Abstract
The N7-methylguanosine cap is a hallmark of the 5′ end of eukaryotic mRNAs and is required for gene expression. Loss of the cap was believed to lead irreversibly to decay. However, nearly a decade ago it was discovered that mammalian cells contain enzymes in the cytoplasm that are capable of restoring caps onto uncapped RNAs. In this review, we summarize recent advances in our understanding of cytoplasmic RNA recapping and discuss the biochemistry of this process and its impact on regulating and diversifying the transcriptome. Although most studies focus on mammalian RNA recapping, we also highlight new observations for recapping in disparate eukaryotic organisms, with the trypanosome recapping system appearing to be a fascinating example of convergent evolution. We conclude with emerging insights into the biological significance of RNA recapping and prospects for the future of this evolving area of study.
Graphical/Visual Abstract and Caption
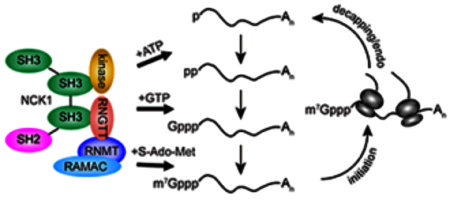
Cytoplasmic recapping is catalysed by a complex of enzymes that restores the m7G cap onto decapped or endonucleolytically cleaved mRNAs. Recapping maintains the translation of some mRNAs and has impacts on transcriptome and proteome complexity.
Introduction
The 5′ end of all eukaryotic mRNAs is defined by a cap structure consisting of N7-guanosine (m7G) joined by a 5′,5′-triphosphate linkage to the first transcribed nucleotide. The m7GpppN cap structure (where N is the first transcribed nucleotide) was first identified in the 1970s through studies of viral RNAs and later as a common feature of all eukaryotic mRNAs (Furuichi, 2015; Furuichi et al., 1975; Furuichi, Morgan, Muthukrishnan, & Shatkin, 1975; Furuichi et al., 1975; Moss, 2017; Wei & Moss, 1975; Wei, Gershowitz, & Moss, 1975). A number of recent reviews have covered the nuclear steps involved in cotranscriptional capping of pre-mRNAs (Cowling, 2010; Ghosh & Lima, 2010; Ramanathan, Robb, & Chan, 2016) and the role of the cap in downstream processes including translation, mRNA turnover, and nonsense-mediated mRNA decay (Topisirovic, Svitkin, Sonenberg, & Shatkin, 2011; Grudzien-Nogalska & Kiledjian, 2017).
Prior to 2009 it was thought that loss of the cap, either by specific decapping enzymes or as a consequence of endonuclease decay, led to rapid transcript loss by the 5′ exonuclease XRN1. This thinking began to change with the concurrent publication of two papers that year. One of these presented the first evidence for the existence of capped ends downstream of transcription start sites and within spliced exons (Fejes-Toth et al., 2009). The other, from our lab, described a cytoplasmic pool of capping enzyme (RNGTT, RNA guanylyltransferase and 5’-phosphatase) in complex with a kinase that together convert RNA with a 5′-monophosphate end to a molecule with a GpppN 5′ terminus (Otsuka, Kedersha, & Schoenberg, 2009). That same year we wrote a review summarizing what was known at the time about the function of RNA recapping and its relationship to transcriptome complexity (Schoenberg & Maquat, 2009). In the intervening years much has changed, with discoveries driven by insights into the biochemistry of cytoplasmic capping, high throughput sequencing, improvements in proteomics, and single molecule approaches for studying translation. Here we summarize these findings as they relate to RNA recapping and its role in transcriptome and proteome complexity.
The cap: canonical nuclear synthesis and functions in mRNA metabolism
The canonical nuclear capping pathway modifies the 5′ end of every RNA polymerase II (Pol II) transcript, including all pre-mRNAs, miRNA precursors, long non-coding RNAs (lncRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs). Nuclear capping of the nascent transcript is catalyzed by RNGTT bound to the phosphorylated C-terminal domain of Pol II (Ho, C. K., Sriskanda, V., McCracken, S., Bentley, D., Schwer, B., & Shuman, S., 1998; Yue et al., 1997; Martinez-Rucobo et al., 2015). Capping starts with conversion of the emergent 5′-triphosphate end to a diphosphate, a process that is catalyzed by the N-terminal triphosphatase domain of RNGTT. Next, the guanylyltransferase domain transfers GMP bound covalently to the ε amino group of lysine 294 to create a G-capped RNA. N7 methylation by RNA guanine-7 methyltransferase (RNMT, also called cap methyltransferase) completes the synthesis of cap 0, the basic cap structure. Cap 0 is sufficient for recognition by cap-binding effector proteins (Calero et al., 2002; Marcotrigiano, Gingras, Sonenberg, & Burley, 1997; Niedzwiecka et al., 2002). Additional cap modifications are generated by 2’-O-methylation of the first (cap 1) and second (cap 2) transcribed nucleotide by CMTR1 and CMTR2, respectively (Belanger, Stepinski, Darzynkiewicz, & Pelletier, 2010; Werner et al., 2011). These additional methylations function to enhance translation (Kuge, Brownlee, Gershon, & Richter, 1998) and to mark an RNA as “self” to evade innate immunity responses against viral RNAs (Hyde & Diamond, 2015; Schuberth-Wagner et al., 2015).
Cap-binding proteins participate in every step of mRNA metabolism including nuclear processing (Gonatopoulos-Pournatzis & Cowling, 2014; Izaurralde et al., 1994; Schwer & Shuman, 1996; Flaherty, Fortes, Izaurralde, Mattaj, & Gilmartin, 1997; Gilmartin, McDevitt, & Nevins, 1988), export (Jarmolowski, Boelens, Izaurralde, & Mattaj, 1994; Visa, Izaurralde, Ferreira, Daneholt, & Mattaj, 1996), translation (Lee, Kranzusch, Doudna, & Cate, 2016; Tcherkezian et al., 2014; Lahr et al., 2017; Philippe, Vasseur, Debart, & Thoreen, 2018; Topisirovic et al., 2011), microRNA silencing (Chapat et al., 2017), nonsense-mediated mRNA decay (Hosoda, Kim, Lejeune, & Maquat, 2005) and ultimately mRNA turnover (Schoenberg & Maquat, 2012). A detailed discussion of the interplay between the cap and cap-binding proteins is beyond the scope of this review. What is relevant is the interplay between decapping enzymes and cap quality control mechanisms (Grudzien-Nogalska & Kiledjian, 2017). Most decapping enzymes are members of the Nudix (nucleoside diphosphate linked to another moiety X) family of enzymes, the best known and most characterized being DCP2. DCP2 binds RNA and cleaves between the α and β phosphates to generate m7Gpp and RNA with a 5′-monophosphate end. As noted above, 5′-monophosphate RNA is susceptible to degradation by XRN1 (Schoenberg & Maquat, 2012), but can also serve as substrate for recapping. There are 22 genes encoding Nudix family proteins in the human genome, but to date only DCP2, NUDT3 and NUDT16 have been shown to catalyze mRNA decapping in vivo (Song, Li, & Kiledjian, 2010). However, in vitro decapping activity has been observed for many other Nudix family proteins, with different specificities and products that may be relevant to mRNA recapping. For example, NUDT3, NUDT12, and NUDT15 are capable of cleaving between the β and γ phosphates of the cap 0 structure to yield RNA with a 5′-diphosphate end (Song, Bail, & Kiledjian, 2013). RNA with a 5′ diphosphate is resistant to degradation by XRN1 (Fujimura & Esteban, 2010) and may serve as a direct substrate for cytoplasmic recapping (Figure 1A).
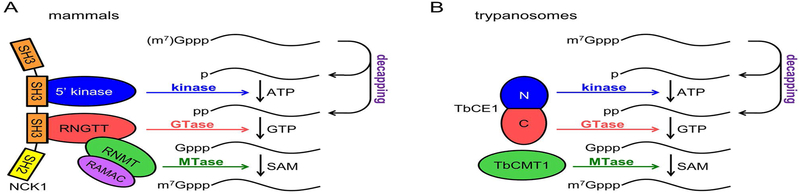
Figure 1. Biochemistry of RNA recapping.
