as HEK 293T cells. Knockdown of endogenous TRIM43 using an shRNA with a different targeting sequence than the shRNAs used in our screen, led to efficient KSHV lytic gene expression and production of infectious virions in infected HEK 293T cells (Supplementary Fig. 2a,b). Depletion of TRIM43 also enhanced KSHV lytic replication in SLK cells harboring recombinant KSHV.219 (iSLK.219) (Supplementary Fig. 2c). To determine if TRIM43 specifically inhibits herpesviral infection, we next examined the effect of silencing TRIM43 on the replication of HSV-1, Ad, VSV, and encephalomyocarditis virus (EMCV). We also tested seven additional TRIM proteins (TRIM25, TRIM42, TRIM45, TRIM46, TRIM47, TRIM65 and TRIM66) that had demonstrated pronounced effects on KSHV reactivation in our shRNA screen (Fig. 1b). siRNA-mediated depletion of endogenous TRIM43 led to a significant increase in HSV-1 replication. In contrast, knocking down TRIM43 had no or a marginal effect on the replication of Ad, VSV or EMCV (Fig. 1h and Supplementary Fig. 2d–i), whereas depletion of the other seven TRIM proteins affected the replication of multiple viruses, and each TRIM possessed a unique restriction profile (Fig. 1h), strengthening the conclusion that TRIM43 specifically restricts herpesviruses. In further support of this hypothesis, silencing of endogenous TRIM43 also induced the spontaneous reactivation of EBV from latently-infected cells and enhanced lytic replication of HCMV, as indicated by the enhanced expression of the immediate-early/early genes, BMRF1 and Zta or immediate-early-1 (IE-1), respectively (Fig. 1i–k and Supplementary Fig. 2j). Finally, we asked the inverse question of whether ectopic expression of TRIM43 suppresses herpesviral infection, and whether this activity is dependent on its RING E3 ligase activity, which is known to play an important role in the antiviral activity of other TRIM proteins5, 10, 11. Ectopic expression of wild-type (WT) TRIM43 suppressed HSV-1 replication, while expression of a mutant of TRIM43 which has the RING-finger domain deleted (TRIM43ΔRING), did not have an inhibitory effect on HSV-1 replication (Supplementary Fig. 2k). Collectively, these results demonstrate that TRIM43 suppresses the reactivation or lytic replication of multiple herpesviruses, and further suggest that the TRIM43 antiviral activity is dependent on its RING E3 ligase activity.
TRIM43 is induced by DUX4 during herpesvirus infection as part of a germline-specific transcriptional program
As TRIM43 is a completely uncharacterized protein we first tested its expression pattern in different human tissues (Supplementary Fig. 3a). We found ubiquitous but very low mRNA expression of TRIM43 in all human tissues tested, with the exception of the brain, thymus and testis, which showed higher expression. We next used laser-scanning confocal microscopy to assess the localization of TRIM43 protein in primary human foreskin fibroblast (HFF) cells that were either left uninfected, or infected with HSV-1 (Fig. 2a). We observed extremely low expression of endogenous TRIM43 protein in uninfected cells; TRIM43 was detectable only at high magnification, where the protein appeared as one or two small puncta per cell. However, TRIM43 protein expression was profoundly upregulated in HSV-1-infected cells, showing punctate expression throughout the cytoplasm (Fig. 2a). UV-inactivated HSV-1 did not induce TRIM43 expression, suggesting that TRIM43 upregulation is dependent on active viral replication (Fig. 2a).
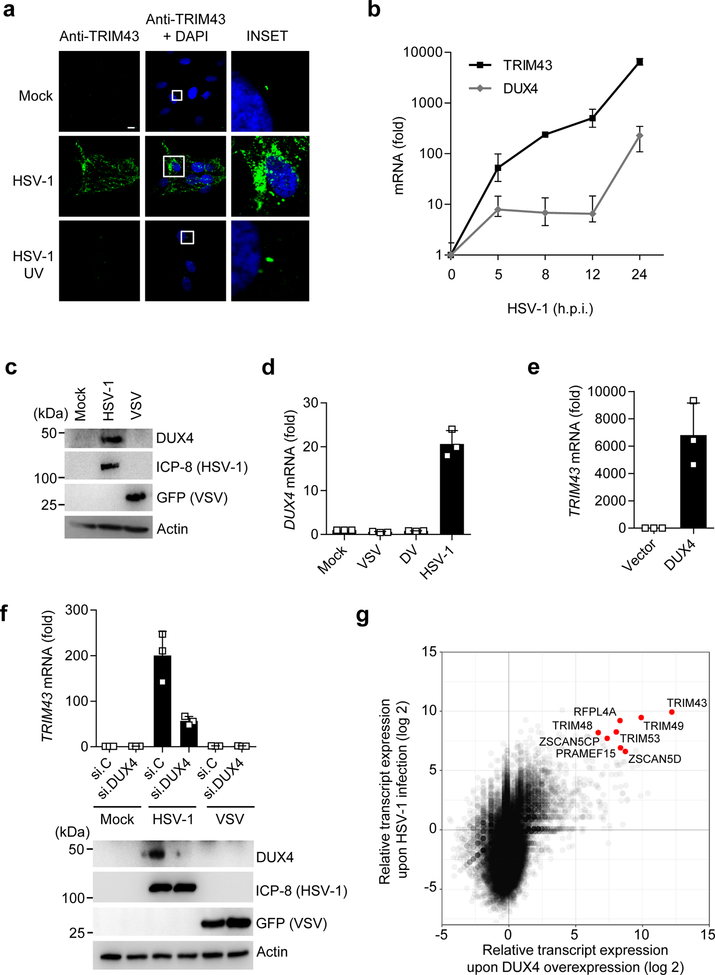
Fig. 2. TRIM43 is induced upon herpesvirus infection as part of a DUX4-dependent germline transcriptional program.
a, TRIM43 protein expression in primary HFF cells that were either mock-infected, or infected with live HSV-1 or UV-inactivated HSV-1 (HSV-1 UV) (both MOI 1) for 16 h, determined by IF using an anti-TRIM43 antibody. DAPI, nuclei (blue). Scale bar, 20 μm. b, TRIM43 and DUX4 transcripts in HFF cells infected with HSV-1 (MOI 1) for the indicated times, determined by qRT-PCR. Values are presented as fold induction (normalized to 18S rRNA) relative to uninfected control cells (0 h). c, Endogenous DUX4 protein expression in HFF cells that were infected with HSV-1 (MOI 5) or VSV-GFP (MOI 0.05) for 18 h, or that remained uninfected (mock), determined by immunoblot (IB) analysis using anti-DUX4 antibody. IB of HSV-1 infected cell protein 8 (ICP-8) and GFP (VSV-GFP) served as controls. d, DUX4 transcripts in Huh7 cells infected with VSV-GFP (MOI 0.05), HSV-1 (MOI 5), or DV (MOI 1) for 18 h, assessed by qRT-PCR. Values were normalized and calculated as in (b). e, TRIM43 transcripts in HEK 293T cells transfected with empty vector or a plasmid encoding human DUX4, determined by qRT-PCR at 36 h post-transfection. f, Upper: TRIM43 transcripts in HFF cells that were transfected with non-targeting control siRNA (si.C) or DUX4-specific siRNA (si.DUX4) for 24 h and subsequently infected with HSV-1 (MOI 0.1) or VSVGFP (MOI 0.05) for 14 h, assessed by qRT-PCR. Values were normalized and calculated as in (b). Lower panel: Knockdown of endogenous DUX4 was confirmed by IB with an anti-DUX4 antibody. IB of ICP-8 (HSV-1) and GFP (VSV-GFP) served as infection controls. g, RNAseq of HEK 293T cells that were either mock-treated, transfected with a plasmid encoding human DUX4 for 18 h, or infected with HSV-1 (MOI 10) for 18 h. Scatter blot of global gene expression in DUX4-expressing vs. HSV-1-infected cells, relative to mock-treated cells, is shown. Data represent mean and s.d. of n = 3 (biological replicates) (b, d–f). Data are representative of three (a–f) or two (g) independent experiments.
To provide insight into the mechanism of TRIM43 transcriptional upregulation, we first asked whether TRIM43 gene expression is type I interferon (IFN)-inducible, as is the case for many other TRIM proteins4, 12. Treatment with IFN-α or infection by Sendai virus (SeV), known to robustly trigger IFN induction, failed to enhance TRIM43 transcript levels, while the mRNA levels of IFN-stimulated gene 15 (ISG15) were strongly upregulated (Supplementary Fig. 3b). TRIM43 was also robustly upregulated by HSV-1 infection in Vero cells (Supplementary Fig. 3c), which are known to be defective in IFN-α/β signaling13, further supporting that herpesvirus-induced TRIM43 expression is independent of type I IFNs.
Previous reports indicated that TRIM43 is efficiently expressed during early embryogenesis but minimally expressed in adult tissues14–17. Moreover, TRIM43 gene expression was found to be robustly upregulated in cells from patients with facioscapulohumeral muscle dystrophy (FSHD); in this disease, TRIM43 was shown to be a target gene of the germline transcription factor double homeobox 4 (DUX4), whose aberrant expression causes FSHD18–20. Therefore, we asked whether DUX4 gene expression is induced by herpesvirus infection, and whether TRIM43 expression is mediated by DUX4 in herpesvirus-infected cells. DUX4 transcripts were profoundly upregulated over the course of HSV-1 infection, which correlated closely with the induction of TRIM43 mRNA (Fig. 2b). DUX4 protein was also readily detectable in HSV-1-infected cells, where it localized to the cell nucleus, but was not detectable in uninfected cells (Fig. 2c and Supplementary Fig. 3d). In a pattern similar to that of herpesvirus-specific induction of TRIM43 expression (Fig. 1c–g), DUX4 was highly expressed only during herpesvirus infection, whereas its expression was minimal during infection with VSV or DV (Fig. 2d). To examine if DUX4 is sufficient for the induction of TRIM43, we ectopically expressed DUX4 in HEK 293T cells, which do not detectably express DUX4 under normal (uninfected) conditions, and found that ectopic expression of DUX4 robustly induced TRIM43 transcripts (Fig. 2e). Next, we depleted endogenous DUX4 in HSV-1-infected cells using specific siRNA (Fig. 2f). TRIM43 mRNA expression was highly upregulated by HSV-1 infection in cells transfected with non-targeting control siRNA (si.C), whereas knockdown of DUX4 markedly reduced TRIM43 mRNA levels in infected cells. These results indicate that DUX4 is necessary and sufficient to drive TRIM43 gene expression.
