Abstract
The skeletogenic gene regulatory network (GRN) of sea urchins and other echinoderms is one of the most intensively studied transcriptional networks in any developing organism. As such, it serves as a pre-eminent model of GRN architecture and evolution. This review summarizes our current understanding of this developmental network. We describe in detail the most comprehensive model of the skeletogenic GRN, one developed for the euechinoid sea urchin Strongylocentrotus purpuratus, including its initial deployment by maternal inputs, its elaboration and stabilization through regulatory gene interactions, and its control of downstream effector genes that directly drive skeletal morphogenesis. We highlight recent comparative studies that have leveraged the euechinoid GRN model to examine the evolution of skeletogenic programs in diverse echinoderms, studies that have revealed both conserved and divergent features of skeletogenesis within the phylum. Lastly, we summarize the major insights that have emerged from analysis of the structure and evolution of the echinoderm skeletogenic GRN and identify key, unresolved questions as a guide for future work.
Keywords: development, evolution, gene regulatory network, transcriptional network, echinoderm, sea urchin, morphogenesis, skeletogenesis, primary mesenchyme
I. Introduction
The process by which a single cell gives rise to a multicellular organism is encoded in the genome (Peter and Davidson, 2015). A central challenge of biology is to explain how information contained in the genomic sequence (which is inherently one-dimensional in nature) is read out during embryogenesis, ultimately producing the three-dimensional anatomy characteristic of an organism. It is well established that although all cells in the embryo contain the same genome, they progressively acquire distinct properties by expressing different subsets of genes. Differential gene expression involves diverse regulatory mechanisms, but during metazoan development, transcriptional regulation plays a pivotal role (Spitz and Furlong, 2012; Andrey and Mundlos, 2017). Any comprehensive model of development must explain how distinct domains of differential gene transcription arise in the early embryo, how they become progressively refined, and how they control embryo anatomy.
Gene regulatory networks (GRNs) have emerged as a valuable tool for studying the genetic control and evolution of development (Levine and Davidson, 2005; Ettensohn, 2013; Peter and Davidson, 2015). At their core, GRNs represent interactions among regulatory genes (i.e., genes that encode transcription factors). The sum total of these interactions determines the regulatory state of a cell, which can be thought of as the ensemble of functional transcription factors present in the cell at a given time (Peter, 2017). GRNs are typically represented as wiring diagrams that describe functional interactions (which can be direct or indirect) among regulatory genes (Fig. 1). Any single representation cannot capture the dynamic nature of developmental GRNs but a series of embryonic stage-specific diagrams can do so to a first approximation. Because cell-cell signals play a critically important role in regulating embryonic cell fates and because they act to modulate cell regulatory states, genes associated with cell-cell signaling (i.e., genes encoding ligands, receptors, and signal transduction components) are also sometimes incorporated into GRNs. The acquisition of cell identities during development can be interpreted as the deployment of distinct GRNs in different cells or territories of the early embryo (Fig. 2).
Fig. 1-. A hypothetical developmental Gene Regulatory Network (GRN).

GRNs are typically depicted using circuit diagrams like the one shown here. Because GRNs are dynamic, such diagrams are either snapshots of network topology at a single developmental stage or time-averaged views (as here). In this simplified GRN, localized maternal inputs activate early regulatory (transcription factor-encoding) genes (Regulatory Genes A and B), which activate the expression of late regulatory genes (Regulatory Genes C-E). Positive and negative interactions among the regulatory genes in the network are depicted by arrows and bars, respectively. The sum total of these gene interactions determines the suite of transcription factors present in a cell at any particular stage of development (the cell “regulatory state”) and ultimately specifies the cellular phenotype. One of the consequences of the cell regulatory state is the activation of non-transcription factor-encoding (“effector”) genes that carry out cell type-specific developmental functions. These include signaling genes, which play critically important roles in developmental processes, and genes that regulate morphogenetic cell behaviors. Sets of effector genes often share regulatory controls (Effector Gene Sets 1 and 2). Bent arrows projecting from each gene symbol indicate transcription.
Fig. 2-. Establishment of cell identities via the deployment of distinct GRNs in territories of the early embryo.
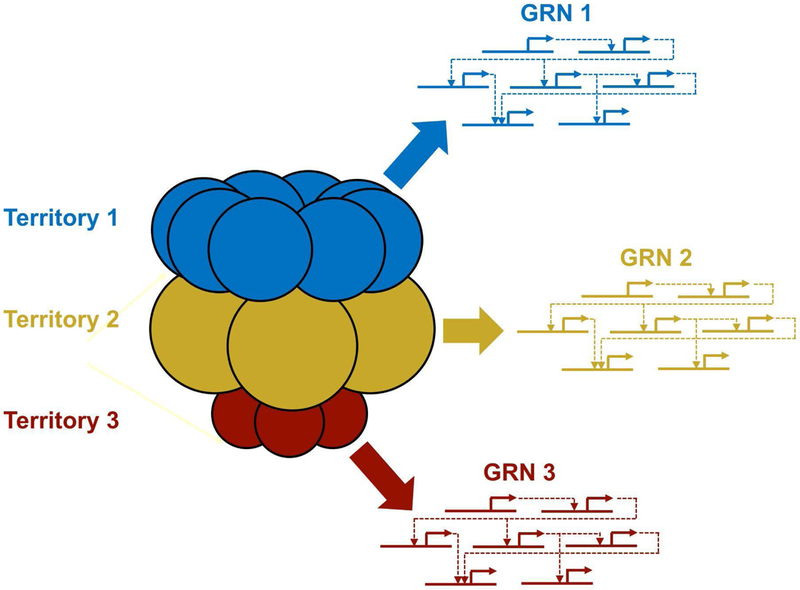
The early embryo can be thought of as comprising several multi-cellular territories (colored blue, gold, and red in this diagram). A different gene regulatory program (i.e., a distinct GRN) is deployed in each territory (colored arrows and circuit diagrams), endowing the constituent cells with a distinct identity.
The final readout of development is anatomy; therefore, regulatory networks have much greater explanatory power if they can be linked to effector genes that control the cellular processes that shape embryonic tissues (Lyons et al., 2012; Ettensohn, 2013). The cell-level properties that directly drive tissue morphogenesis (e.g., cell adhesion, shape, motility, proliferation, etc.) are regulated by effector genes that are controlled by the same transcriptional networks that specify cell identity. Therefore, the most comprehensive GRN models include linkages between regulatory genes and downstream genes that perform such morphogenetic functions. By means of such linkages, GRNs provide a logical framework for understanding how morphology is encoded in the genome; i.e., for explaining the connection between genotype and (morphological) phenotype.
The insight that developmental anatomy is controlled by GRNs provides a conceptual basis for examining changes in genome sequence that underlie the evolution of morphology. Comparative studies of GRN architecture across organisms can reveal conserved features, providing evidence of ancient regulatory systems or of homologous structures, as well as novel network circuitry that has arisen during the independent evolution of animal taxa (Rebeiz et al., 2015; Rebeiz and Tsiantis, 2017). By comparing the structure of GRNs in appropriate developmental model systems that span a range of evolutionary distances, it may be possible to reconstruct the evolutionary changes in the genome that have led to the appearance of new embryonic cell types, new embryonic structures, and new morphologies.
Sea urchins and other echinoderms are well-suited to gene regulatory network analysis, and detailed GRNs have been constructed for many territories of the early embryo (Smith, 2008; Ettensohn, 2009; Ben-Tabou and Davidson, 2009; Peter and Davidson, 2015; Arnone et al., 2016; Martik et al., 2016). Among these, the GRN specifying the embryonic skeleton is among the most comprehensive and, arguably, the most illuminating with respect to the evolution of development. Below, we first briefly introduce the process of skeleton formation in sea urchin embryos. Second, we examine in detail the architecture of the GRN that operates in embryonic skeletogenic cells. Lastly, we discuss recent studies that have leveraged this network to examine the evolution of skeletogenesis within echinoderms.
II. Skeletogenesis in Sea Urchins
All adult echinoderms have an endoskeleton composed of calcite, a crystalline form of calcium carbonate. The most prominent calcified structures of the adult are the test, spines, and structures associated with Aristotle’s lantern- the feeding apparatus of the animal (Veis, 2011; Stock, 2014). In most sea urchins (and in most echinoderms), the adult form arises through maximal, indirect development; i.e, through the metamorphosis of a free-swimming, feeding larva that has a morphology radically different from that of the adult. Indirect development is usually considered to be ancestral within echinoderms, and is certainly ancestral among echinoids, although direct development has arisen independently many times (Emlet, 1985; Raff, 1987; Smith, 1997).
For those echinoderms that produce a larval skeleton, including sea urchins, the skeleton establishes the angular shape of the larva and influences its swimming, orientation and feeding (Strathmann, 1971; Pennington and Strathmann, 1990; Hart and Strathmann, 1994; Strathman and Grunbaum, 2006). The elongated arms of the larva are supported by skeletal rods and decorated with ciliated cells that move food toward the mouth. Echinoderm larvae that have relatively long arms clear algae from the seawater more rapidly than do larvae with short arms (Strathman, 1971). In addition, sea urchin larvae regulate skeletal growth in response to food availability; they form relatively short arms when food is abundant and longer arms when food is scarce (Boidron-Metairon, 1988; Hart and Strathman, 1994; Miner, 2007). When food is abundant, dopamine-based signaling slows the growth of skeletal rods that support the larval arms (Adams et al., 2011).
In euechinoid sea urchins (the largest subclass), the embryonic founder cells of the skeletogenic lineage arise at the 32-cell stage, when four large micromeres form at the vegetal pole of the embryo (Fig. 3). These cells undergo several additional mitotic divisions and their descendants are transiently incorporated into the epithelial wall of the blastula. Later in development, the 32–64 large micromere descendants undergo a spectacular sequence of morphogenetic behaviors (see reviews by Ettensohn et al., 1997; Wilt and Ettensohn, 2007; Ettensohn, 2013; McIntyre et al., 2014). At the mesenchyme blastula stage, they undergo an epithelial-to-mesenchymal transition (EMT) and ingress into the blastocoel, after which they are referred to as primary mesenchyme cells (PMCs). During gastrulation, PMCs extend filopodia and migrate along a thin basal lamina that lines the blastocoel wall. Their migration is guided by ectoderm-derived cues that direct the cells to adopt a ring-like pattern near the equator of the embryo, within which two ventrolateral clusters of PMCs form. As the PMCs migrate, their filopodia fuse, creating a pseudopodial cable that joins the cells in a single, syncytial network. The calcite-based rods that form the skeleton are secreted within this pseudopodial cable, beginning with the deposition of a small, tri-radiate spicule rudiment in each ventrolateral PMC cluster at the mid-gastrula stage. The three radii of each spicule rudiment subsequently elongate and branch in a stereotypical manner (Okazaki, 1975a; Guss and Ettensohn, 1997), eventually producing the elaborate endoskeleton of the early pluteus larva. When the larva begins to feed, it has two pairs of elongated arms- the anterolateral and postoral arms.
Fig. 3-. Skeletogenesis in euechinoids.
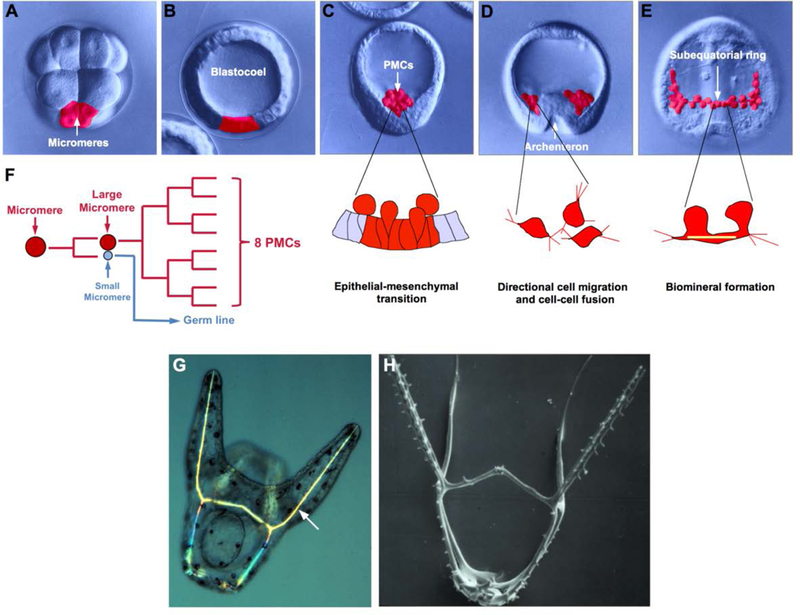
Panels A-E show living embryos (L. variegatus) viewed with differential interference contrast optics. Cells of the micromere-large micromere-PMC lineage are pseudo-colored red. A) 16-cell stage. B) Blastula. C) Mesenchyme blastula. D) Early gastrula. E) Late gastrula. Major PMC morphogenetic behaviors characteristic of specific developmental stages are also illustrated. F) PMC lineage. Each of the four micromeres divides unequally, giving rise to a large and small micromere. Each large micromere divides 3 times to produce 8 PMCs (in some species there is one additional round of division). G) Living, pre-feeding pluteus larva (L. variegatus) viewed with partially crossed polarizers. Skeletal rods (arrow) appear bright due to their birefringence. H) Biomineralized endoskeleton of a pre-feeding pluteus larva (Dendraster excentricus), with all cellular material removed.
While cells of the large micromere-PMC lineage construct the entire embryonic (pre-feeding) skeleton, many additional skeletal elements (the dorsal arch, posterodorsal rods, and preoral rods) arise after the larva begins to feed, and other mesodermal cells participate in the formation of these structures (Yajima et al., 2007). Eventually, the feeding larva undergoes metamorphosis, a major transformation during which the juvenile sea urchin emerges from a primordium known as the echinus rudiment. During this process, most larval structures (including most larval skeletal elements) are lost. During the late feeding stage, some future adult skeletal elements (e.g., certain genital and terminal plates that will be incorporated into the test on the aboral side of the animal) grow from the proximal tips of larval rods (Emlet, 1985; Gosselin and Jangoux, 1998). Most adult skeletal structures, however, arise de novo within the echinus rudiment, and are produced by fusogenic, mesenchymal cells of unknown lineage (Kniprath, 1974; Märkel et al., 1986).
III. The PMC Gene Regulatory Network of Sea Urchins
A). Cell-Autonomous Activation by Maternal Factors
Embryological studies, primarily involving micromere transplantation and recombination experiments, originally showed that the skeletogenic lineage is autonomously specified during early cleavage (see references in Ettensohn et al., 1997). Furthermore, when micromeres are isolated from 16-cell stage embryos and cultured in unsupplemented sea water, they divide, become motile, fuse, and sometimes produce small calcareous granules, although these never elongate (Okazaki 1975b; Hodor and Ettensohn, 1998). There has been no large-scale analysis of the gene expression program of isolated micromeres cultured under these conditions, but at least some biomineralization genes are activated cell-autonomously (Page and Benson, 1992), strongly suggesting that essential, early regulatory genes are as well.
