Abstract
Spinophilin is the most abundant protein phosphatase 1 targeting protein in the postsynaptic density of dendritic spines. Spinophilin associates with myriad synaptic proteins to regulate normal synaptic communication; however, the full complement of spinophilin interacting proteins and mechanisms regulating spinophilin interactions are unclear. Here we validate an association between spinophilin and the scaffolding protein, disks large-associated protein 3 (SAP90/PSD-95 associated protein 3; SAPAP3). Loss of SAPAP3 leads to obsessive-compulsive disorder (OCD)-like behaviors due to alterations in metabotropic glutamate receptor (mGluR) signaling. Here we report that spinophilin associates with SAPAP3 in the brain and in a heterologous cell system. Moreover, we have found that expression or activation of group I mGluRs along with activation of the mGluR-dependent kinase, protein kinase C β, enhances this interaction. Functionally, global loss of spinophilin attenuates amphetamine-induced hyperlocomotion, a striatal behavior associated with dopamine dysregulation and OCD. Together, these data delineate a novel link between mGluR signaling, spinophilin, and SAPAP3 in striatal pathophysiology.
Keywords: Phosphatases, Signaling, Striatum, Scaffolding Proteins
Graphical Abstract
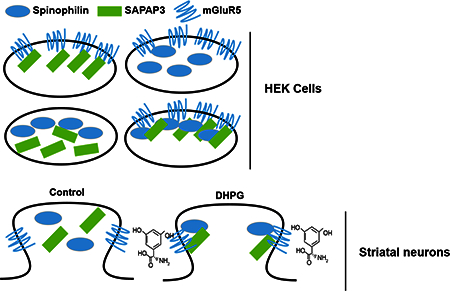
Introduction
Spinophilin knockout mice have deficits in long-term depression (LTD) in the striatum and hippocampus (Allen et al., 2006; Di Sebastino et al., 2016) and behavioral pathologies, such as enhanced cocaine-induced conditioned place preference (Allen et al., 2006) and increased sedation in response to α2 adrenergic agonists (Lu et al., 2010). As spinophilin is the major protein phosphatase 1 (PP11) targeting protein in the PSD (Colbran et al., 1997), many of spinophilin’s functions are thought to be due to alterations in spinophilin-dependent PP1 targeting. Using a global proteomics analysis of spinophilin immunoprecipitates, we have previously reported that spinophilin binding to multiple classes of proteins are diminished following dopamine depletion in the striatum (Hiday et al., 2017), an animal model of Parkinson disease (PD). One protein that was detected in this screen to potentially have a decreased association with spinophilin was SAP90/PSD-95 associated protein 3 (SAPAP3).
SAPAP3 is implicated in obsessive-compulsive disorder (OCD) pathologies as knockout of this protein leads to excessive grooming in mice, causing them to develop facial lesions (Welch et al., 2007). These alterations are due to increases in α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) internalization and alterations in synaptic plasticity (Chen et al., 2011; Wan et al., 2011). Furthermore, this internalization is caused by excessive metabotropic glutamate receptor (mGluR) 5-dependent activity and plasticity (Chen et al., 2011; Wan et al., 2011). Interestingly, others have found that in addition to SAPAP3, spinophilin associates with mGluR5 and loss of spinophilin enhances mGluR endocytosis (Di Sebastino et al., 2016). Moreover, treatment with the group I mGluR agonist, dihydroxyphenylglycine (DHPG), fails to elicit LTD in hippocampus of spinophilin KO animals. However, the mechanisms regulating the spinophilin/SAPAP3 complex are unknown. Here we identify SAPAP3 as a spinophilin interacting protein whose association with spinophilin is regulated by mGluR signaling. Specifically, mGluR expression and downstream activation of protein kinase C appear to play a role. Moreover, we find that SAPAP3 also regulates spinophilin binding to mGluR5. Functionally, we observed that a loss of spinophilin attenuates amphetamine-dependent locomotor sensitization, a model of dopamine-dependent striatal dysfunction that overlaps with compulsive behaviors. Together these data shed mechanistic light on the association of spinophilin with SAPAP3 and mGluR5 and delineate a novel potential role for spinophilin in striatal pathophysiology.
MATERIALS AND METHODS
Animals.
Dissected striatum for immunoprecipitations from WT and spinophilin KO animals were performed from female 2-month old mice. Dissected forebrains for immunoprecipitations from WT and SAPAP3 KO animals were performed from male (N=2, 1 WT and 1 KO) and female (N=2, 1 WT and 1 KO) 1-month old mice. Whole brains from these mice were a kind gift of Dr. Nicole Calakos (Duke University School of Medicine, Durham, NC). Striatal slices for DHPG studies were prepared from female, 1.5–3.5-month old mice. For amphetamine-sensitization studies, 3 – 5.5-month old male spinophilin KO (B6N(Cg)-Ppp1r9btml.1(KOMP)Vlcg/J N=6) or male (N=4) or female (N=2) control, WT littermate mice were used. The KO and WT mice from above had previously been subjected to rotarod analysis (Edler et al., 2018). All animal experiments were conducted under the guidelines of the Guide for the Care and Use of Laboratory Animals (ΝIH) and approved by the Indiana University-Purdue University School of Science Institutional Animal Care and Use Committee.
Generating DNA constructs.