Our data showed that herpesvirus infection upregulates TRIM43 gene expression in a manner that is dependent on DUX4, which is a germline transcription factor that is usually only expressed during early development, or in FSHD patients, which carry genetic modifications of the DUX4 gene locus19, 21. Comparative global transcriptome analysis of HEK 293T cells transfected with DUX4 or infected with HSV-1 showed that the transcript levels of TRIM43 and also several other known DUX4-target genes (e.g., members of the PRAMEF and ZSCAN protein families15–18, 20) are highly upregulated by HSV-1 infection or DUX4 expression (Fig. 2g and Supplementary Fig. 4a,b). Quantitative real-time PCR analysis confirmed that these DUX4-target genes are also markedly upregulated upon KSHV reactivation (Supplementary Fig. 4c). Together, these results indicate that herpesvirus infection triggers TRIM43 gene expression as part of a DUX4-dependent germline-specific transcriptional program.
TRIM43 regulates the stability of Pericentrin and thereby centrosomal integrity
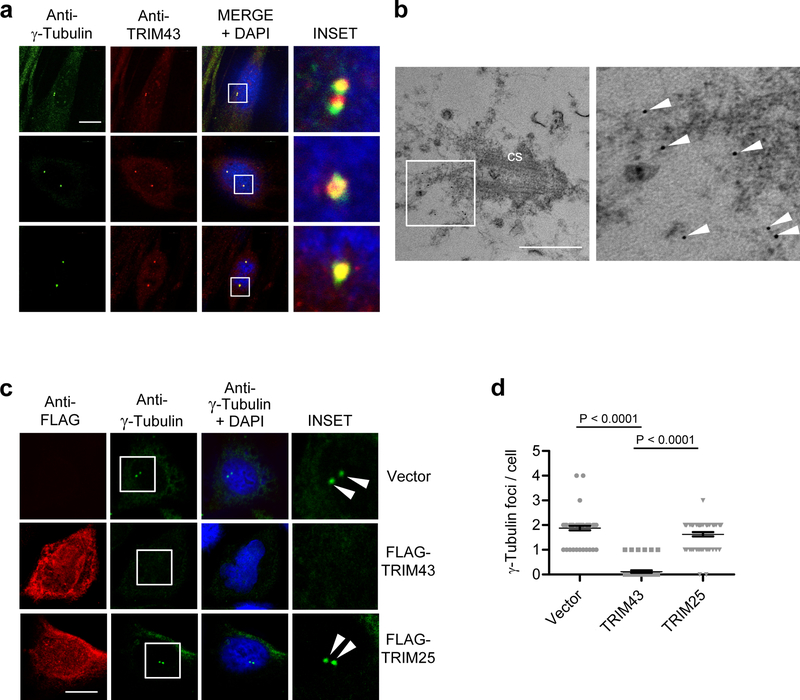
To elucidate the mechanism behind TRIM43-mediated herpesvirus restriction, we began by characterizing the localization of TRIM43, which under normal (uninfected) conditions appears as either one or two puncta per cell (Fig. 2a). Confocal microscopy analysis revealed that the TRIM43 puncta co-stained with the centrosomal marker γ-tubulin, suggesting that TRIM43 localizes to the centrosome (Fig. 3a). Of note, siRNA-mediated depletion of TRIM43 abolished its centrosomal staining pattern, ruling out unspecific staining of centrosomes by the polyclonal TRIM43 antibody (Supplementary Fig. 5a). Electron microscopy analysis confirmed that TRIM43 localization was restricted to the centrosome in uninfected cells, where it localized to the pericentrosomal material (Fig. 3b and Supplementary Fig. 5b). Furthermore, we observed that ectopic expression of TRIM43, but not TRIM25, which has a similar domain structure as TRIM43 but does not localize to centrosomes11, led to a near-complete loss of centrosomal γ−tubulin staining. Ectopic expression of TRIM43 also led to a loss of staining for the centrosomal proteins centrobin and Sas-6 (Supplementary Fig. 5c–e), which prompted us to hypothesize that TRIM43 controls herpesviral reactivation/lytic replication by modulating the structural integrity and/or function of the centrosome.
Fig. 3. TRIM43 localizes to centrosomes and regulates centrosomal integrity.
a, IF analysis of endogenous TRIM43 and γ-Tubulin in HFF cells, determined by confocal microscopy using anti-TRIM43 and anti-γ-Tubulin antibodies. Nuclei were stained with DAPI (blue). Scale bar, 20 μm. b, Electron microscopy analysis of HFF cells after immunogold labeling and staining with anti-TRIM43 antibody and protein A coupled to 5 nm gold particles. Arrows indicate TRIM43. cs, centrosome. Scale bar, 500 nm. c, HeLa cells were transfected with empty vector, or plasmids encoding FLAG-tagged TRIM43 or TRIM25. 36 h later, cells were immunostained with anti-γ−Tubulin (green) and anti-FLAG (red) antibodies and subjected to IF analysis. Nuclei, DAPI (blue). Scale bar, 20 μm. d, Quantification of the number of γ-Tubulin foci per cell from (c) for 50 cells. Data represent mean and s.d. of n = 50, and statistical significance was calculated by unpaired two-tailed t-test. Data are representative of three (a,c) or two (b) independent experiments.
Indeed, mass spectrometry (MS) analysis of affinity-purified TRIM43ΔRING showed that it interacted with several proteins that are important for centrosome function or structural integrity: the key structural centrosomal proteins Pericentrin (PCNT) and γ-tubulin22, 23, as well as the cohesin subunits, SMC1, SMC3, and RAD21, which are involved in centrosome disengagement24 (Supplementary Fig. 5f). Importantly, we used in this experiment the E3 ligase-defective TRIM43ΔRING mutant because we postulated that this would allow us to purify putative TRIM43 substrates that would otherwise have been degraded.
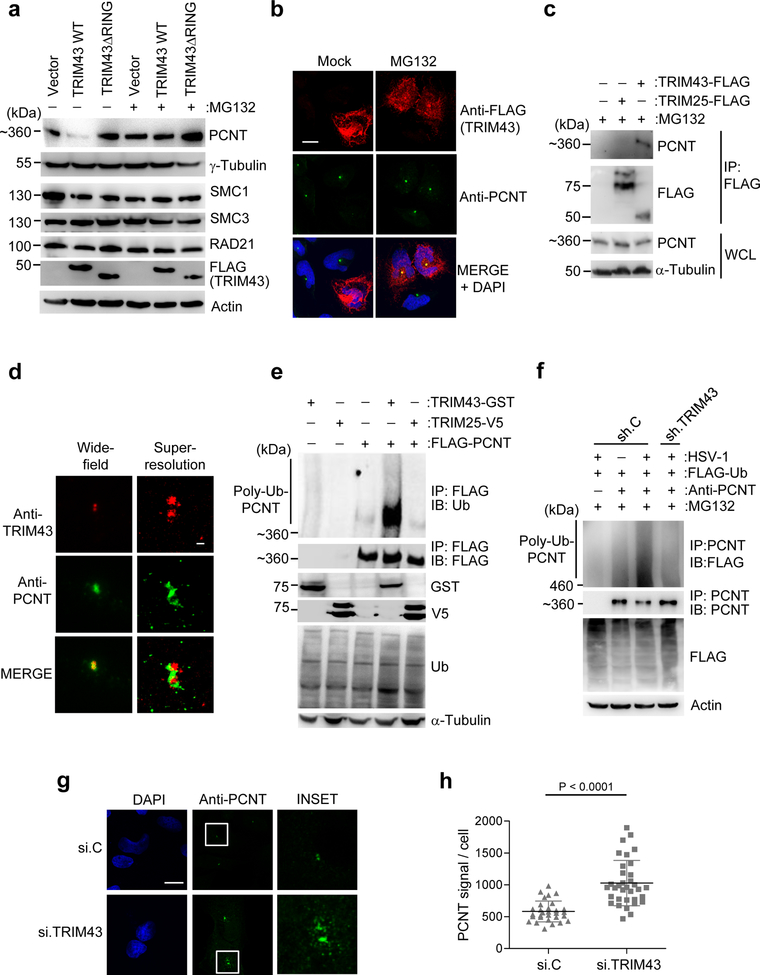
Therefore, we next asked whether TRIM43 E3 ligase activity triggers the degradation of the centrosomal proteins identified by our MS analysis. We found that ectopic expression of FLAG-TRIM43 WT effectively induced the degradation of endogenous PCNT (Fig. 4a,b and Supplementary Fig. 6a) but did not trigger the degradation of γ-tubulin, SMC1, SMC2, or RAD21 (Fig. 4a). Treatment with the proteasome inhibitor MG132 rescued TRIM43-induced degradation of PCNT (Fig. 4a,b and Supplementary Fig. 6a). In contrast to WT TRIM43, ectopic expression of the catalytically-inactive TRIM43ΔRING mutant did not induce PCNT degradation, but instead, led to an increase in PCNT protein abundance (Fig. 4a and Supplementary Fig. 6a), suggesting a dominant negative effect of TRIM43ΔRING. Kinetics analysis of PCNT protein abundance in HSV-1-infected or KSHV-reactivated cells showed a gradual loss of PCNT protein, which closely correlated with increased TRIM43 protein expression (Supplementary Fig. 6b and 7a). In contrast, VSV or Ad infection did not lead to changes in PCNT protein abundance (Supplementary Fig. 7b,c). These data suggested that TRIM43 interacts with PCNT at the centrosome and regulates its protein stability during herpesvirus infection.