A. Model of recapping in mammals. Multiple decapping enzymes can convert unmethylated (GpppN) and/or methylated (m7GpppN) capped ends to mono- and diphosphate substrates for recapping. 5′-monophosphate RNA is phosphorylated by an ATP-dependent kinase to produce 5′-diphosphate RNA. Guanylation by the guanylyltransferase (GTase) domain of RNGTT yields the GpppN cap structure that is then methylated at the N7 position by the RNMT-RAMAC heterodimeric cap methyltransferase. The 5′-monophosphate RNA kinase and RNGTT bind to adjacent SH3 domains of NCK1, and RNMT-RAMAC is recruited to RNGTT through the C-terminal catalytic domain of RNMT. B. Model of recapping in trypanosomes. Trypanosome decapping enzymes producing both mono- and diphosphate ends from m7GpppN caps have been described. Conversion of 5′-monophosphate RNA to the GpppN cap structure is catalyzed by the bifunctional enzyme TbCE1, which has an N-terminal kinase domain and a C-terminal GTase domain. The methyltransferase TbCMT1 is proposed to catalyze N7-methylation.
Several of the Nudix hydrolases preferentially degrade unmethylated or improperly methylated caps (Song et al., 2013; Grzela et al., 2018). As such they likely function to limit expression to mRNAs with properly modified caps. The unrelated DXO (decapping exoribonuclease) family of proteins also functions in cap surveillance. The initial evidence for cap quality control came from the discovery that yeast Rai1, a pyrophosphoryl hydrolyase, forms a heterodimer with the nuclear 5′-exonuclease Rat1 to form a complex that selectively degrades mRNAs with improperly methylated caps (Jiao et al., 2010). A related protein Dxo1 acts similarly to the Rai1/Rat1 heterodimer to degrade mRNAs with improperly methylated caps, and the mammalian ortholog of this (DXO) also degrades pre-mRNAs with improperly methylated caps (Jiao, Chang, Kilic, Tong, & Kiledjian, 2013). DXO shows similar activity in vitro against mRNAs with methylated and unmethylated caps, and recent results indicate its primary function lies in the ability to degrade NAD-capped mRNAs (Jiao et al., 2017). NAD capping is beyond the scope of this article and is the subject of a recent review (Kiledjian, 2018).
Early evidence of cytoplasmic recapping
The view that uncapped 5′ ends are irreversibly destined for exonucleolyic decay was challenged by early studies of nonsense-mediated decay (NMD). In erythroid cells of a β-thalassemic mouse model, decay intermediates from nonsense codon-containing β-globin mRNA were found that lacked portions of their 5′ ends but were surprisingly stable (Lim, Mullins, Chen, Gross, & Maquat, 1989). These decay intermediates were polyadenylated like their parent mRNAs and were only detected in the cytoplasm (Lim et al., 1989; Stevens et al., 2002). Loss of parent mRNA levels upon transcription inhibition was coincident with the accumulation of the decay intermediates, supporting their generation as a cytoplasmic process independent of transcription (Lim & Maquat, 1992). Further analysis of these decay intermediates revealed their 5′ ends had been modified by a cap or cap-like structure (Lim, Sigmund, Gross, & Maquat, 1992). Supporting this conclusion, anti-cap antibody bound both decay intermediates and parent mRNA, and both were competitively eluted with m7G. We later confirmed these findings, showing that the decay intermediates bind to recombinant cap-binding protein EIF4E and that they could be eluted with m7GDP. These decay intermediates were resistant to degradation by XRN1, and they could be decapped in vitro by DCP2 under conditions specific to m7G caps (Otsuka et al., 2009). NMD occurs in the cytoplasm (Trcek, Sato, Singer, & Maquat, 2013), and later work showed that the capped β-globin decay intermediates are generated by action of the SMG6 endonuclease (Mascarenhas, Dougherty, & Schoenberg, 2013). Prior to 2009 there were other hints that RNAs might be recapped in the cytoplasm, one of the most notable being the observation that the downstream product of antisense cleavage of hepatitis B virus RNA was stable and translated to generate an N-terminally truncated protein product (Thoma et al., 2001). The cleaved transcript retained an intact poly(A) tail, and at the time the authors speculated that it may have undergone some form of cytoplasmic recapping.
Recapping enzymes and cofactors
The first mechanistic insight into how a cap could be synthesized on a 5′-end-processed, cytoplasmic RNA came with the observation that RNGTT is not restricted to the nucleus (Otsuka et al., 2009; Wen, Yue, & Shatkin, 1998). While RNGTT is a predominantly nuclear enzyme, it is also present in the cytoplasm of multiple mammalian cell lines, including U2OS, Huh7, MEL, Cos-1, HEK293 and RH-30 (Otsuka et al., 2009; Mukherjee, Bakthavachalu, & Schoenberg, 2014; Thul et al., 2017). Drosophila also has a population of RNGTT (mRNA-cap) that localizes to the cytoplasm (Chen et al., 2017b).
A common feature among the diverse eukaryotic cap synthesis reactions is the presence of the necessary enzymatic activities in either a single polypeptide or in a complex of interacting proteins. The latter (termed a metabolon (Srere, 1987)) facilitates efficient substrate channeling and limits the accumulation of reaction intermediates. For example, mammalian RNGTT is a bifunctional triphosphatase-guanylyltransferase whereas the same reactions in yeast are catalyzed by the Cet1p:Ceg1p heterodimer (Ho et al., 1998). The trypanosome nuclear capping enzyme TbCGM1 is a bifunctional guanylyltransferase-methyltransferase (Takagi, Sindkar, Ekonomidis, Hall, & Ho, 2007), while the trifunctional capping enzymes of poxviruses, African swine fever virus, and mimivirus each contain triphosphatase, guanylyltransferase, and methyltransferase activities in a single polypeptide (Benarroch, Smith, & Shuman, 2008; Kyrieleis, Chang, de la Peña, Shuman, & Cusack, 2014). Consistent with this theme, cytoplasmic capping in mammalian cells is accomplished by a metabolon consisting of RNGTT, a 5′-monophosphate kinase that generates a diphosphate substrate for RNGTT guanylyltransferase activity (Otsuka et al., 2009), and guanine-N7 cap methyltransferase (RNMT) (Trotman, Giltmier, Mukherjee, & Schoenberg, 2017) (Figure 1A).
Initial evidence for an mRNA recapping metabolon came from experiments in which immunoprecipitated cytoplasmic RNGTT was able to add GMP onto RNA with a 5′-monophosphate end but not the same RNA with a 5′ hydroxyl (Otsuka et al., 2009). Although RNGTT is 68.5 kDa, the enzymatic activities needed to add GMP onto 5’-monophosphate RNA sedimented at ~140 kDa on glycerol gradients, suggesting that RNGTT might be associated with other proteins. Importantly, RNGTT coprecipitated with an RNA kinase activity that was capable of transferring the γ phosphate of γ-[32P]ATP onto 5′-monophosphate RNA but not onto the same RNA with a 5′-hydroxyl end.
In the process of categorizing portions of cytoplasmic RNGTT that function in recapping, we noted that modifications to the C-terminus impaired the ability of immunoprecipitated protein to transfer [32P]GMP from α-[32P]GTP onto 5′-monophosphate RNA in vitro (Mukherjee et al., 2014). The very C-terminus of RNGTT is a proline-rich sequence that was predicted to function as a ligand for binding by one or more SH3-domain-containing proteins, notably NCK1 (NCK adapter protein 1). consists of three SH3 domains and a C-terminal SH2 domain. Mutational analyses of RNGTT and NCK1 showed that RNGTT binds through its proline-rich C-terminus to the third SH3 domain of NCK1. Importantly, the as-of-yet unidentified 5′-monophosphate kinase binds to the second SH3 domain, presumably through a proline-rich peptide sequence of its own (Mukherjee et al., 2014). These findings established NCK1 as a scaffold upon which the cytoplasmic capping complex assembles (Figure 1A).
Multiple lines of evidence showed recapped transcripts undergo N7 methylation to generate the same m7G caps found on nuclear capped RNAs. RNMT is the only mammalian enzyme known to catalyze the N7-methylation of G-capped RNA (Cowling, 2010), and like RNGTT, it was thought to be restricted to the nucleus. RNMT functions as a heterodimer with RNMT-associated miniprotein (RAMAC, formerly known as RAM), which binds RNA and stimulates RNMT activity (Gonatopoulos-Pournatzis, Dunn, Bounds, & Cowling, 2011). Biochemical fractionation and immunofluorescence identified cytoplasmic pools of both RNMT and RAMAC, and RNA interference and co-immunoprecipitation showed that these two proteins catalyze cytoplasmic cap methylation in the same manner as nuclear cap methylation (Trotman et al., 2017). Gel filtration identified RNMT in a complex with RNGTT and NCK1, and this was confirmed by proximity-dependent biotinylation by BirA*-tagged cytoplasmic RNGTT. In vitro and in vivo binding experiments agreed with an earlier report of direct interaction between RNMT and RNGTT (Pillutla, Yue, Maldonado, & Shatkin, 1998) and showed that the C-terminal methyltransferase domain of RNMT binds to the C-terminal guanylyltransferase domain of RNGTT. Together, these findings describe a cytoplasmic capping metabolon that contains all activities necessary to regenerate cap 0 on uncapped RNA.