The templates for the constructs used were: human spinophilin (Hiday et al., 2017); rat SAPAP3 (Uniprot ID P97838; Addgene 40217, Cambridge, MA); mouse mGluR5 (Uniprot ID Q3UVX5; Transomic Technology, Huntsville, AL BC144727); human ΡKCβ (Uniprot ID P05771; Transomic Technology BC036472); human PP1α (Uniprot ID P62136; Transomic Technology BC001888); and rat PP1γ1 (Uniprot ID P63088; (Carmody et al., 2008)). PCR products were generated from these template DNAs and inserted into pDONR221 (ThermoFisher, Waltham MA) for Gateway cloning into either pcDNA3.1/nV5-DEST (SAPAP3) or modified pcDNA3.1 vectors containing an HA-tag (spinophilin) His-tag (mGluR5), or myc-tag (ΡΚCβ, PP1α, PP1γ1, mGluR5) in place of the V5 tag. Spinophilin fragments and F451A mutant were generated from the full-length construct. PCRs (cloning or mutagenesis) were performed with Q5 Hot Start high-fidelity TAQ polymerase (New England Biolabs, Ipswich, MA), VAPRase (Vanderbilt Antibody Protein Resource, Vanderbilt University, Nashville TN), or a site-directed mutagenesis kit (QuickChange II, Agilent Technologies, Santa Clara, CA). All constructs and mutations were sequence validated (Genewiz, South Plainfield, NJ).
Mammalian cell protein lysis.
Human embryonic kidney 293FT cells (HEK293; ThermoFisher) were transfected in 25 cm2 flat bottom flasks with each relevant DNA construct in 500 μL serum-free Dulbecco’s modified Eagle’s medium using PolyJet reagent (SignaGen Laboratories Rockville, MD) in a 3:1 (reagent volume:DNA mass) ratio. Empty vector DNAs were used to transfect equal DNA concentrations. Cells were incubated overnight, medium was aspirated from each flask, and cells were washed one time with cold phosphate-buffered saline (PBS). HEK293 cells were then suspended in a KCl homogenization buffer (150 mM KCl, 50 mM Tris-HCl pH 7.5, 1 mM DTT, 2 mM EDTA, 1% Triton X-100) or RIPA homogenization buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.5, 1 mM EDTA, 1% NP-40, 1% Deoxycholate) containing protease inhibitors (ThermoFisher or Bimake, Houston, TX) and phosphatase inhibitors (20 mM sodium fluoride, 20 mM sodium orthovanadate, 20 mM β-glycerophosphate, and 10 mM sodium pyrophosphate; MilliporeSigma, St. Louis, MO or ThermoFisher).
PKCβ activation.
Prior to harvesting, HEK293 cells were incubated with 200 nM Phorbol 12-myristate 13 acetate (TPA; Tocris Bioscience, Minneapolis, MN) or vehicle (dimethyl sulfoxide, ThermoFisher) for 30 minutes. Following activation, cells were processed as above.
Slice culture pharmacology.
Slices (400 μm) containing the striatum were made in ice-cold sucrose (206 mM sucrose, 2.5 mM KCl, 0.5 mM CaCl2, 7 mM MgCl2, 1.2 mM NaH2PO4, 26 mM NaHC03, 5 mM glucose, 5 mM HEPES.) using a VT1200S vibratome (Leica Biosystems, Buffalo Grove, IL). Slices were allowed to equilibrate for 20 minutes in artificial cerebrospinal fluid (aCSF; 130 mM NaCl, 3.5 mM KCl, 1.25 mM NaH2PO4, 1.5 mM MgSO4-7 H2O, 2 mM CaCl2, 24 mM NaHCO3, 10 mM glucose) prior to treatment. Four striatal slices were generated per mouse and two were treated with vehicle and two with DHPG. Slices were incubated in a slice chamber at 30°C for 30 minutes in aCSF with vehicle or 50 μΜ DHPG (Tocris Bioscience). Following incubation, striata were dissected out of the slices and spinophilin and SAPAP3 were immunoprecipitated as described below. Two pooled slices were used to generate each N and each animal was its own control.
Immunoprecipitation assays
HEK293 cell lysates, striatal slices, or dissected striatum were homogenized, sonicated, and centrifuged at 10,000 xg for 10 min at 4°C. HEK293 cell lysates were lysed in KCl or RIPA buffer, whereas brain lysates were generated in RIPA or low-ionic (2 mM Tris, 2 mM EDTA, 1 mM DTT, protease and phosphatase inhibitors as above) buffer. Lysates were added to microfuge tubes with appropriate primary antibody: goat spinophilin antibody (SC-14774, Santa Cruz Biotechnology, Dallas Texas), SAPAP3 antibody (ABN325, MilliporeSigma), mGluR5 antibody (AB5675, MilliporeSigma), HA antibody (A190–107A Bethyl Laboratories), HA-magnetic beads (88836, ThermoFisher), c-myc antibody (A190–104A, Bethyl Laboratories), goat IgG Isotype Control (02–6202, ThermoFisher), or rabbit IgG Isotype control (10500C, ThermoFisher) and incubated at 4°C overnight with rotation. Protein G magnetic beads were then added to the samples and incubated for 2 h at 4°C with rotation. Beads were then washed three times with immunoprecipitation buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 0.5% (v/v) Triton X-100) using a magnet. Washed beads were incubated with 2X Laemmli sample buffer, heated at 70°C for 10 minutes, separated by SDS-PAGE, and immunoblotted.
Immunoblotting.