Fig. 4. TRIM43 mediates ubiquitination-dependent proteasomal degradation of PCNT.
a, IB analysis of indicated proteins in HEK 293T cells transfected with empty vector, FLAG-tagged TRIM43 WT or ΔRING for 24 h, and then mock-treated or treated with MG132 for 5 h. Expression of transfected FLAG-TRIM43 constructs was confirmed by IB with anti-FLAG. b, IF analysis of endogenous PCNT in HeLa cells transfected with FLAG-TRIM43 for 24 h and then mock-treated or treated with MG132 for 5 h, determined by immunostaining with an anti-PCNT antibody. TRIM43 expression was confirmed by staining with anti-FLAG. Nuclei, DAPI (blue). Scale bar, 20 μm. c, Binding of endogenous PCNT and FLAG-tagged WT TRIM43 in transiently transfected and MG132-treated HEK 293T cells, determined by co-IP with anti-FLAG and IB with anti-PCNT. Loading control, anti-α-Tubulin. d, Complex formation of endogenous PCNT and TRIM43 in HFF cells, determined by IF using anti-PCNT and anti-TRIM43 and imaged by GSD-TIRF microscopy. Widefield pictures (upper panels, 160 × objective) or superresolution pictures (lower panels). Scale bar, 200 nm. e, Ubiquitination of FLAG-PCNT in transiently transfected HEK 293T cells co-transfected with empty vector, TRIM43-GST, or TRIM25-V5 and treated with MG132 for 5 h, determined by IP with anti-FLAG and IB with anti-ubiquitin (Ub). Whole cell lysates (WCL) were immunoblotted with anti-Ub, anti-GST, anti-V5 and anti-α-Tubulin (loading control). f, Ubiquitination of endogenous PCNT in HEK 293T cells expressing FLAG-tagged Ub and either control shRNA (sh.C) or TRIM43-specific shRNA (sh.TRIM43) and infected with HSV-1 (MOI 1) for 16 h, determined by IP with anti-PCNT. Cells were further treated with MG132 for 4 h to limit PCNT degradation. g, PCNT protein abundance in HFF cells that were transfected with TRIM43-specific siRNA (si.TRIM43) or non-targeting control siRNA (si.C) for 40 h, determined by IF using an anti-PCNT antibody. Nuclei, DAPI (blue). Scale bar, 20 μm. h, Quantification of PCNT fluorescence intensity per cell (n = 30) from (g). Data represent mean and s.d. of n = 30, and statistical significance was calculated by unpaired two-tailed t-test. Data are representative of three (a–c, e–g) or two (d) independent experiments.
In support of an interaction between TRIM43 and PCNT, co-IP experiments showed that endogenous PCNT interacted with FLAG-TRIM43, but not FLAG-TRIM25 (Fig. 4c). Ground state depletion combined with total internal reflection fluorescence (GSD-TIRF) super-resolution microscopy indicated that endogenous TRIM43 and PCNT form a complex at the centrosome (Fig. 4d). We next tested whether TRIM43 mediates PCNT degradation through direct ubiquitination by examining the effect of ectopically-expressed TRIM43 on PCNT ubiquitination by immunoblot. TRIM43, but not TRIM25, expression strongly enhanced PCNT polyubiquitination (Fig. 4e). Conversely, whereas endogenous PCNT ubiquitination was increased upon HSV-1 infection, silencing of TRIM43 diminished PCNT ubiquitination in infected cells (Fig. 4f). Depletion of endogenous TRIM43 led to a strong increase in the abundance (both signal intensity and numbers) of PCNT puncta, further supporting that TRIM43 regulates PCNT protein stability via degradative ubiquitination (Fig. 4g,h). These data indicate that TRIM43 regulates the structural integrity of centrosomes by interacting with PCNT and inducing its proteasomal degradation.
Herpesvirus restriction by TRIM43 is dependent on Pericentrin degradation
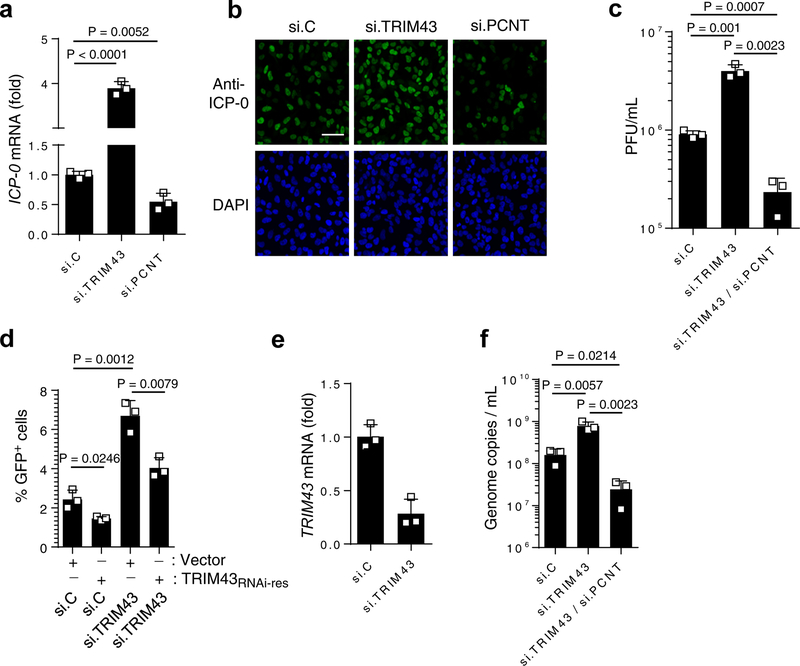
To address the question whether the ability of TRIM43 to degrade PCNT is responsible for TRIM43’s antiviral activity, we used siRNA to deplete PCNT, thus mimicking the effects of TRIM43-mediated PCNT degradation. In parallel, we also silenced TRIM43, which leads to higher stability of PCNT and is therefore expected to have the opposite effect on virus replication. Knockdown of PCNT significantly decreased (by ~ 50%) the mRNA and protein expression of HSV-1 Infected cell protein-0 (ICP-0), which is indicative of repressed lytic replication (Fig. 5a,b), while it had no effect on Ad or VSV replication (Supplementary Fig. 7d,e). In contrast, silencing TRIM43 markedly enhanced ICP-0 mRNA and protein abundance as well as HSV-1 titers (Fig. 5a–c). Importantly, ectopic expression of a siRNA-resistant TRIM43 (TRIM43RNAi-res) construct in cells depleted of TRIM43 reversed the effect of TRIM43 knockdown on HSV-1 replication, ruling out off-target effects of the siRNA (Fig. 5d,e). Moreover, while knockdown of TRIM43 enhanced HSV-1 replication and KSHV reactivation, double-knockdown of TRIM43 and PCNT reversed this effect, strongly suggesting that TRIM43’s antiviral activity is mediated by PCNT (Fig. 5c,f). Of note, ectopic expression of TRIM43 did not lead to changes in the cell cycle, ruling out that TRIM43’s effect on PCNT leads to defective mitosis and thereby affects herpesvirus infection (Supplementary Fig. 8a). These results indicate that TRIM43-mediated restriction of herpesvirus replication is dependent on its ability to degrade PCNT.
Fig. 5. Herpesvirus restriction by TRIM43 is dependent on PCNT degradation.
a, HSV-1 ICP-0 transcripts in HEK 293 cells that were transfected with the indicated siRNAs and 36 h later infected with HSV-1 (MOI 0.5) for 1 h, determined by qRT-PCR. b, HSV-1 ICP-0 protein expression in HeLa cells that were transfected with non-targeting control siRNA (si.C), or siRNAs specific for TRIM43 or PCNT (si.TRIM43 or si.PCNT) for 24 h and then infected with HSV-1 (MOI 0.5) for 3 h, determined by IF using an anti-ICP-0 antibody. Nuclei, DAPI (blue). Scale bar, 100 μm. c, HSV-1 titers in the supernatants of HFF cells transfected with the indicated siRNAs and infected with HSV-1 for 48 h. d, GFP-positive HEK293T cells, which were transfected with the indicated siRNAs together with either empty vector or an RNAi-resistant TRIM43 plasmid (TRIM43RNAi-res) for 36 h and then infected with HSV-1-GFP, determined by FACS at 16 hpi. e, TRIM43 knockdown efficiency for (d), determined by qPCR. f, Genome copies of KSHV in the supernatants of iSLK.219 cells transfected with the indicated siRNAs for 36 h and then treated with doxycyclin (dox) for 48 h, determined by qPCR of orf26. Pfu, plaque forming units. Data represent mean and s.d. of n = 3 (a, c–f), and statistical significance was calculated by unpaired two-tailed t-test. Data are representative of three (a–f) independent experiments.