Trypanosomes have a nuclear guanylyltransferase-methyltransferase, TbCGM1, and a predominantly cytoplasmic guanylyltransferase, TbCE1. Strikingly, TbCE1 has a novel 5′-monophosphate RNA kinase domain at its N-terminus, making it the only enzyme identified to date with this activity (Ignatochkina, Takagi, Liu, Nagata, & Ho, 2015). TbCE1 kinase activity is magnesium-dependent and can use ATP or dATP as a phosphate donor. Of the 5′-monophosphate RNA substrates tested, TbCE1 was most effective in recapping the spliced leader (SL) sequence present on all Trypanosome mRNAs. It is unclear whether recapping activity is stimulated by SL sequence or is simply more efficient on substrate RNAs with a 5′-terminal G or A. As a bifunctional kinase-guanylyltransferase, TbCE1 is the first dedicated cytoplasmic recapping enzyme to be described. Another trypanosome cytoplasmic protein, TbG5-IP, has a domain structure similar to TbCE1, probably resulting from a gene duplication event (Freire et al., 2014). TbG5-IP interacts with the translation initiation factors TbEIF4G5 and TbEIF4E6, raising the possibility of translational control through an additional, similar recapping enzyme (Freire et al., 2014; Freire, Sturm, Campbell, & de Melo Neto, 2017). It has also been speculated that an additional standalone guanine-N7 methyltransferase, TbCMT1, functions to methylate cytoplasmically recapped RNAs in trypanosomes (Hall & Ho, 2006; Ignatochkina et al., 2015). The trypanosome recapping machinery is summarized in Figure 1B. The fact that TbCE1-related proteins are limited to kinetoplastids (NCBI BLAST analysis) suggests a different evolutionary origin for recapping by these organisms compared to higher eukaryotes.
Other enzymes have been described that may function in recapping or in generating substrates for recapping. The earliest example was a 5′-monophosphate RNA kinase from vaccinia viral cores that can yield di- or triphosphate ends (Spencer, Loring, Hurwitz, & Monroy, 1978). The identity of this enzyme remains unknown despite the limited number (~200) of protein-coding genes that have been annotated in vaccinia (Moss, 2017). Cytoplasmic recapping has not been reported in yeast, but the yeast L-A virus synthesizes RNA transcripts with a 5′ diphosphate, and the ATP-dependent generation of 5′-diphosphate ends from GMP-primed transcripts points to the possibility of 5′-monophosphate RNA kinase activity (Fujimura & Esteban, 2010). As noted above, mammalian cells have decapping enzymes that generate 5′-diphosphate ends (Song et al., 2013). The trypanosome decapping enzyme TbALPH1 can also generate products with 5′ diphosphate (Kramer, 2017). 5′-diphosphate ends are resistant to XRN1 activity (Fujimura & Esteban, 2010), and transcripts bearing 5′-diphosphate ends may undergo cytoplasmic recapping independent of the need to be converted from a 5′-monophosphate terminus (Figure 1B).
Recapping target RNAs and functional consequences of recapping
RNGTT functions in both nuclear and cytoplasmic capping and one of the challenges to studying cytoplasmic capping was the development of a way to inhibit this process in vivo without disrupting nuclear capping. This was achieved by development a catalytically-inactive form of RNGTT that is restricted to the cytoplasm (Otsuka et al., 2009). This form of RNGTT has the HIV Rev nuclear export sequence at the N-terminus and is missing the four amino acids that function as a nuclear localization sequence. Changing the active site lysine 294 in the guanylylation domain to alanine resulted in a catalytically-inactive protein, termed K294A. Overexpression of K294A inhibits cytoplasmic capping in a dominant-negative manner, most likely through competitive disruption of the cytoplasmic capping complex. Uncapped forms of recapping target mRNAs were identified by their susceptibility to in vitro degradation by XRN1, and were grouped into three classes based on their appearance with respect to K294A overexpression (Mukherjee et al., 2012). 2666 mRNAs were identified as having ‘natively’ uncapped forms in the absence of K294A expression. These mRNAs are relatively unstable and they are stabilized by XRN1 knockdown. A separate group of 675 mRNAs was identified for which uncapped forms appeared only in K294A-expressing cells. This “capping-inhibited” class likely represents transcripts that undergo rapid recapping whenever an uncapped form appears in the cytoplasm. A third group of 835 “common” transcripts were found to be uncapped in both K294A-expressing and uninduced cells; these mRNAs are natively uncapped to some degree but have an increased proportion that appears uncapped upon inhibiting cytoplasmic recapping.
As noted above cyclic decapping and recapping, or “cap homeostasis”, serves to regulate the stability of natively uncapped mRNAs (Mukherjee et al., 2012). It also impacts translation, particularly of mRNAs in the “common” and “capping-inhibited” pool. Normally these mRNAs sediment with translating polysomes; however, when cytoplasmic capping is blocked, their resulting uncapped forms redistribute to non-translating mRNPs (Figure 2A). This raised the question whether uncapped mRNAs in non-translating mRNPs retained poly(A) tails of sufficient length to enable their return to the translating pool following cytoplasmic recapping. Recent transcriptome-wide studies of polyadenylation showed little correlation between translation efficiency and poly(A) tail length in somatic cells (Zheng & Tian, 2014; Park, Yi, Kim, Chang, & Kim, 2016). Inhibition of cytoplasmic capping has no impact on global poly(A) length distribution across the transcriptome (Kiss et al., 2016). More importantly, poly(A) tails of similar length were present on capped forms of recapping targets on translating polysomes and uncapped forms of the same mRNAs in non-translating mRNP complexes. These observations support a model of cap homeostasis wherein stable uncapped mRNAs retain a translationally competent poly(A) tail, thus enabling these transcripts to be stored in a state that is primed for rapid reentry into the translating pool upon recapping (Figure 2B). Recent advances in live-cell imaging of translation events have revealed sporadic on-off “bursting” kinetics (Wu, Eliscovich, Yoon, & Singer, 2016; Yan, Hoek, Vale, & Tanenbaum, 2016) that could be explained by cycles of decapping and recapping regulating translation in a temporal manner (Iwasaki & Ingolia, 2016).
Figure 2. Translational control by cap homeostasis.
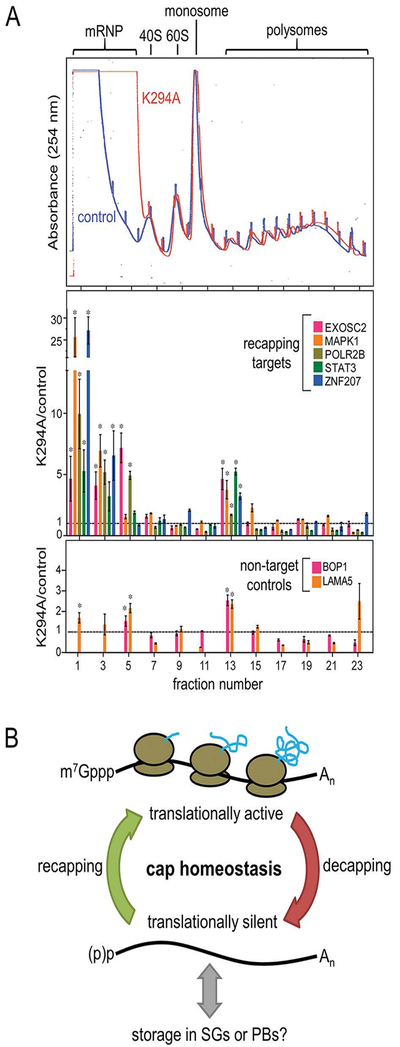
A. Polysome profiling shows inhibition of recapping by overexpression of a catalytically inactive, cytoplasmically restricted form of RNGTT (K294A) increases transcript accumulation in non-translating messenger ribonucleoprotein complexes (mRNPs, top panel). In the middle and lower panels individual fractions were assayed by RT-qPCR for recapping target and non-target mRNAs. Inhibition of cytoplasmic capping results in the redistribution of target mRNAs into non-translating mRNPs whereas non-target controls remain essentially unchanged. This figure is adapted from Figure 5 in (Mukherjee et al., 2012) and is reproduced here in accordance with Creative Commons License CC-BY. B. Model of cap homeostasis. Because the majority of translation initiation is cap-dependent, the translatability of an mRNA can be regulated by cycles of decapping and recapping. Such cap homeostasis is independent of changes in poly(A) tail length. Translationally silent mRNAs can be stored in stress granules (SGs) and P-bodies (PBs), though it remains to be elucidated how cap homeostasis intersects with RNA granule formation.
