Abstract
Enantiomeric molecular evaluations remain an enormous challenge for current analytical techniques. To date, derivatization strategies and long separation times are generally required in these studies, and the development and implementation of new approaches are needed to increase speed and distinguish currently unresolvable compounds. Herein, we describe a method using chiral cyclodextrin adducts and structures for lossless ion manipulations (SLIM) and serpentine ultralong path with extended routing (SUPER) ion mobility (IM) to achieve rapid, high resolution separations of d and l enantiomeric amino acids. In the analyses, a chiral cyclodextrin is added to each sample. Two cyclodextrins were found to complex each amino acid molecule (i.e. potentially sandwiching the amino acid in their cavities) and forming host-guest noncovalent complexes that were distinct for each d and l amino acid pair studied and thus separable with IM in SLIM devices. The SLIM was also used to accumulate much larger ion populations than previously feasible for evaluation and therefore allow enantiomeric measurements of higher sensitivity, with gains in resolution from our ultralong path separation capabilities, than previously reported by any other IM-based approach.
Keywords: chiral separations, cyclodextrins, enantiomers, ion mobility-mass spectrometry, structures for lossless ion manipulations
Introduction
One of the most challenging tasks in the field of bioanalytical separations is separating and evaluating enantiomers (non-superimposable stereoisomers) [1–4]. Enantiomers have identical physical and chemical properties, but the mirror-image isomers may exhibit dramatically different, and potentially undesirable, biological or physiological properties. To date, performing enantiomeric separations involves derivatization or use of a chiral environment [1–5]. Chemical derivatizations are used to transform enantiomers into diastereomers or molecules having different physical and chemical properties, enabling their separation [3, 5–6]. However, this approach can be quite difficult when dealing with small sample sizes or when accurate quantitation is desired. The introduction of a chiral environment provides a much simpler approach, but unfortunately the interactions between the introduced chiral molecules and enantiomeric analytes are not always sufficient to achieve the separation needed for characterization. In order to facilitate enantiomer-related applications, such as in the food, pharmaceutical, and agricultural industries, new assays, and especially ones that provide high resolution and high sensitivity, are desired [1–4].
When dealing with the separation of underivatized d/l enantiomeric pairs with chiral liquid chromatography (LC) chiral stationary phases are often used [2–3, 7–8]. Of the wide array of commercially available chiral LC stationary phases, cyclodextrin-based phases continue to be popular for targeted applications [2–3, 7–12]. Cyclodextrins are cyclic oligosaccharides comprised of D-glucose and thus are inherently chiral. Cyclodextrins have also shown some utility as chiral mobile phase additives in both LC and capillary electrophoresis, but have not been as effective as when used in the LC stationary phase [3, 5, 13–18].
While chiral LC analysis approaches presently dominate for enantioseparations, they are often limited by their speed, as well as the lack of sufficient resolution and sensitivity. Rapid ion mobility-mass spectrometry (IM-MS) analyses provide an attractive alternative to LC-MS methods [4, 19]. In IM-MS, analyte ions are separated by their mobilities as well as mass-to-charge in milliseconds to seconds [4, 19]. While enantiomers can be separated based on their interactions with a chiral stationary phase, they pose a challenge for IM separations since they have identical mobilities [20]. Chiral ion interactions must be induced for gas-phase IM enantiomeric separations to potentially resolve d and l enantiomers. To date, most IM enantioseparations rely on the formation of chiral non-covalent complexes, i.e. where the d or l analyte complexes with other chiral molecules [21–28]. These approaches have utilized transition metal cations in conjunction with chiral amino or nucleic acids to form unique interactions with the enantiomeric molecule of interest and potentially providing sufficient differences in the mobilities of the resulting non-covalent complexes to allow their resolution [21–28]. Chiral drift gas modifiers (e.g. S-(+)-2-butanol) have also been previously used for enantiomer IM separations at atmospheric pressure, but have also been difficult to implement [29–30]. Additionally, these approaches have not yet been replicated at lower pressures (which dominates in commercially available IM implementations with MS due to their greater sensitivity), potentially because of differences in ion-molecule clustering and related interactions at the different pressures.