Immunoblotting was performed as previously described (Hiday et al., 2017). Briefly, inputs (0.5% −2%) or immunoprecipitates (25% of the immunoprecipitate) were separated by SDS-PAGE and blotted using antibodies to spinophilin, SAPAP3, mGluR5, PP1α (SC-7482; Santa Cruz Biotechnology), ΡΡ1γ1 (SC-6108; Santa Cruz Biotechnology), Phospho-PKC Substrate Motif antibody (6967, Cell Signaling Technology), HA-tag (as above or rabbit HA (SC-805, Santa Cruz Biotechnology)), c-myc tag (as above or mouse c-myc (SC-40, Santa Cruz Biotechnology)). Appropriate infrared secondary antibodies were used (Donkey anti goat, donkey anti rabbit, or donkey anti mouse conjugated to Alexa Fluor 690 or 780; ThermoFisher and Jackson Immunologicals) and imaging and fluorescence intensity measurements were made using Image Studio (LI-COR Biosciences, Lincoln, NE) set to automatic to ensure no saturated pixels. Linearity of signal intensity was validated from 125 ng −78.3 μg (spinophilin) and from 250 ng to 156.6 μg (SAPAP3 and mGluR5) of total lysate loaded (Figure S1). For HEK293 cell immunoblotting experiments, each transfection was performed in 25 cm2 flasks isolated on separate days and/or from a separate 75cm2 parent flask. Therefore, each transfection set is an independent experiment. Each HEK293 immunoblot was analyzed on a separate gel (see Quantitation and Statistics, below). For slice studies, 2–3 sets of gels were run with an N of 3–4 on each gel. Samples were normalized to the control values within each gel to allow for comparisons across gels.
Mammalian cell imaging
HEK293 cells were plated in 6-well plates on 18 mm x 18mm x 170nm (LxHxW) coverslips (Carl Zeiss Microscopy, Thornwood, NY; 474030–9000-000). Cells were stained with V5 rabbit antibody (SC-83849 Santa Cruz Biotechnology; for SAPAP3 detection), spinophilin goat antibody, and Myc mouse antibody (for mGluR5 detection). Primary antibodies were used at a 1:500 concentration. Secondary antibodies were a Donkey anti rabbit Alexa-Fluor 546 antibody, Donkey anti goat Alexa-Fluor 488, and a Donkey anti mouse Alexa-Flour 647 antibody (ThermoFisher; A10040, A11055, A31571, respectively). Secondary antibodies were used at a 1:2000 dilution. No signal was observed in non-transfected cells imaged using similar parameters (data not shown). Imaging was performed on a Zeiss 710 confocal microscope (Carl Zeiss Microscopy) using Zeiss Elyra PS.1 lasers in structured illumination microscopy (SIM) mode (we thank Dr. Teresa Mastracci at the Indiana Biosciences Research Institute for use of the Zeiss 710 microscope). Image processing was performed on Carl Zeiss Zen 2012 SP2 software, version 11.03.190 (Carl Zeiss Microscopy). Imaging parameters were set for each channel to ensure no saturated pixels. Single optical slices from different fields of view from two transfection conditions using SIM were performed.
Amphetamine sensitization.
For amphetamine-induced behavioral sensitization, WT or whole-body spinophilin KO mice (above) were given an intraperitoneal (i.p.) injection of 3.0 mg/kg of d-amphetamine (Sigma) or the saline volume equivalent to the control groups. There was an N of 3 in each group (WT saline, WT d-amphetamine, spinophilin KO saline, spinophilin KO d-amphetamine). d-Amphetamine was administered once every 24 hours for five consecutive days. Immediately after each i.p. injection, mice were placed in a VersaMax Animal Activity Monitoring System (Accuscan Instruments Inc., Columbus, OH). The 40 × 40 cm test chamber was equipped with evenly spaced intersecting photocell beams along the walls of the test chamber. Interruptions in intersecting photocell beams detected locomotor activity. The test chambers were sound-attenuating in that each 40 × 40 cm test chamber is situated in a 53 cm across x 58 cm deep x 40 cm high chamber with a house light and fan for ventilation and background noise. The VersaMax apparatus is interfaced with a Dell computer and the software is programed to track consecutive photocell beam interruptions that could be translated into distance traveled (cm). Data was collected in 1-minute intervals for 60 minutes.
Quantitation and statistics.
To normalize immunoprecipitates, the fluorescence intensity of the co-immunoprecipitated protein in the immunoprecipitate was divided by the fluorescence intensity of the immunoprecipitated protein in the immunoprecipitate as well as the intensity of the co-immunoprecipitated protein in the lysate. This allows for normalization of immunoprecipitation in the different conditions as well as normalizes any protein expression differences. To compare across multiple gels, the control and treated normalized value was divided by the control normalized value, to give a control value of 1. For comparison of a single control and treated group, a 1-column t-test comparing the treated group to a theoretical value of 1 was performed. For comparison of DHPG-treated striatal slices, 2–3 sets of gels were run with an N of 3–4 on each gel and a paired t-test was utilized. For TPA activation and ΡKCβ expression in HEK293 cells and for open-field, a two-way ANOVA was performed followed by a Tukey multiple comparison test or Sidak multiple comparison test to identify specific differences. Significance values are listed in the individual figures and legends.
RESULTS
Validation of the spinophilin and SAPAP3 protein interaction in mouse brain.