It remains to be determined whether cytoplasmic recapping occurs more frequently at the canonical 5′ end or at downstream sites generated by endonucleolytic cleavage (Mercer et al., 2010; Schmidt et al., 2015) or at sites of pausing by XRN1 (Moon et al., 2015; Charley, Wilusz, & Wilusz, 2018) (Figure 3A). Nonetheless, growing evidence suggests a role for cytoplasmic recapping in generating novel capped ends at downstream sites. A number of methods have been developed for the transcriptome-wide identification of capped sites (Afik et al., 2017; Batut, Dobin, Plessy, Carninci, & Gingeras, 2013; Afik et al., 2017; Djebali et al., 2012; Cartolano, Huettel, Hartwig, Reinhardt, & Schneeberger, 2016; Gu et al., 2012; Machida & Lin, 2014), with cap analysis of gene expression (CAGE (Shiraki et al., 2003)) being the most commonly used. CAGE was originally developed for the annotation of transcription start sites (TSSs); however, its early application identified human exonic CAGE tags downstream of canonical cap sites (Fejes-Toth et al., 2009). Such downstream CAGE tags likely represent the recapping of post-transcriptionally processed mRNAs, as the tags are considered too short to be generated by the splicing of mRNAs produced from alternative TSSs (Fejes-Toth et al., 2009; Le Hir, Gatfield, Izaurralde, & Moore, 2001; Le,H., Izaurralde, Maquat, & Moore, 2000) and do not coincide with the active chromatin marks or Pol II occupancy that would be expected of alternative transcription initiation (Mercer et al., 2010; Mercer et al., 2011). Subsequent analysis concluded that ~72% of human CAGE tags map to actual TSSs, leaving the remaining 30% as possible recapping sites (Djebali et al., 2012). A ligation-based approach identified locations of uncapped 5′ ends in recapping target mRNAs that mapped either exactly to or in the vicinity of downstream CAGE tags (Kiss, Oman, Bundschuh, & Schoenberg, 2015), supporting a role for cytoplasmic recapping in generating novel capped ends. CAGE-based sequencing of vaccinia virus transcripts similarly found many capped 5′ ends that mapped within positive-sense coding sequences (CDSs) (Yang, Bruno, Martens, Porcella, & Moss, 2011). Recapping of these processed ends could potentially be catalyzed by either viral or host enzymes in the cytoplasm.
Figure 3.
Diversification of the transcriptome and proteome by recapping of 5′-end-processed RNAs.A. Possible avenues for the generation of 5′-end-processed RNA recapping substrates for recapping. New 5′ ends can be created by action of endoribonucleases (1) or by inhibition of XRN1 5′-exoribonuclease activity by strong secondary structure (2) or RNA-binding proteins (3). B. Consequences of recapping at downstream sites. A full-length mRNA with a cap at its canonical transcription start site is shown as (1). Recapping of an mRNA with a shortened 5′ UTR (2) can remove regulatory elements. Recapping within the CDS (3) may facilitate translation from downstream initiation codons, producing N-terminally truncated proteins. Noncoding RNAs with regulatory potential can be generated by recapping within the CDS downstream of alternative translation initiation codons (4) or recapping within the 3’ UTR (5).
Downstream capped ends have also been documented in Drosophila. Based on promoter motifs and chromatin immunoprecipitation sequencing data for the transcription factors TBP and TRF2, it is unlikely that these are generated by initiation at alternative TSSs (Hoskins et al., 2011; Ni et al., 2010). Targeted validation by two independent methods, cap-trapping and oligo-capping, confirmed that 10 out of 12 candidate downstream sites indeed represented capped RNAs (Ni et al., 2010). Uncapped mRNAs are also abundant in Drosophila (Machida & Lin, 2014), further pointing to cap homeostasis as a process affecting the Drosophila transcriptome.
Recapping of 5′-end-processed RNAs has implications for diversification of the transcriptome and proteome through the generation of capped transcripts with shortened 5′ untranslated regions (UTRs), shortened CDSs, and noncoding RNAs consisting primarily of 3′ UTR sequences (Figure 3B). Recapping an mRNA with a shortened 5′ UTR would leave the CDS-encoded protein unchanged but could alter its translational regulation by removing structural elements, sites of regulatory protein binding, and/or upstream open reading frames (uORFs) (Leppek, Das, & Barna, 2018; Sendoel et al., 2017; Tamarkin-Ben-Harush, Vasseur, Debart, Ulitsky, & Dikstein, 2017; Hinnebusch, Ivanov, & Sonenberg, 2016). Recapped RNAs with 5′-end-shortened CDSs could presumably be translated from downstream initiation codons to produce N-terminally truncated protein products. Detection of proteoforms corresponding to different transcript isoforms has been hampered by limits in coverage and sensitivity of proteomic methods (Aebersold et al., 2018). Nonetheless, ribosome profiling and N-terminal proteomics studies have provided ample evidence for translation initiating at downstream sites (Kim et al., 2014; Van Damme, Gawron, Van Criekinge, & Menschaert, 2014; Na et al., 2018; Menschaert et al., 2013), with downstream translation initiation estimated to occur on a quarter of human transcripts (Lee et al., 2012). Specific examples of this phenomenon include nonsense-containing CCHCR1 and ABHD14B mRNAs (Jagannathan & Bradley, 2016) and EIF4A1 mRNA with capped ends downstream of the canonical initiation codon (Tamarkin-Ben-Harush et al., 2017). A combination of ribosome profiling and positional proteomics with modulation of cytoplasmic capping will be necessary to determine if recapping of 5′-end-processed mRNAs contributes to translation initiating downstream of canonical start sites. Finally, recapping downstream of initiation codons within the CDS or within the 3’ UTR could produce noncoding RNAs that may regulate the physiology of other mRNAs by acting as “sponges” that sequester miRNAs, RNA-binding proteins, and other trans-acting factors (Mayr, 2017; Mercer et al., 2011).
Biological significance of recapping
The overall biological significance of cytoplasmic recapping remains an open question. Temporal regulation of translation and RNA stability through cap homeostasis may serve important roles in the cell cycle and in the cellular response to stress. Gene ontology (GO) analysis of recapping targets revealed that the “capping-inhibited” class of transcripts was enriched for proteins involved in mitotic cell cycle control (Mukherjee et al., 2012). This is consistent with more recent reports suggesting some level of coordination between cap synthesis and the cell cycle (Aregger & Cowling, 2017; Aregger et al., 2016). Another clue comes from studies characterizing the integrated stress response. In general, cap-dependent translation is repressed by cellular stresses such as endoplasmic reticulum (ER) stress, oxidative stress, and heat shock. Each of these stressors also results in the sequestration of translationally repressed mRNPs in stress granules (SGs (Sheinberger & Shav-Tal, 2017)), which may functionally intersect with translational repression by P-bodies (Stoecklin & Kedersha, 2013; Hubstenberger et al., 2017). The cap status of mRNAs in these granules is unknown, but several observations suggest a possible link between cytoplasmic recapping and the ability of cells to recover from stress. Recovery from arsenite-induced oxidative stress was reduced when cytoplasmic recapping was blocked (Otsuka et al., 2009). A recent study found that only a portion of the transcriptome is sequestered in cytoplasmic RNP granules of stressed cells (Namkoong, Ho, Woo, Kwak, & Lee, 2018). These mRNAs had similar GO terms as mRNAs that were identified in (Mukherjee et al., 2012) as cytoplasmic recapping targets. Of potentially greater significance, results of both studies identified a particular enrichment for mRNAs with AU-rich motifs in their 3’ UTRs. Thus, one role for cytoplasmic recapping may be to facilitate the post-stress resumption of cap-dependent translation.
A recent paper on the Hedgehog signaling pathway raised the fascinating possibility of a role for RNA recapping in animal development (Chen, Zhu, & Sun, 2017a). An siRNA screen of ~7000 genes conserved between Drosophila and mammals found that RNGTT is required for Drosophila wing development. Importantly, wing defects caused by loss of RNGTT were rescued by expressing wild-type RNGTT in a form restricted to the cytoplasm. However, wing defects were not rescued with the same active-site mutant (K294A) used to inhibit cytoplasmic capping in mammalian cells. Overexpression of a cytoplasmic form of catalytically active RNGTT increased hedgehog signaling in mouse 3T3 cells, and hedgehog signaling was reduced by overexpression of the K294A mutant. The dependence of these phenomena on the catalytic activity of cytoplasmic RNGTT is consistent with a role of cap homeostasis in regulating translation and/or mRNA decay during development. Hedgehog signaling is dysregulated in a wide range of human diseases, including cancer and neurodevelopmental disorders (Fattahi, Pilehchian Langroudi, & Akhavan-Niaki, 2018), and future studies into the involvement of cytoplasmic recapping may lead to novel therapeutic strategies.