While previous chiral IM separations have been reported, instrumental challenges in both resolution and sensitivity have limited their utility [31–32]. Recently, structures for lossless ion manipulations (SLIM) IM-MS have shown potential for overcoming the drawbacks present of conventional IM approaches [31–35]. Utilizing traveling wave (TW) IM separations over long serpentine path lengths (SUPER; serpentine ultralong path with extended routing) can overcome the limitations of drift tube IM resolution arising from the heightened voltage needed as the length of the drift tube increases [31–35]. SLIM SUPER IM-MS [33] also enables achieving targeted ultrahigh resolution analyses using a previously described “ion switch” to route the ions through the serpentine IM separation region multiple times prior to MS analysis [32–33]. This capability permits arbitrarily long path length separations to be performed, further increasing the IM resolution achievable [32–33]. However, even with theoretically unlimited resolution IM still cannot separate molecules with the same mobility or cross section, such as d/l amino acid enantiomer pairs (Figure 1). In this regard, numerous studies have probed and modeled the formation of host-guest inclusion complexes for enantiomeric separations [36–44], and, as already noted, the use of cyclodextrins as both chiral stationary phase supports and mobile phase additives [5, 7–18].
Figure 1.
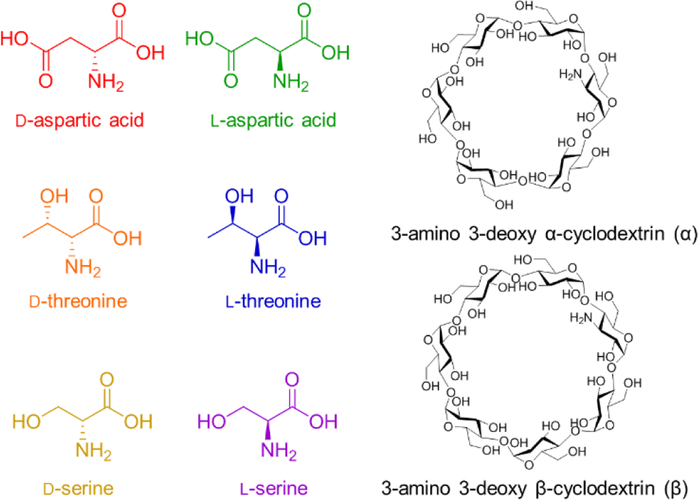
Structures of the d/l amino acid pairs and α/β cyclodextrins used in the SLIM SUPER IM-MS experiments.
In this work we have initially explored and utilized cyclodextrins as chiral complexing agents for enantioseparations in our SLIM SUPER IM-MS platform. Since positive ion mode ESI-MS analyses are more readily and widely used than their negative mode counterpart, a substituted cyclodextrin with a cationic functional group was selected for the studies. The effect of the cyclodextrin cavity size was also of interest. Based on these criteria, we selected 3-amino 3-deoxy α-cyclodextrin (α, six d-glucose ring) and 3-amino 3-deoxy β-cyclodextrin (β, seven d-glucose ring) as our potential chiral hosts for the enantiomeric separations of d/l-aspartic acid, d/l-serine, and d/l-threonine (Figure 1).
Materials and Methods
3-amino 3-deoxy α and β cyclodextrins were purchased from TCI Chemicals (Portland, OR USA). d/l-aspartic acid, d/l-serine, and d/l-threonine were purchased from Sigma Aldrich (Milwaukee, WI USA). All solvents were LC-MS grade and purchased from Sigma Aldrich (Milwaukee, WI USA). All solutions were prepared at a final concentration of 50 μM for each individual component (amino acid and cyclodextrin), either alone or as an enantiomeric (d/l) mixture, in 50/50 (v/v) water/methanol with 0.5% acetic acid (v/v).
A previously described SLIM IM-MS platform was used in all experiments [31, 33]. Solutions were infused for nanoelectrospray ionization (nESI) (+3000 V; 130 °C inlet capillary temperature) for ~10 min each at a flow rate of 0.3 μL/min (<5 μL total sample used). A 60 mTorr positive pressure differential was maintained between the SLIM chamber (2.36 Torr nitrogen gas) and the ion funnel trap region (2.30 Torr). 15 V DC was applied to the SLIM guard electrodes and RF electrode potentials were 360 Vpp at 900 kHz frequency. Traveling waves of 30 Vpp amplitude at 200 m/s were applied. The first SLIM traveling wave section (TW1, 9 m path length) was used as an extended ion accumulation region by halting the traveling wave in the second section [31] for two seconds, before ions were transferred to the second traveling wave section (TW2, 4.5 m) for separation. Longer path separations were performed by the use of an ion switch [33], which can route the ions either to the Agilent 6224 TOF-MS for detection or back to the start of TW1 serpentine path for another 13.5 m of separation (9 m TW1 + 4.5 m TW2) for SLIM SUPER analyses. Compression ratio ion mobility programming [31, 35] (CRIMP) was applied in the same manner as previously described [45]. Briefly, a stuttering (i.e. intermittently applied) TW2 allows for ions to be compressed at the interface between TW1 and TW2. CRIMP is only applied on the second pass of ions around the SLIM module, and its duration was 100 ms (enough time for all ions of interest to be successfully compressed). See the Supporting Information for an example on the benefit of CRIMP for improving both signal-to-noise (S/N) and signal intensity. SLIM data was acquired with the homebuilt Falkor data station software in the unified ion mobility file format (UIMF). From there, files were converted to the Agilent (.d) format, where MassHunter was used to process the data.