The deployment of the PMC GRN in the micromere lineage is entrained by the molecular polarity of the unfertilized egg (Fig. 4). The signaling protein, Dishevelled (Dsh), becomes concentrated in puncta in the vegetal cortex of the egg during oogenesis (Weitzel et al. 2004; Peng and Wikramanayake, 2013). Dsh localization requires N-terminal motifs that have been shown in other cell types to mediate homo-oligomerization, suggesting that the puncta in the oocyte are super-assemblies of Dsh protein (Leonard and Ettensohn, 2007; Bienz, 2014). Mis-expression studies have shown that Dsh is not only concentrated in the vegetal cortex but is also locally activated there, by mechanisms that remain poorly understood (Weitzel et al. 2004; Peng and Wikramanayake, 2013). Vegetally localized/activated Dsh is partitioned predominantly into the micromeres during cleavage, where it stabilizes and promotes the nuclear accumulation of maternal β-catenin (Wikramanayake et al., 1998; Logan et al., 1999; Weitzel et al., 2004; Ettensohn, 2006).
Fig. 4-. Activation of the PMC GRN in euechinoids (S. purpuratus).
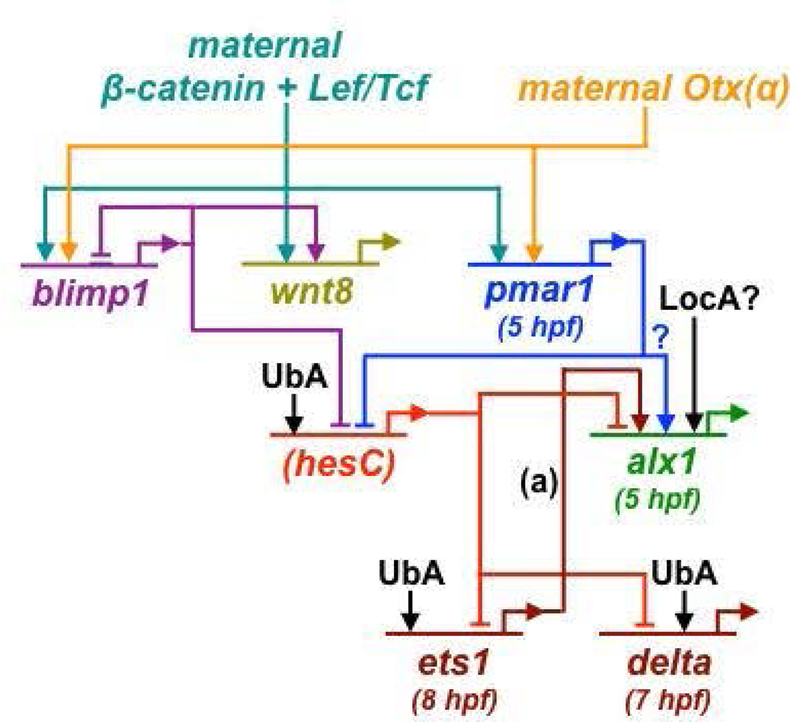
Maternal factors are localized at the vegetal pole, including the maternal protein Dishevelled, which stabilizes β-catenin in the micromeres. β-catenin and maternal Otx(α) activate pmar1, a pivotal initiator of skeletogenic specification. One function of Pmar1 is to represses hesC in the large micromere lineage (indicated by the parentheses surrounding “hesC”), a transient function later assumed by Blimp1. In non-PMC lineages, HesC acts to repress PMC regulatory genes. Transcriptional repression of hesC does not account for the early activation of alx1, however, which may involve hesC-independent functions of Pmar1 (blue question mark) or other unidentified, localized activators (LocA) (see Section II(A) for details). Both Alx1 and Ets1 play pivotal roles in PMC specification and provide positive inputs into many downstream regulatory and effector genes. Delta and Wnt8 are signaling molecules produced by the micromere lineage. Several regulatory genes in the PMC GRN are activated ectopically throughout the embryo following Pmar1 overexpression or HesC knockdown, pointing to ubiquitous activators (UbA). Times shown in parentheses (hours post-fertilization, or hpf) are approximate times of gene activation based on quantitative, high-resolution NanoString time course data (Rafiq and Ettensohn, unpublished observations). Arrows and bars show positive and negative regulatory inputs, respectively. See Sections II(A) and II(B) for references. Notes: (a) The input from zygotic Ets1 into alx1 occurs several hours after alx1 activation.
Another maternal protein, Otx(α), exhibits a polarized distribution in the early embryo. Several different mRNAs and proteins are produced from the single sea urchin otx gene by means of alternative promoter usage and alternative splicing. The single, early isoform, Otx(α), is present maternally as both mRNA and protein, both of which are found in all cells of the early embryo (Li et al., 1997). At the 16-cell stage, however, the Otx(α) protein becomes transiently concentrated selectively in the nuclei of micromeres (Chuang et al., 1996). The mechanism of this micromere-specific protein localization is unknown, although Otx(α) binds to α-actinin and it has been suggested that the micromeres are relatively devoid of actin-based cortical cytoskeleton, freeing Otx(α) to enter the nucleus.
In the micromeres, β-catenin interacts with maternal TCF (Huang et al., 2000; Vonica et al., 2000) to activate a variety of molecular targets. In concert with maternal Blimp1 protein, β-catenin activates wnt8, a gene which encodes a secreted signaling ligand, in the micromere territory by the end of 5th cleavage (Wikramanayake et al., 2004; Minokawa et al., 2005; Smith et al., 2007). Studies using C59, a global inhibitor of Wnt protein secretion, argue against the possibility that secreted Wnt8 acts in an autocrine fashion to reinforce β-catenin nuclearization in the micromere territory (Cui et al., 2014). Maternal Otx(α) acts with β-catenin/TCF to drive the zygotic expression of blimp1 during the sixth cleavage (Smith et al., 2007). Later, accumulation of Blimp1 in the micromeres leads to transcriptional autorepression and a decrease in the expression of both blimp1 and wnt8 in the micromeres. These various gene regulatory processes may play little or no role in the specification and morphogenesis of the large micromere-PMC lineage, but they contribute to a wave of dynamic signaling events in the vegetal region of the embryo, originating from the micromere territory, that regulates the development of non-skeletogenic mesoderm and endoderm (Smith and Davidson, 2008; Materna and Davidson, 2012).
Beta-catenin also interacts with maternal TCF and Otx(α) to activate the zygotic transcription of pmar1/micro1 (Oliveri et al., 2002; Nishimura et al., 2004) specifically in the micromeres at the end of the fourth cleavage. Pmar1, a paired class homeodomain-containing protein, is both necessary and sufficient for the specification of skeletogenic cells (Kitamura et al., 2002; Oliveri et al., 2002, 2003; Nishimura et al., 2004; Yamazaki et al., 2005). Pmar1 functions as a repressor (Oliveri et al., 2002; Yamazaki et al., 2009) and indirectly activates the expression of skeletogenic genes, at least in part by blocking the expression of a second repressor, hesC, a member of the HES (Hairy-Enhancer-of-Split) family (Revilla-i Domingo et al., 2007). hesC mRNA is present ubiquitously in the early embryo but is cleared from the micromere territory by the early blastula stage. Overexpression of Pmar1 results in a global decrease in hesC expression while knockdown of HesC results in an expanded region of expression of delta (Revilla-i Domingo et al., 2007), which encodes a signaling molecule ordinarily restricted to the micromere territory during early development (Sweet et al., 2002). In addition, cis-regulatory analyses of alx1 (Damle and Davidson, 2011), tbr (Wahl et al., 2009) and delta (Smith and Davidson, 2008) have identified putative binding sites for HesC that are required for the repression of these genes in cells other than the skeletogenic lineage, thereby establishing a direct repressive linkage between HesC and these skeletogenic genes. Together, this evidence supports the view that a pmar1/hesC “double-negative gate” controls the expression skeletogenic genes specifically in the micromere lineage; i.e., when the expression of pmar1 is activated in the micromeres, hesC is repressed, thereby allowing the activation of skeletogenic genes. The ectopic activation of skeletogenic genes in non-micromere lineages following experimental perturbation of the pmar1/hesC double-negative system presumably involves the activity of ubiquitous activators, but these proteins have not been identified. Furthermore, in the large micromere-PMC lineage, the double-repression system must work in concert with as yet-unidentified activators that drive the expression of early regulatory genes. Therefore, characterization of the maternally provisioned activators that provide inputs into the first layer of zygotic regulatory genes in the PMC GRN will be very important in order to extend the current model of the network.
Other observations challenge the view that hesC repression is sufficient to fully account for the earliest deployment of the PMC GRN. It has been shown in two different species that at the time when alx1 and delta are first activated specifically in the large micromere territory, levels of hesC mRNA are uniform across the embryo; i.e., selective clearing of hesC mRNA from the large micromere territory does not occur until later in development (Sharma and Ettensohn, 2010). Thus, the initial spatial restriction of alx1 and delta transcription cannot be explained solely by the localized transcriptional repression of hesC and additional mechanisms must be involved. It is possible that at early cleavage stages, HesC activity is inhibited specifically in the micromere territory by post-transcriptional mechanisms, that Pmar1 can act through a repressor distinct from HesC, or that Pmar1-independent mechanisms are involved. One caveat regarding the published experimental data showing that hesC binding sites are required to restrict the expression of alx1 and delta is that the spatial expression patterns of the relevant transcriptional reporters have been analyzed only at relatively late developmental stages, not at the initial stages of large micromere-specific gene expression (Smith and Davidson, 2008; Damle and Davidson, 2011). Another issue is that several early genes in the PMC do not appear to be highly sensitive to HesC-mediated repression. A structure-function analysis of Pmar1 identified one mutant construct (N-HD-A-C) that downregulated hesC mRNA levels throughout the embryo and resulted in an expansion of delta expression but did not significantly expand the expression of alx1, tbr or ets1 (Yamazaki et al., 2009). More recently, Yamazaki and Minokawa (2016) have re-visited the effects of HesC morpholino knockdown using two different sea urchin species (Hemicentrotus pulcherrimus and Scaphechinus mirabilis). They report that, while mis-expression of Pmar1 efficiently converts all cells to PMC-like fate as previously reported, morpholino-based HesC knockdowns had much more modest consequences, with effects on gene expression that varied among genes (for example, the expression domains of alx1 and tbr were only slightly expanded when assayed at the mid- and late blastula stages). These various findings argue strongly that, while the localized transcriptional repression of hesC plays a significant role in restricting the domain of expression of skeletogenic genes, other mechanisms are responsible for the initial deployment of the PMC GRN specifically in the large micromere-PMC lineage.
The early specification of micromeres is dependent upon unequal cell division (Langelan and Whiteley, 1985; Sharma and Ettensohn, 2010). The cell division that produces micromeres is a consequence of the displacement of the nuclei of the four vegetal blastomeres of the 8-cell stage embryo towards the vegetal pole prior to nuclear envelope breakdown and the close association of one pole of their mitotic spindles with the vegetal cortex (Schroeder, 1987; Dan and Tanaka, 1990; Holy and Schatten, 1991). These events rely on maternally derived components of G-protein signaling (Voronina and Wessel, 2006). Pharmacological agents have been used to inhibit the vegetal positioning of the mitotic spindles, thereby equalizing cleavage and producing embryos that lack micromeres (Tanaka, 1976; Langelan and Whiteley, 1985; see also Kominami and Takaichi, 1998). Equally-cleaving embryos show a striking reduction in the development of the skeleton. Sharma and Ettensohn (2010) showed that the zygotic expression of two early markers, alx1 and delta, but not that of pmar1, was reduced in such embryos. This suggests that unequal cleavage is not required in order to concentrate sufficient Dsh or β-catenin to activate pmar1, but may be required in order to concentrate sufficient Pmar1 (or other proteins) to activate downstream targets such as alx1.
Each micromere undergoes an additional round of unequal cell division at the 5th cleavage, thereby producing one large daughter cell (large micromere) and one small daughter cell (small micromere). The four large micromeres are the founder cells of the PMC lineage; they undergo an additional 3 or 4 rounds of cell division, depending upon the species, and give rise exclusively to skeletogenic PMCs. The small micromeres, in contrast, contribute to the germ line (Yajima and Wessel, 2011). The skeletogenic program is deployed in the large, but not the small, daughter cells of the micromeres, despite the presence of pmar1 mRNA in both cells. This is likely due to a global repression of gene expression in the small micromeres which involves both transcriptional and translational mechanisms (Swartz et al., 2014; Oulhen et al., 2017).
B). Alx1 and Ets1: Key Early Transcription Factors
Alx1 and Ets1 play pivotal roles in PMC specification and morphogenesis. The alx1 gene, which encodes a homeodomain protein, is the first regulatory gene activated specifically in the large micromere-PMC lineage and its expression is restricted to this lineage throughout embryogenesis (Ettensohn et al., 2003). In S. purpuratus, alx1 is activated in the four large micromeres in the first cell cycle after these cells are born. Knockdown of alx1 diverts large micromere progeny into non-PMC fates and entirely blocks skeletogenesis, while overexpression of alx1 converts macromere descendants to a skeletogenic fate (Ettensohn et al., 2003; Ettensohn et al., 2007). RNA-seq analysis of Alx1 morphants has shown that this transcription factor provides positive inputs into almost half of the ~400 genes differentially expressed by PMCs (and an even larger fraction of the highly expressed genes in this set), pointing to the pivotal role of Alx1 in establishing PMC identity (Rafiq et al., 2012; 2014). Targets of Alx1 include several regulatory genes expressed selectively in PMCs (e.g., alx4, fos, nk7, and foxB); therefore, the extent to which Alx1 controls PMC effector genes indirectly via its effects on the expression of intermediary transcription factors remains unclear. Alx1 also appears to function as an auto-regulator, promoting a rise in alx1 expression when present at low levels but functioning as an auto-repressor at high concentrations (Ettensohn et al., 2003; Damle and Davidson, 2011).
One of the functions of Alx1 is to repress the deployment of alternative transcriptional programs in the large micromere territory. Ordinarily, this territory is surrounded by prospective non-skeletogenic mesoderm (prospective pigment and blastocoelar cells) (Ruffins and Ettensohn, 1996). The domain of expression of pigment cell markers, including the key regulatory gene gcm, expands into the large micromere territory in Alx1 morphants (Oliveri et al., 2008). In addition, gene expression profiling of Alx1 morphants reveals increases in the levels of expression of several regulatory genes associated with blastocoel cell specification, suggesting that this transcriptional program is also ectopically expressed in the large micromere territory, although this has not been confirmed by WMISH analysis (Rafiq et al., 2014).