Pericentrin degradation by TRIM43 induces nuclear lamina alterations
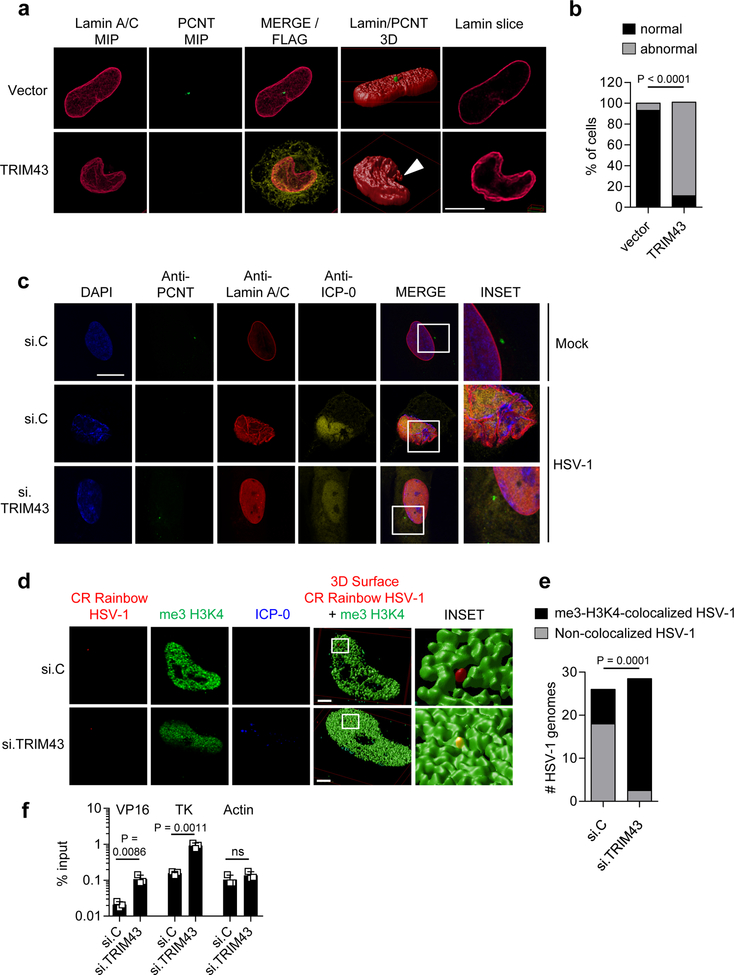
We next sought to determine the molecular mechanism by which TRIM43-mediated PCNT degradation affects the reactivation/lytic replication of herpesviruses, which replicate in the host cell nucleus. It is well established that the centrosome is associated with the nuclear envelope by microtubules and LINC (linker of nucleoskeleton and cytoskeleton) proteins25, 26. As such, centrosomal proteins such as PCNT can influence nuclear positioning and the architecture of the nuclear envelope27, 28. As the nuclear lamina has been shown to influence not only cellular chromatin but also viral chromatin, we postulated that TRIM43, through its regulation of PCNT stability, induces nuclear lamina alterations that may affect the transcriptional activity of viral chromatin and thereby modulate viral reactivation/lytic replication. Ectopic expression of TRIM43, which induces PCNT degradation, led to a range of morphology changes of the nuclear lamina, such as lamina invaginations and ruffling, and even complete breakdown of the nuclear lamina when TRIM43 was highly expressed (Fig. 6a,b and Supplementary Fig. 8b,c). In contrast, TRIM25 expression had no effect on nuclear envelope integrity, whereas TRIM43ΔRING expression induced ‘thickening’ of the nuclear lamina (Supplementary Fig. 8b), suggesting that the lamin alterations induced by TRIM43 are dependent on its E3 ligase activity. Notably, the observed nuclear lamina changes were however not mediated by TRIM43-induced proteasomal degradation of lamin A/C or lamin B, as the overall abundance of lamin proteins remained unchanged upon TRIM43 overexpression (Supplementary Fig. 8d). In concordance with our hypothesis that TRIM43-mediated PCNT degradation induces lamin alterations, depleting endogenous PCNT resulted in very similar lamina changes (e.g., lamina invaginations/ruffling, donut-shaped nuclei, and extranuclear lamin) (Supplementary Fig. 9a–c), which is consistent with published results27, and further strengthens that the effect of TRIM43 on nuclear lamina integrity is dependent on PCNT. Along these lines, HSV-1 infection led to the loss of PCNT, as previously reported29, as well as nuclear lamina alterations (Fig. 6c) similar to those observed in cells that ectopically express TRIM43 (Fig. 6a). In contrast, VSV or Ad infection, which did not trigger TRIM43 upregulation or PCNT degradation (Supplementary Fig. 7b,c), did not induce nuclear lamina changes (Supplementary Fig. 10a). Next, we asked whether the nuclear lamina alterations observed in HSV-1-infected cells are mediated by TRIM43. Strikingly, HSV-1-induced alterations of lamina morphology were reversed by knocking down endogenous TRIM43 (Fig. 6c). Consistent with this, TRIM43 depletion also rescued the degradation of endogenous PCNT in HSV-1-infected cells (Fig. 6c). Together, these data indicate that herpesvirus-induced TRIM43 expression leads to degradation of PCNT and the loss of centrosomal integrity, ultimately inducing alterations of the nuclear lamina.
Fig. 6. TRIM43 regulates nuclear lamina integrity and thereby the association of viral chromatin with transcriptionally-active host chromatin.
a, Nuclear lamina and PCNT abundance in HeLa cells transfected with empty vector or FLAG-tagged WT TRIM43 for 36 h, by IF analysis using anti-Lamin A/C (red) and anti-PCNT (green). FLAG-TRIM43 expression was confirmed by anti-FLAG staining (yellow). Stacks of confocal images were subjected to 3D surface rendering MIP: maximum intensity projection. Arrow: extranuclear lamin. Scale bar, 10 μm. b, Quantification of n = 50 cells showing nuclear lamina changes from (a). c, HFF cells were transfected with si.C or si.TRIM43. 24 h later, cells were mock-treated or infected with HSV-1 (MOI 0.5) for 16 h, followed by immunostaining using anti-PCNT (green) and anti-Lamin A/C (red). Virus infection was confirmed by co-staining with anti-ICP-0 antibody (yellow). Nuclei, DAPI (blue). Scale bar, 30 μm. d, Co-localization of HSV-1 genomes with me3-H3K4 chromatin using CRISPRainbow (CRainbow). HeLa cells expressing the CRainbow components dCAS9, a HSV-1 genome-specific sgRNA and a sgRNA-binding blue-fluorescent protein (shown in red), were transfected with si.C or si.TRIM43. 24 h later, cells were infected with HSV-1 (MOI 1) for 1.5 h, followed by immunostaining with antibodies specific for me3-H3K4 (green) and ICP-0 (blue). Stacks of confocal images were subjected to 3D surface rendering. e, Quantification of the co-localization of HSV-1 genomes with me3-H3K4 for the experiment shown in (d). n = 25 cells. Scale bar, 3 μm. f, Chromatin-IP (ChIP) analysis of HFF cells transfected with the indicated siRNAs for 48 h and then infected with HSV-1 (MOI 1) for 5 h using an anti-me3-H3K4 antibody. ChIP DNA was quantified by qPCR with promoter-specific primers to viral genes VP16 and TK as well as cellular actin (control). Data were normalized to the level of input DNA and presented as percentage of input. Data represent mean and s.d. of n = 3 (f), and statistical significance was calculated using two-tailed chi-square test (b), two-way ANOVA test (e), or unpaired two-tailed t-test (f). Data are representative of three (a, c–e) or two (f) independent experiments.
TRIM43-induced nuclear lamina alterations suppress transcriptionally-active viral chromatin states and thereby inhibit herpesvirus replication
Lamina-associated domains typically serve as anchoring platforms for cellular heterochromatin. In the case of viral chromatin, however, lamin A/C is required for targeting viral genomic DNA to the periphery of the cell nucleus30. As such, in Lmna -/- cells, viral replication compartments are reduced in size, which results in diminished virion production due to increased viral heterochromatin and transcriptional silencing of viral genes30. Therefore, we next asked whether TRIM43-mediated nuclear lamina changes influence viral chromatin and thereby viral lytic replication/reactivation. To test this hypothesis, we adapted the recently developed CRISPRainbow technology31 to visualize the chromatin of HSV-1 genomes in cells that were transfected with either non-targeting control siRNA, or TRIM43-specific siRNA. (Fig. 6d,e). Briefly, we designed a guide RNA (gRNA) with sequence-specificity for HSV-1 repeat DNA that is fused to MS2 RNA-binding sites. Co-transfection of this gRNA along with a construct encoding a catalytically-defective mutant of CRISPR-associated protein 9 (dCas9) and a construct encoding MS2-RNA-binding domains fused to blue fluorescent protein (BFP) allows for recruitment of dCas9 and BFP to HSV-1 DNA and visualization of viral genomic DNA. Importantly, BFP foci co-localized extensively with ICP-8, an HSV-1-encoded single-stranded DNA binding protein that binds to replicating viral genomes, which confirmed that our system allows for specific visualization of viral genomes and not cellular chromatin (Supplementary Fig. 10b). As compared to HSV-1 genomes in cells transfected with non-targeting siRNA, the co-localization of HSV-1 genomes with the euchromatic marker histone H3 tri-methyl lysine 4 (H3K4me3) was markedly enhanced in cells depleted of endogenous TRIM43 (Fig. 6d,e). Cells depleted of TRIM43 also showed a significantly enhanced association of the euchromatic marker H3K4me3 with the promoters of the viral genes VP16 and thymidine kinase (TK) as compared to cells transfected with control siRNA (Fig. 6f). Collectively, these data demonstrate that TRIM43, through degradation of PCNT, induces nuclear lamina alterations that suppress transcriptionally-active viral chromatin states and thereby viral lytic replication/reactivation.
Discussion
It has become evident that members of the TRIM protein family play a critical role in the antiviral host response. Here we reveal a role for TRIM proteins in modulating KSHV reactivation. We found that silencing specific TRIM proteins in cells latently infected with KSHV induced spontaneous virus reactivation, suggesting that these TRIM proteins play a role in suppressing KSHV lytic replication.
Among the TRIM proteins identified by our screen, the previously uncharacterized TRIM43 protein was distinguished by its profound upregulation upon infection with both KSHV and other herpesviruses and its ability to restrict a broad range of herpesviruses belonging to the α-, β-, and γ-herpesvirus subfamilies. Furthermore, TRIM43 was also upregulated in herpesvirus-associated diseases in humans, including γ-herpesvirus-associated tumors. As such, TRIM43 and its transcription factor DUX4 could potentially serve as biomarkers of active herpesviral replication and pathogenesis. Moreover, it is tempting to speculate that differences in steady-state levels of endogenous TRIM43 due to cell type or cell differentiation stage could determine whether the virus undergoes active lytic replication or establishes latency in a particular cell.
In contrast to many TRIM proteins whose expression is type-I IFN-inducible, we found that the upregulation of TRIM43 during herpesvirus infection was not dependent on IFN but was instead mediated by the germline-specific transcription factor DUX4, which is itself transcriptionally upregulated during herpesviral infection via an unknown mechanism. In the absence of infection, we saw minimal to no expression of DUX4 in fully differentiated adult human cells. We further observed that DUX4 expression and herpesvirus infection transcriptionally induced a similar set of genes, some of which encode additional TRIM proteins. Intriguingly, based on genome-wide transcriptional analysis, the DUX4-dependent TRIM proteins that are upregulated during herpesviral infection are minimally or not at all expressed in normal (uninfected) differentiated cells, and their activities have yet to be characterized. Further investigation is needed to determine how herpesvirus infection upregulates DUX4 expression and how other DUX4-induced genes affect herpesviral replication. Moreover, future studies will need to determine the physiological relevance of TRIM43 in other DUX4-regulated cellular processes and diseases, such as regulation of zygote gene activation16 and FSHD19.