Results and Discussion
Prior to the attempted enantiomer separations on our SLIM SUPER IM-MS platform, several questions were formulated. First, would the chiral non-covalent complexes form between the amino acids and the cyclodextrins and if so, would their stoichiometry be different for the enantiomeric pairs? Second, would our SLIM and traveling wave conditions be “gentle” enough to not perturb or fragment these non-covalent complexes? Third, would the complexes survive several passes through the SLIM module ion path? Finally, would this approach be amenable in achieving faster and higher resolution separations than existing techniques?
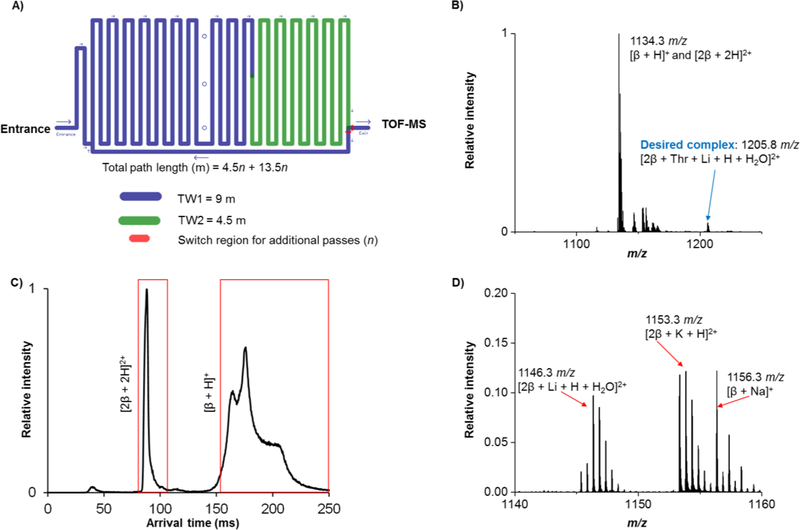
In order to answer the question regarding complex formation, a mixture of d/l-threonine and 3-deoxy 3-amino β-cyclodextrin (β) was assessed after ‘in-SLIM’ ion accumulation for 2 seconds followed by a 4.5 meter path length IM separation (Figure 2A). It is important to note that the total path length for all our separations will be of the form 4.5 + 13.5n (where n is the number of passes) since traveling wave 1 (TW1, 9 m) is used as an ion accumulation region for the first pass. The high resolution mass spectrum for the mixture (Figure 2B) showed both the protonated monomer and doubly protonated dimer of β-cyclodextrin were present. More importantly, the desired complex between the d/l-Thr and β formed. Interestingly, other m/z values were also observed (Figure 2D) due to complexes with other cations such as sodium, potassium, etc. None of these cations were added intentionally, indicating that they were either present in the solvents used or inherently present in the commercially purchased reagents. We also note that in some instances the isotope peaks due to complexes with similar m/z overlap. This further illustrates the benefit of long-pass SLIM SUPER IM separations for separating the species based upon their mobilities, and improving the confidence of identification.
Figure 2.
Analysis of the cyclodextrin and d/l-Thr mixture with SLIM SUPER IM-MS. A) The cartoon depiction of our SLIM module consisting of a 9 m traveling wave 1 (TW1) section and 4.5 m traveling wave 2 (TW2) section, along with an ion switching to direct ions for additional passes through the TW1 and TW2 regions. B) Mass spectrum of an equimolar mixture of d/l-Thr with β cyclodextrin after a 2 sec in-SLIM accumulation step and a 4.5 m IM separation. C) Mobility distribution of m/z 1134.3, highlighting the difference in conformational flexibility of the β monomer versus the rigidity of the β dimer. D) Other identified complexes include m/z 1146.3: [2β + Li + H + H2O]2+, m/z 1153.3: [2β + K + H]2+, m/z 1156.3: [β + Na]+ and [2β + 2Na]2+.