Damle and Davidson (2011) have argued that zygotically expressed Ets1, expressed selectively in the large micromere-PMC lineage as a consequence of the pmar1/hesC double-negative gate, is the primary activator of alx1 expression in the large micromere lineage. As noted above, the role of HesC-mediated repression in alx1 activation is unclear, as alx1 is expressed specifically in the large micromere territory before there is any local depletion of hesC mRNA in that region (Sharma and Ettensohn, 2010). Moreover, while there is no doubt that Ets1 provides important positive inputs into alx1 at post-cleavage stages (Ettensohn et al., 2003; Oliveri et al., 2008; Damle and Davidson, 2011), the hypothesis that zygotic Ets1 is responsible for the initial activation of alx1 in the large micromere lineage is inconsistent with several lines of experimental evidence. First, a dominant negative form of Ets1 that effectively blocks alx1 expression at late developmental stages has no effect on the initial accumulation of alx1 mRNA in the large micromere territory (Sharma and Ettensohn, 2010). Second, immunostaining studies in two different species show that Ets1 protein does not accumulate in the nuclei of large micromere descendants until the blastula stage, several hours after alx1 is first expressed (Sharma et al., 2010; Yajima et al., 2010). Third, mis-expression of Ets1 throughout the embryo fails to induce the ectopic expression of alx1, although it converts many cells to a migratory (mesenchymal) phenotype (Kurokawa et al., 1999; Röttinger et al., 2004; Koga et al., 2010; Sharma and Ettensohn, 2010). Lastly, high-resolution Nanostring studies of alx1 and ets1 expression using intron-specific probes show that zygotic expression of ets1 follows, rather than precedes, that of alx1 and therefore zygotic Ets1 protein is not present when alx1 is first activated (Rafiq and Ettensohn, unpublished observations). These findings indicate that zygotically produced Ets1 is required for the maintenance, but not for the activation, of alx1 expression. The mechanisms that underlie the lineage-specific transcription of alx1, a critically important regulatory gene in the PMC GRN, remain to be elucidated.
The ets1 gene, one of several ETS family genes in sea urchins, is expressed maternally as well as zygotically. Both ets1 mRNA and Ets1 protein are maternally supplied and distributed uniformly throughout the early embryo (Kurokawa et al., 1999; Yajima et al., 2010; Rizzo et al., 2006). Zygotic transcription begins during late cleavage and is restricted to the skeletogenic lineage until the late mesenchyme blastula stage, when ets1 is also expressed in presumptive blastocoelar cells (Kurokawa et al., 1999; Yajima et al., 2010; Flynn et al., 2011). The role of maternally-derived Ets1 is unclear, as only the zygotic protein is detected in cell nuclei (Sharma and Ettensohn, 2010; Yajima et al., 2010). Knockdown of Ets1 or forced expression of a dominant negative form of the protein blocks PMC specification and skeletogenesis (Kurokawa et al., 1999; Oliveri et al., 2008; Sharma and Ettensohn, 2010). As noted above, overexpression of ets1 transforms most cells of the embryo into mesenchymal cells. Ets1, like Alx1, provides positive inputs into a large fraction of PMC effector genes (Rafiq and Ettensohn, 2012; 2014). Some of these inputs are very likely to be direct, based on the identification of essential, consensus ETS-binding sites in the cis-regulatory control regions of several PMC effector gene (Yamasu and Wilt, 1999; Amore and Davidson, 2006; Yajima et al., 2010), although several other ETS family proteins are also expressed by PMCs (Zhu and Ettensohn, 2001; Rizzo et al., 2006) that might bind to these sequences. Ets1 also controls the expression of numerous regulatory genes (including alx1, alx4, dri, erg, fos, foxB, foxO, mef2, nk7, and smad2/3) and may therefore regulate downstream effectors by indirect mechanisms (Oliveri et al., 2008; Rafiq et al., 2014).
The Raf/MEK/ERK (MAPK) pathway plays a critical role in the ingression of PMCs into the blastocoel at the mesenchyme blastula stage, and this is effect is mediated by Ets1. Ets1 contains a single consensus site for phosphorylation by the MAP kinase, ERK, as well as a predicted ERK docking site (Röttinger et al., 2004). ERK is transiently activated in presumptive PMCs just prior to ingression, and PMCs fail to ingress in embryos treated with the MEK inhibitor U0126, although gastrulation occurs normally otherwise (Fernandez-Serra et al., 2004; Röttinger et al., 2004). MAPK signaling is also required for PMC specification, as MEK inhibition downregulates the expression of several skeletogenic regulatory genes (Röttinger et al., 2004). These include alx1 and tbr, which require MEK signaling to maintain (but not to activate) their expression (Sharma and Ettensohn, 2010; 2011). Surprisingly, the MAPK pathway is activated cell autonomously in the large micromere-lineage (Röttinger et al., 2004). This pathway appears to act entirely via the phosphorylation of Ets1, a conclusion consistent with the finding that almost ¾ of all PMC effector genes affected by U1026 are also affected by Ets1 knockdown (Röttinger et al., 2004; Rafiq et al., 2014). Although Alx1 also contains a predicted MAP kinase phosphorylation site, this site is not required for the embryonic function of Alx1 (Khor and Ettensohn, 2017).
C). The Progression and Stabilization of PMC Specification
Many other regulatory genes are expressed selectively in the large micromere-PMC lineage. These include alx4, dri, erg, fos, jun, foxB, foxN2/3, foxO, hex, mitf, nfkbil1L, nk7, nurr1, smad1/5/8, smad2/3, tbr, tel, and tgif (Zhu et al., 2001; Fuchikami et al., 2002; Rizzo et al., 2006; Oliveri et al., 2008; Barsi, 2014; Rafiq et al., 2012; 2014). In most cases (with nk7 being one exception), these genes are also expressed zygotically in other embryonic territories or show ubiquitous, maternal expression. All are activated in the PMC lineage by the late blastula stage, prior to overt PMC morphogenesis, although their precise temporal patterns of expression vary (Materna et al., 2010).
The best characterized of these genes is tbr. In sea urchins, maternal stockpiles of tbr RNA and protein are distributed to all cells of the early embryo, but zygotic transcription is entirely restricted to PMCs (Croce et al., 2001; Fuchikama et al., 2002). Tbr protein is predominantly cytoplasmic until the blastula stage, when it becomes concentrated in the nuclei of presumptive PMCs (Fuchikama et al., 2002). In this regard, Tbr protein resembles Ets1; i.e., in both cases the cytoplasmic localization of the ubiquitous, maternal protein suggests that this form may not regulate transcription. Zygotic tbr expression is dependent upon β-catenin (Fuchikami et al., 2002) and tbr is activated ectopically in response to pmar1 misexpression or perturbation of HesC function (Oliveri et al., 2002; Wahl et al., 2011). The early, positive drivers of tbr expression in the large micromere-PMC lineage, however, are unknown. Late (post-ingression) regulation by ETS family proteins (probably Ets1) via one CRM (the γ(2) module) of the tbr cis-regulatory apparatus has been demonstrated (Wahl et al., 2011). Downstream, tbr is only weakly connected to the PMC GRN. It provides inputs into a much smaller number of downstream targets than either alx1 or ets1 (Oliveri et al., 2008; Rafiq et al., 2012). This is consistent with the finding that in tbr morphants, PMC migration, fusion, and patterning are not affected, although skeletogenesis is perturbed (Fuchikami et al., 2002; Oliveri et al., 2008). As discussed below, tbr was only recently co-opted into the PMC GRN of echinoids (Hinman et al., 2007; Gao and Davidson, 2008), and the limited connectivity of tbr may reflect the recent recruitment of this gene into the network.
With respect to other PMC regulatory genes, overexpression of pmar1 or a form of cadherin that interferes with β-catenin function have shown that erg, foxN2/3, hex, tel, and tgif are all downstream of β-catenin and pmar1 (Oliveri et al., 2008; see also Rho and McClay, 2011, with respect to foxN2/3) while foxO, jun, and mitf showed less pronounced changes in expression. The other PMC regulatory genes listed above have not been tested, but it seems likely that many are also downstream of β-catenin and pmar1. Rafiq et al., (2014) used morpholino knockdowns of Alx1 and Ets1 and RNA-seq gene expression profiling at the mesenchyme blastula stage to show that Ets1 provides positive inputs into most PMC regulatory genes (alx1, alx4, dri, fos, foxB, foxO, mef2, nfkbil1L, nk7, and smad2/3), while Alx1 provides positive inputs into a subset of the Ets1-regulated genes (alx4, dri, fos, foxB, nfkbil1L, and nk7) and negatively regulates its own expression (Ettensohn et al., 2003; Damle and Davidson, (2011). There is some evidence of a direct input from Ets1 into dri (Amore and Davidson, 2006), but it is unknown whether Ets1 regulates its other targets directly, indirectly, or through a combination of both mechanisms. These Alx1 knockdown data are entirely consistent with those of Oliveri et al. (2008), who also identified inputs from Alx1 into dri and foxB (alx4, cebpa, fos, and nk7 were not tested). On the other hand, while there is substantial agreement in these two studies with respect to regulatory genes that receive inputs from Ets1, there are also differences. Most notably, Oliveri et al. (2008) reported significant changes in the expression of erg, tgif, and hex following Ets1 knockdown, while Rafiq and co-workers (Rafiq et al., 2014) did not detect such changes. It is not clear whether these differences are due to different significance thresholds, slight differences in the developmental stages used, or other factors.
Oliveri et al. (2008) carried out morpholino knockdowns of nine of the regulatory genes expressed selectively in the large micromere-PMC lineage. They found evidence of a variety of regulatory interactions among these genes- primarily positive feedback loops that serve to enhance and maintain gene expression. One prominent example is a set of mutual regulatory interactions among erg, hex, and tgif that reinforces the expression of all three genes. Such feedback loops may convert the transient expression of pmar1 into a more stable regulatory state in the skeletogenic lineage and buffer against initial variation in the level of expression of these genes. Since the genes involved in these feedback loops are co-dependent, loss of expression of even one gene severely affects the expression of all genes in the circuit, and this has a catastrophic effect on the expression of downstream genes. This probably imposes an evolutionary constraint on the network, making rewiring of the circuit difficult without completely losing its function (Peter and Davidson, 2015).
The possible role of the transcriptional repressor, snail, in the PMC GRN is currently unsettled. Wu and McClay (2007), working with Lytechinus variegatus, provided evidence that this regulatory gene acts downstream of alx1 to regulate PMC ingression. On the other hand, Oliveri et al., (2008) concluded that, in S. purpuratus, snail is irrelevant with respect to early PMC specification because it is expressed at an extremely low level until gastrulation. One gene expression study (Barsi et al., 2014) reported a substantial enrichment of Sp-sna in PMCs at the mesenchyme blastula stage, as reported in L. variegatus, while another did not (Rafiq et al., 2014). In addition, the latter study reported that levels of Sp-sna mRNA increased in Alx1 morphants, in contrast to the findings in L. variegatus. It remains unclear whether the role of snail is different in the two species or whether technical differences in these studies account for the apparent variability.
From the above discussion, it is apparent that not all the possible interactions among PMC regulatory genes have been explored, and our understanding of the architecture of this layer of the PMC GRN is undoubtedly incomplete. Nevertheless, it is very likely that all regulatory genes expressed selectively by PMCs have now been identified, with the exception of Zn-finger genes, which have not been carefully analyzed. In addition, due largely to the work of Oliveri et al., (2008), many regulatory interactions among these genes are known. Our current view of the interactions among PMC regulatory genes is shown in Fig. 5.
Fig. 5-. Regulatory gene interactions (S. purpuratus).

Tiers of regulatory genes are shown top-to-bottom in their approximate order of activation. Genes in gray box represent regulatory genes expressed selectively by PMCs that currently have no regulatory linkages to any other genes in the PMC GRN. Experimental evidence for the regulatory linkages shown comes primarily from Oliveri et al. (2008) and Rafiq et al. (2014), with additional information from Wahl et al., (2009), Sharma and Ettensohn (2010), Damle and Davidson (2011), Rho and McClay (2011), and Rafiq et al. (2012). Notes: (a) MAPK signaling, activated autonomously in the PMC lineage by unknown mechanisms, is required for Ets1 function. (b) Inputs from Ets1 and Tgif into alx1 occur several hours after alx1 activation. (c) The input from Ets1 into tbr occurs late (post-ingression). (d) The input from Tbr into foxN2/3 is extrapolated from data for L. variegatus. (e) Inputs from Hex and Tgif into foxO are evident at 24 hpf but not at 18 hpf. See Fig. 1 for description of symbols.
D). Activation of Skeletogenic Effector Genes
Historically, interest in the development of PMCs was spurred in no small part by their spectacular morphogenetic behaviors, which include EMT (ingression), directional cell migration, cell-cell fusion, and biomineral deposition (see reviews by Ettensohn et al., 1997; Wilt and Ettensohn, 2007; McIntyre et al., 2014). With the elucidation of the transcriptional network deployed by these cells, there is now an opportunity to develop a comprehensive explanation for their behaviors. More broadly, analysis of the PMC GRN provides a paradigm for linking cell specification to morphogenesis (Lyons et al., 2012; Ettensohn, 2013).
Genome-wide transcriptome profiling has identified several hundred effector genes expressed selectively by PMCs (Rafiq et al., 2014; Barsi et al., 2014). The vast majority of these genes, even those associated directly with biomineralization (see below) are expressed prior to the first overt morphogenetic activity of PMCs (ingression) and several hours before the onset of biomineral deposition, which begins at the mid-gastrula stage. As noted above (Section IIB) Alx1 provides positive inputs (direct or indirect) into almost half of the effector genes differentially expressed by PMCs, and Ets1 regulates only a slight smaller fraction (40%), pointing to the prominent roles of these two transcription factors in driving PMC behaviors (Fig. 6). Moreover, there is striking overlap between the targets of Alx1 and Ets1; 85% of Ets1 targets are also regulated by Alx1, and 73% of all Alx1 targets are also regulated by Ets1. On the whole, more than a third of all effector genes are co-regulated by these two transcription factors. This fraction is even higher if one considers only the most highly expressed effector genes; of the 100 most abundant, PMC-specific mRNAs, almost 2/3 are positively regulated by both Ets1 and Alx1, including most of the effectors discussed in detail below. The effector genes that are co-regulated by Ets1 and Alx1 show a characteristic temporal pattern of expression; most are exclusively zygotically expressed and exhibit a strong spike in expression between the late blastula and mid-gastrula stages (Rafiq et al., 2014). The mechanism of co-regulation appears to be a widespread feedforward circuit with the structure: Ets1>Alx1, Ets1+Alx1>effector gene. Ets1 positively regulates Alx1 (Ettensohn et al., 2003 Oliveri et al., 2008) and also appears to have direct inputs into the effector genes cyclophilin, msp130, msp103L, and sm30 (Yamasu and Wilt, 1999; Amore and Davidson, 2006; Oliveri et al., 2008). Moreover, a recent genome-wide analysis of chromatin accessibility in PMCs showed a substantial enrichment of candidate Ets1 and Alx1 binding sites in PMC enhancers, suggesting that a feedforward circuit might regulate a large fraction of effector genes (Shashikant et al., 2018).