Mouse striata were homogenized and spinophilin and SAPAP3 immunoprecipitations were performed from the lysates. These isolated immunoprecipitates were separated by SDS-PAGE and immune complexes were analyzed via western blot. Spinophilin immunoprecipitates contained SAPAP3 whereas SAPAP3 immunoprecipitates contained spinophilin (Fig. 1A). Less co-immunoprecipitation was observed in IgG controls (Fig. 1A). To further validate this association, we utilized spinophilin KO animals as a control. As with IgG control there was more SAPAP3 (1.5 – 3-fold) in the WT when compared to the KO control. This was evaluated using two different homogenization buffers. We next evaluated SAPAP3 association with spinophilin in SAPAP3 KO mice. Like spinophilin KO mice, there was an ~3-fold greater association of spinophilin and SAPAP3 in the WT compared to SAPAP3 KO mice (Fig. 1C). Therefore, while there is some non-specific binding of SAPAP3 to the spinophilin antibody and spinophilin to the SAPAP3 antibody, these data suggest a selective interaction between these two proteins in striatum.
Figure 1. Spinophilin and SAPAP3 interact in striatal lysates.
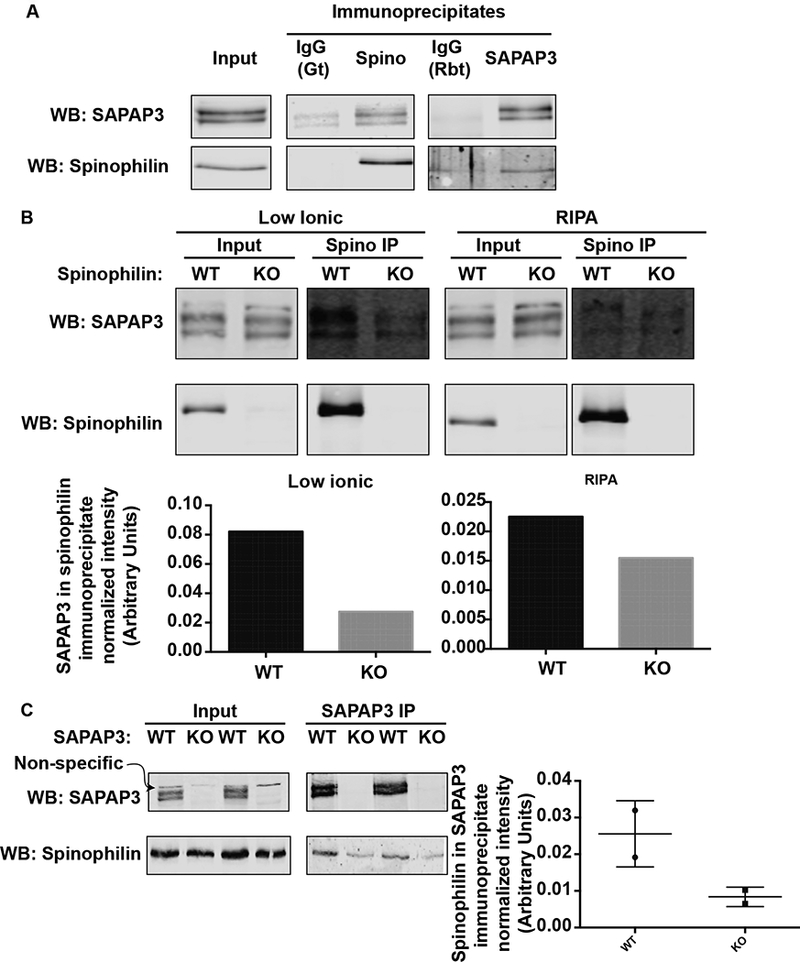
Striata from WT, spinophilin KO, or SAPAP3 KO mice were homogenized in RIPA (1A, 1B) or low-ionic (1B, 1C) buffer and immunoprecipitated with goat or rabbit IgG, goat spinophilin, or SAPAP3 rabbit antibody as detailed. Immunoprecipitates were immunoblotted with the same antibodies.
Mechanisms regulating the spinophilin, SAPAP3, mGluR5 complex.
It was recently shown that spinophilin can anchor to Group I mGluRs to inhibit their endocytosis (Sebastiano et al., 2016). We observed SAPAP3 in spinophilin immunoprecipitates (Fig. 1) confirming our previous proteomics data (Hiday et al., 2017). The above data suggest that spinophilin associates with SAPAP3 and group I mGluRs. SAPAP3 has been suggested to play a role in diminishing mGluR5-driven silencing of AMPARs in striatal medium spiny neurons (MSNs) (Wan et al., 2011); however, if mGluRs modulate the association between spinophilin and SAPAP3 is unknown. In agreement with Figure 1, we observed an association between spinophilin and SAPAP3 in a heterologous cell system when both spinophilin and SAPAP3 were expressed (Fig. 2A). In addition to spinophilin associating with SAPAP3 in heterologous cells, upon co-expression of mGluR5, there was a dramatic increase in the interaction between spinophilin and SAPAP3 (Fig. 2B).
Figure 2. mGluR5 and SAPAP3 coordinate spinophilin binding in a heterologous cell system.