Future prospects and concluding remarks
Research over the past decade has established RNA recapping as a novel mechanism of gene expression control; however, many questions remain unanswered. One of the biggest challenges is to define the rules for the selection of recapped sites. Such selectivity could be achieved through preferential activity of any of the recapping enzymes or by biases in decapping, exonuclease, and/or endonuclease activities. Sequence motifs provide possible insights into this selectivity. Motif analyses found only a slight preference for a guanosine at presumptive novel 5′ ends in mouse (Mercer et al., 2010), but in Drosophila, 59% of the most abundant signals in downstream capped read clusters mapped to genomic TC dinucleotides (Ni et al., 2010). This finding implies that recapping frequently occurs after processing events that target UC dinucleotides in fruit flies. It also remains unknown whether RNA secondary structure is important for recapping. XRN1 exonuclease activity is inhibited by strong secondary structures (Charley et al., 2018; Moon et al., 2015; Muhlrad, Decker, & Parker, 1994), so 5′-monophosphate ends at such structures may be preferentially stabilized and targeted for recapping. The presence of RNA-binding proteins could similarly cause XRN1 to stall (Figure 3A). Insight into any such rules could guide the development of reporter mRNAs as new tools to aid studies of recapping. Further, does cap homeostasis depend on or influence epitranscriptomic marks such as pseudouridine or N6-methyladenosine (m6A)? N6-methylation of adenosine at the cap-proximal nucleotide is one of the most prevalent internal modifications in the transcriptome, and its presence can modulate sensitivity to decapping by DCP2 (Mauer et al., 2016). This warrants investigating whether m6A influences the selection of RNAs for recapping.
Another major hurdle regarding mammalian recapping is elucidating the identity of the 5′-monophosphate RNA kinase. Is it a previously uncharacterized protein? Or does a known protein possess this as-of-yet uncharacterized activity? Additionally, considering the discovery that the trypanosome TbCE1 kinase domain is highly similar to the adenylate kinase family of proteins (Ignatochkina et al., 2015), nucleoside monophosphate kinases (NMPKs) are obvious candidates for investigation. The chemistry of any previously unrecognized 5′-monophosphate RNA kinase activity would presumably be analogous to NMPKs, and given the abundance of NMPK genes in mammals (nine different adenylate kinases, two different uridylate/cytidylate kinases, one guanylate kinase, and one thymidylate kinase (Panayiotou, Solaroli, & Karlsson, 2014)), it is conceivable that some NMPKs may act upon 5′-monophosphate RNA ends to generate diphosphate substrates for the guanylyltransferase activity of RNGTT. Methods capable of detecting 5′-diphosphate ends have recently been developed (Ettwiller, Buswell, Yigit, & Schildkraut, 2016; Luciano, Vasilyev, Richards, Serganov, & Belasco, 2017) and have shown that 5′-diphosphate mRNAs are surprisingly common in bacteria. Do the transcriptomes of eukaryotic species contain prevalent 5′-diphosphate RNAs as well?
Accumulating evidence suggests that recapping may target diverse species of RNA other than mRNA. m7G caps have been observed on tRNA precursors (Ohira & Suzuki, 2016), and because these RNAs are synthesized by Pol III instead of Pol II, it appears unlikely that pre-tRNA capping occurs through the canonical co-transcriptional pathway. Capped pre-tRNAs in yeast were most prevalent on those that had undergone splicing, and because tRNA splicing occurs on the cytoplasmic surface of mitochondria in yeast, it will be interesting to learn where this pre-tRNA capping occurs. Human pre-tRNAs can also undergo capping, as demonstrated by recovery with anti-cap antibody and the mapping of CAGE tags to the 5′ regions of mammalian tRNA genes (Ohira & Suzuki, 2016). Furthermore, analyses of RNAs shorter than 200 nucleotides have uncovered the existence of short, capped RNAs that appear to be derived from post-transcriptional processing of tRNAs, mature mRNAs, and snoRNAs (Abdelhamid et al., 2014; Fejes-Toth et al., 2009; Mercer et al., 2010). In C. elegans, caps have also been reported on snoRNAs and pre-miRNAs processed from larger primary transcripts (Chen et al., 2017a; Xiao et al., 2012). It is thus likely that the cytoplasmic capping machinery plays a role in the metabolism of RNAs beyond those with protein-coding potential.
Another outstanding question is whether the caps added in the cytoplasm are distinguishable from those added in the nucleus. Cap 1 and cap 2 ribose methylations would be preserved following recapping at canonical 5′ ends but could potentially be absent upon recapping at downstream sites. The cap 1 ribose methyltransferase CMTR1 is mostly nuclear and binds to the C-terminal repeat domain of Pol II, suggesting that cap 1 methylation is co-transcriptional and restricted to the nucleus (Belanger et al., 2010; Haline-Vaz, Silva, & Zanchin, 2008). In the absence of ribose methylation, caps added at downstream sites may targeted for translational repression by IFIT proteins, since these bind cap 0 more tightly than cap 1 (Johnson et al., 2018; Kumar et al., 2014). Such regulation could be further mediated by IFIT2, which binds AU-rich elements that are prevalent in recapping target RNAs (Fensterl & Sen, 2015; Fensterl & Sen, 2015; Mukherjee et al., 2012; Yang et al., 2012). Cap 2 methyltransferase CMTR2 is present in the nucleus and cytoplasm and is capable of modifying both cap 0 and cap 1 (Werner et al., 2011). The activity of this enzyme is greater in cytoplasmic extracts than in nuclear extracts of HeLa cells (Langberg & Moss, 1981), making it conceivable that CMTR2 catalyzes ribose methylation of recapped RNAs.
Also to be addressed is the relationship of cytoplasmic capping to proteome complexity. As noted above there is a large body of evidence for downstream capped ends and a growing body of proteomic evidence for alternative N-termini. These need to be reconciled, and the most straightforward way will be through ribosome profiling and N-terminal proteomics of matching samples from cells with normal cytoplasmic capping and cells in which this process is blocked. Another open question is whether RNA recapping is a regulated process. NCK1 is an adapter protein that transduces signaling from receptor tyrosine kinases to other cellular moieties, such as the actin cytoskeleton (Fawcett et al., 2007). The identification of NCK1 as a scaffold for assembling the cytoplasmic capping complex raises the possibility that recapping might be modulated by the binding of tyrosine phosphorylated protein(s) to the C-terminal SH2 domain or by binding of a proline-rich motif to the first SH3 domain (Figure 1A). Cap methylation by RNMT-RAMAC is a regulated process (Cowling, 2010), and cytoplasmic cap methylation could be similarly subject to the same regulatory events as nuclear cap methylation. Cap methylation is also an attractive target for expanding the study of cytoplasmic capping to more cell lines and to animal studies. In (Trotman et al., 2017) cytoplasmic cap methylation was blocked by overexpression of a catalytically-inactive form of RNMT. This construct lacks N-terminal nuclear localization sequences but retains the ability to bind to RNGTT and RAMAC, and its overexpression resulted in a significant decrease in the steady-state levels of known recapping targets without affecting the matching controls. This effect is most likely due to degradation by cytoplasmic cap surveillance enzymes that target RNAs with unmethylated caps (Song et al., 2013; Jiao et al., 2013; Grzela et al., 2018). This approach should make it possible to more broadly identify cytoplasmic capping targets simply by changes in their steady-state levels by RNA-Seq.
Finally, most of the work cited here used immortalized cell lines, and while there is still much to learn about the process of RNA recapping, the field is poised to move into addressing the larger questions of how this process relates to cellular and organismal function. For example, some viruses encode their own capping enzymes and “snatch” caps from host RNAs (Ferron, Decroly, Selisko, & Canard, 2012), but do any RNA viruses hijack the cytoplasmic capping machinery (Snijder, Decroly, & Ziebuhr, 2016)? The identity of such a process could make recapping a therapeutic target. What role, if any, does RNA recapping play in development? One can envision a role for regulated downstream recapping in generating protein isoforms with specific functional domains that appear at some stages but not at others. Similarly, does cytoplasmic recapping play a role in initiation or progress of cancer? Bioinformatics analysis of cytoplasmic capping targets yielded numerous examples for initiation downstream of internal CAGE tags that could result in expression of proteins lacking an N-terminal regulatory domain. There already is good evidence for a link between RNA granules (e.g., stress granules), RNA recapping, and the ability of cells to recover from stress. RNA granules feature prominently in a number of neurodegenerative diseases such as ALS (Ito, Hatano, & Suzuki, 2017), and nothing is known about if and/or how recapping might play into this. Lastly, is recapping a strategy used by neurons to control localized translation (Rangaraju, Tom Dieck, & Schuman, 2017; Kapur, Monaghan, & Ackerman, 2017), particularly around synapses? With so much left to discover, it is our hope that insights from these and other fields will continue to reveal new understanding as to how cytoplasmic recapping impacts RNA biology.