Our work has unveiled a mechanism of herpesviral inhibition in which a TRIM E3 ligase controls viral lytic gene expression by regulating the structure of host cell centrosomes and thereby integrity of the nuclear lamina. As such, TRIM43 provides an attractive target for addressing latent herpesvirus reservoirs. Chemical inhibition or RNA-based depletion of TRIM43 may allow for reactivation of human herpesviruses from latent reservoirs, which could be exploited for therapeutic intervention against herpesvirus-associated diseases.
Methods
Cell Culture and Viruses.
Primary human foreskin fibroblasts (HFF), HeLa, A549, Vero, HEK 293T and HEK 293 cells (all purchased from ATCC) as well as Huh7 (provided by M. Farzan, Scripps Institute Florida), were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Thermo Fisher Scientific) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific), 2 mM GlutaMAX (Thermo Fisher Scientific), and 1% (v/v) penicillin-streptomycin (Thermo Fisher Scientific) under standard culture conditions. iSLK and iSLK.219 cells (harboring recombinant (r) KSHV.219) were kindly provided by Don Ganem (University of California) and maintained in DMEM supplemented with 10% (v/v) FBS, 2 μg/mL puromycin (Sigma) and 250 μg/mL G418 (Sigma). AKBM cells32 (provided by Maaike Ressing, Leiden University Medical Center, The Netherlands) and BJAB cells (provided by Jae Jung, University of Southern California) were cultured in Roswell Park Memorial Institute medium (RPMI, Thermo Fisher Scientific) supplemented with 10% (v/v) FBS, 2 mM GlutaMAX, and 1% (v/v) penicillin-streptomycin. AGS-EBV cells33 (kindly provided by Nancy Raab-Traub, University of North Carolina, Chapel Hill) were cultured in Ham’s F12 medium (Thermo Fisher Scientific) supplemented with 10% (v/v) FBS, 2 mM GlutaMAX, 1% (v/v) penicillin-streptomycin, and 500 μg/mL G418 (Sigma).
HEK 293.rKSHV.219 cells were generated by infecting HEK 293 cells with rKSHV.2198, followed by selection with 1 μg/mL puromycin for at least 7 days. Lytic replication of KSHV was induced in iSLK.219 cells by plating cells in the absence of antibiotics and by adding 900 μM sodium butyrate (Sigma) and 1 μg/mL doxycycline (Sigma). Supernatants from induced iSLK cells containing infectious virus were harvested 3 d post-induction, and viral stocks were stored in aliquots at −80 °C until use.
Cell lines from ATCC were authenticated by ATCC and were not validated further in our laboratory. Cell lines that were obtained and validated by other groups were not further authenticated. All cell lines used in this study have been regularly tested for potential mycoplasma contamination by PCR or using the MycoAlert Kit (Lonza).
Herpes simplex virus 1 (HSV-1, strain KOS) was kindly provided by David Knipe (Harvard University). HSV-1-GFP (strain 17) was generously provided by Benedikt Kaufer (Freie Universität Berlin, Germany), and adenovirus 12 (Ad12) was a gift from Walter Doerfler (University of Erlangen-Nuremberg, Germany). Ad-GFP (type 5) was obtained from Vector Biolabs (#1060). For infection experiments with HCMV, the laboratory strain AD169 was used. VSV-GFP was a gift from Sean Whelan (Harvard University) and SeV (Cantell strain) was purchased from Charles River Laboratories. DV serotype 2 (strain 16681), propagated in Aedes albopictus C6/36 cells (provided by Michael Farzan, Scripps Institute Florida), has been previously described34. EMCV (strain EMC) was purchased from ATCC.
TRIM shRNA Screen for KSHV Reactivation.
A custom-made TRIM shRNA library cloned into the pGIPZ lentiviral vector was purchased from Open Biosystems (173795 TRIM ReArray). To individually silence the 62 TRIM proteins, a pool of 3 shRNAs per TRIM protein with different targeting sequences were used. Lentiviral particles were generated by transfecting the respective pGIPZ plasmids encoding TRIM-specific shRNAs together with the lentiviral packaging plasmids psPAX2 and the VSV-G envelope expressing plasmid pMD2.G (both provided by Didier Trono; Addgene plasmids #12260 and #12259, respectively) into HEK 293T cells using Lipofectamine and Plus reagent (Thermo Fisher Scientific). Forty-eight hours later supernatants were harvested, filtered using a 0.2 μm filter to remove cellular debris, and then stored at −80 °C until use. HEK 293.rKSHV.219 cells harboring the rKSHV.219 genome were seeded into 24-well plates (~50,000 cells per well). The KSHV strain rKSHV.219 is a recombinant virus strain that contains a green fluorescent protein (GFP) under control of a CMV-immediate early promoter and a red fluorescent protein (RFP) under control of the viral PAN-promoter that allows monitoring of lytic replication. Cells transduced with pooled supernatants containing three different lentivirally-packaged shRNAs for each TRIM protein were daily monitored for RFP expression using a Nikon TiE fluorescent microscope, and then harvested at 96 h post-transduction by detaching them using 0.1% EDTA (in PBS, pH 7.4). Cells were pelleted by centrifugation at 300 × g for 5 min, and then fixed with 2% (w/v) paraformaldehyde (PFA, in PBS) for 10 min, followed by washing cells twice with PBS. RFP-positive cells were analyzed by FACS analysis using a LSRII flow cytometer equipped with a 546 nm-laser (BD Biosciences), and data were analyzed using FlowJo (Tree Star) software. The screen was performed in 3 biological replicates for each sample.
Reagents, Plasmids and Transfections.
TRIM43A cDNA was purchased from Thermo Fisher (MGC Human TRIM43 Sequence-Verified cDNA, Cat# MHS6278–202755530) and cloned into the pEF-BOS vector (containing an N-terminal FLAG tag) using NotI and SalI restriction sites, or into the pEBG vector (containing N-terminal glutathione S-transferase [GST]) using BamHI and ClaI. pEF-BOS-FLAG-TRIM43 was further used as a PCR template to generate FLAG-TRIM43 ΔRING (amino acids 53–446). Plasmid pCS2-mkg-DUX4, which encodes untagged human DUX4, was a gift from Stephen Tapscott (Addgene plasmid # 2115635). The plasmids encoding TRIM25-V5 and FLAG-TRIM25 have been described previously11, 36. pLKO.1-sh.NT (Sense sequence: CCTAAGGTTA AGTCGCCCTC GCTCGAGCGA GGGCGACTTA ACCTTAGG) and pLKO.1-sh.TRIM43 (Cat# RHS3979–9601504) were purchased from Open Biosystems. FLAG-PCNT was kindly provided by Kunsoo Rhee (Seoul National University, Korea) and has been described previously37, 38. To generate the TRIM43RNAi-res plasmid, the following silent mutations (indicated in lower case) were introduced into the TRIM43 sequence targeted by the four individual siRNAs included in the Dharmacon siRNA SMARTpool. aCAacGctcgTGGGTtAAg (target sequence 1; bp 636–654); GaACgCAccGtCAgACgAA (target sequence 2; bp 281–299); TtTAtcGttTaAAtaGaTT (target sequence 3; bp 836–854); and CaGTgtTaCAcAAaGAgGA (target sequence 4; bp 563–581). The TRIM43RNAi-res-coding sequence was synthesized as an IDT gBlock and then cloned into pEF-Bos between BamHI and NotI with an N-terminal FLAG-tag using the Gibson Assembly method. All constructs were sequenced to verify 100% agreement with the original sequence.
Transfections were performed using the calcium phosphate method, GenJet (SignaGen Laboratories), or Lipofectamine and Plus reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. A goat F(ab’)2 fragment directed against human IgG (MP Biomedicals LLC) was used at a concentration of 0.1 mg/mL for 24 h to induce EBV reactivation in AKBM cells. Recombinant IFNα2 was obtained from PBL Biomedical Laboratories, and MG132 was purchased from Sigma (M8699). Protease inhibitor cocktail was obtained from Sigma (P2714) and used at 1:500.
Virus Replication or Reactivation upon siRNA-mediated TRIM Knockdown.
For determining HSV-1, Ad5-GFP, VSV-GFP and EMCV replication upon transient knockdown of selected TRIM proteins (data shown in Fig. 1h and Supplementary Fig. 2d–i), Huh7 (used for HSV-1 or VSV-GFP) or HEK 293T (used for Ad5-GFP and EMCV) cells were seeded into 12-well plates (~5 × 105 cells per well) and, the next day, transfected with non-targeting control siRNA (si.C) or TRIM-specific siRNA (all purchased from Dharmacon) as described in detail below. At 36 h post-transfection, cells were infected with HSV-1 (MOI 0.1), Ad5-GFP (MOI 0.1), EMCV (MOI 1), or VSV-GFP (MOI 0.01). At 36 h (for HSV-1), 72 h (for Ad5-GFP), 24 h (for VSV-GFP), or 8 h (for EMCV) post-infection, cells were harvested by detaching them using 0.1% EDTA (in PBS, pH 7.4), and then pelleted by centrifugation at 300 × g for 5 min. Cells were fixed with 2% (w/v) PFA (in PBS) for 15 min, followed by washing cells twice with PBS. Ad5-GFP and VSV-GFP replication was assessed by determining GFP-positive cells using FACS analysis (LSRII flow cytometer, BD Biosciences).
To determine HSV-1 replication in TRIM-depleted cells, cells were incubated with blocking/permeabilization buffer (5% [v/v] FBS and 0.1% [w/v] Saponin in PBS), followed by staining with primary (anti-ICP-8; 1:1,000) and secondary antibody (anti-rabbit-Alexa 488 (1:400) on ice in blocking/permeabilization buffer for 30 min each. Percentage of ICP-8-positive cells was determined on a LSRII flow cytometer (BD Biosciences), and data were analyzed using FlowJo (Tree Star) and FCS Express 3 softwares (De Novo Software).
EMCV replication was determined by TCID50 (median tissue culture infective dose) assay. Briefly, supernatants from TRIM-depleted cells that had been infected with EMCV were serially diluted and then added onto Vero cells. Twenty-four hours later, wells with dead cells were counted to determine the infectious titer according to the Reed-Muench method39.