At a first glance, it was surprising that the most abundant β:Thr complexes contained two cyclodextrin molecules (i.e. a 2:1 host-guest inclusion complex) as opposed to only one. Additionally, it was interesting that no 1:1 host-guest inclusion complex was detectable. To investigate why this was the preferred stoichiometric ratio, we performed a 4.5 m SLIM IM-MS separation of the singly protonated β monomer and the doubly protonated β dimer (for an 18 m separation, see the Supporting Information). In Figure 2C, it was observed that the mobility distribution of the [2β + 2H]2+ existed as a much narrower ion mobility peak, potentially indicating the presence of only a single conformation and thus some level of structural rigidity for its dimer form. The mobility distribution of the [β + H]+, however, existed as an extremely broad peak with some evident substructure, possibly indicating the presence of multiple structural or ion conformations. Alternatively, the large difference in peak widths could be attributed to the presence of multiple unresolved species (potentially a mobile protonation site) in its monomer form. We envision future applications to exclusively probe, and theoretically model, the mobility distributions, and potential conformer ensembles, of various cyclodextrin species depending on their substituents. Since the amino acids are significantly smaller in size and molecular weight than the cyclodextrins, the mobility distribution, or overall structure of the host-guest complex, might be expected to be dominated by the preferred conformation of the cyclodextrin. In other words, we might expect that the cyclodextrin amino acid complex to adopt a cyclodextrin dimer conformation (2:1 host-guest complex) that is potentially more rigid and stable than the cyclodextrin monomer (1:1 ratio). Consistent with these observations, no other complex stoichiometries (e.g., 1:1 or 1:2) were observed for any of the other amino acids assessed in this study.
While [2β + Thr + Li + H + H2O]2+ was the dominant complex formed between d/l-Thr and β, other doubly charged complexes having protonated, sodiated, and potassiated forms were also observed, again at low levels, and presumably due to the trace levels of these salts in the solutions. Since complexation was possible between β and M (where M is the enantiomer analyte), we explored the IM separation parameters in our SLIM IM-MS platform to see if we could tease apart the individual d/l enantiomers for aspartic acid, serine, and threonine. All SLIM SUPER IM-MS separations for this study were performed using a 58.5 m path length. In this, the 9 m first TW region was used to accumulate ions in lieu of the ion funnel trap (for gains in sensitivity [31]) and separation was performed for 4.5 m on the first pass, followed by four additional passes (13.5 m each) for a 58.5 m total path [(4.5 m) + (4) × (13.5 m)]. CRIMP [31, 35] was applied only on the second pass (as previously described [45]) for increases in both S/N and signal intensity.
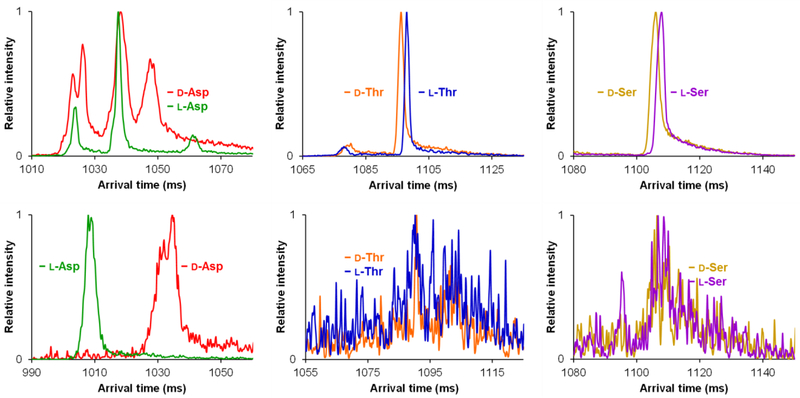
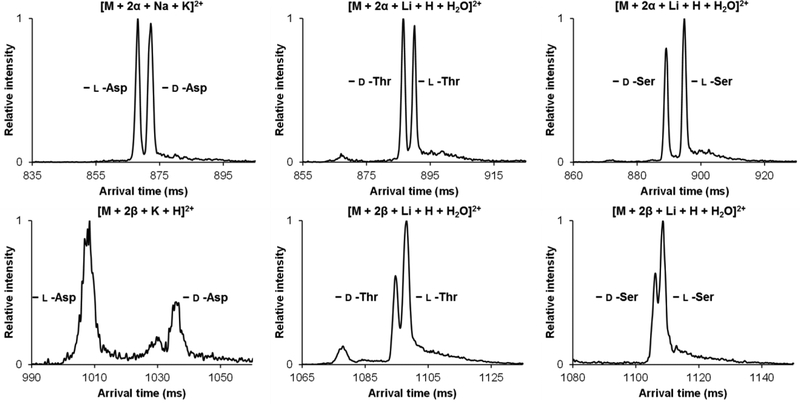