Fig. 6-. Regulatory inputs into PMC effector genes (S. purpuratus).

More than 400 effector genes are expressed selectively by PMCs (Rafiq et al., 2014), of which only a small subset is shown here. The effector genes shown encode members of the P16 (p16, p16rel1, and p16rel2) and MSP130 families (msp130, msp130rel1, msp130rel2, and msp130rel3), matrix metalloproteases (mmp16, mtmmpb, mtmmpd, and mtmmpe), spicule matrix proteins (sm20, sm21, sm27, sm29, sm49, sm50, and C-lectin), signaling receptors (tgfbr2 and vegfr-Ig10), an Ig-domain adhesion protein (kirrelL), and several other proteins. A large number of effector genes are regulated positively by both Ets1 and Alx1 (Rafiq et al., 2014). Regulatory inputs into msp130, sm27 (= pm27), sm50, vegfr-Ig10, frp, and cyp1 are from Oliveri et al. (2008) with additional information from Amore et al., (2006). Additional tbr targets were identified by Rafiq et al., (2012). The developmental functions of several of these effectors are discussed in Section IID. For interactions among the regulatory genes shown here, see Fig. 4.
PMC effector genes have diverse functions, but a large fraction are genes associated directly with the secretion of the skeleton. The biomineralized skeleton is composed primarily of calcite, within which small amounts of secreted proteins are incorporated. Although these secreted proteins make up less than 0.1% of the mass of the biomineral, they play an important role in controlling its mechanical properties and growth (Wilt and Ettensohn, 2007). In the sea urchin, the most abundant of these secreted proteins are the spicule matrix proteins, a family of 17 closely related proteins, each of which contains a single C-type lectin domain and a variable number of proline/glycine-rich repeats (Livingston et al., 2006). One important function of spicule matrix proteins is to regulate the conversion of amorphous calcium carbonate (ACC), a precursor of calcite, to a crystalline state (Gong et al., 2012). The genes that encode spicule matrix proteins, like the members of most of the other families of biomineralization gene discussed below, are clustered in the genome, strongly suggesting that they expanded through gene duplication.
Members of the MSP130 family of cell-surface glycoproteins are among the most highly expressed effector genes in PMCs. MSP130 proteins are also expressed selectively in biomineralizing tissues in other marine phyla, pointing to a conserved role (Ettensohn, 2014; Szabó and Ferrier, 2015). The biochemical functions of MSP130 proteins are not well understood, but these proteins regulate the internalization of calcium, which appears to enter PMCs primarily via endocytosis (see references in Wilt and Ettensohn, 2007; Ettensohn, 2014; Vidavsky et al., 2016; Killian and Wilt, 2017). Three type I transmembrane proteins specifically expressed by PMCs, P16 (a member of a small family of related proteins), and the two related proteins, P59A and P58B, are each required for skeletal growth (Cheers and Ettensohn, 2005; Adomako-Ankomah and Ettensohn, 2011). A PMC-specific, GPI-anchored carbonic anhydrase may be involved in biomineral remodeling (Livingston et al., 2006; Mitsunaga et al., 1986; Zito et al., 2015). Non-fibrillar collagens produced by PMCs provide an essential substrate for the cells, although they do not appear to be a structural component of the biomineral, as they are in vertebrates (Livingston et al., 2006; Wessel et al., 1991). Otopetrin (otop2L), expressed selectively by PMCs, is the ortholog of a vertebrate protein essential for the development of otoliths/otoconia, extracellular calcium carbonate-containing crystals that mediate vestibular mechanosensory function. PMCs also express a suite of matrix metalloprotease genes, arranged in tandem on a single chromosome, which probably encode the metalloprotease activities required for spiculogenesis in vivo and in vitro (Roe et al., 1989; Ingersoll and Wilt, 1998). Lastly, a PMC-specific TgfbrtII receptor was recently shown to be required for biomineral deposition (Sun and Ettensohn, 2017).
Biomineralization proteins regulate skeletal growth in complex ways (Killian and Wilt, 2008). Moreover, heritable variation in the expression of these proteins may play an important role in evolutionary changes in skeletal morphology. Garfield et al. (2013) analyzed population-level variation in the expression of genes in the embryonic skeletogenic GRN of S. purpuratus. They identified variability that was attributable to parent-of-origin effects and that was associated with variation in larval skeletal morphology. Moreover, they found that the early GRN buffers this variation, while variation in skeletal morphology is primarily attributable to differences in effector gene expression.
PMC effectors have also been identified that control morphogenetic processes other than biomineralization. After PMCs ingress, they migrate directionally within the blastocoel by means of filopodia. VEGF3, a signaling molecule secreted by specific regions of the overlying ectoderm, provides critically important guidance cues that are recognized by a cognate, PMC-specific receptor, VEGFR-Ig10 (Duloquin et al., 2007; Adomako-Ankomah and Ettensohn, 2013). The role of VEGF3 in directing PMC migration can be distinguished from its second important function, the local maintenance within the PMC syncytium of the expression of genes required for skeletal growth (Section IIE, below). At the same time that PMCs are migrating directionally within the blastocoel in response to VEGF3, their filopodia contact one another and fuse, leading to the formation of a pseudopodial cable that links the cells in an extensive syncytial network (see Section IC). KirrelL, a PMC-specific member of the Ig-domain superfamily of cell adhesion proteins, is required for PMC filopodial contacts to result in cell-cell fusion (Ettensohn and Dey, 2017).
In only three cases have the cis-regulatory elements that control PMC effector genes been analyzed in detail. These studies have focused on two spicule matrix genes, sm50 (Makabe et al., 1995; Otim, 2017) and sm30 (Frudakis and Wilt, 1995; Yamasu and Wilt, 1999), and cyclophilin/cyp1 (Amore and Davidson, 2006). In general, these studies have provided evidence of both positive and negative regulatory inputs and have pointed to direct regulation by ETS family proteins. A more complete understanding of the direct transcriptional inputs into PMC effector genes would clearly be facilitated by the high-throughput identification of CRMs that control the expression of these genes. Recently, the genome-wide chromatin accessibility profile of PMCs has been compared to that of other cells in the embryo and has led to the identification of hundreds of PMC CRMs (Shashikant et al., 2018). The experimental and computational analysis of this large collection of regulatory elements will undoubtedly reveal new features of GRN architecture upstream of PMC effector genes and link the specification layers of the network to PMC morphogenesis is a more robust manner.
E). A Developmental Shift in Regulatory Mode: Signal-Dependent Control
As described above (Section IIIA), the skeletogenic lineage is autonomously specified by means of maternal factors. The cell-autonomous phase of PMC specification produces prospective skeleton-forming cells that, at least based upon qualitative WMISH analysis, appear to be homogeneous with respect to their programs of gene expression. After gastrulation begins, however, migratory PMCs come under the influence of localized signals that emanate from the adjacent embryonic ectoderm. The immediate effect of these signals is to maintain high levels of effector gene expression in only those PMCs (or, more properly, PMC cell bodies) nearest the source of the signals, while expression declines elsewhere. As a consequence, by the late gastrula stage, the PMC syncytium is a mosaic of distinct subdomains of gene expression. The localization of mRNAs within the PMC syncytium is likely stabilized by their limited diffusion and results in the non-uniform distribution of proteins encoded by these mRNAs (Gross et al., 2003; Harkey et al., 1995; Urry et al., 2000; Wilt et al., 2008). Significantly, regions of high PMC effector gene expression are intimately associated with sites of active skeletogenesis; for example, most effector genes are expressed at high levels in the ventrolateral PMC clusters where the two skeletal primordia form and later in the clusters of PMCs associated with the growing tips of skeletal rods that support the larval arms (Harkey et al., 1992; Guss and Ettensohn, 1997; Sun and Ettensohn, 2014). These observations strongly suggest that the signal-dependent pattern of gene expression within the PMC syncytium underlies the stereotypical pattern of skeletal rod growth.
Multiple ectoderm-derived cues regulate skeletogenesis (reviewed by Adomako-Ankomah and Ettensohn, 2014; McIntyre et al., 2014). The best characterized of these is VEGF3, which, in addition to its role in directing PMC migration, maintains the expression of many biomineralization genes and enhances skeletal growth (Duloquin et al., 2007, Adomako-Ankomah and Ettensohn, 2013; Sun and Ettensohn, 2014). At post-gastrula stages, VEGF3 expression is tightly associated with sites of active biomineral deposition, but only on the ventral (oral) side of the embryo, where the skeletal primordia and the larval arms form. A separate, unidentified signal acts on the opposite side of the embryo to enhance effector gene expression in the scheitel, a region of active biomineral deposition at the posterior tips of the body rods (Sun and Ettensohn, 2014). Recombinant sea urchin VEGF3 enhances the deposition of skeletal elements by cultured PMCs, demonstrating that this protein affects the cells directly (Knapp et al., 2012). Remarkably, the direction and pattern of skeletal rod branching are also sensitive to levels of VEGF3, suggesting that local differences in vegf3 expression might regulate the stereotypical pattern of skeletal rod branching observed in vivo. In the embryo, VEGF3 signaling is facilitated by ventrally-localized, sulfated proteoglycans (Fujita et al., 2010; Piacentino et al., 2016). One of the consequences of VEGF3 signaling is to maintain expression of the cognate receptor, VEGFR-Ig10, in neighboring domains of the PMC syncytium (Duloquin et al., 2007).
The mechanism by which signaling through VEGF3 and VEGFR-Ig10 impinges on the PMC GRN is poorly understood, but is of great interest in light of the evolutionary conservation of this regulatory mechanism (see Section III). Because many effector genes are sensitive to VEGF/VEGFR signaling (Adomako-Ankomah and Ettensohn, 2013; Sun and Ettensohn, 2014), one hypothesis is that this signaling pathway impinges on a key regulatory gene in the PMC GRN; i.e., one that controls many downstream effectors. VEGF3 appears to act through the ERK/MAPK pathway, as the effects of MEK inhibition on PMC gene expression at late developmental stages mimic those of VEGFR inhibition (Sun and Ettensohn, 2014). As discussed above (Section IIB), Ets1 provides positive inputs into many PMC effector genes; moreover, early in development this protein is directly activated by ERK (Röttinger et al., 2004). Ets1 may therefore serve as a critically important link between VEGF/VEGFR signaling and the expression of skeletogenic genes, although this has not been tested directly.
Two other signaling pathways have been implicated in the control of skeletal morphogenesis; TGF-β signaling and FGF signaling. The possible regulation of PMC effector gene expression by these pathways has not been explored in detail, and it should be noted that both pathways (and even VEGF signaling) could modulate skeletal growth by transcription-independent mechanisms. A PMC-specific, Type II TGF-β receptor, encoded by tgfbrtII (a target of both Ets1 and Alx1), is required for biomineral deposition and likely acts by binding TGF-β sensu stricto (Sun and Ettensohn, 2016). Piacentino et al., (2015) also found that TGF-β signaling regulates skeletal development and showed that the formation of anterior skeletal elements is particularly sensitive to signaling through this pathway. A PMC-specific FGF receptor, encoded by fgfr2, is also downstream of Ets1 and Alx1 and is selectively expressed in regions of the PMC syncytium associated with skeletal growth (Röttinger et al., 2008; Rafiq et al., 2014). The likely ligand, FGFA, shows a dynamic pattern of expression; it is expressed by the equatorial ectoderm cells and by all PMCs at the early gastrula stage but becomes restricted to the gut and to subdomains of the PMC syncytium associated with skeletal growth at post-gastrula stages (Röttinger et al., 2008; Adomako-Ankomah Adomako-Ankomah and Ettensohn, 2013). The role of FGF signaling in PMC morphogenesis is controversial and may differ among sea urchin species (reviewed by Adomako-Ankomah and Ettensohn, 2014). Despite these unresolved issues, the general pattern that emerges is the following: among those effectors that are activated in the PMC lineage during the initial, cell-autonomous phase of GRN deployment are signaling receptors that will play a pivotal role in regulating skeletal growth and patterning during the second, signal-dependent phase. During and after gastrulation, localized ectodermal cues maintain the transcriptional network selectively at sites of active skeletal growth. This localized up-regulation of the GRN includes the expression of the signaling receptors themselves, which reinforces local variations in effector gene expression and skeletal growth within the PMC syncytium.
IV. Evolution of the Skeletogenic GRN in Echinoderms
A). Echinoderm Phylogeny
All extant deuterostomes are grouped into three phyla: Chordata, Hemichordata and Echinodermata. Echinoderms and hemichordates, collectively referred to as ambulacarians, diverged from one another more than 500 million years ago (Erwin et al., 2011). Evolutionary relationships within the echinoderms have been well resolved through numerous morphological and molecular phylogenetic analyses (Cannon et al., 2014; Telford et al., 2014; Reich et al., 2015; Thuy and Stohr, 2016; Miller et al., 2017) (Fig. 7). Echinoderms are grouped into five classes: crinoids (sea lilies and feather stars), asteroids (sea stars), ophiuroids (brittle stars), holothuroids (sea cucumbers) and echinoids (sea urchins and sand dollars). Most extant sea urchin species are members of the euechinoid subclass, while a much smaller number belong to the cidaroid subclass. Among indirect-developing species, the larvae of sea urchins and brittle stars form an elaborate endoskeleton, holothuroid larvae form a highly reduced skeleton, and asteroid and crinoid larvae lack any larval endoskeleton. It should be noted that no crinoids with a feeding larva have been described.
Fig. 7-. Echinoderm phylogeny.

Asterozoan topology, the consensus view of relationships within echinoderms, is shown. Hemichordata is included as the nearest outgroup. Branch lengths are not drawn to scale. Images of representative adult and larval morphologies are shown (not all images correspond to the species listed as examples). Reprinted from Cary, G. A. and Hinman, V. F. (2017) Echinoderm development and evolution in the post-genomic era. Dev Biol 427:203–211, with permission from Elsevier. Photo credits: Adult Euechinoidea and Cidaroid are © Ann Cutting, Caltech; Holothuroidea is © Richard Ling/www.rling.com; Asteroidea is © Jerry Kirkhart, Los Osos, CA; Ophiuroidea is © Hans Hillewaert; Crinoidea is © NOAA Okeanos Explorer Program, INDEX-SATAL 2010; and Hemichordata is © Moorea Biocode / calphotos.berkeley.edu 4444 4444 0513 0997. Cidaroidea larval image is adapted from Bennett et al. (2012), all other whole (SEM) images of echinoderm and tornaria larvae are © T.C. Lacalli and T.H.J. Gilmour (University of Saskatchewan n.d.).