HCMV (strain AD169) replication was determined via IE-1 fluorescence as described in detail previously40. Briefly, HFF cells transiently transfected with control non-targeting siRNA or TRIM43-specific siRNA were infected with HCMV at the indicated MOI. Twenty-four hours later, cells were fixed and stained with a monoclonal antibody (p63–27) directed against IE-141. The number of IE-1-positive cells was determined and used to calculate viral titers, expressed as IE protein-forming units (IEU).
EBV reactivation in latently-infected AGS-EBV cells transfected with non-targeting control siRNA or TRIM43-specific siRNA, was determined by quantitative real-time PCR analysis of viral BMRF1 gene expression using forward primer 5’-CAACACCGCACTGGAGAG-3’, reverse primer 5’-GCCTGCTTCACTTTCTTGG-3’, and probe 5’-AGGAAAAGGACATCGTCGGAGGC-3’ on a 7500 Fast Real-Time PCR System (Thermo Fisher Scientific), or by immunoblot (IB) analysis of EBV Zta protein using an anti-Zta antibody (1:500, BZ1, Santa Cruz).
HSV-1 Plaque Assays.
Viral stocks were serially diluted in PBS containing 0.5 mM MgCl2, 0.9 mM CaCl2, 0.1% dextrose, and 5% (v/v) heat-inactivated FBS. Vero cells, seeded into 6- or 12-well plates, were incubated with virus dilutions for 1 h at 37°C. The inoculum was then replaced with MEM containing 5% (v/v) heat-inactivated FBS and 1.5% (w/v) carboxymethylcellulose and incubated at 37°C for 4 d. Cells were fixed using methanol for 5 min and then stained for 30 min with crystal violet stain (Sigma) diluted in 20% (v/v) ethanol. Cells were rinsed with water, dried, and plaques counted.
SiRNA-mediated Knockdown of TRIM proteins, PCNT and DUX4.
HEK 293T cells were seeded into 24- or 12-well plates (~ 2.5 or 5 × 105 cells per well). The next day, cells were transfected with 60 nM or 120 nM of gene-specific siRNAs using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s instructions. SiRNAs targeting TRIM43 (siGENOME SMARTpool M-007127-01-0050), PCNT (siGENOME SMARTpool M-012172-01-0005), DUX4 (SMARTpool Lincode R-188080-00-0005), TRIM4 (siGENOME SMARTpool M-007101-00-0005), TRIM23 (siGENOME SMARTpool M-006523-00-0005), TRIM25 (siGENOME SMARTpool M-006585-00-0005), TRIM42 (siGENOME SMARTpool M-007173-00-0005), TRIM45 (siGENOME SMARTpool M-007073-01-0005), TRIM46 (siGENOME SMARTpool M-007071-01-0005), TRIM47 (siGENOME SMARTpool M-007106-02-0005), TRIM65 (siGENOME SMARTpool M-018490-01-0005) and TRIM66 (siGENOME SMARTpool M-026772-01-0005) as well as the non-targeting control siRNA (siGenome Control Pool D-001206-14-50) were purchased from Dharmacon. At 48 h post-transfection, knockdown efficiency of endogenous TRIM proteins, DUX4, or PCNT was determined by qRT-PCR or immunoblot (IB) analysis, as described below.
Analysis of TRIM43 mRNA Expression in Human Tissues.
The Human Total RNA Master Panel II (Clontech Cat# 636643) was used to determine the absolute number of TRIM43 transcripts in different human tissues. qPCR was performed in one step using 20 ng of the individual cDNA library samples and the SuperScript III Platinum Kit (Thermo Fisher Scientific) on a 7500 Fast Real-Time PCR Machine (Thermo Fisher Scientific) according to the manufacturer’s protocols. A TaqMan probe for the TRIM43 gene was acquired as premixed Taqman MGB assay (Thermo Fisher Scientific, #4351370) and added to the reaction. A standard of known concentration was serially diluted, which served to determine the absolute number of TRIM43 transcripts in the individual tissues.
Electron Microscopy.
HFF cells were grown in 6-well plates on glass cover glasses (Marienfeld Superior). Twenty-four hours later, cells were fixed with 4% (w/v) PFA (in PBS) for 5 min and then permeabilized with 0.1% (v/v) Triton X-100 for 3 min. Cells were blocked with 5% (v/v) bovine serum (in PBS) for 1 h and then stained with anti-TRIM43 antibody (Abcam 80460; diluted 1:50 in 5% (v/v) bovine serum [in PBS]) over night at 4 °C. Cells were extensively washed with PBS and then stained with 5 nm gold particles coupled to protein A for 1 h at room temperature (RT). Cells were washed and then fixed with 1.25 % glutaraldehyde and 0.015% picric acid in 50 mM sodium cacodylate buffer (pH 7.4) at RT for 30 min. After washing with sodium cacodylate buffer (50 mM, pH 7.4), cell layers were embedded in Epon for 24 – 48 h at 40 °C. Perfused cells were cut ultrathin (Reichert-Ultracut-S) and then transferred into a formvar/carbon-coated copper grid for imaging. All images were acquired using the Tecnai G2 Spirit BioTWIN (FEI) at the Electron Microscopy Facility of Harvard, Boston.
Pull-down Assay, Co-immunoprecipitation, and Immunoblot Analysis.
HEK 293T cells were lysed in RIPA buffer (150 mM NaCl, 0.5 – 1% [v/v] NP-40, 1% [w/v] deoxycholic acid (DOC), 0.01% [v/v] SDS, 20 mM Tris pH 8.0) or NP-40 buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 1% [v/v] NP-40, protease inhibitor cocktail [Sigma], and 10 μg/mL proteasome inhibitor MG132), and cell debris pelleted by centrifugation at 13,000 rpm for 20 min at 4 °C. FLAG pull-downs, Co-IP, and western blot analyses were performed as previously described11. Elution of immunoprecipitated proteins was performed by heating samples in Laemmli SDS sample buffer at 95 °C for 5 min. For immunoblotting, the following primary antibodies were used: anti-FLAG (M2; 1:2,000; Sigma), anti-HA (1:2,000; clone HA-7; Sigma), anti-ICP-8 (1:5,000, kindly provided by David Knipe, Harvard), anti-Zta (1:500, BZ1, Santa Cruz), anti-V5 (1:5,000, R960–25, Novex), anti-GST (1:5,000, Sigma), anti-PCNT (1:1,000, clone 28144, Abcam), anti-Lamin A/C Antibody (clone 636, Santa Cruz Biotech), anti-γ-Tubulin (1:1,000, clone GTU-88, Abcam), anti-DUX4 (1:1,000, clone 9A12, Millipore), anti-SMC1a (1:500, rabbit polyclonal, Bethyl antibodies) anti-SMC3 (1:500, ab9263, Abcam), anti-RAD21 (1:500, 05–908, Millipore), anti-α-Tubulin (1:1,000, rabbit polyclonal, Genscript), anti-Ubiquitin (1:500, clone P4D1, Santa Cruz Biotech) and anti-β-Actin (1:10,000; Abcam or AC-15, Sigma).
To determine endogenous PCNT ubiquitination upon HSV-1 infection of cells depleted of TRIM43, HEK 293T cells were transduced with lentivirally-packaged non-targeting control shRNA or TRIM43-specific shRNA and then selected using puromycin (1 μg/mL) at 72 h post-transduction. Cells were transiently transfected with FLAG-tagged ubiquitin, and 24 h later, either infected with HSV-1 (MOI 1) for 16 h, or left uninfected. Cells were further treated with MG132 (50 μM) for 4 h to limit the degradation of PCNT triggered by HSV-1 infection. Cells were lysed in NP40 buffer (300 mM NaCl, 10 mM Tris HCl pH 8.0, 1% [v/v] NP40, and protease inhibitor cocktail [Sigma]), and cell debris pelleted by centrifugation at 10,000 rpm for 10 min at 4 °C. Lysates were incubated with anti-PCNT-coupled G magnetic beads to immunoprecipitate PCNT.
Large-scale Protein Purification and Mass Spectrometry Analysis.
To identify cellular interaction partners of TRIM43, twelve 10cm-dishes of HEK 239T cells (~ 1 × 107 cells per dish) were each transfected with 15 μg of pEF-BOS empty vector, pEF-BOS-FLAG-TRIM43 ΔRING, or pEF-BOS-FLAG-TRIM25 (control). Thirty-six hours later, cells were lysed with Triton X-100 buffer (150 mM NaCl, 5 % EDTA, 1% [v/v] Triton X-100, pH 7.5, protease inhibitor cocktail [Sigma]), followed by centrifugation at 13,000 rpm for 20 min at 4 °C. Lysates were pre-cleared using 100 μl Sepharose® 4B (Sigma-Aldrich) to remove cell debris and then mixed with a ~50% slurry of anti-FLAG M2 Affinity Gel (Sigma) and incubated for 4 h at 4 °C. After extensive washing of the beads with lysis buffer, bound proteins were eluted by heating samples in 2× Laemmli SDS sample buffer for 5 min at 95 °C and subsequently separated on a NuPAGE 4 – 12% Bis-Tris gradient gel (Thermo Fisher Scientific). To stain co-immunoprecipitated proteins, a Silverquest™ staining kit (Thermo Fisher Scientific) was used according to the manufacturer’s instruction. Bands that were specifically present in the FLAG-TRIM43 ΔRING sample, but not in the empty vector and FLAG-TRIM25 samples, were excised and analyzed by ion-trap mass spectrometry at the Harvard Taplin Biological Mass Spectrometry Facility, Boston.
Confocal Microscopy.