Figure 3 displays the overlaid, individual, d/l amino acid separations as their various β host-guest inclusion complexes, with the top panel showing the [M + 2β + Li + H + H2O]2+ complexes and the bottom panel the [M + 2β + K + H]2+ complexes. Both the d- and l-Asp lithium cation-based complexes can be seen to adopt multiple unique ion conformations, which are only detectable after the present extremely long path length separations (i.e. the separations are not sufficient to be observable using conventional e.g. 1 m drift tube IM separations [20, 32–34]). Even so, the overlaid mobility spectra for d/l-Asp show several co-arriving or unresolved conformer mobility peaks. d/l-Thr and d/l-Ser, however, both showed the potential for resolution as their overlaid individual standards based on their respective [M + 2β + Li + H + H2O]2+ host-guest complexes. As their potassium cation-based β complexes, only d/l-Asp could clearly be individually resolved from one another, with a broader peak suggesting at least two ion conformations being present for the d-Asp complex. For example, a mobility peak presumed to be in a single conformation is ~4–5 ms in width in these measurements, and any peaks significantly wider than this (i.e. the d/l-Asp β-complexes which are ~10 ms in width) could potentially be due to the presence of multiple species/conformations, especially since their arrival times are similar to those of the other amino acid cyclodextrin complexes. However, both d/l-Thr and d/l-Ser were unable to form the [M + 2β + K + H]2+ inclusion complexes with significant abundance in this work. We hypothesize that this could be potentially be due to the larger cavity size of the β inhibiting complex formation for the smaller amino acids. Since all complexes could form as lithiated-water adduct versions, it can be assumed that the role of the lithium cation in conjunction with a water molecule can overcome the large size of the β cavity.
Figure 3.
Individual overlaid d- and l-amino acid SLIM SUPER IM separations with β cyclodextrin (58.5 m). Top: [M + 2β + Li + H + H2O]2+. Bottom: [M + 2β + K + H]2+. M is the amino acid noted in each case.
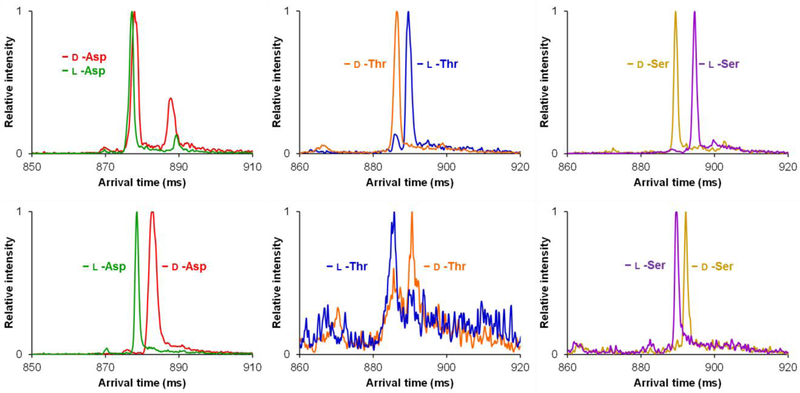
In order to further interrogate the role of cyclodextrin cavity size on enantiomeric separations of amino acids, we decreased the cavity size from seven to six D-glucoses by moving from β- to α-cyclodextrin as the chiral host. Similar to the host-guest complexes formed with β, the α complexes also showed preference for dimers (2α). Additionally, the same adducts (see Figure 3 with β) were observed to form when α was used. Figure 4 illustrates the overlaid individual d/l amino acid SLIM SUPER IM separations (58.5 m) as their [M + 2α + Li + H + H2O]2+ (top) and [M + 2α + K + H]2+ (bottom) host-guest complexes. Similar to the β complexes, the lithiated complexes with α resulted in multiple ion conformations being present for both d and l-Asp, with no resolution between the two enantiomers even after a 58.5 m separation. Conversely, d/l-Thr and d/l-Ser exhibited much more resolution as compared to when β was used. This points to the role of cavity size having a significant effect on both overall resolution and sensitivity. When their respective potassiated adducts were surveyed, it was observed that all three d/l amino acid pairs could be resolved in a 58.5 m path length separation. However, the overlaid individual separation intensities for d/l-Thr and d/l-Ser were noticeably noisier than d/l-Asp, potentially indicating a less preferred complex formation than with lithiated versions.