B). Skeletogenesis in Cidaroids
Cidaroids are extremely valuable for comparative studies as they represent the closest outgroup to the euechinoids. The morphology of adult cidaroids has remained similar to that of the few sea urchin species that survived the Permian-Triassic extinction (a major bottleneck in echinoderm evolution), while euechinoid morphologies have diversified greatly since that time. Cidaroid embryos form variable numbers of micromeres and lack an early-ingressing, skeletogenic mesenchyme (Schroeder, 1981; Emlet, 1988; Wray and McClay 1988; Yamazaki et al., 2012). Only after the archenteron has invaginated to a considerable extent do mesenchyme cells, including skeletogenic cells, delaminate from its tip. Micromeres give rise to at least some of the skeletogenic mesenchyme (Wray and McClay 1988), although the precise relationship between the cleavage pattern of vegetal blastomeres and the specification of skeletogenic cells has not been analyzed in detail. Skeletogenic mesenchyme cells migrate directionally to form ventrolateral clusters and deposit a skeleton later in development.
Recent studies using two different cidaroid species, Prionocidaris baculosa (Yamazaki et al., 2014) and Eucidaris tribuloides (Erkenbrack and Davidson, 2015) have revealed both similarities and differences with respect to the euechinoid program of skeletogenesis. Alx1 is activated early and specifically in the micromere lineage (Yamazaki et al., 2014 and see Fig. S1 in Erkenbrack and Davidson, 2015), and experimental knockdown shows the protein plays an essential role in skeletogenesis. In cidaroids, as in euechinoids, alx1 expression is dependent on the maternally-driven, polarized stabilization of β-catenin (Erkenbrack and Davidson, 2015) but the intervening regulatory steps are unknown. The ets1 gene is not expressed maternally in cidaroids as it is in euechinoids, but ets1 is expressed zygotically initially in the micromere lineage and later in non-skeletogenic mesenchyme, similar to the zygotic pattern of expression in euechinoids, and ets1 appears to have a conserved function as a driver of genes required for EMT and skeletogenesis. In E. tribuloides, Ets1 expression is extinguished in the micromere lineage prior to invagination, but this does not appear to be the case in P. baculosa. Expression of delta commences very early and is initially confined to the micromere lineage, as in euechinoids. Tbr is initially activated selectively in micromeres but, in contrast to euechinoids, expression later expands to non-skeletogenic mesoderm. Most strikingly, the double-negative gate does not exist in either cidaroid species. In the presumptive skeletogenic cells, hesC is co-expressed with delta, ets1, and alx1 and therefore does not appear to act as a repressor of these genes. Furthermore, despite considerable effort, an ortholog of pmar1 has not been identified. The double-negative gate arose more than 220 million years ago and is ubiquitous among modern echinoids (Yamazaki et al., 2010; Yamazaki and Minokawa, 2015; Thompson et al., 2017). Although absent from modern cidaroids, it has been difficult to establish whether the last common ancestor of all echinoids (cidaroids + euechinoids) utilized this regulatory circuitry (Thompson et al., 2017). The double-negative gate is absent from holothuroid and asteroid embryos, supporting the view that it evolved specifically within the echinoid lineage (McCauley et al., 2012; Thompson et al., 2017). In contrast to the double-negative gate, the regulation of the skeletogenic pathway by ectodermally-derived VEGF appears to be conserved in cidaroids (Gao et al., 2005; Erkenbrack and Petsios, 2017). In addition, similarities in the expression patterns of downstream components of the network indicate that common regulatory mechanisms are at work.
C). Skeletogenesis in Holothuroids
The embryos of sea cucumbers, the closest living relatives of sea urchins, do not produce micromeres during early cleavage. Nevertheless, skeletogenic mesenchyme cells ingress from the vegetal plate prior to invagination, migrate directionally, and construct a highly reduced embryonic skeleton consisting of a small spicule located in the posterior region of the embryo (Koga et al., 2010). McCauley et al. (2012) examined the expression of several mesodermal regulatory genes in Parastichopus parvimensis and found that orthologs of ets1, erg, foxN2/3, tbr and tgif are all expressed in the presumptive mesodermal territory in the central region of the vegetal plate at the blastula stage. Strikingly, alx1 is initially expressed in just four cells (presumably the founder cells of the skeletogenic lineage) within this territory, and knockdown of alx1 confirms that this gene is essential for skeletogenesis. The early ingression of the skeletogenic mesenchyme in P. parvimensis, also seen in all euechinoids, suggests that this may have been a developmental character of the common ancestor of sea urchins and sea cucumbers, a hypothesis which implies that the delayed ingression of skeletogenic cells in cidaroids is derived. Whether a highly simplified embryonic skeleton was ancestral to sea urchins + sea cucumbers or whether there has been a reduction of the skeleton in the holothuroid lineage is not possible to discern. At present, the mechanisms that underlie the activation of the skeletogenic network and the expression of downstream skeletal effector genes in holothuroids remain largely unexplored although, as noted above, the double-negative gate appears not operate in this taxon.
D). Skeletogenesis in Ophiuroids
Like sea cucumbers, brittle stars do not form micromeres at the vegetal pole during early development, yet skeletogenic mesenchyme cells ingress early in development, before the archenteron begins to invaginate. As in euechinoids, these cells are numerous and form an extensive larval skeleton that is initiated at two ventrolateral sites. A recent analysis of the Amphiura filiformis skeletogenic GRN has revealed several differences compared to the S. purpuratus PMC GRN, most notably with respect to the activation of the network (Dylus et al., 2016). The spatio-temporal expression of the likely ortholog of pmar1 in A. filiformis (Afi-pplx1) is similar to that Sp-pmar1, but Afi-Pplx1 lacks eh1 motifs that are required for the repressive function of Sp-pmar1. Moreover, Afi-pplx1 is co-expressed with Afi-hesC and therefore is unlikely to repress this gene. Similarly, co-expression of Afi-hesC with Afi-tbr, Afi-ets1/2 and Afi-delta suggests that repressive functions of hesC observed in sea urchins are not conserved in brittle stars. As gene knockdown studies have not been carried out in A. filiformis, it remains possible that Afi-pplx1 regulates the brittle star skeletogenic GRN, but by mechanisms that are independent of hesC. In addition to its distinct mechanism of activation, the brittle star skeletogenic GRN differs from that of euechinoids with respect to downstream regulatory gene interactions. For example, hex, erg and tgif, genes that are thought to engage in an interlocking regulatory loop in sea urchins, exhibit a reversed order of activation in A. filiformis, suggesting differences in their activating inputs. Furthermore, the expression patterns of Afi-foxB and Afi-dri do not support a potential role for these genes in skeletogenesis in brittle star embryos.
E). Skeletogenesis in Asteroids
Sea star embryos lack micromeres, skeletogenic mesenchyme, and embryonic skeletal elements. Numerous non-skeletogenic mesenchyme cells migrate into the blastocoel but only after after archenteron invagination is complete. Orthologs of several genes associated with mesoderm specification in sea urchins (including genes expressed by, but not restricted to, the skeletogenic mesoderm) are also expressed by the prospective mesoderm of sea stars (Hinman et al., 2007; McCauley et al., 2010). Hex, ets1, tbr, erg, tgif and foxN2/3 orthologs are first expressed in the central vegetal plate (prospective mesoderm) of sea star blastulae and then in distinct endodermal and mesodermal territories by the mid-gastrula stage. The mesodermal territory is likely established by a recursively wired circuit consisting of erg, hex and tgif, activated by tbr. The regulatory interactions between erg, hex, and tgif consist primarily of positive feedback loops that serve to ensure stable and robust expression of these genes. This subcircuit is conserved in sea stars and sea urchins as part of an ancient, mesoderm-specification network, although sea star embryos lack skeletogenic mesoderm. This conclusion is supported by the expression patterns of erg, hex, and tgif in cidaroids, which are consistent with the view that this subcircuit also has a broad, mesodermal function in that clade (Erkenbrack et al., 2016). Significantly, alx1 is expressed only at extremely low levels during embryonic development in sea stars, but is robustly expressed in skeletogenic centers of the adult rudiment (Gao and Davidson, 2008; Koga et al., 2016). Remarkably, forced expression of sea urchin alx1 in sea star embryos stimulates the expression of several biomineralization-related genes that are also regulated positively by alx1 in sea urchins, including p19, p16, carbonic anhydrase (can1), and lamG/egff2 (Koga et al., 2016). These findings are consistent with the view that heterochronic changes in alx1 expression played an important role in the evolutionary origin of the larval skeleton, as discussed below.
F). Skeletogenesis in Crinoids
Adult crinoids, like all adult echinoderms, have an extensive endoskeleton. Embryonic development has been described in detail in relatively few species, and in all these cases the embryo gives rise to a non-feeding larva that lacks skeletal elements. To date, there have been no studies of the gene regulatory basis of skeletogenesis in crinoids.
G). Insights from Comparative GRN Studies
Given what we know about the structure and deployment of the skeletogenic network in various echinoderm clades, we can begin to consider how skeletogenesis has evolved within the phylum (for recent reviews, see Koga et al., 2014, and Cary and Hinman, 2017). Insights that are emerging from this experimental model have important implications for our broader understanding of the evolution of developmental programs.
1). Co-option of an ancestral, adult program of skeletogenesis
The presence of a calcite-based endoskeleton in all adult echinoderms strongly suggests that this was a trait of their last common ancestor. This conclusion is supported by the recent demonstration of widespread, calcified biomineralized elements within (or beneath) the epidermis of adult hemichordates (Cameron and Bishop, 2012), the nearest outgroup to echinoderms. Although a calcitic endoskeleton is common to all adult echinoderms, embryonic skeletal elements are not found in all taxa. They are missing in the most basal group, crinoids (although, as noted above, no maximally indirect developing crinoids have been described) and in hemichordates, suggesting that the embryonic skeleton is an evolutionary novelty within echinoderms.
It has been widely hypothesized that the embryonic skeleton of sea urchins arose via co-option of the adult skeletogenic program (Gao and Davidson, 2008). According to this model, there has been a shift in the deployment of the ancestral GRN from the skeletogenic cells of the adult sea urchin to the micromeres of the early embryo. This would not only have required a heterochronic change but also shift in the lineage of cells that deploy the network, as micromeres do not contribute to adult skeletogenic structures, while macromere-derived mesenchyme cells do (Yajima, 2007). The co-option model is consistent with numerous similarities in the programs of gene expression in the skeletogenic cells of embryos and adults, including the expression of several key regulatory genes (e.g., alx1, ets1, and hex) and many effector genes (Gao et al., 2005; Gao and Davidson, 2008; Mann et al., 2008, Mann et al., 2010; Czarkwiani et al., 2013; Koga et al., 2016). Whether such a co-option occurred in a single evolutionary step, or perhaps two (first from the adult to post-feeding larval skeletogenic cells, then to the micromeres of the cleavage stage embryo), is unknown.
2). Possible examples of developmental drift
Despite the many similarities, there are also several differences in gene usage between the adult and embryonic skeletogenic programs in sea urchins. With respect to effector genes, for example, distinct members of the sm30 gene family are expressed in embryos and adults (Livingston et al., 2006). With respect to regulatory genes, Gao and Davidson (2008) did not detect expression of tbr, tel, foxO, or foxB in adult skeletogenic centers, at least at the stages examined, although all these genes are components of the embryonic GRN. Thus, although most key elements of the skeletogenic network are conserved, there are subtle differences as well. These differences may be examples of what has been called “developmental systems drift”, a term that has been put forward to describe stochastic evolutionary changes in homologous developmental pathways that do not result in major phenotypic changes (True and Haag, 2001; see also Halfon, 2017). Developmental systems drift may also contribute to taxon-specific differences in the embryonic skeletogenic GRN. For example, foxB, tbr, and dri, are components of the skeletogenic GRN in sea urchins but not in brittle stars (Czarkwiani et al., 2013; Dylus et al., 2016). Notably, in sea urchins all three of these regulatory genes provide inputs into relatively few effector genes compared with the cardinal regulators, ets1 and alx1. Other evidence supports the view that tbr has undergone substantial evolutionary modifications with respect to its network connectivity (Hinman et al., 2007). Variations in regulatory gene usage such as these likely reveal peripheral features of the echinoderm skeletogenic GRN that are not tightly constrained and therefore subject to drift.
3). Possible convergent evolution of larval skeletons
The formation of an extensive embryonic endoskeleton in two well-separated taxa, ophiuroids and echinoids, affords an opportunity to explore what appears to be an example of the convergent evolution of a complex structure. It is important to note that the alternative scenario, i.e, that the embryonic skeleton arose only once and was lost secondarily in sea stars (and reduced in sea cucumbers) cannot be rigorously excluded (Morino et al., 2012; 2016). Nevertheless, evidence that the initial deployment of the skeletogenic network in brittle stars is not associated unequal cell division, the latter a process critical to PMC specification in echinoids (Sharma and Ettensohn, 2010), or with the double-negative gate (Dylus et al., 2016) suggests that the adult skeletogenic program may have been co-opted separately in the two taxa. To further clarify this issue, it will be of considerable interest to elucidate the inputs into early skeletogenic regulatory genes (such as alx1) in brittle stars. As noted above, there is strong evidence that the double-negative gate alone is insufficient to account for the activation of alx1 in sea urchins (Sharma and Ettensohn, 2010), and further work may uncover common regulatory mechanisms in these two taxa.
4). Layering of cell specification pathways
A unique feature of modern-day euechinoid development is the precocious specification of the large micromeres, the founder cells of the PMC lineage. The specification of these cells relies, in part, on conserved gene networks that play a more general role in mesoderm specification in all echinoderms and even outside the phylum. One such mechanism is the vegetal targeting and activation of Dishevelled, which underlies the accumulation of nuclear β-catenin in the vegetal-most blastomeres of the cleavage stage embryo (Ettensohn, 2006). In all echinoderms that have been studied (and in other phyla) this axial patterning system is linked to the activation of regulatory genes that play a conserved role in the early specification of mesoderm and endoderm. This mechanism can therefore be viewed as part of a basal mesoderm specification network which likely includes the regulatory genes ets1, hex, erg, tgif, and foxN2/3, as well as the signaling gene delta. These genes are expressed in the vegetal plate of every echinoderm that has been studied, including sea stars, which lack a skeletogenic mesenchyme (see discussion and references in McCauley et al., 2012 and Dylus et al., 2016). Some part of this pan-mesodermal specification network probably also specifies a general “mesenchymal” (i.e., migratory) state. Evidence of such a pan-mesenchymal state comes also from the many similarities in the programs of gene expression of PMCs and migratory, non-skeletogenic mesenchyme in euechinoid sea urchins (Ettensohn et al., 2007; Rafiq et al., 2012) and from the highly conserved role of ets1 and MAPK signaling in the ingression of both skeletogenic and non-skeletogenic mesenchyme among echinoderms (Kurokawa et al., 1999; Röttinger et al., 2004; Serra et al., 2004; Koga et al., 2010).