Acknowledgments
This work was supported by the National Institutes of Health grant GM084177 to DRS. JBT was supported by National Institutes of Health training grant GM08512 and a predoctoral fellowship from The Ohio State University Center for RNA Biology. The content is solely the responsibility of the authors and does not necessarily represent the official views of The Ohio State University or the National Institutes of Health.
Contributor Information
Jackson B. Trotman, Department of Biological Chemistry and Pharmacology, Center for RNA Biology, The Ohio State University, Columbus, OH 43210, jbtrot@gmail.com.
*Daniel R. Schoenberg, Department of Biological Chemistry and Pharmacology, Center for RNA Biology, The Ohio State University, Columbus, OH 43210, schoenberg, 3@osu.edu.
References
- Abdelhamid RF, Plessy C, Yamauchi Y, Taoka M, de Hoon M, Gingeras TR, . . . Carninci P (2014). Multiplicity of 5’ cap structures present on short RNAs. PLoS One, 9(7), e102895. doi:10.1371/journal.pone.0102895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, . . . Zhang B (2018). How many human proteoforms are there. Nat Chem Biol, 14(3), 206–214. doi:10.1038/nchembio.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afik S, Bartok O, Artyomov MN, Shishkin AA, Kadri S, Hanan M, . . . Kadener S (2017). Defining the 5΄ and 3΄ landscape of the Drosophila transcriptome with Exo-seq and RNaseH-seq. Nucleic Acids Res, 45(11), e95. doi:10.1093/nar/gkx133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aregger M, & Cowling VH (2017). Regulation of mRNA capping in the cell cycle. RNA Biol, 14(1), 11–14. doi:10.1080/15476286.2016.1251540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aregger M, Kaskar A, Varshney D, Fernandez-Sanchez ME, Inesta-Vaquera FA, Weidlich S, & Cowling VH (2016). CDK1-cyclin B1 activates RNMT, coordinating mRNA cap methylation with G1 phase transcription. Mol. Cell, 61(5), 734–746. doi:10.1016/j.molcel.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, & Gingeras TR (2013). High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res, 23(1), 169–180. doi:10.1101/gr.139618.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger F, Stepinski J, Darzynkiewicz E, & Pelletier J (2010). Characterization of hMTr1, a human cap1 2’O-ribose methyltransferase. J. Biol. Chem doi:10.1074/jbc.M110.155283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch D, Smith P, & Shuman S (2008). Characterization of a trifunctional mimivirus mRNA capping enzyme and crystal structure of the RNA triphosphatase domain. Structure, 16(4), 501–512. doi:10.1016/j.str.2008.01.009 [DOI] [PubMed] [Google Scholar]
- Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, & Cerione RA (2002). Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol, 9(12), 912–917. doi:10.1038/nsb874 [DOI] [PubMed] [Google Scholar]
- Cartolano M, Huettel B, Hartwig B, Reinhardt R, & Schneeberger K (2016). cDNA library enrichment of full length transcripts for SMRT long read sequencing. PLoS One, 11(6), e0157779. doi:10.1371/journal.pone.0157779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapat C, Jafarnejad SM, Matta-Camacho E, Hesketh GG, Gelbart IA, Attig J, . . . Sonenberg N (2017). Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc Natl Acad Sci U S A, 114(21), 5425–5430. doi:10.1073/pnas.1701488114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charley PA, Wilusz CJ, & Wilusz J (2018). Identification of phlebovirus and arenavirus RNA sequences that stall and repress the exoribonuclease XRN1. J Biol Chem, 293(1), 285–295. doi:10.1074/jbc.M117.805796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhu D, & Sun Y (2017a). Cap-seq reveals complicated miRNA transcriptional mechanisms in C elegans and mouse. Quantitative Biology. doi:10.1007/s40484-017-0123-4 [Google Scholar]
- Chen P, Zhou Z, Yao X, Pang S, Liu M, Jiang W, . . . Zhang Q (2017b). Capping enzyme mRNA-cap/RNGTT regulates Hedgehog pathway activity by antagonizing protein kinase A. Sci Rep, 7(1), 2891. doi:10.1038/s41598-017-03165-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH (2010). Regulation of mRNA cap methylation. Biochem J, 425(2), 295–302. doi:10.1042/BJ20091352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, . . . Gingeras TR (2012). Landscape of transcription in human cells. Nature, 489(7414), 101–108. doi:10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwiller L, Buswell J, Yigit E, & Schildkraut I (2016). A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome. BMC Genomics, 17, 199. doi:10.1186/s12864-016-2539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattahi S, Pilehchian Langroudi M, & Akhavan-Niaki H (2018). Hedgehog signaling pathway: Epigenetic regulation and role in disease and cancer development. J Cell Physiol. doi:10.1002/jcp.26506 [DOI] [PubMed] [Google Scholar]
- Fawcett JP, Georgiou J, Ruston J, Bladt F, Sherman A, Warner N, . . . Pawson T (2007). Nck adaptor proteins control the organization of neuronal circuits important for walking. Proc. Natl. Acad. Sci USA, 104(52), 20973–20978. doi:10.1073/pnas.0710316105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, . . . Gingeras TR (2009). Post-transcriptional processing generates a diversity of 5’-modified long and short RNAs. Nature, 457, 1028–1032. doi:10.1038/nature07759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterl V, & Sen GC (2015). Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol, 89(5), 2462–2468. doi:10.1128/JVI.02744-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Decroly E, Selisko B, & Canard B (2012). The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res, 96(1), 21–31. doi:10.1016/j.antiviral.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, & Gilmartin GM (1997). Participation of the nuclear cap binding complex in pre-mRNA 3’ processing. Proc.Natl.Acad.Sci.USA PNAS, 94(22), 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire ER, Malvezzi AM, Vashisht AA, Zuberek J, Saada EA, Langousis G, . . . Campbell DA (2014). Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot Cell, 13(7), 896–908. doi:10.1128/EC.00071-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire ER, Sturm NR, Campbell DA, & de Melo Neto OP (2017). The role of cytoplasmic mRNA cap-binding protein complexes in Trypanosoma brucei and other Trypanosomatids. Pathogens, 6(4). doi:10.3390/pathogens6040055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, & Esteban R (2010). Yeast double-stranded RNA virus L-A deliberately synthesizes RNA transcripts with 5’-diphosphate. J Biol Chem, 285(30), 22911–22918. doi:10.1074/jbc.M110.138982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y (2015). Discovery of m7G-cap in eukaryotic mRNAs. Proc Jpn Acad Ser B Phys Biol Sci, 91(8), 394–409. doi:10.2183/pjab.91.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Muthukrishnan S, & Shatkin AJ (1975). Reovirus messenger RNA contains a methylated, blocked 5’-terminal structure: m-7G(5’)ppp(5’)G-MpCp-. Proc Natl Acad Sci U S A, 72(1), 362–366. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/1054511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, & Darnell JE (1975). Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A, 72(5), 1904–1908. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/1057180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, & Lima CD (2010). Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA, 1(1), 152–172. doi:10.1002/wrna.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM, McDevitt MA, & Nevins JR (1988). Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev, 2(5), 578–587. [DOI] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, & Cowling VH (2014). Cap-binding complex (CBC). Biochem J, 457(2), 231–242. doi:10.1042/BJ20131214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Dunn S, Bounds R, & Cowling VH (2011). RAM/Fam103a1 is required for mRNA cap methylation. Mol. Cell, 44(4), 585–596. doi:10.1016/j.molcel.2011.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien-Nogalska E, & Kiledjian M (2017). New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip Rev. RNA, 8(1). doi:10.1002/wrna.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzela R, Nasilowska K, Lukaszewicz M, Tyras M, Stepinski J, Jankowska-Anyszka M, . . . Darzynkiewicz E (2018). Hydrolytic activity of human Nudt16 enzyme towards dinucleotide cap analogs and short capped oligonucleotides. RNA. doi:10.1261/rna.065698.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte DJ, & Mello CC (2012). CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell, 151(7), 1488–1500. doi:10.1016/j.cell.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haline-Vaz T, Silva TC, & Zanchin NI (2008). The human interferon-regulated ISG95 protein interacts with RNA polymerase II and shows methyltransferase activity. Biochem Biophys Res Commun, 372(4), 719–724. doi:10.1016/j.bbrc.2008.05.137 [DOI] [PubMed] [Google Scholar]