Figure 4.
Individual overlaid d- and l-amino acid SLIM SUPER IM separations with α cyclodextrin (58.5 m). Top: [M + 2α + Li + H + H2O]2+. Bottom: [M + 2α + K + H]2+. M is the amino acid noted.
In an effort to confirm the observations described in this work for individual enantiomers run separately, we utilized mixtures of both d/l enantiomers to assess their true separability, as shown in Figure 5 with α (top panel) and β (bottom panel). The analyses of mixtures is of importance, as relying on individual standards run separately may be misleading in determining the true resolution of an enantiomeric pair or artifacts due to potential experimental variations (e.g. pressure) causing small variations in arrival times. All three amino acid pairs could be distinguished in the analyses, and the highest resolution separations always occurred with the α-cyclodextrin rather than the β-cyclodextrin, indicating that cavity size plays a key role in initial complex formation, as well as in increasing overall separation. d/l-Asp was the only enantiomeric pair examined that was unable to be separated as its lithium-based adducts (Figures 3 and 4), illustrating the effect of cations on the formation of an energetically favorable host-guest inclusion complex. d/l-Asp was, however, successfully separated as its [M + 2α + Na + K]2+. Figure 5 clearly demonstrates the ability to resolve mixtures of enantiomers, rather than relying solely on arrival times of individual species. We tentatively attribute the observed peak tailing for certain complexes to lower mobility (i.e. slower) species interconverting with higher mobility ones (i.e. the main peak observed).
Figure 5.
High-resolution enantiomeric SLIM SUPER IM separations of the d/l mixtures of amino acids with either α cyclodextrin (top) or β cyclodextrin (bottom) after 58.5 m of IM separation.
Concluding Remarks
Herein a non-covalent cyclodextrin complexation strategy is described for the separation of d/l amino acid enantiomeric pairs using a new SLIM SUPER IM-MS platform. To the best of our knowledge, this is the first demonstration of cyclodextrins used as chiral hosts for the IM separation of enantiomeric amino acid mixtures. Additionally, this methodology provides much higher resolution than any previous IM-based enantiomer separations technique, largely permitted by the ultralong path length capabilities of our SLIM SUPER IM-MS system. All d/l amino acid separations presented in this work were performed using 58.5 m path length separations, with arrival times ranging from 800 ms to 1200 ms. These separation times are significantly faster than chromatographic approaches (min to hours) and provide an exciting alternative to these slower techniques. We note that targeted chromatographic approaches have utility when a researcher is interested in collecting an enantiopure sample, and while highly sensitive MS-based approaches are destructive in nature, the ability to use nESI conditions with low flow rates, etc. affords sample size requirements much lower than other analytical techniques (<10 nanograms of sample in the present work, and potentially much less). In this work, the observed increase in sensitivity is largely due to our ability to use a massive ion accumulation region in the SLIM, as opposed to ion introduction with the ion funnel trap [31] used previously for drift tube IM-MS.
In the analyses of the cyclodextrin and amino acid complexes, we found that the smaller cavity size of α cyclodextrin complex is preferred over its β counterpart. It was also evident that the presence of certain cations in the overall host-guest complex either assist or hinder the enantiomeric separation resolution achieved. Interestingly, regardless of what cyclodextrin was used with the lithium cation complexes, the d- enantiomer for both Ser and Thr arrived earlier than l-, indicating that the d-complex is more compact in nature. Previous literature has demonstrated the utility of lithium-water adduction to individually discriminate amongst certain monosaccharides [46–47]. This supports our findings that cation and water molecule addition can also be beneficial for other analytes. We emphasize the importance of the cyclodextrin in providing the chiral environment necessary for these enantiomer separations, and that insight into the observed stoichiometric ratio of complex formation (2:1 cyclodextrin host to amino acid guest) was provided from the observed mobility features for the β monomer versus β dimer (Figure 1). Here we infer that the presence of two cyclodextrins plays an important role in the encapsulation of the amino acid, and potentially increases the overall structural rigidity of the formed complex. With the combination of cyclodextrin-based complexation and the high-resolution capabilities of our SLIM SUPER IM-MS platform, we envision other enantiomers could be resolved with this or a similar strategy.