Layered onto this basal “mesoderm” or “mesenchymal” specification pathway are additional gene regulatory mechanisms that are unique to the large micromere-PMC lineage and endow these cells with the distinctive cell behaviors and biosynthetic activities associated with skeletogenesis. As discussed above, micromere specification is dependent upon the unequal cell division that results from the displacement of the nuclei of the four vegetal blastomeres of the 8-cell stage embryo toward the vegetal pole. One critically important consequence of this unequal cell division may be to concentrate Pmar1 or other regulatory factors at sufficiently high levels in the micromeres to activate early regulatory genes such as alx1. The evolution of unequal cleavage in euechinoids must have involved the redistribution of cortical protein complexes that regulate spindle positioning. How this might have occurred in concert with changes in transcriptional regulatory processes (e.g, the invention of Pmar1 and its function in PMC specification) is an intriguing, open question.
5). Alx1 as a pivotal regulator of skeletogenesis and a model of transcription factor evolution
Recent studies have highlighted the pivotal role of alx1 as a primary driver of skeletogenic specification in all echinoderms. In all clades that form embryonic skeletons- euchinoids (including non-camarodont species; see Yamazaki and Minokawa, 2015), cidaroids, holothuroids, and ophiuroids- alx1 is expressed only by the skeletogenic lineage and is activated early in the specification process, prior to ingression. In all three clades in which functional studies have been performed (euechinoids, cidaroids, and holothuroids), alx1 has been shown to play an essential role in skeletogenic specification (Ettensohn et al., 2003; McCauley et al., 2012; Erkenbrack and Davidson, 2015.) As noted above, in euechinoids, alx1 has positive inputs into almost half of all genes selectively expressed by PMCs, highlighting its role as a primary determinant of PMC identity. Moreover, even in sea stars, which lack an embryonic skeleton entirely, alx1 is expressed selectively in the skeletogenic centers of the adult rudiment (Gao and Davidson, 2008: Koga et al., 2016). These observations establish alx1 as a pivotal component of the skeletogenic network.
The alx1 gene arose early in echinoderm evolution via a gene duplication event and Alx1 secondarily acquired the robust, skeletogenic function that it currently exhibits in all echinoderms (Koga et al., 2016; Khor and Ettensohn, 2017). Recent work has shown that the acquisition of this new regulatory function was associated with the exonization of a short (41 amino acid) motif located between the homeodomain of Alx1 and its C-terminus (Khor and Ettensohn, 2017). Remarkably, experimental insertion of this motif is sufficient to confer robust skeletogenic function on Alx4, the paralogue of Alx1. Thus, Alx1 provides a particularly striking example of a specific evolutionary change in transcription factor sequence that has led to a major shift in developmental function (see Lynch and Wagner, 2008). Another transcription factor in the PMC GRN, Tbr, has also undergone significant protein sequence changes, as evidenced by subtly different DNA binding preferences of the sea star and sea urchin orthologs both in vivo and in vitro (Cheatle-Jarvela et al., 2014; Cary et al., 2017).
6). Rapid evolution of effector genes
A final insight from comparative studies on the skeletogenic GRN is that effector genes evolve more rapidly than core regulatory machinery. Thus, although a core set of transcription factors has been implicated in skeletogenesis across the phylum, recent transcriptomic and proteomic studies have revealed surprising variation in biomineralization-related effector genes, such as those of the MSP130 and spicule matrix protein families, and point to recent, lineage-specific expansions (Seaver and Livingston, 2015; Flores and Livingston, 2017; Dylus et al., 2018). Indeed, effector gene duplication appears to have been rampant during the evolution of echinoderm biomineralization (Livingston et al., 2006; Adomako-Ankomah and Ettensohn, 2011; Rafiq et al., 2012; Rafiq et al., 2014; Ettensohn, 2014; Ettensohn and Dey, 2017). The flexibility in activation mechanisms described earlier, and the rapid evolution of effector genes, generally support the view that the periphery of a developmental GRN is less constrained than its regulatory core (see also Davidson and Erwin, 2006; Dylus et al., 2018).
V. Future Directions
A). The PMC GRN of Euechinoids
The skeletogenic GRN of euechinoid embryos is highly detailed and serves as a model of GRN architecture and as a gold-standard for comparisons with other taxa. Even in the case of this well-developed GRN, however, important questions remain unanswered. Progress on the following fronts will enhance the value of the euechinoid skeletogenic network as a general model of GRN architecture and, at the same time, increase its utility for evolutionary studies:
(1) The mechanism by which the GRN is initially deployed in the large micromeres is not fully understood; for example, questions remain concerning the role of HesC-mediated repression, the precise function of unequal cell division, the identity of the postulated ubiquitous transcription factors that activate early regulatory genes, and the mechanism by which the MAPK pathway is activated cell-autonomously in the PMC lineage.
(2) Recent transcriptome profiling studies have led to the identification of additional transcription factors expressed selectively by PMCs (e.g., Nk7, Alx4, Mitf, and others), but these have not yet been integrated into the network.
(3) An overarching issue is that, although much of the circuitry of the network has been revealed, in the vast majority of cases it is not known whether regulatory interactions are direct or indirect. Additional experimental dissection of PMC CRMs, by a variety of approaches, will clarify the fine structure of the network. In addition, temporal changes in the architecture of the GRN are poorly understood, mostly because conditional knockdowns of regulatory genes at late developmental stages, which might reveal developmental changes in network circuitry, have not been carried out.
(4) The mechanisms by which ectoderm-derived signals such as VEGF impinge on the network are unknown, as are the mechanisms by which the skeletogenic network is activated in non-micromere lineages after larval feeding commences (these may be prove to be related mechanisms).
(5) Undoubtedly, many critically important effector genes, and their precise roles in PMC behavior and skeletal morphogenesis, have yet to be uncovered.
B). Comparative Studies
Comparative studies of the skeletogenic GRN in echinoderms will continue to provide important new insights concerning the gene regulatory programs that underlie embryonic development and morphological evolution. Because the skeleteogenic network appears to have been transferred to different developmental addresses in a modular fashion, it follows that some of the most pressing questions concern evolutionary modifications to the mechanisms that activate the network:
For those taxa that lack micromeres yet form an early ingressing, skeletogenic mesenchyme (brittle stars and sea cucumbers), a central challenge is to elucidate the mechanisms that underlie the early, lineage-specific activation of alx1.
With respect to cidaroid sea urchins, it will be important to determine whether the variable, unequal cell division of vegetal blastomeres bears any causal relationship to skeletogenic specification. The late ingression of skeletogenic mesenchyme during cidaroid embryogenesis is also of interest; this may be a consequence of a delayed activation of the network, or there may be other regulatory mechanisms at work that slow the progression of the network once deployed
The mechanisms that control the activation of the skeletogenic GRN in the adult rudiment of any echinoderm are entirely unknown, but presumably reflect the most ancient regulatory mechanisms.
Striking evolutionary changes in the timing of the activation of the skeletogenic GRN have been documented in direct developing euechinoids (Parks et al., 1998; Israel et al., 2016), but the mechanisms are not well understood.
A different question concerns the extent to which the control of skeletogenesis by VEGF and VEGFR-Ig10 is strictly conserved in all developmental contexts throughout the phylum, and the intriguing, related problem of how evolutionary changes in the developmental expression of VEGF and other signaling ligands in the ectoderm occurred in parallel with changes in the developmental expression of the cognate receptors (see Morino et al., 2012; 2016).
Further analysis of skeletogenesis in hemichordates, the closest outgroup to the echinoderms, will be valuable in highlighting possible features of the ancestral program in echinoderms.
VI. Acknowledgements
This work was supported by grants from the National Science Foundation (IOS-1656580) and National Institutes of Health (R24OD023046) to C.A.E.
VII. References
- Adams DK, Sewell MA, Angerer RC, Angerer LM. 2011. Rapid adaptation to food availability by a dopamine-mediated morphogenetic response. Nat Commun 2:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adomako-Ankomah A, Ettensohn CA. 2011. P58-A and P58-B: novel proteins that mediate skeletogenesis in the sea urchin embryo. Dev Biol 353:81–93. [DOI] [PubMed] [Google Scholar]
- Adomako-Ankomah A, Ettensohn CA. 2013. Growth factor-mediated mesodermal cell guidance and skeletogenesis in the sea urchin embryo. Development 140:4212–4225. [DOI] [PubMed] [Google Scholar]
- Adomako-Ankomah A, Ettensohn CA. 2014. Growth factors and early mesoderm morphogenesis: insights from the sea urchin embryo. Genesis 52:158–172. [DOI] [PubMed] [Google Scholar]
- Akasaka K, Frudakis TN, Killian CE, George NC, Yamasu K, Khaner O, Wilt FH. 1994. Genomic organization of a gene encoding the spicule matrix protein SM30 in the sea urchin Strongylocentrotus purpuratus. J Biol Chem 269:20592–20598. [PubMed] [Google Scholar]
- Amore G, Davidson EH. 2006. cis-Regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev Biol 293:555– 564. [DOI] [PubMed] [Google Scholar]
- Andrey G, Mundlos S. 2017. The three-dimensional genome: regulating gene expression during pluripotency and development. Development 144:3646–3658. [DOI] [PubMed] [Google Scholar]
- Arnone MI, Andrikou C, Annunziata R. 2016. Echinoderm systems for gene regulatory studies in evolution and development. Curr Opin Genet Dev 39:129–137. [DOI] [PubMed] [Google Scholar]
- Barsi JC, Tu Q, Calestani C, Davidson EH. 2015. Genome-wide assessment of differential effector gene use in embryogenesis. Development 142:3892–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsi JC, Tu Q, Davidson EH. 2014. General approach for in vivo recovery of cell type-specific effector gene sets. Genome Res 24:860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH. 2009. Experimentally based sea urchin gene regulatory network and the causal explanation of developmental phenomenology. Wiley Interdiscip Rev Syst Biol Med 1:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M 2014. Signalosome assembly by domains undergoing dynamic head-to-tail polymerization. Trends Biochem Sci 39:487–495. [DOI] [PubMed] [Google Scholar]
- Boidron-Metairon IF. 1988. Morphological plasticity in laboratory-reared echinoplutei of Dendraster excentricus (Eschscholtz) and Lytechinus variegatus (Lamarck) in response to food conditions. J Exp Mar Biol Ecol 119:31–41. [Google Scholar]
- Cannon JT, Kocot KM, Waits DS, Weese DA, Swalla BJ, Santos SR, Halanych KM. 2014. Phylogenomic resolution of the hemichordate and echinoderm clade. Curr Biol 24:2827–2832. [DOI] [PubMed] [Google Scholar]
- Cameron CB, Bishop CD. 2012. Biomineral ultrastructure, elemental constitution and genomic analysis of biomineralization-related proteins in hemichordates. Proc Biol Sci 279:3041–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary GA, Cheatle Jarvela AM, Francolini RD, Hinman VF. 2017. Genome-wide use of high- and low-affinity Tbrain transcription factor binding sites during echinoderm development. Proc Natl Acad Sci U S A 114:5854–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary GA, Hinman VF. 2017. Echinoderm development and evolution in the post- genomic era. Dev Biol 427:203–211. [DOI] [PubMed] [Google Scholar]
- Cheatle-Jarvela AM, Brubaker L, Vedenko A, Gupta A, Armitage BA, Bulyk ML, Hinman VF. 2014. Modular evolution of DNA-binding preference of a Tbrain transcription factor provides a mechanism for modifying gene regulatory networks. Mol Biol Evol 31:2672–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA. 2005. P16 is an essential regulator of skeletogenesis in the sea urchin embryo. Dev Biol 283:384–396. [DOI] [PubMed] [Google Scholar]
- Chuang CK, Wikramanayake AH, Mao CA, Li X, Klein WH. 1996. Transient appearance of Strongylocentrotus purpuratus Otx in micromere nuclei: cytoplasmic retention of SpOtx possibly mediated through an alpha-actinin interaction. Dev Genet 19:231–237. [DOI] [PubMed] [Google Scholar]
- Croce J, Lhomond G, Lozano JC, Gache C. 2001. ske-T, a T-box gene expressed in the skeletogenic mesenchyme lineage of the sea urchin embryo. Mech Dev 107:159–162. [DOI] [PubMed] [Google Scholar]
- Czarkwiani A, Dylus DV, Oliveri P. 2013. Expression of skeletogenic genes during arm regeneration in the brittle star Amphiura filiformis. Gene Expr Patterns 13:464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle S, Davidson EH. 2011. Precise cis-regulatory control of spatial and temporal expression of the alx-1 gene in the skeletogenic lineage of S. purpuratus. Dev Biol 357:505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan K, Tanaka Y. 1990. Attachment of one spindle pole to the cortex in unequal cleavage. Ann N Y Acad Sci 582:108–119. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Cameron RA, Ransick A. S. 1998. Specification of cell fate in the sea urchin embryo: summary and some proposed mechanisms. Development 125:3269–3290. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311:796–800. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, et al. 2002. A genomic regulatory network for development. Science 295:1669–1178. [DOI] [PubMed] [Google Scholar]
- Duloquin L, Lhomond G, Gache C. 2007. Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134:2293–2302. [DOI] [PubMed] [Google Scholar]
- Dylus DV, Czarkwiani A, Blowes LM, Elphick MR, Oliveri P. 2018. Developmental transcriptomics of the brittle star Amphiura filiformis reveals gene regulatory network rewiring in echinoderm larval skeleton evolution. Genome Biol 19:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dylus DV, Czarkwiani A, Stangberg J, Ortega-Martinez O, Dupont S, Oliveri P. 2016. Large-scale gene expression study in the ophiuroid Amphiura filiformis provides insights into evolution of gene regulatory networks. Evodevo 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlet RB. 1985. Crystal axes in recent and fossil adult echinoids indicate trophic mode in larval development. Science 230:937–940. [DOI] [PubMed] [Google Scholar]
- Emlet RB. 1988. Larval form and metamorphosis of a “primitive” sea urchin, Eucidaris thouarsi (Echinodermata: Echinoidea: Cidaroidea), with implications for developmental and phylogenetic studies. Biol Bull 174:4–19. [DOI] [PubMed] [Google Scholar]