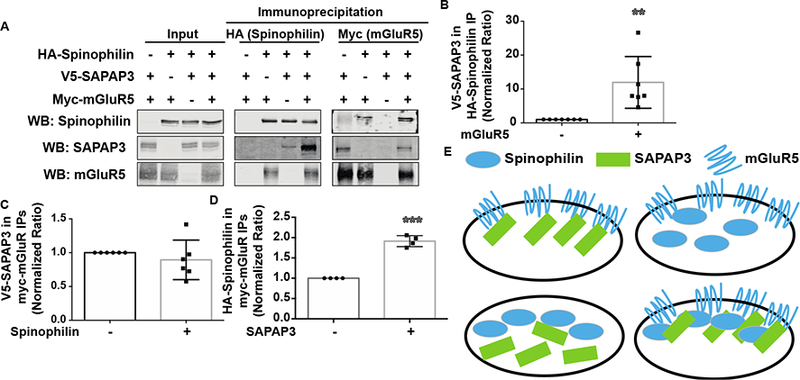
HEK293 cells were transfected with HA-tagged spinophilin, V5-tagged SAPAP3, and Myc-tagged mGluR5. A. Transfected lysates were immunoprecipitated with HA or Myc antibodies and immunoblotted with spinophilin, SAPAP3, and mGluR5 antibodies. B. Overexpression of mGluR5 significantly increased the amount of SAPAP3 in spinophilin immunoprecipitates. C. Overexpression of spinophilin had no significant effect on the association of SAPAP3 with mGluR5. D. SAPAP3 overexpression significantly enhanced the association of spinophilin with mGluR5. E. Schema of SAPAP3/mGluR5-dependent bridging of spinophilin. ** p<0.01, *** p <0.001.
To further explore mechanisms modulating the interactions between spinophilin, SAPAP3, and mGluR5, we evaluated SAPAP3 in mGluR5 immunoprecipitates in the absence or presence of spinophilin. Spinophilin appeared to have no effect on SAPAP3 binding to mGluR5 (Fig. 2C). However, SAPAP3 expression enhanced the association of spinophilin with mGluR5 (Fig. 2D). These data suggest that SAPAP3 and mGluR5 enhance spinophilin association with mGluR5 and SAPAP3, respectively.
While the above data suggest that spinophilin associates with SAPAP3 and mGluR5, it is unclear if all three proteins reside in the same area of the cell. We used high-resolution, SIM imaging to evaluate the localization of the transfected proteins. There were areas of co-localization of all 3 proteins (Fig. S2; white arrowheads). Moreover, histograms plotting pixel signal intensity by frequency in each of the 3 channels show areas of single protein expression (red, green, or blue), areas of overlap between SAPAP3 and spinophilin (yellow), areas of overlap between SAPAP3 and mGluR5 (Purple), areas of overlap between spinophilin and mGluR5 (Cyan), and areas where all three proteins co-localize (grey). Images from two different transfections and their corresponding histograms are shown in Fig. S2, whereas additional images are shown in Fig. S3. These data, along with the co-immunoprecipitation data suggest at least indirect, and possibly direct, interactions between the 3 proteins. However, while we show multiple cells with a range of transfection efficiencies, these data are limited based on image resolution and experimental design. Specifically, while SIM imaging has high resolution, the laser intensity may lead to detection outside the image plane (e.g. out of focus blurring). Moreover, overexpression of any protein may lead to areas of colocalization.
SAPAP3 and mGluR5 bind different domains on spinophilin
The above data suggest that SAPAP3 and mGluR5 enhance spinophilin binding to each individual component. We next delineated the regions on spinophilin that associate with SAPAP3 and mGluR5. We expressed deletion mutants of HA-tagged spinophilin (Fig. 3A) with SAPAP3 and mGluR5. Previous studies demonstrated that spinophilin associates with the PDZ ligand of group I mGluRs (Di Sebastino et al., 2016); however, where on spinophilin mGluR5 binds was not elucidated. mGluR5 bound to full-length and spinophilin containing amino acids 1–670, but did not bind to amino acids 1–460 or 1–301 on spinophilin (Fig. 3B). These data suggest that mGluR5 binds to spinophilin in the region containing amino acids 1–670 (containing the F-actin binding domain, receptor binding domain, and PDZ domain). To narrow this region down, we utilized a second set of fragments and found that the domain containing 151–670, but not 441–670, of spinophilin bound mGluR5 (Fig. 3C). This suggests that spinophilin binding to mGluR5 requires both the receptor binding domain and PDZ domain. Whereas SAPAP3 bound to full-length spinophilin, its association with spinophilin lacking the coiled-coil domain was abrogated (Fig. 3B). However, binding to additional fragments, including the 665–817 fragment was highly variable (data not shown). This may suggest that SAPAP3 binding to spinophilin requires the coiled-coil region, but this region is not sufficient for binding.
Figure 3. Mapping the domains on spinophilin that interact with SAPAP3 and mGluR5.