Supplementary Material
Acknowledgements
This work was partially supported by the National Institute of General Medical Sciences (P41 GM103493), the National Institute of Environmental Health Sciences of the NIH (R01 ES022190), and the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory. Experiments were performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle under contract DE-AC05-76RL0 1830 for the DOE.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References:
- 1.Lorenz H; Seidel-Morgenstern A, Angew. Chem. Int. Ed. Engl 2014, 53, 1218–50. [DOI] [PubMed] [Google Scholar]
- 2.Chankvetadze B, J. Chromatogr. A 2012, 1269, 26–51. [DOI] [PubMed] [Google Scholar]
- 3.Ward TJ; Ward KD, Anal. Chem 2012, 84, 626–35. [DOI] [PubMed] [Google Scholar]
- 4.Enders JR; McLean JA, Chirality 2009, 21 Suppl 1, E253–64. [DOI] [PubMed] [Google Scholar]
- 5.Scriba GK; Harnisch H; Zhu Q, Methods Mol. Biol 2016, 1483, 277–99. [DOI] [PubMed] [Google Scholar]
- 6.Patel AV; Kawai T; Wang L; Rubakhin SS; Sweedler JV, Anal. Chem 2017, 89, 12375–12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ai F; Wang Y; Chen H; Yang Y; Tan TT; Ng SC, Analyst 2013, 138, 2289–94. [DOI] [PubMed] [Google Scholar]
- 8.Xiao Y; Ng SC; Tan TT; Wang Y, J. Chromatogr. A 2012, 1269, 52–68. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J; Tang J; Tang W, TrAC Trends in Analytical Chemistry 2015, 65, 22–29. [Google Scholar]
- 10.Tang W; Ng SC, Nat. Protoc 2007, 2, 3195–200. [DOI] [PubMed] [Google Scholar]
- 11.O’Keeffe F; Shamsi SA; Darcy R; Schwinte P; Warner IM, Anal. Chem 1997, 69, 4773–82. [DOI] [PubMed] [Google Scholar]
- 12.Hinze WL; Riehl TE; Armstrong DW; DeMond W; Alak A; Ward T, Anal. Chem 1985, 57, 237–242. [Google Scholar]
- 13.Escuder-Gilabert L; Martin-Biosca Y; Medina-Hernandez MJ; Sagrado S, J. Chromatogr. A 2014, 1357, 2–23. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y; Zhou J; Liu Y; Tang J; Tang W, Electrophoresis 2014, 35, 2744–51. [DOI] [PubMed] [Google Scholar]
- 15.Dai Y; Wang S; Zhou J; Liu Y; Sun D; Tang J; Tang W, J. Chromatogr. A 2012, 1246, 98–102. [DOI] [PubMed] [Google Scholar]
- 16.Cucinotta V; Contino A; Giuffrida A; Maccarrone G; Messina M, J. Chromatogr. A 2010, 1217, 953–67. [DOI] [PubMed] [Google Scholar]
- 17.de Boer T; de Zeeuw RA; de Jong GJ; Ensing K, Electrophoresis 2000, 21, 3220–39. [DOI] [PubMed] [Google Scholar]
- 18.Wang F; Khaledi MG, Electrophoresis 1998, 19, 2095–100. [DOI] [PubMed] [Google Scholar]
- 19.Gabelica V; Marklund E, Curr. Opin. Chem. Biol 2018, 42, 51–59. [DOI] [PubMed] [Google Scholar]
- 20.Dodds JN; May JC; McLean JA, Anal. Chem 2017, 89, 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaye MM; Nagy G; Clemmer DE; Pohl NL, Anal Chem 2016, 88, 2335–44. [DOI] [PubMed] [Google Scholar]
- 22.Hilderbrand AE; Myung S; Clemmer DE, Anal. Chem 2006, 78, 6792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chouinard CD; Cruzeiro V. c. W. D.; Roitberg AE; Yost RA, J. Am. Soc. Mass Spectrom 2017, 28, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mie A; Jornten-Karlsson M; Axelsson BO; Ray A; Reimann CT, Anal. Chem 2007, 79, 2850–8. [DOI] [PubMed] [Google Scholar]