- Hall MP, & Ho CK (2006). Characterization of a Trypanosoma brucei RNA cap (guanine N-7) methyltransferase. RNA, 12(3), 488–497. doi:10.1261/rna.2250606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, & Sonenberg N (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science, 352(6292), 1413–1416. doi:10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Sriskanda V, McCracken S, Bentley D, Schwer B, & Shuman S (1998). The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem, 273(16), 9577–9585. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9545288 [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, . . . Celniker SE (2011). Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res, 21(2), 182–192. doi:10.1101/gr.112466.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, & Maquat LE (2005). CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol, 12, 893–901. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Bénard M, Souquere S, Ernoult-Lange M, Chouaib R, . . . Weil D (2017). P-Body purification reveals the condensation of repressed mRNA regulons. Mol Cell, 68(1), 144–157. e5. doi:10.1016/j.molcel.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Hyde JL, & Diamond MS (2015). Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation. Virology, 479–480, 66–74. doi:10.1016/j.virol.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatochkina AV, Takagi Y, Liu Y, Nagata K, & Ho CK (2015). The messenger RNA decapping and recapping pathway in Trypanosoma. Proc. Nat.l Acad. Sci. USA, 112(22), 6967–6972. doi:10.1073/pnas.1424909112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito D, Hatano M, & Suzuki N (2017). RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Sci Transl Med, 9(415). doi:10.1126/scitranslmed.aah5436 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, & Ingolia NT (2016). PROTEIN TRANSLATION. Seeing translation. Science, 352(6292), 1391–1392. doi:10.1126/science.aag1039 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, & Mattaj IW (1994). A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell, 78(4), 657–668. [DOI] [PubMed] [Google Scholar]
- Jagannathan S, & Bradley RK (2016). Translational plasticity facilitates the accumulation of nonsense genetic variants in the human population. Genome Res, 26(12), 1639–1650. doi:10.1101/gr.205070.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, & Mattaj IW (1994). Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol, 124(5), 627–635. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7509815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Doamekpor SK, Bird JG, Nickels BE, Tong L, Hart RP, & Kiledjian M (2017). 5’ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell, 168(6), 1015–1027.e10. doi:10.1016/j.cell.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chang JH, Kilic T, Tong L, & Kiledjian M (2013). A mammalian pre-mRNA 5’ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell, 50, 104–115. doi:10.1016/j.molcel.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, & Kiledjian M (2010). Identification of a quality-control mechanism for mRNA 5’-end capping. Nature, 467(7315), 608–611. doi:10.1038/nature09338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B, VanBlargan LA, Xu W, White JP, Shan C, Shi PY, . . . Amarasinghe GK (2018). Human IFIT3 modulates IFIT1 RNA binding specificity and protein stability. Immunity, 48(3), 487–499.e5. doi:10.1016/j.immuni.2018.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE, & Ackerman SL (2017). Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron, 96(3), 616–637. doi:10.1016/j.neuron.2017.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M (2018). Eukaryotic RNA 5’-End NAD+ Capping and DeNADding. Trends Cell Biol, 28, 454–464. doi:10.1016/j.tcb.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, . . . Pandey A (2014). A draft map of the human proteome. Nature, 509(7502), 575–581. doi:10.1038/nature13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Oman K, Bundschuh R, & Schoenberg DR (2015). Uncapped 5’ ends of mRNAs targeted by cytoplasmic capping map to the vicinity of downstream CAGE tags. Febs Lett, 589, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Oman KM, Dougherty JA, Mukherjee C, Bundschuh R, & Schoenberg DR (2016). Cap homeostasis is independent of poly(A) tail length. Nucleic Acids Res, 44(1), 304–314. doi:10.1093/nar/gkv1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S (2017). The ApaH-like phosphatase TbALPH1 is the major mRNA decapping enzyme of trypanosomes. PLoS Pathog, 13(6), e1006456. doi:10.1371/journal.ppat.1006456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge H, Brownlee GG, Gershon PD, & Richter JD (1998). Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res, 26(13), 3208–3214. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9628920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Sweeney TR, Skabkin MA, Skabkina OV, Hellen CU, & Pestova TV (2014). Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5’-terminal regions of cap0-, cap1- and 5’ppp- mRNAs. Nucleic Acids Res, 42(5), 3228–3245. doi:10.1093/nar/gkt1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrieleis OJ, Chang J, de la Peña M, Shuman S, & Cusack S (2014). Crystal structure of vaccinia virus mRNA capping enzyme provides insights into the mechanism and evolution of the capping apparatus. Structure, 22(3), 452–465. doi:10.1016/j.str.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr RM, Fonseca BD, Ciotti GE, Al-Ashtal HA, Jia JJ, Niklaus MR, . . . Berman AJ (2017). La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife, 6. doi:10.7554/eLife.24146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langberg SR, & Moss B (1981). Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2’-)-methyltransferases from HeLa cells. J Biol Chem, 256(19), 10054–10060. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/7275966 [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, & Moore MJ (2001). The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J, 20(17), 4987–4997. doi:10.1093/emboj/20.17.4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H,H, Izaurralde E, Maquat LE, & Moore MJ (2000). The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. The EMBO Journal EMBO J., 19(24), 6860–6869. Retrieved from http://wwwembojorg/cgi/content/full/19/24/6860URLS_ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, & Cate JH (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature, 536, 96–99. doi:10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, & Qian SB (2012). Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A, 109(37), E2424–32. doi:10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, & Barna M (2018). Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol, 19(3), 158–174. doi:10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Mullins JJ, Chen CM, Gross KW, & Maquat LE (1989). Novel metabolism of several βo_-thalassemic β-globin mRNAs in the erythroid tissues of transgenic mice. EMBO J, 8(9), 2613–2619. Retrieved from PM:2573525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, & Maquat LE (1992). Human beta-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5’ termini. EMBO J, 11, 3271–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Sigmund CD, Gross KW, & Maquat LE (1992). Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol. Cell. Biol, 12, 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Vasilyev N, Richards J, Serganov A, & Belasco JG (2017). A novel RNA phosphorylation state enables 5’ end-dependent degradation in Escherichia coli. Mol Cell, 67(1), 44–54.e6. doi:10.1016/j.molcel.2017.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida RJ, & Lin YY (2014). Four methods of preparing mRNA 5’ end libraries using the Illumina sequencing platform. PLoS One, 9(7), e101812. doi:10.1371/journal.pone.0101812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, & Burley SK (1997). Cocrystal structure of the messenger RNA 5’ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell, 89, 951–961. [DOI] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJ, Hemann M, Herzog F, . . . Cramer P (2015). Molecular basis of transcription-coupled pre-mRNA capping. Mol Cell, 58(6), 1079–1089. doi:10.1016/j.molcel.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Mascarenhas R, Dougherty JA, & Schoenberg DR (2013). SMG6 cleavage generates metastable decay intermediates from nonsense-containing β-globin mRNA. PLoS One, 8, e74791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, . . . Jaffrey SR (2016). Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature, 371–375. doi:10.1038/nature21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C (2017). Regulation by 3’-Untranslated Regions. Annu Rev Genet, 51, 171–194. doi:10.1146/annurev-genet-120116-024704 [DOI] [PubMed] [Google Scholar]
- Menschaert G, Van Criekinge W, Notelaers T, Koch A, Crappé J, Gevaert K, & Van Damme P (2013). Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol Cell Proteomics, 12(7), 1780–1790. doi:10.1074/mcp.M113.027540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, . . . Mattick JS (2010). Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res, 20(12), 1639–1650. doi:10.1101/gr.112128.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Wilhelm D, Dinger ME, Soldà G, Korbie DJ, Glazov EA, . . . Mattick JS (2011). Expression of distinct RNAs from 3’ untranslated regions. Nucleic Acids Res, 39(6), 2393–2403. doi:10.1093/nar/gkq1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJ, Keene JD, . . . Wilusz J (2015). XRN1 stalling in the 5’ UTR of Hepatitis C virus and Bovine Viral Diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog, 11(3), e1004708. doi:10.1371/journal.ppat.1004708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B (2017). Investigating Viruses during the Transformation of Molecular Biology. J Biol Chem, 292(10), 3958–3969. doi:10.1074/jbc.X117.778712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, & Parker R (1994). Deadenylation of the unstable mRNA encoded by the yeast mfa2 gene leads to decapping followed by 5’->3’ digestion of the transcript. Genes and Development Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Mukherjee C, Bakthavachalu B, & Schoenberg DR (2014). The cytoplasmic capping complex assembles on adapter protein NCK1 bound to the proline-rich C-terminus of mammalian capping enzyme. PLoS Biol, 12(8), e1001933. doi:10.1371/journal.pbio.1001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, & Schoenberg DR (2012). Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep, 2, 674–684. doi:10.1016/j.celrep.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na CH, Barbhuiya MA, Kim MS, Verbruggen S, Eacker SM, Pletnikova O, . . . Pandey A (2018). Discovery of noncanonical translation initiation sites through mass spectrometric analysis of protein N termini. Genome Res, 28(1), 25–36. doi:10.1101/gr.226050.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, & Lee JH (2018). systematic characterization of stress-induced RNA granulation. Mol Cell, 70(1), 175–187. e8. doi:10.1016/j.molcel.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, . . . Zhu J (2010). A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods, 7(7), 521–527. doi:10.1038/nmeth.1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, . . . Stolarski R (2002). Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5’ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol, 319(3), 615–635. doi:10.1016/S0022-2836(02)00328-5 [DOI] [PubMed] [Google Scholar]