- Erkenbrack EM, Davidson EH. 2015. Evolutionary rewiring of gene regulatory network linkages at divergence of the echinoid subclasses. Proc Natl Acad Sci U S A 112:4075–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkenbrack EM, Petsios E. 2017. A conserved role for VEGF signaling in specification of homologous mesenchymal cell types positioned at spatially distinct developmental addresses in early development of sea urchins. J Exp Zool B Mol Dev Evol 328:423–432. [DOI] [PubMed] [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. 2011. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334:1091–1097. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA. 2006. The emergence of pattern in embryogenesis: regulation of beta- catenin localization during early sea urchin development. Sci STKE 361:pe48. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA. 2009. Lessons from a gene regulatory network: echinoderm skeletogenesis provides insights into evolution, plasticity and morphogenesis. Development 136:11–21. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA. 2013. Encoding anatomy: developmental gene regulatory networks and morphogenesis. Genesis 51:383–409. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA. 2014. Horizontal transfer of the msp130 gene supported the evolution of metazoan biomineralization. Evol Dev 16:139–48. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Dey D. 2017. KirrelL, a member of the Ig-domain superfamily of adhesion proteins, is essential for fusion of primary mesenchyme cells in the sea urchin embryo. Dev Biol 421:258–270. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Guss KA, Hodor PG, Malinda KM. 1997. The morphogenesis of the skeletal system of the sea urchin embryo. In: Reproductive Biology of Invertebrates, Progress in Developmental Biology 7:225–265. [Google Scholar]
- Ettensohn CA, Illies MR, Oliveri P, De Jong DL. 2003. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development 130:2917–2928. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Kitazawa C, Cheers MS, Leonard JD, Sharma T. 2007. Gene regulatory networks and developmental plasticity in the early sea urchin embryo: alternative deployment of the skeletogenic gene regulatory network. Development 134:3077–3087. [DOI] [PubMed] [Google Scholar]
- Fernandez-Serra M, Consales C, Livigni A, Arnone MI. 2004. Role of the ERK-mediated signaling pathway in mesenchyme formation and differentiation in the sea urchin embryo. Dev Biol 268:384–402. [DOI] [PubMed] [Google Scholar]
- Flores RL, Livingston BT. 2017. The skeletal proteome of the sea star Patiria miniata and evolution of biomineralization in echinoderms. BMC Evol Biol 17:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn CJ, Sharma T, Ruffins SW, Guerra SL, Crowley JC, Ettensohn CA. 2011. High-resolution, three-dimensional mapping of gene expression using GeneExpressMap (GEM). Dev Biol 357:532–540. [DOI] [PubMed] [Google Scholar]
- Frudakis TN, Wilt F. 1995. Two cis elements collaborate to spatially repress transcription from a sea urchin promoter. Dev Biol 172:230–241. [DOI] [PubMed] [Google Scholar]
- Fuchikami T, Mitsunaga-Nakatsubo K, Amemiya S, Hosomi T, Watanabe T, et al. 2002. T- brain homologue (HpTb) is involved in the archenteron induction signals of micromere descendant cells in the sea urchin embryo. Development 129:5205–5216. [DOI] [PubMed] [Google Scholar]
- Fujita K, Takechi E, Sakamoto N, Sumiyoshi N, Izumi S, Miyamoto T, Matsuura S, Tsurugaya T, Akasaka K, Yamamoto T. 2010. HpSulf, a heparan sulfate 6-O-endosulfatase, is involved in the regulation of VEGF signaling during sea urchin development. Mech Dev 127:235–245. [DOI] [PubMed] [Google Scholar]
- Gao F, Thompson JR, Petsios E, Erkenbrack E, Moats RA, Bottjer DJ, Davidson EH. 2015. Juvenile skeletogenesis in anciently diverged sea urchin clades. Dev Biol 400:148–58. [DOI] [PubMed] [Google Scholar]
- Gao F, Davidson EH. 2008. Transfer of a large gene regulatory apparatus to a new [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfield DA, Runcie DE, Babbitt CC, Haygood R, Nielsen WJ, Wray GA. 2013. The developmental address in echinoid evolution. Proc Natl Acad Sci USA 105:6091–6096.impact of gene expression variation on the robustness and evolvability of a developmental gene regulatory network. PLoS Biol 11:e1001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong YU, Killian CE, Olson IC, Appathurai NP, Amasino AL, Martin MC, Holt LJ, Wilt FH, Gilbert PU. 2012. Phase transitions in biogenic amorphous calcium carbonate.Proc Natl Acad Sci USA 109:6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin P, Jangoux M. 1998. From competent larva to exotrophic juvenile: a morphofunctional study of the perimetamorphic period of Paracentrotus lividus (Echinodermata, Echinoida). Zoomorphology 118:31–43. [Google Scholar]
- Gross JM, Peterson RE, Wu SY, McClay DR. 2003. LvTbx2/3: a T-box family transcription factor involved in formation of the oral/aboral axis of the sea urchin embryo. Development 130:1989–1899. [DOI] [PubMed] [Google Scholar]
- Guss KA, Ettensohn CA. 1997. Skeletal morphogenesis in the sea urchin embryo: regulation of primary mesenchyme gene expression and skeletal rod growth by ectoderm-derived cues. Development 124:1899–1908. [DOI] [PubMed] [Google Scholar]
- Halfon MS. 2017. Perspectives on gene regulatory network evolution. Trends Genet 33:436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkey MA, Klueg K, Sheppard P, Raff RA. 1995. Structure, expression, and extracellular targeting of PM27, a skeletal protein associated specifically with growth of the sea urchin larval spicule. Dev Biol 168:549–566. [DOI] [PubMed] [Google Scholar]
- Harkey MA, Whiteley HR, Whiteley AH. 1992. Differential expression of the msp130 gene among skeletal lineage cells in the sea urchin embryo: a three dimensional in situ hybridization analysis. Mech Dev 37:173–184. [DOI] [PubMed] [Google Scholar]
- Hart MW, Strathmann RR. 1994. Functional consequences of phenotypic plasticity in echinoid larvae. Biol Bull 186:291–299. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen A, and Davidson EH. 2007. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol 312:584–595. [DOI] [PubMed] [Google Scholar]
- Hodor PG, Ettensohn CA. 1998. The dynamics and regulation of mesenchymal cell fusion in the sea urchin embryo. Dev Biol 199:111–24. [DOI] [PubMed] [Google Scholar]
- Holy J, Schatten G. 1991. Differential behavior of centrosomes in unequally dividing blastomeres during fourth cleavage of sea urchin embryos. J Cell Sci 98:423–31. [DOI] [PubMed] [Google Scholar]
- Huang L, Li X, El-Hodiri HM, Dayal S, Wikramanayake AH, Klein WH. 2000. Involvement of Tcf/Lef in establishing cell types along the animal-vegetal axis of sea urchins. Dev Genes Evol 210:73–81. [DOI] [PubMed] [Google Scholar]
- Illies MR, Peeler MT, Dechtiaruk AM, Ettensohn CA. 2002. Identification and developmental expression of new biomineralization proteins in the sea urchin Strongylocentrotus purpuratus. Dev Genes Evol 212:419–431. [DOI] [PubMed] [Google Scholar]
- Israel JW, Martik ML, Byrne M, Raff EC, Raff RA, McClay DR, Wray GA. 2016. Comparative developmental transcriptomics reveals rewiring of a highly conserved gene regulatory network during a major life history switch in the sea urchin genus Heliocidaris. PLoS Biol 14:e1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingersoll EP, Wilt FH. 1998. Matrix metalloproteinase inhibitors disrupt spicule formation by primary mesenchyme cells in the sea urchin embryo. Dev Biol 196:95–106. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502. [DOI] [PubMed] [Google Scholar]
- Khor JM, Ettensohn CA. 2017. Functional divergence of paralogous transcription factors supported the evolution of biomineralization in echinoderms. Elife 6 pii: e32728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian CE, Wilt FH. 2008. Molecular aspects of biomineralization of the echinoderm endoskeleton. Chem Rev 108:4463–4474. [DOI] [PubMed] [Google Scholar]
- Killian CE, Wilt FH. 2017. Endocytosis in primary mesenchyme cells during sea urchin larval skeletogenesis. Exp Cell Res 359:205–214. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Nishimura Y, Kubotera N, Higuchi Y, Yamaguchi M. 2002. Transient activation of the micro1 homeobox gene family in the sea urchin (Hemicentrotus pulcherrimus) micromeres. Dev Genes Evol 212:1–10. [DOI] [PubMed] [Google Scholar]
- Knapp RT, Wu CH, Mobilia KC, Joester D. 2012. Recombinant sea urchin vascular endothelial growth factor directs single-crystal growth and branching in vitro. J Am Chem Soc 134:17908–17911. [DOI] [PubMed] [Google Scholar]
- Kniprath E 1974. Ultrastructure and growth of the sea urchin tooth. Calcif Tissue Res 14:211–228. [DOI] [PubMed] [Google Scholar]
- Koga H, Fujitani H, Morino Y, Miyamoto N, Tsuchimoto J, Shibata TF, Nozawa M, Shigenobu S, Ogura A, Tachibana K, Kiyomoto M, Amemiya S, Wada H. 2016. Experimental approach reveals the role of alx1 in the evolution of the echinoderm larval skeleton. PLoS One 11:e0149067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Matsubara M, Fujitani H, Miyamoto N, Komatsu M, Kiyomoto M, Akasaka K, Wada H. 2010. Functional evolution of Ets in echinoderms with focus on the evolution of echinoderm larval skeletons. Dev Genes Evol 220:107–115. [DOI] [PubMed] [Google Scholar]
- Koga H, Morino Y, Wada H. 2014. The echinoderm larval skeleton as a possible model system for experimental evolutionary biology. Genesis 52:186–192. [DOI] [PubMed] [Google Scholar]
- Kominami T, Takaichi M. 1998. Unequal divisions at the third cleavage increase the number of primary mesenchyme cells in sea urchin embryos. Dev Growth Differ 40:545–53. [DOI] [PubMed] [Google Scholar]
- Kurokawa D, Kitajima T, Mitsunaga-Nakatsubo K, Amemiya S, Shimada H, et al. 1999. HpEts, an ets-related transcription factor implicated in primary mesenchyme cell differentiation in the sea urchin embryo. Mech Dev 80:41–52. [DOI] [PubMed] [Google Scholar]
- Langelan RE, Whiteley AH. 1985. Unequal cleavage and the differentiation of echinoid primary mesenchyme. Dev Biol 109:464–475. [DOI] [PubMed] [Google Scholar]
- Leonard JD, Ettensohn CA. 2007. Analysis of dishevelled localization and function in the early sea urchin embryo. Dev Biol 306:50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Davidson EH. 2005. Gene regulatory networks for development. Proc Natl Acad Sci USA 102:4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chuang CK, Mao CA, Angerer LM, Klein WH. 1997. Two Otx proteins generated from multiple transcripts of a single gene in Strongylocentrotus purpuratus. Dev Biol 187:253–66. [DOI] [PubMed] [Google Scholar]
- Livingston BT, Killian CE, Wilt F, Cameron A, Landrum MJ, Ermolaeva O, Sapojnikov V, Maglott DR, Buchanan AM, Ettensohn CA. 2006. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev Biol 300:335–348. [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. 1999. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development 126:345– 57. [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Wagner GP. 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62:2131–54. [DOI] [PubMed] [Google Scholar]
- Lyons DC, Kaltenbach SL, McClay DR. 2012. Morphogenesis in sea urchin embryos: linking cellular events to gene regulatory network states. Wiley Interdiscip Rev Dev Biol 1:231–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makabe KW, Kirchhamer CV, Britten RJ, Davidson EH. 1995. Cis-regulatory control of the SM50 gene, an early marker of skeletogenic lineage specification in the sea urchin embryo. Development 121:1957–1970. [DOI] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M. 2008a. The sea urchin (Strongylocentrotus purpuratus) test and spine proteomes. Proteome Sci 6:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Poustka AJ, Mann M. 2008b. In-depth, high-accuracy proteomics of sea urchin tooth organic matrix. Proteome Sci 6:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Wilt FH, Poustka AJ. 2010. Proteomic analysis of sea urchin (Strongy- locentrotus purpuratus) spicule matrix. Proteome Sci 8:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Märkel K, Roeser U, Mackenstedt U, Klostermann M. 1986. Ultrastructural investigation of matrix-mediated biomineralization in echinoids (Echinodermata, Echinoidea). Zoomorphology 106:232–243. [Google Scholar]
- Martik ML, Lyons DC, McClay DR. 2016. Developmental gene regulatory networks in sea urchins and what we can learn from them. F1000Res 5:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Davidson EH. 2012. A comprehensive analysis of Delta signaling in pre-gastrular sea urchin embryos. Dev Biol 364:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Nam J, Davidson EH. 2010. High accuracy, high-resolution prevalence measurement for the majority of locally expressed regulatory genes in early sea urchin development. Gene Expr Patterns 10:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley BS, Weideman EP, Hinman VF. 2010. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol 340:200–208. [DOI] [PubMed] [Google Scholar]
- McCauley BS, Wright EP, Exner C, Kitazawa C, Hinman VF. 2012. Development of an embryonic skeletogenic mesenchyme lineage in a sea cucumber reveals the trajectory of change for the evolution of novel structures in echinoderms. Evodevo 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre DC, Lyons DC, Martik M, McClay DR. 2014. Branching out: origins of the sea urchin larval skeleton in development and evolution. Genesis 52:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellott DO, Thisdelle J, Burke RD. 2017. Notch signaling patterns neurogenic ectoderm and regulates the asymmetric division of neural progenitors in sea urchin embryos. Development 144:3602–3611. [DOI] [PubMed] [Google Scholar]
- Miller AK, Kerr AM, Paulay G, Reich M, Wilson NG, Carvajal JI, Rouse GW. 2017. Molecular phylogeny of extant Holothuroidea (Echinodermata). Mol Phylogenet Evol 111:110–131. [DOI] [PubMed] [Google Scholar]
- Miner BG. 2007. Larval feeding structure plasticity during pre-feeding stages of echinoids: not all species respond to the same cues. J Exp Mar Biol Ecol 343:158–165. [Google Scholar]
- Minokawa T, Wikramanayake AH, Davidson EH, 2005. cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev Biol 288:545–558. [DOI] [PubMed] [Google Scholar]
- Mitsunaga K, Akasaka K, Shimada H, Fujino Y, Yasumasu I, Numanoi H. 1986. Carbonic anhydrase activity in developing sea urchin embryos with special reference to calcification of spicules. Cell Differ 18:257–262. [DOI] [PubMed] [Google Scholar]
- Morino Y, Koga H, Tachibana K, Shoguchi E, Kiyomoto M, Wada H. 2012. Heterochronic activation of VEGF signaling and the evolution of the skeleton in echinoderm pluteus larvae. Evol Dev 14:428–436. [DOI] [PubMed] [Google Scholar]
- Morino Y, Koga H, Wada H. 2016. The conserved genetic background for pluteus arm development in brittle stars and sea urchin. Evol Dev 18:89–95. [DOI] [PubMed] [Google Scholar]
- Nam J, Davidson EH. 2012. Barcoded DNA-tag reporters for multiplex cis-regulatory analysis. PLoS One 7:e35934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Sato T, Morita Y, Yamazaki A, Akasaka K, Yamaguchi M. 2004. Structure, regulation, and function of micro1 in the sea urchin Hemicentrotus pulcherrimus. Dev Genes Evol 214:525–536. [DOI] [PubMed] [Google Scholar]
- Okazaki K 1975a. Normal development to metamorphosis In: Czihak G, editor. The Sea Urchin Embryo- Biochemistry and Morphogenesis. Springer-Verlag: Berlin: Ch. 7, pp 177–232. [Google Scholar]
- Okazaki K 1975b. Spicule formation by isolated micromeres of the sea urchin embryo. Am Zool 15:567–581. [Google Scholar]