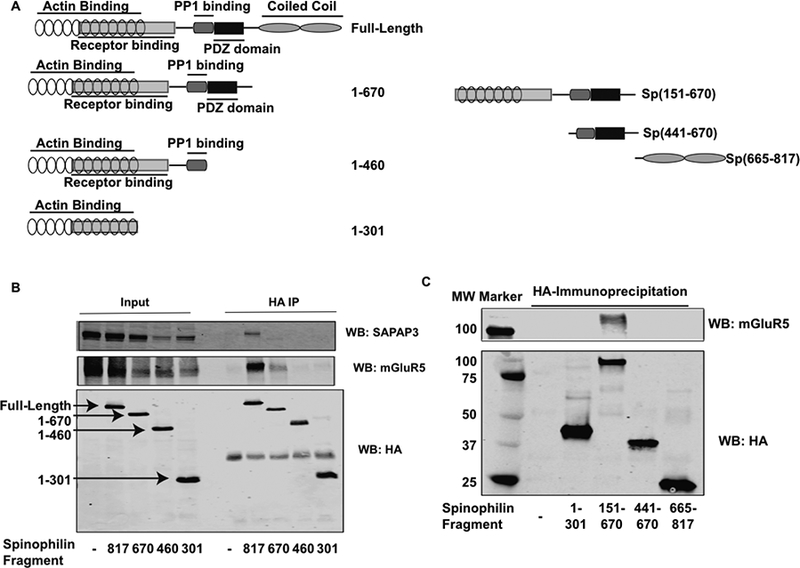
HEK293 cells were transfected with SAPAP3 and mGluR5 along with different deletion mutants of spinophilin. A. Schematic representation of spinophilin deletion mutants. Numbers refer to amino acids on different spinophilin fragments. B. HEK293 cells were transfected with SAPAP3 and mGluR5 alone or in the presence of different HA-tagged spinophilin deletion mutants. HEK293 cell lysates were immunoprecipitated with an HA antibody and immunoprecipitates were immunoblotted for SAPAP3, mGluR5, and HA. C. A different set of HA spinophilin fragments were immunoprecipitated and immunoblotted for HA and mGluR5. Experiments are representative of 3 independent trials.
PKCβ regulates the spinophilin and SAPAP3 interaction.
While our data suggest that SAPAP3 binding to spinophilin is regulated by mGluR5, the mechanisms by which mGluRs modulate spinophilin/SAPAP3 binding is unclear. mGluR5 is a Gq-coupled GPCR. Given this, it was of interest to explore the effects of protein kinase C beta (PKCβ), an mGluR5-activated second messenger, on the spinophilin/SAPAP3 interaction. Among the many PKC isoforms, ΡKCβ was selected among others due to its high striatal expression observed in the Allen Brain Atlas (Lein et al., 2007). To this end, PKCP was overexpressed with spinophilin and SAPAP3 in HEK293 cells. Endogenous levels of ΡKCβ are undetectable (Fig. 4A) and others suggest that ΡKCβ is not expressed in HEK293 cells (Ludowyke et al., 2006). Mock transfected or ΡKCβ transfected cells were treated with either vehicle or the phorbol ester, TPA, for 30 minutes prior to processing. Following treatment, HA-tagged spinophilin was immunoprecipitated and immune complexes were analyzed via western blot (Fig. 4B). In evaluating SAPAP3 expression, a Two-Way ANOVA comparison found a significant effect of activation (F(1,24) = 52.55; P<0.0001), PKC expression (F(l,24) = 8.491); P = 0.0076), and an interaction between variables (F(1,24) = 9.635; P = 0.0048). A Tukey multiple comparison’s test revealed no effect of overexpression of PKCβ in the non-active conditions on SAPAP3 expression. However, activation of endogenous PKC decreased SAPAP3 expression and overexpression of PKCβ under the TPA condition further decreased this expression (Fig. 4B). This decrease was not due to cell death or regulation of the expression plasmid as ponceau stains were equal and there was no difference in the expression of spinophilin (Fig. 4B). In evaluating the spinophilin/SAPAP3 interaction, a Two-Way ANOVA comparison found a significant effect of both PKC expression (F(l,28)=24.97; PO.OOOl) and activation (F(l,28)=4.613 P0.0405) with no interaction effect (F(l,28) = 3.279; P0.0809). A Tukey post-hoc test found a trend for a significant difference in interaction upon expression of PKCβ in the non-activated state (P=0.1338) and a significant effect of PKCβ in the activated state (P = 0.0003). Moreover, activation of overexpressed PKCβ further increased the association compared to PKCβ expression alone P=0.0429; Fig. 4C). Given the TPA-dependent decrease in SAPAP3 protein levels, the increase in association may be due to technical floor/ceiling effects; however, there was a significant PKCβ effect by ANOVA and a trend for an increased association between spinophilin and SAPAP3 when PKCβ was overexpressed (in the absence of TPA activation), a condition that did not alter SAPAP3 expression. These data suggest that activation of PKCβ decreases SAPAP3 expression but enhances the spinophilin/SAPAP3 association.
Figure 4. Activation of PKC𝛃 enhanced spinophilin/SAPAP3 interaction.