- Ohira T, & Suzuki T (2016). Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat. Chem. Biol, 12, 648–655. doi:10.1038/nchembio.2117 [DOI] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, & Schoenberg DR (2009). Identification of a cytoplasmic complex that adds a cap onto 5’-monophosphate RNA. Mol. Cell. Biol, 29, 2155–2167. doi:10.1128/MCB.01325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayiotou C, Solaroli N, & Karlsson A (2014). The many isoforms of human adenylate kinases. Int J Biochem Cell Biol, 49, 75–83. doi:10.1016/j.biocel.2014.01.014 [DOI] [PubMed] [Google Scholar]
- Park JE, Yi H, Kim Y, Chang H, & Kim VN (2016). Regulation of poly(A) tail and translation during the somatic cell cycle. Mol Cell, 62(3), 462–471. doi:10.1016/j.molcel.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Philippe L, Vasseur JJ, Debart F, & Thoreen CC (2018). La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res, 46(3), 1457–1469. doi:10.1093/nar/gkx1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillutla RC, Yue Z, Maldonado E, & Shatkin AJ (1998). Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem, 273(34), 21443–21446. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=9705270 [DOI] [PubMed] [Google Scholar]
- Ramanathan A, Robb GB, & Chan SH (2016). mRNA capping: biological functions and applications. Nucleic Acids Res, 7511–7526. doi:10.1093/nar/gkw551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, Tom Dieck S, & Schuman EM (2017). Local translation in neuronal compartments: how local is local. EMBO Rep, 18(5), 693–711. doi:10.15252/embr.201744045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SA, Foley PL, Jeong DH, Rymarquis LA, Doyle F, Tenenbaum SA, . . . Green PJ (2015). Identification of SMG6 cleavage sites and a preferred RNA cleavage motif by global analysis of endogenous NMD targets in human cells. Nucleic Acids Res, 43(1), 309–323. doi:10.1093/nar/gku1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, & Maquat LE (2009). Re-capping the message. Trends Biochem. Sci, 34(9), 435–442. doi:10.1016/j.tibs.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, & Maquat LE (2012). Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet, 13(4), 246–259. doi:10.1038/nrg3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner AM, Zillinger T, Goldeck M, . . . Schlee M (2015). A Conserved Histidine in the RNA Sensor RIG-I Controls Immune Tolerance to N1–2’O-Methylated Self RNA. Immunity, 43(1), 41–51. doi:10.1016/j.immuni.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, & Shuman S (1996). Conditional invactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA, 2, 574–583. [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, . . . Fuchs E (2017). Translation from unconventional 5’ start sites drives tumour initiation. Nature, 541(7638), 494–499. doi:10.1038/nature21036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinberger J, & Shav-Tal Y (2017). mRNPs meet stress granules. FEBS Lett, 591(17), 2534–2542. doi:10.1002/1873-3468.12765 [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, . . . Hayashizaki Y (2003). Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA, 100(26), 15776–15781. doi:10.1073/pnas.2136655100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Decroly E, & Ziebuhr J (2016). The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv Virus Res, 96, 59–126. doi:10.1016/bs.aivir.2016.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Li Y, & Kiledjian M (2010). Multiple mRNA decapping enzymes in mammalian cells. Mol. Cell, 40(3), 423–432. doi:10.1016/j.molcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Bail S, & Kiledjian M (2013). Multiple Nudix family proteins possess mRNA decapping activity. RNA, 19(3), 390–399. doi:10.1261/rna.037309.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer E, Loring D, Hurwitz J, & Monroy G (1978). Enzymatic conversion of 5’-phosphate-terminated RNA to 5’-di- and triphosphate-terminated RNA. Proc. Natl. Acad. Sci. USA, 75(10), 4793–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere PA (1987). Complexes of sequential metabolic enzymes. Annu. Rev. Biochem, 56, 89–124. doi:10.1146/annurev.bi.56.070187.000513 [DOI] [PubMed] [Google Scholar]
- Stevens A, Wang Y, Bremer K, Zhang J, Hoepfner R, Antoniou M, . . . Maquat LE (2002). Beta-globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc. Natl. Acad. Sci. USA, 99(20), 12741–12746. Retrieved from PM:12242335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, & Kedersha N (2013). Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol, 768, 197–211. doi:10.1007/978-1-4614-5107-5_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Sindkar S, Ekonomidis D, Hall MP, & Ho CK (2007). Trypanosoma brucei encodes a bifunctional capping enzyme essential for cap 4 formation on the spliced leader RNA. J Biol Chem, 282(22), 15995–16005. doi:10.1074/jbc.M701569200 [DOI] [PubMed] [Google Scholar]
- Tamarkin-Ben-Harush A, Vasseur JJ, Debart F, Ulitsky I, & Dikstein R (2017). Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. Elife, 6. doi:10.7554/eLife.21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, & Roux PP (2014). Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5’TOP mRNA translation. Genes Dev, 28(4), 357–371. doi:10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma C, Hasselblatt P, Kock J, Chang SF, Hockenjos B, Will H, . . . Offensperger WB (2001). Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligodeoxynucleotides. Mol. Cell, 8(4), 865–872. [DOI] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, . . . Lundberg E (2017). A subcellular map of the human proteome. Science, 356(6340). doi:10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Svitkin YV, Sonenberg N, & Shatkin AJ (2011). Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA, 2(2), 277–298. doi:10.1002/wrna.52 [DOI] [PubMed] [Google Scholar]
- Trcek T, Sato H, Singer RH, & Maquat LE (2013). Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev, 27, 541–551. doi:10.1101/gad.209635.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman JB, Giltmier AJ, Mukherjee C, & Schoenberg DR (2017). RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res, 45(18), 10726–10739. doi:10.1093/nar/gkx801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Gawron D, Van Criekinge W, & Menschaert G (2014). N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men. Mol Cell Proteomics, 13(5), 1245–1261. doi:10.1074/mcp.M113.036442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, & Mattaj IW (1996). A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J.Cell.Biol, 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, & Moss B (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell, 4, 379–386. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/164293 [DOI] [PubMed] [Google Scholar]
- Wei CM, & Moss B (1975). Methylated nucleotides block 5’-terminus of vaccinia virus messenger RNA. Proc Natl Acad Sci U S A, 72(1), 318–322. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/164018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Yue Z, & Shatkin AJ (1998). Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc. Natl. Acad. Sci. USA, 95(21), 12226–12231. Retrieved from PM:9770468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Purta E, Kaminska KH, Cymerman IA, Campbell DA, Mittra B, . . . Bujnicki JM (2011). 2’-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res, 39(11), 4756–4768. doi:10.1093/nar/gkr038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, & Singer RH (2016). Translation dynamics of single mRNAs in live cells and neurons. Science, 352(6292), 1430–1435. doi:10.1126/science.aaf1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Wang Y, Luo H, Liu L, Wei G, Chen X, . . . Chen R (2012). A differential sequencing-based analysis of the C. elegans noncoding transcriptome. RNA, 18(4), 626–639. doi:10.1261/rna.030965.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, & Tanenbaum ME (2016). Dynamics of Translation of Single mRNA Molecules In Vivo. Cell, 165(4), 976–989. doi:10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Bruno DP, Martens CA, Porcella SF, & Moss B (2011). Genome-wide analysis of the 5’ and 3’ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J Virol, 85(12), 5897–5909. doi:10.1128/JVI.00428-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Liang H, Zhou Q, Li Y, Chen H, Ye W, . . . Liu Y (2012). Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res, 22(9), 1328–1338. doi:10.1038/cr.2012.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, & Shatkin AJ (1997). Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase. Proc. Natl. Acad. Sci. USA, 94(24), 12898–12903. Retrieved from http://www.pnas.org/cgi/content/abstract/94/24/12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, & Tian B (2014). Sizing up the poly(A) tail: insights from deep sequencing. Trends Biochem. Sci, 39, 255–257. doi:10.1016/j.tibs.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]