- Oliveri P, Carrick DM, Davidson EH. 2002. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol 246:209–228. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH, McClay DR. 2003. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol 258:32–43. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. 2008. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA 105:5955–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otim O 2017. An empirical model of Onecut binding activity at the sea urchin SM50 C-element gene regulatory region. Int J Dev Biol 61:537–543. [DOI] [PubMed] [Google Scholar]
- Oulhen N, Swartz SZ, Laird J, Mascaro A, Wessel GM. 2017. Transient translational quiescence in primordial germ cells. Development 144:1201–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page L, Benson S. 1992. Analysis of competence in cultured sea urchin micromeres. Exp Cell Res 203: 305–311. [DOI] [PubMed] [Google Scholar]
- Parks AL, Parr BA, Chin JE, Leaf DS, Raff RA. 1988. Molecular analysis of heterochronic changes in the evolution of direct developing sea urchins. J Evol Biol 1:27–44. [Google Scholar]
- Peng CJ, Wikramanayake AH. 2013. Differential regulation of disheveled in a novel vegetal cortical domain in sea urchin eggs and embryos: implications for the localized activation of canonical Wnt signaling. PLoS One 8:e80693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington JT, Strathmann RR. 1990. Consequences of the calcite skeletons of planktonic echinoderm larvae for orientation, swimming, and shape. Biol Bull 179:121–133. [DOI] [PubMed] [Google Scholar]
- Peter IS. 2017. Regulatory states in the developmental control of gene expression. Brief Funct Genomics 16:281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH., 2015. Genomic Control Process: Development and Evolution. Academic Press. [Google Scholar]
- Piacentino ML, Ramachandran J, Bradham CA. 2015. Late Alk4/5/7 signaling is required for anterior skeletal patterning in sea urchin embryos. Development 142:943–952. [DOI] [PubMed] [Google Scholar]
- Piacentino ML, Zuch DT, Fishman J, Rose S, Speranza EE, Li C, Yu J, Chung O, Ramachandran J, Ferrell P, Patel V, Reyna A, Hameeduddin H, Chaves J, Hewitt FB, Bardot E, Lee D, Core AB, Hogan JD, Keenan JL, Luo L, Coulombe-Huntington J, Blute TA, Oleinik E, Ibn-Salem J, Poustka AJ, Bradham CA. 2016. RNA-Seq identifies SPGs as a ventral skeletal patterning cue in sea urchins. Development 143:703–714. [DOI] [PubMed] [Google Scholar]
- Raff RA. 1987. Constraint, flexibility, and phylogenetic history in the evolution of direct development in sea urchins. Dev Biol 119:6–19. [DOI] [PubMed] [Google Scholar]
- Rafiq K, Cheers MS, Ettensohn CA. 2012. The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development 139:579–590. [DOI] [PubMed] [Google Scholar]
- Rafiq K, Shashikant T, McManus CJ, Ettensohn CA. 2014. Genome-wide analysis of the skeletogenic gene regulatory network of sea urchins. Development 141:950– 961. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Patel NH, Hinman VF. 2015. Unraveling the tangled skein: the evolution of transcriptional regulatory networks in development. Annu Rev Genomics Hum Genet 16:103–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich A, Dunn C, Akasaka K, Wessel G. 2015. Phylogenomic analyses of Echinodermata support the sister groups of Asterozoa and Echinozoa. PLoS One 10:e0119627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-i Domingo R, Minokawa T, Davidson EH. 2004. R11: a cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev Biol 274:438–451. [DOI] [PubMed] [Google Scholar]
- Revilla-i Domingo R, Oliveri P, Davidson EH. 2007. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA 104:12383–12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho HK, McClay DR. 2011. The control of foxN2/3 expression in sea urchin embryos and its function in the skeletogenic gene regulatory network. Development 138:937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. 2006. Identification and developmental expression of the ets gene family in the sea urchin (Strongylocentrotus purpuratus). Dev Biol 300:35–48. [DOI] [PubMed] [Google Scholar]
- Roe JL, Park HR, Strittmatter WJ, Lennarz WJ. 1989. Inhibitors of metalloendoproteases block spiculogenesis in sea urchin primary mesenchyme cells. Exp Cell Res 181:542–550. [DOI] [PubMed] [Google Scholar]
- Röttinger E, Besnardeau L, Lepage T. 2004. A Raf/MEK/ERK signaling pathway is required for development of the sea urchin embryo micromere lineage through phosphorylation of the transcription factor Ets. Development 131:1075–1087. [DOI] [PubMed] [Google Scholar]
- Röttinger E, Saudemont A, Duboc V, Besnardeau L, McClay D, Lepage T. 2008. FGF signals guide migration of mesenchymal cells, control skeletal morphogenesis [corrected] and regulate gastrulation during sea urchin development. Development 135:353–365. [DOI] [PubMed] [Google Scholar]
- Ruffins SW, Ettensohn CA. 1996. A fate map of the vegetal plate of the sea urchin (Lytechinus variegatus) mesenchyme blastula. Development 122:253–263. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. 1987. Fourth cleavage of sea urchin blastomeres: microtubule patterns and myosin localization in equal and unequal cell divisions. Dev Biol 124:9–22. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. 1981. Development of a “primitive” sea urchin (Eucidaris tribuloides): irregularities in the hyaline layer, micromeres, and primary mesenchyme. Biol Bull 161:141–151. [Google Scholar]
- Seaver RW, Livingston BT. 2015. Examination of the skeletal proteome of the brittle star Ophiocoma wendtii reveals overall conservation of proteins but variation in spicule matrix proteins. Proteome Sci 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma T, Ettensohn CA. 2010. Activation of the skeletogenic gene regulatory network in the early sea urchin embryo. Development 137:1149–1157. [DOI] [PubMed] [Google Scholar]
- Shashikant T, Khor JM, Ettensohn CA. 2018. Global analysis of primary mesenchyme cell cis-regulatory modules by chromatin accessibility profiling. BMC Genomics, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J 2008. A protocol describing the principles of cis-regulatory analysis in the sea urchin. Nat Protoc 3:710–718. [DOI] [PubMed] [Google Scholar]
- Smith AB. 1997. Echinoderm larvae and phylogeny. Annu Rev Ecol Syst 28:219–241. [Google Scholar]
- Smith J, Davidson EH. 2008. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc Natl Acad Sci USA 105:20089–20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Theodoris C, Davidson EH. 2007. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science 318:794–797. [DOI] [PubMed] [Google Scholar]
- Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13:613–626. [DOI] [PubMed] [Google Scholar]
- Stock SR. 2014. Sea urchins have teeth? A review of their microstructure, biomineralization, development and mechanical properties. Connect Tissue Res 55:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathmann RR. 1971. The behavior of planktotrophic echinoderm larvae: mechanisms, regulation, and rates of suspension feeding. J Exp Mar Biol Ecol 6:109–160. [Google Scholar]
- Strathmann RR, Grunbaum D. 2006. Good eaters, poor swimmers: compromises in larval form. Int Comp Biol 46:312–322. [DOI] [PubMed] [Google Scholar]
- Summerton J, Weller D. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev 7:187–195. [DOI] [PubMed] [Google Scholar]
- Sun Z, Ettensohn CA. 2014. Signal-dependent regulation of the sea urchin skeletogenic gene regulatory network. Gene Expr Patterns 16:93–103. [DOI] [PubMed] [Google Scholar]
- Sun Z, Ettensohn CA. 2017. TGF-β sensu stricto signaling regulates skeletal morphogenesis in the sea urchin embryo. Dev Biol 421:149–160. [DOI] [PubMed] [Google Scholar]
- Swartz SZ, Reich AM, Oulhen N, Raz T, Milos PM, Campanale JP, Hamdoun A, Wessel GM. 2014. Deadenylase depletion protects inherited mRNAs in primordial germ cells. Development 141:3134–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet HC, Gehring M, Ettensohn CA. 2002. LvDelta is a mesoderm-inducing signal in the sea urchin embryo and can endow blastomeres with organizer-like properties. Development 129:1945–1955. [DOI] [PubMed] [Google Scholar]
- Szabó R, Ferrier DE. 2015. Another biomineralising protostome with an msp130 gene and conservation of msp130 gene structure across Bilateria. Evol Dev 17:195–197. [DOI] [PubMed] [Google Scholar]
- Tanaka Y 1976. Effects of surfactants on the cleavage and further development of the sea urchin embryo. I. The inhibition of micromere formation at the fourth cleavage. Develop Growth Differ 18:113–122. [DOI] [PubMed] [Google Scholar]
- Telford MJ, Lowe CJ, Cameron CB, Ortega-Martinez O, Aronowicz J, et al. 2014. Phylogenomic analysis of echinoderm class relationships supports Asterozoa. Proc Biol Sci 281:pii: 20140479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Erkenbrack EM, Hinman VF, McCauley BS, Petsios E, Bottjer DJ. 2017. Paleogenomics of echinoids reveals an ancient origin for the double-negative specification of micromeres in sea urchins. Proc Natl Acad Sci U S A 114:5870–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy B, Stöhr S. 2016. A new morphological phylogeny of the Ophiuroidea (Echinodermata) accords with molecular evidence and renders microfossils accessible for cladistics. PLoS One 11:e0156140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Haag ES. 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 3:109–19. [DOI] [PubMed] [Google Scholar]
- Tu Q, Cameron RA, Davidson EH. 2014. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev Biol 385:160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH. 2012. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Res 22:2079–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry LA, Hamilton PC, Killian CE, Wilt FH. 2000. Expression of spicule matrix proteins in the sea urchin embryo during normal and experimentally altered spiculogenesis. Dev Biol 225:201–213. [DOI] [PubMed] [Google Scholar]
- Veis A 2011. Organic matrix-related mineralization of sea urchin spicules, spines, test and teeth. Front Biosci 16:2540–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidavsky N, Addadi S, Schertel A, Ben-Ezra D, Shpigel M, Addadi L, Weiner S. 2016. Calcium transport into the cells of the sea urchin larva in relation to spicule formation. Proc Natl Acad Sci U S A 113:12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A, Weng W, Gumbiner BM, Venuti JM. 2000. TCF is the nuclear effector of the beta-catenin signal that patterns the sea urchin animal-vegetal axis. Dev Biol 217:230–243. [DOI] [PubMed] [Google Scholar]
- Voronina E, Wessel GM. 2006. Activator of G-protein signaling in asymmetric cell divisions of the sea urchin embryo. Dev Growth Differ 48:549–557. [DOI] [PubMed] [Google Scholar]
- Wahl ME, Hahn J, Gora K, Davidson EH, Oliveri P. 2009. The cis-regulatory system of the tbrain gene: Alternative use of multiple modules to promote skeletogenic expression in the sea urchin embryo. Dev Biol 335:428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel HE, Illies MR, Byrum CA, Xu R, Wikramanayake AH, Ettensohn CA. 2004. Differential stability of beta-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development 131:2947–2956. [DOI] [PubMed] [Google Scholar]
- Wessel GM, Etkin M, Benson S. 1991. Primary mesenchyme cells of the sea urchin embryo require an autonomously produced, nonfibrillar collagen for spiculogenesis. Dev Biol 148:261–272. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Huang L, Klein WH. 1998. Beta-catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc Natl Acad Sci USA 95:9343–9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. 2004. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis 39:194–205. [DOI] [PubMed] [Google Scholar]
- Wilt FH, Ettensohn CA. 2007. The morphogenesis and biomineralization of the sea urchin larval skeleton. In: Handbook of Biomineralization: Biological Aspects and Structure Formation (ed. Bauerlein E), pp.183–210. [Google Scholar]
- Wilt FH, Killian CE, Hamilton P, Croker L. 2008. The dynamics of secretion during sea urchin embryonic skeleton formation. Exp Cell Res 314:1744–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, McClay DR. 1988. The origin of spicule-forming cells in a ‘primitive’ sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development 103:305–15. [DOI] [PubMed] [Google Scholar]
- Wu SY, McClay DR. 2007. The Snail repressor is required for PMC ingression in the sea urchin embryo. Development 134:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M 2007. A switch in the cellular basis of skeletogenesis in late-stage sea urchin larvae. Dev Biol 307:272–281. [DOI] [PubMed] [Google Scholar]
- Yajima M, Umeda R, Fuchikami T, Kataoka M, Sakamoto N, Yamamoto T, Akasaka K. 2010. Implication of HpEts in gene regulatory networks responsible for specification of sea urchin skeletogenic primary mesenchyme cells. Zoolog Sci 27:638–646. [DOI] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. 2011. Small micromeres contribute to the germline in the sea urchin. Development 13:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasu K, Wilt FH. 1999. Functional organization of DNA elements regulating SM30alpha, a spicule matrix gene of sea urchin embryos. Dev Growth Differ 41:81–91. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Furuzawa Y, Yamaguchi M. 2010. Conserved early expression patterns of micromere specification genes in two echinoid species belonging to the orders clypeasteroida and echinoida. Dev Dyn 239:3391–3403. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Kawabata R, Shiomi K, Amemiya S, Sawaguchi M, Mitsunaga-Nakatsubo K, Yamaguchi M. 2005. The micro1 gene is necessary and sufficient for micromere differentiation and mid/hindgut-inducing activity in the sea urchin embryo. Dev Genes Evol 215:450–459. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Ki S, Kokubo T, Yamaguchi M. 2009. Structure-function correlation of micro1 for micromere specification in sea urchin embryos. Mech Dev 126:611–623. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Kidachi Y, Minokawa T. 2012. “Micromere” formation and expression of endomesoderm regulatory genes during embryogenesis of the primitive echinoid Prionocidaris baculosa. Dev Growth Differ 54:566–78. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Kidachi Y, Yamaguchi M, Minokawa T. 2014. Larval mesenchyme cell specification in the primitive echinoid occurs independently of the double-negative gate. Development 141:2669–2679. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Minokawa T. 2015. Expession patterns of mesenchyme specification genes in two distantly related echinoids, Glyptocidaris crenularis and Echinocardium cordatum. Gene Expr Patterns 17:87–97. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Minokawa T. 2016. Roles of hesC and gcm in echinoid larval mesenchyme cell development. Dev Growth Differ 58:315–326. [DOI] [PubMed] [Google Scholar]
- Zhu X, Mahairas G, Illies M, Cameron RA, Davidson EH, Ettensohn CA. 2001. A large-scale analysis of mRNAs expressed by primary mesenchyme cells of the sea urchin embryo. Development 128:2615–2627. [DOI] [PubMed] [Google Scholar]
- Zito F, Koop D, Byrne M, Matranga V. 2015. Carbonic anhydrase inhibition blocks vskeletogenesis and echinochrome production in Paracentrotus lividus and Heliocidaris tuberculata embryos and larvae. Dev Growth Differ 57:507–514. [DOI] [PubMed] [Google Scholar]