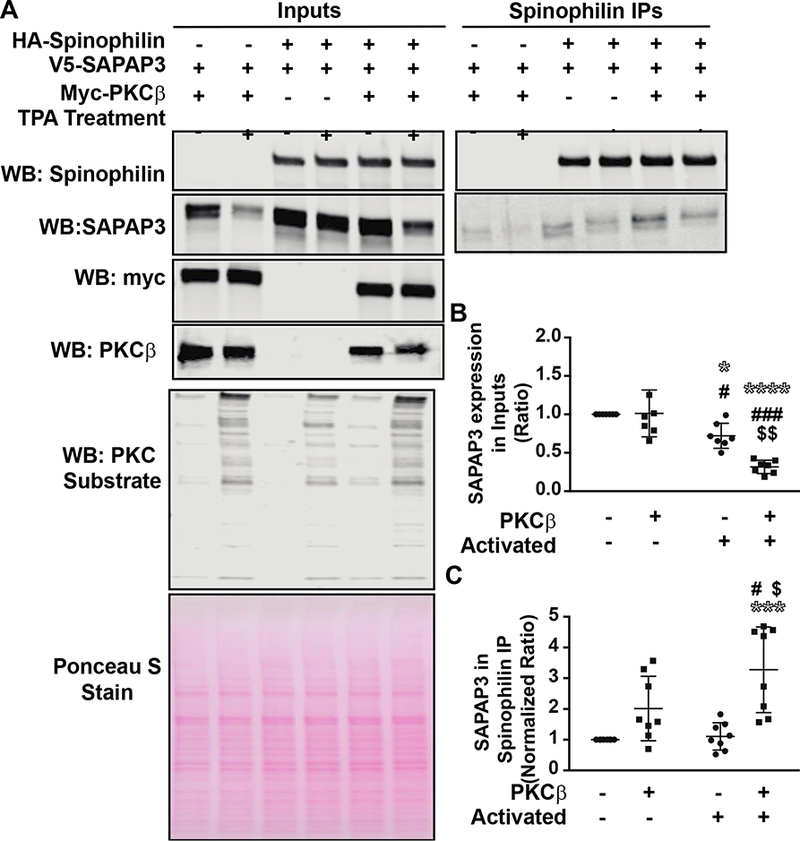
HEK293 cells were transfected with spinophilin and SAPAP3 in the absence or presence of PKCβ. The following day, HEK293 cells were treated with 200 nM TPA or vehicle for 30 minutes. A. Lysates were immunoprecipitated with an HA spinophilin antibody and lysates and immunoprecipitates were stained with Ponceau S and immunoblotted for spinophilin, SAPAP3, PKCβ, and PKC substrate substrate antibody. B. Activation of PKC and overexpression of PKCβ along with activation of PKC significantly decreased the expression of SAPAP3 compared to: no overexpression or activation (* symbol), PKCβ overexpression only (# symbol), or activation only ($ symbol). C. Activation of PKC along with overexpression of PKCβ significantly increased the association between spinophilin and SAPAP3. *,#,$ p<0.05, $$ p<0.01, ***, ###p <0.001, **** p<0.0001.
Overexpression of PP1α decreases the interaction between spinophilin and SAPAP3
Given the above results suggesting that PKC-specific phosphorylation events play an important role on the spinophilin/SAPAP3 interaction, we were further interested to investigate the role a specific phosphatase has on the protein interaction. Specifically, the importance of PP1α, promiscuous phosphatase targeted to substrates by spinophilin, was tested (Fig. 5A). Results showed that in contrast to PKCβ, PP1α dramatically attenuated the spinophilin/SAPAP3 interaction (Fig. 5B). As mGluR5 enhances spinophilin binding to SAPAP3, we wanted to determine if overexpression of PP1α modulated the association of spinophilin with mGluR5. As with SAPAP3, overexpression of PP1α decreased binding of spinophilin with mGluR5 (Fig. 5C, 5D). One possible explanation for overexpression of PP1α regulating the spinophilin/SAPAP3/mGluR5 association is that PP1α competitively displaces mGluR5 and/or SAPAP3 from spinophilin. To test this, we transfected WT or a PPl-deficient binding mutant (F451A) of spinophilin in the presence of SAPAP3 and mGluR5 and immunoblotted for spinophilin, SAPAP3, and mGluR5 (Fig. 5E). These studies were performed in the absence of overexpressed PP1. WT spinophilin had greater endogenous (HEK293 cell) PP1 binding than F451A mutant spinophilin (Fig. 5E). If PP1 were competing for spinophilin binding to SAPAP3 and mGluR5, we would expect to see increased binding of mutant spinophilin; however, we observed no change in binding of an F451A mutant spinophilin to either SAPAP3 or mGluR5 (Fig. 5F, 5G). These data demonstrate that PP1α expression decreases spinophilin binding to both mGluR5 and SAPAP3 and that this decreased association is not due to competition between spinophilin and PP1 binding. This suggests a role for PP1 activity on modulating the association of spinophilin with mGluR5 and SAPAP3.
Figure 5. Overexpression of PP1𝛂 decreased spinophilin binding to mGluR5 and SAPAP3.
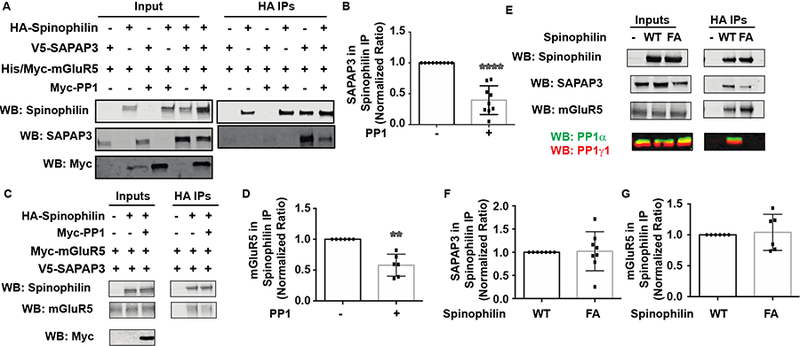
HEK293 cells were transfected with SAPAP3 and mGluR5 and/or spinophilin in the absence or presence of PPlα. A. HA immunoprecipitates and/or lysates were immunoblotted with spinophilin, SAPAP3 and Myc antibodies. B. Overexpression of PP1 significantly decreased SAPAP3 association with spinophilin. C. HA immunoprecipitates and/or lysates were immunoblotted with spinophilin, mGluR5, and Myc antibodies. D. Overexpression of PP1 decreased mGluR association with spinophilin. E. WT or F451A (FA) mutant spinophilin were transfected with SAPAP3 and mGluR5. Mutant spinophilin did not bind endogenous PP1, but had no effect on the association of spinophilin with SAPAP3 (F) or mGluR5 (G). **P<0.01, ****P<0.0001.