- 25.Domalain V; Hubert-Roux M; Tognetti V; Joubert L; Lange CM; Rouden J; Afonso C, Chem. Sci 2014, 5, 3234–3239. [Google Scholar]
- 26.Zhang JD; Mohibul Kabir KM; Lee HE; Donald WA, Int. J. Mass Spectrom 2018, 428, 1–7. [Google Scholar]
- 27.Zheng X; Zhang X; Schocker NS; Renslow RS; Orton DJ; Khamsi J; Ashmus RA; Almeida IC; Tang K; Costello CE; Smith RD; Michael K; Baker ES, Anal. Bioanal. Chem 2017, 409, 467–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y; Dodds ED, Anal. Chem 2013, 85, 9728–35. [DOI] [PubMed] [Google Scholar]
- 29.Kulyk K; Rebrov O; Ryding M; Thomas RD; Uggerud E; Larsson M, J. Am. Soc. Mass Spectrom 2017, 28, 2686–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi P; Wu C; Matz LM; Clowers BH; Siems WF; Hill HH Jr., Anal. Chem 2006, 78, 8200–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng L; Garimella SVB; Hamid AM; Webb IK; Attah IK; Norheim RV; Prost SA; Zheng X; Sandoval JA; Baker ES; Ibrahim YM; Smith RD, Anal. Chem 2017, 89, 6432–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng L; Ibrahim YM; Baker ES; Aly NA; Hamid AM; Zhang X; Zheng X; Garimella SVB; Webb IK; Prost SA; Sandoval JA; Norheim RV; Anderson GA; Tolmachev AV; Smith RD, ChemistrySelect 2016, 1, 2396–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng L; Webb IK; Garimella SVB; Hamid AM; Zheng X; Norheim RV; Prost SA; Anderson GA; Sandoval JA; Baker ES; Ibrahim YM; Smith RD, Anal. Chem 2017, 89, 4628–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng L; Ibrahim YM; Hamid AM; Garimella SV; Webb IK; Zheng X; Prost SA; Sandoval JA; Norheim RV; Anderson GA; Tolmachev AV; Baker ES; Smith RD, Anal. Chem 2016, 88, 8957–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garimella SV; Hamid AM; Deng L; Ibrahim YM; Webb IK; Baker ES; Prost SA; Norheim RV; Anderson GA; Smith RD, Anal. Chem 2016, 88, 11877–11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Y; Wei J; Costello CE; Lin C, Journal of The American Society for Mass Spectrometry 2018, 29, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha S; Roy A; Roy K; Roy MN, Sci. Rep 2016, 6, 35764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saha S; Ray T; Basak S; Roy MN, New J.Chem 2016, 40, 651–661. [Google Scholar]
- 39.Qi Y; Geib T; Volmer DA, J. Am. Soc. Mass Spectrom 2015, 26, 1143–9. [DOI] [PubMed] [Google Scholar]
- 40.Roy MN; Roy MC; Roy K, RSC Advances 2015, 5, 56717–56723. [Google Scholar]
- 41.Subramaniam P; Mohamad S; Alias Y, Int. J. Mol. Sci 2010, 11, 3675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kralj B; Smidovnik A; Kobe J, Rapid Commun. Mass Spectrom 2009, 23, 171–80. [DOI] [PubMed] [Google Scholar]
- 43.Lebrilla CB, Acc. Chem. Res 2001, 34, 653–61. [DOI] [PubMed] [Google Scholar]
- 44.Grigorean G; Ramirez J; Ahn SH; Lebrilla CB, Anal. Chem 2000, 72, 4275–81. [DOI] [PubMed] [Google Scholar]
- 45.Chouinard CD; Nagy G; Webb IK; Garimella SVB; Baker ES; Ibrahim YM; Smith RD, Anal. Chem 2018, DOI:10.1021/acs.analchem.8b02990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell MT; Chen D; Wallbillich NJ; Glish GL, Anal. Chem 2017, 89, 10504–10510. [DOI] [PubMed] [Google Scholar]
- 47.Campbell MT; Chen D; Glish GL, J. Am. Soc. Mass Spectrom 2017, 28, 1420–1424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.