HeLa cells or HFF cells (~ 2.5 × 105 cells per sample) were seeded onto cover glasses (Marienfeld Superior). The next day, cells were transfected with plasmid DNA (usually 1 μg DNA per sample) using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. At 24 – 36 h post-transfection, cells were fixed with 2% (w/v) PFA (in PBS) for 10 min, permeabilized with 0.1% (v/v) Triton X-100, and then blocked with 5% (v/v) bovine serum (in PBS) for 1 h at RT. Cell preparation and confocal microscopy analysis were performed as previously described11. For immunostaining, cells were incubated with the following antibodies usually 1 h at room temperature: anti-TRIM43 (1:400; Abcam 80460), anti-DUX4 (1:4,000; clone 9A12, Millipore), anti-ICP-8 (1:1,000; kindly provided by David Knipe, Harvard), anti-PCNT (1:400; clone 28144, Abcam), anti-γ-Tubulin (1:400; clone GTU-88, Abcam), anti-FLAG (1:1,000; M2, Sigma), anti-Lamin A/C Antibody (clone 636, Santa Cruz Biotech), anti-Lamin B1 (clone B10, Santa Cruz Biotech), anti-centrobin (1:500; Sigma), anti-SAS-6 (1:400; clone G1, Santa Cruz). This was followed by incubation for 1 h at room temperature with the secondary antibodies donkey anti-rabbit Alexa Fluor 594, donkey anti-mouse Alexa Fluor 488, donkey anti-mouse Alexa Fluor 555, donkey anti-rabbit Alexa Fluor 555, donkey anti-rabbit Alexa Fluor 488, donkey anti-mouse Alexa Fluor 647, or donkey anti-rabbit Alexa Fluor 647 (all purchased from Thermo Fisher Scientific). Nuclei were stained with DAPI (Sigma). The samples were imaged using an Olympus IX8I confocal microscope or a Leica TCS SP5 confocal microscope, with 405 nm, 488 nm, 543 nm or 633 nm laser lines, scanning each channel separately under image capture conditions that eliminated channel overlap. The Fiji distribution of ImageJ was used for image analysis and quantification. For 3D reconstruction of images, stacks of confocal images from single cells imaged using a Leica TCS SP5 microscope were deconvoluted and subjected to 3D surface rendering with Huygens Professional Software (Scientific Volume Imaging).
Quantitative Real-Time PCR (qRT-PCR).
Total RNA was extracted from cells using the HP Total RNA Kit (OMEGA bio-tek) according to the manufacturer’s instructions. Reverse transcription and qRT-PCR was performed in one step using 0.1–1 μg of RNA as template using the SuperScript III Platinum One-Step qRT-PCR kit with ROX (Thermo Fisher Scientific) on a 7500 or 7500 Fast Real-Time PCR Machine (Thermo Fisher Scientific) according to the manufacturer’s protocols. Primers/probes for GAPDH, HPRT, 18S rRNA, DUX4, TRIM43, TRIM4, TRIM8, TRIM15, TRIM23, TRIM33, TRIM34, TRIM41, TRIM42, TRIM43, TRIM45, TRIM46, TRIM47, TRIM54, TRIM65, TRIM66, TRIM69 were acquired as premixed master mixes (IDT or Thermo Fisher Scientific/ABI) and added to the reaction. For detection of HSV-1 ICP-0, forward primer 5’-CGGACACGGAACTGTTCGA-3’, reverse primer 5’-CGCCCCCGCAACTGC-3’, and probe 5’-/56-FAM/CCCCATCCA/ZEN/CGCCCTG/3IABKFQ/−3’ (all purchased from IDT) were used. Expression levels for each target gene were calculated by normalizing values to cellular GAPDH, HPRT or 18S rRNA. Fold induction of each target gene relative to mock-infected or mock-treated cells was calculated using the comparative CT Method (ΔΔCT Method). Quantification of KSHV viral genome copies with primers specific for KSHV orf26 (orf26-fwd primer: 5’-GCTAGCAGTGCTACCCCCATT-3; orf26-rev primer 5’-GGTCAAATCCGTTGGATTCG-3 and orf26 probe: FAM / AGCCGAAAGGATTCCACCATTGTGC / TAMRA, all purchased from IDT) was performed as described previously42.
TRIM43 Expression Analysis in BAL Samples.
Anonymized broncheoalveolar lavage (BAL) samples for analysis of TRIM43 transcript expression were selected from the sample repository of the diagnostics department of the Institute for Clinical and Molecular Virology at the University Hospital Erlangen (Erlangen, Germany). Exclusively samples that were no longer needed for diagnostic purposes and assigned for disposal were used. Patient samples that were tested positive for multiple viruses were excluded from the analysis, as well as samples with less than 120,000 albumin transcript copies in the initial nucleic acid extraction to allow for efficient RNA extraction. In total 34 BAL samples from patients with pulmonary infections were analyzed, from which 18 BAL samples were positive for HSV-1 DNA by qPCR, and 16 BAL samples were negative for HSV-1 DNA by qPCR. Total nucleic acids from 400 μL of BAL sample were extracted on an automated Qiagen EZ1 analyzer using the Qiagen EZ1 virus mini kit v2.0 according to the manufacturer’s instructions, and phage M2-RNA added as an extraction control. Extracted nucleic acids were eluted in 60 μL ultrapure water. 6 μL of 10 × PCR buffer (New England Biolabs) was added to the 60 μL of eluted nucleic acids, and DNA was removed by incubation with 1 U of RNAse-free DNAse (Epicentre) for 15 min at 37 °C. DNAse was inactivated at 70 °C for 15 min. qRT-PCR was performed using 8 μL of each sample in a 20 μL reaction with a 2× universal probe One-Step RT-qPCR Kit (New England Biolabs) and primer/probe mixes for HPRT (IDT, Cat# Hs.PT.58v.45621572) and TRIM43 (Thermo Fisher Scientific, # 4351370) spanning two exons to exclude amplification of genomic DNA on a ABI 7500 real time PCR system. Relative induction of TRIM43 transcripts was calculated by normalizing values to HPRT values using 1/2^(CtTRIM43-CtHPRT). In samples negative for TRIM43 mRNA, the Ct-values were set to 45 (45 PCR cycles). Statistical significance was calculated using a Mann-Whitney U test (*p < 0.05).
Next Generation RNA-Sequencing.
HEK 293T cells, seeded into 12-well plates (~500,000 cells per well), were mock-treated, transfected with 2 μg DNA of pCS2-mkg-DUX4 or infected with HSV-1 (MOI 10) for 18 h. Cells were harvested in ice-cold PBS, and total cellular RNA was extracted using standard phenol/chloroform extraction. Purified RNA was converted to a DNA library and sequenced at the Harvard Biopolymers Facility (Boston, MA). Up to 1,000 ng total RNA was used in the Wafergen (formerly IntegenX) PrepX RNA-Seq Library Kit (Cat# 400039) and placed into the Wafergen Apollo, and run using a standard protocol. PCR was performed on samples using indexed primers according to Wafergen instructions and then cleaned using the Wafergen Apollo. The resulting samples were run on an Agilent 2200 Tape Station on a D1000 High Sensitivity Tape with ladder provided to assess the integrity and amount of DNA. DNA libraries were run in a qPCR assay with SYBR green KAPA SYBR FAST Universal 2X qPCR Master Mix (Kapa Biosystems, Cat# KK4602) and primers to the P5 and P7 regions of the adapters. Diluted PhiX was used for the standard curve to determine concentration. Libraries passing the quality control were subjected to Illumina HiSeq 2500 sequencing. A local Galaxy server was used to process the RNAseq data43. Raw sequence reads were quality trimmed using TRIM Galore! (Cutadapt44 and FastQC45 wrapper) and then aligned to the human genome using Tophat246 using standard settings. Htseq-count was used to calculate the induction of human transcripts, and values were normalized using total count normalization47. Of note, the transcripts of the three TRIM43 gene copies found in humans (TRIM43A, TRIM43B and TRIM43C), were analyzed. Data were visualized in R48 using the ggplot2 package. The RNAseq data have been deposited in NCBI’s GEO database (#GSE101435).
Immunohistochemical Analysis of TRIM43 Protein in KS Tissues.
Kaposi’s sarcoma (KS) tissues were obtained from skin biopsies of 17 HIV-1-infected individuals with AIDS in the context of medically required diagnostic or therapeutic procedures. Uninvolved tissues of the skin of five of the patients as well as cerebellum tissue of an HIV-1-negative person were obtained as controls. The use of these post-diagnostic tissues was approved by the ethics committee of the University Hospital Erlangen, Germany.
Staining of PFA-fixed, paraffin-embedded tissue sections for TRIM43 and KSHV LANA-1 were performed as previously described49. A rabbit polyclonal antibody directed against human TRIM43 (ABCAM, ab80460, 1:50) and a rat monoclonal antibody against LANA-1 (Tebu-bio, Cat# 13-210-100, 1:250) were used as primary antibodies. Primary antibody binding was detected using the respective Vectastain Elite ABC Kits (Vector Laboratories). For both antibodies, the citrate buffer pH 6.0 (DakoCytomation) was used for the antigen retrieval. The sections were developed using NovaRed Substrate (Vector Laboratories), counterstained with Hematoxylin Gill III (Merck, Darmstadt, Germany), and analyzed using a DM6000 B microscope (Leica).
CRISPRainbow Labeling of HSV-1 Genomes.
A small guide RNA (sgRNA) directed against the repeats of HSV-1 was designed (5’-GTTACCTGGGACTGTGCGGT-3’) and cloned into pLH-sgRNA1–2XMS2, which was a kind gift from Thoru Pederson (Addgene #7538931). Plasmids pHAGE-TO-MCP-3XBFPnls (Addgene# 7538331), pHAGE-TO-dCas9 (Addgene# 7538131, and pLH-sgHSV1–2XMS2 were co-transfected into HeLa cells. 24 h post-transfection, cells were re-seeded onto cover slips (~ 50,000 cells per sample) and then transfected with either control non-targeting siRNA and TRIM43-specific siRNA (both Dharmacon). 24 h later, cells were infected with HSV-1 (MOI 1) for 1.5 h. Cells were incubated with primary antibodies specific for me3-H3K4 (rabbit polyclonal, Abcam, 1:500) and viral ICP-0 (mouse monoclonal, clone 11060, Santa Cruz, 1:400) for 1 h at room temperature, followed by incubation for 1 h at room temperature with the secondary antibodies donkey anti-rabbit Alexa Fluor 488 and donkey anti-mouse Alexa Fluor 647 (both purchased from Thermo Fisher Scientific). Stacks of confocal images from single cells imaged using a Leica TCS SP5 microscope were deconvoluted and subjected to 3D surface rendering with Huygens Professional Software (Scientific Volume Imaging) to visualize co-localization of HSV-1 genomes with me3-H3K4.
ChIP Assay.
HFF cells, seeded into 10 cm-dishes (~ 1 × 106 cells per dish), were transfected with the indicated siRNAs at a concentration of 120 nM using Lipofectamine 2000 (Thermo Scientific) according to the manufacturer’s instructions. Thirty-six hours later, cells were infected with HSV-1 (KOS strain; MOI of 1) for 5 h. Cells were fixed in 1% formaldehyde for 10 min and fixation quenched by addition of glycine (final concentration of 0.125 M). Cell pellets were washed twice with ice-cold PBS containing protease inhibitor cocktail (Cell Signaling Technologies), resuspended in 300 μL of sonication buffer (10 mM Tris [pH 8.0], 0.25% (w/v) SDS, 2 mM EDTA), and then sonicated using a Bioruptor (Diagenode). Samples were diluted 1:1.5 in equilibration buffer (10 mM Tris, 233 mM NaCl, 1.66 % (w/v) Triton X-100, 0.166% (w/v) Sodium Deoxycholate, 1 mM EDTA, protease inhibitor cocktail), centrifuged at 14,000 rpm for 10 min, and then supernatants transferred into new tubes. Chromatin was immunoprecipitated by incubation with 5 μL of anti-trimethyl-histone3-lysine4 antibody (rabbit polyclonal, ab8580, Abcam) overnight at 4 °C, followed by incubation with 25 μL Protein ADynabeads (Thermo Scientific) for 2.5 h. Beads were washed twice each with RIPA low salt buffer (containing 140 mM NaCl), RIPA high salt buffer (containing 500 mM NaCl), and RIPA LiCl buffer (containing 250 mM LiCl). Beads were then washed once in 10 mM Tris (pH 8.0), and chromatin eluted in 50 μL elution buffer (10 mM Tris [pH 8.0], 0.4% (w/v) SDS, 5 mM EDTA, 300 mM NaCl, and 2 μl Proteinase K) by incubation for 1 h at 65 °C and then 6 h at 55°C. Eluted DNA was purified using Agencourt AMPureXP Beads (Beckmann Coulter) in a 1.8 × volume according to the manufacturer’s instructions. DNA was subjected to SYBR green qRT-PCR analysis using the following forward (fwd) and reverse (rev) primers: VP16-fwd: 5’-CGGATTGGGAAACAAAGGCACGCAACGCC-3’; VP16-rev: 5’-TTGAGGTCTTCGTCTGTG-3’; TK-fwd: 5’-ATGCTGCCCATAAGGTATCG-3’; TK-rev: 5’-GTAATGACAAGCGCCCAGAT-3’; ICP8-fwd: 5’-CCACGCCCACCGGCTGATGAC-3’; ICP8-rev: 5’-TGCTTACGGTCAGGTGCTCCG-3’; ICP27-fwd: 5’-CCACGGGTATAAGGACATCCA-3’; ICP27-rev: 5’-GGATATGGCCTCTGGTGGTG-3’; Actin-fwd: 5’-CGGGAAATCGTGCGTGACATTAAG-3’; Actin-rev: 5’-GAACCGCTCATTGCCAATGGTGAT-3’. Quantitative PCR was performed using the Luna SYBR 2× Master Mix (New England Biolabs) on an ABI 7500 machine (Thermo Fisher Scientific). Values were calculated using the ΔCt method and normalized to input DNA.
Statistical Analysis.
The P values for the TRIM shRNA screen were calculated using an unpaired two-tailed Student’s t-test. Three times each 10,000 cells per samples from biological replicates were quantified. A P value of <0.05 was considered statistically significant.
For analysis of TRIM43 expression in patient samples, statistical significance was calculated using a Mann-Whitney U test. A P value of <0.05 was considered statistically significant.
For the data shown in Figure 6b, statistical significance was calculated using a two-tailed chi-square test. A P value of <0.05 was considered statistically significant.
For all other experiments, an unpaired two-tailed Student’s t-test was used to compare differences between two unpaired experimental groups in all cases. A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
We thank Maria Ericsson from the Harvard Electron Microscopy Facility, Boston, for assistance with sample preparation and electron microscopy, and Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard) for mass spectrometry analysis. We are grateful to Gabriele Förtsch (Division of Molecular and Experimental Surgery, University Hospital Erlangen, Germany) for excellent technical assistance and Dr. Roland Coras (Department of Neuropathology, University Hospital Erlangen, Germany) for cerebellum tissue used as a staining control. This study was supported by the US National Institutes of Health grants R21 AI118509, R01 AI087846 and R01 AI127774 (to M.U.G.), and grants from the German Research Foundation CRC 796, TP B1 and EN423/5–1 (to A.E.), CRC796 and STA357/7–1 (to T.S.), FOR 2438/subproject 2 (to M.S), FU 949/1–1 and FU 949/2–1 (to F.F.), and SP 1600/1–1 (to K.M.J.S.). F.F. was further supported by a Marie Skłodowska-Curie Individual Fellowship from the European Union’s Framework Program for Research and Innovation Horizon 2020 (2014–2020) under the Grant Agreement No. 703896, and the Interdisciplinary Center for Clinical Research Erlangen (IZKF, J57). M.A.Z. received support by NIH training grant T32 GM007183. A.E. also received funding by IZKF, A66.
Footnotes
Data Availability.
The data that support the findings of this study are available from the corresponding author upon request. The RNAseq data from this study are deposited in NCBI GEO under accession code GSE101435. Supplementary figures and tables are available in the Supplementary Information file. Complete western blot images of all figures in the manuscript are provided in Supplementary Figure 11.
Competing financial interests
The authors declare no competing financial interest.
References
- 1.Corey L & Wald A Maternal and neonatal herpes simplex virus infections. N Engl J Med 361, 1376–1385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore PS & Chang Y Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer 10, 878–889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dittmer DP & Damania B Kaposi sarcoma-associated herpesvirus: immunobiology, oncogenesis, and therapy. The Journal of clinical investigation 126, 3165–3175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozato K, Shin DM, Chang TH & Morse HC 3rd TRIM family proteins and their emerging roles in innate immunity. Nature reviews. Immunology 8, 849–860 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajsbaum R, Garcia-Sastre A & Versteeg GA TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol 426, 1265–1284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernardi R & Pandolfi PP Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nature reviews. Molecular cell biology 8, 1006–1016 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Scherer M & Stamminger T Emerging Role of PML Nuclear Bodies in Innate Immune Signaling. J Virol 90, 5850–5854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vieira J & O’Hearn PM Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 325, 225–240 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Duggal NK & Emerman M Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12, 687–695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Tipper C & Sodroski J Role of TRIM5alpha RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J Virol 85, 8116–8132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gack MU et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446, 916–920 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Carthagena L et al. Human TRIM gene expression in response to interferons. PLoS One 4, e4894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desmyter J, Melnick JL & Rawls WE Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 2, 955–961 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanghellini I, Falco G, Lee SL, Monti M & Ko MS Trim43a, Trim43b, and Trim43c: Novel mouse genes expressed specifically in mouse preimplantation embryos. Gene Expr Patterns 9, 595–602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickson PG et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49, 925–934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Iaco A et al. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49, 941–945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiddon JL, Langford AT, Wong CJ, Zhong JW & Tapscott SJ Conservation and innovation in the DUX4-family gene network. Nat Genet 49, 935–940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng LN et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22, 38–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemmers RJ et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreboeuf M et al. DUX4 and DUX4 downstream target genes are expressed in fetal FSHD muscles. Hum Mol Genet 23, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Daxinger L, Tapscott SJ & van der Maarel SM Genetic and epigenetic contributors to FSHD. Curr Opin Genet Dev 33, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doxsey SJ, Stein P, Evans L, Calarco PD & Kirschner M Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76, 639–650 (1994). [DOI] [PubMed] [Google Scholar]
- 23.Dictenberg JB et al. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol 141, 163–174 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schockel L, Mockel M, Mayer B, Boos D & Stemmann O Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol 13, 966–972 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Starr DA A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci 122, 577–586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK & Wilson KL The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6, 21–31 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Verstraeten VL et al. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc Natl Acad Sci U S A 108, 4997–5002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gundersen GG & Worman HJ Nuclear positioning. Cell 152, 1376–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasdeloup D, Labetoulle M & Rixon FJ Differing effects of herpes simplex virus 1 and pseudorabies virus infections on centrosomal function. J Virol 87, 7102–7112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva L, Cliffe A, Chang L & Knipe DM Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog 4, 1000071 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma H et al. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat Biotechnol 34, 528–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ressing ME et al. Impaired Transporter Associated with Antigen Processing-Dependent Peptide Transport during Productive EBV Infection. The Journal of Immunology 174, 6829–6838 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Marquitz AR, Mathur A, Shair KHY & Raab-Traub N Infection of Epstein–Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proceedings of the National Academy of Sciences 109, 9593–9598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan YK & Gack MU A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat Immunol 17, 523–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snider L et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 18, 2414–2430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pauli EK et al. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal 7, 2004577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Lee K & Rhee K PLK1 regulation of PCNT cleavage ensures fidelity of centriole separation during mitotic exit. Nat Commun 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S & Rhee K Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One 9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed LJ & Muench H A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. American journal of epidemiology 27, 493–497 (1938). [Google Scholar]
- 40.Lorz K et al. Deletion of open reading frame UL26 from the human cytomegalovirus genome results in reduced viral growth, which involves impaired stability of viral particles. Journal of virology 80, 5423–5434 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreoni M, Faircloth M, Vugler L & Britt WJ A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. Journal of virological methods 23, 157–167 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Full F et al. Kaposi’s sarcoma associated herpesvirus tegument protein ORF75 is essential for viral lytic replication and plays a critical role in the antagonization of ND10-instituted intrinsic immunity. PLoS pathogens 10, e1003863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afgan E et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44, W3–W10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17 (2011). [Google Scholar]
- 45.Andrews S FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2017).
- 46.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dillies MA et al. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Briefings in bioinformatics 14, 671–683 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Team, R.C. (R Foundation for Statistical Computing, Vienna, Austria; 2013). [Google Scholar]
- 49.Naschberger E et al. Matricellular protein SPARCL1 regulates tumor microenvironment-dependent endothelial cell heterogeneity in colorectal carcinoma. The Journal of clinical investigation 126, 4187–4204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.