In vivo regulation of spinophilin SAPAP3 interaction by mGluR signaling
Whereas mGluR expression and PKC activity modulate spinophilin/SAPAP3 association in HEK293 cells, the role of mGluR activation in regulating the spinophilin/SAPAP3 association in striatum is unclear. To begin to test the role of mGluR activation on modulating the spinophilin/SAPAP3 association in neurons, we treated striatal slices from adult, wild-type mice with 50 μΜ DHPG for 30 minutes. Following treatment, striata were dissected from the slice and homogenized. Subsequently, spinophilin and SAPAP3 were immunoprecipitated and immunoblotted for spinophilin and SAPAP3 (Fig. 6A). DHPG treatment on slice cultures caused a trend towards an increased association in the spinophilin/SAPAP3 interaction (Fig. 6A). Taken together these data suggest that mGluR5 activation may enhance the interaction of spinophilin with SAPAP3 in striatal slices (Fig. 6B); however, there is high variability associated with these assays.
Figure 6. Association of spinophilin with SAPAP3 following DHPG activation of mGluRs.
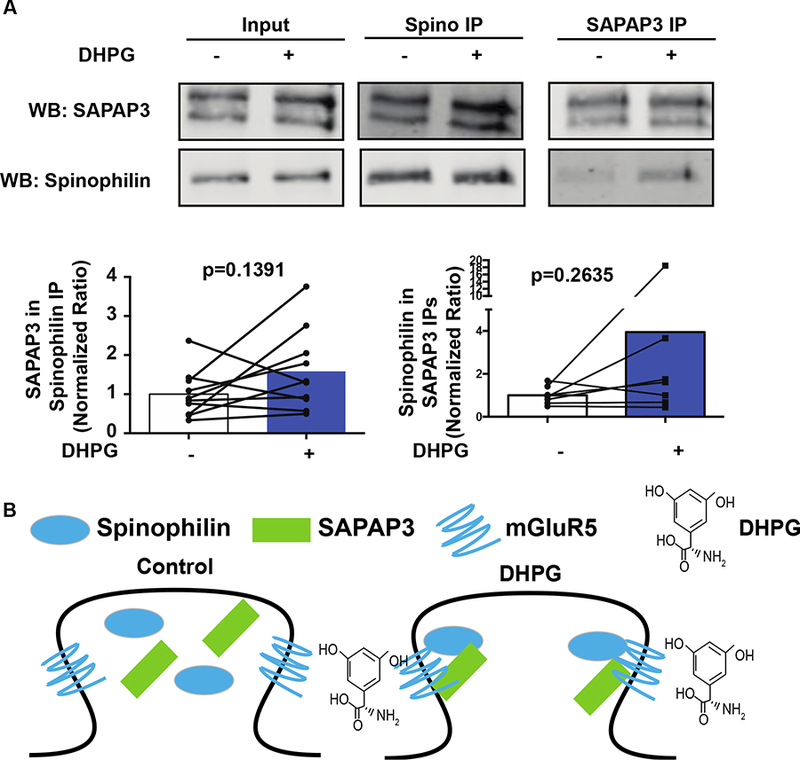
Striatal slices were incubated with DHPG or vehicle for 30 minutes. Following incubation, striata were dissected and immunoprecipitated for spinophilin and SAPAP3. A. Immunoblots of control and treated striatal lysates and spinophilin and SAPAP3 immunoprecipitates. B. Activation of group I mGluRs led to a trend towards an increase in the association of spinophilin with SAPAP3.
Loss of spinophilin attenuates amphetamine-induced locomotor sensitization.
SAPAP3 KO mouse models of OCD have alterations in monoamine release and/or metabolism (Wood et al., 2018). Moreover, blockade of mGluR5 signaling attenuates amphetamine-induced hyperactivity (Gill et al., 2012). Conversely, loss of spinophilin enhances responses to cocaine-induced conditioned place preference (Allen et al., 2006). However, the role of spinophilin in modulating amphetamine-dependent locomotor sensitization has not been evaluated. We treated WT or spinophilin KO mice with amphetamine and measured locomotor activity in an open field assay. A Two-Way ANOVA demonstrated a trend for a significant Day effect (F (4,16) = 2.468; P = 0.0867), a significant genotype effect (F(1,4) = 8.827; P = 0.0411), and a significant interaction effect (F(4,16) = 18.80; P<0.0001). Moreover, a Tukey post-hoc test comparing the means within each genotype across days shows a significant increase in locomotion in WT amphetamine treated animals at days 2 −5 compared to day 1. In contrast, there is no difference in locomotor activity in the spinophilin KO mice at days 2–4, and a significant decrease at day 5, compared to day 1 (Fig. 7). Furthermore, a Sidak post-hoc test across genotypes of amphetamine-treated animals at each day revealed significant differences in locomotion between WT and KO animals at days 3–5. These data suggest that loss of spinophilin prevents amphetamine-induced locomotor sensitization.
Figure 7. Spinophilin KO mice do not undergo amphetamine-dependent sensitization.
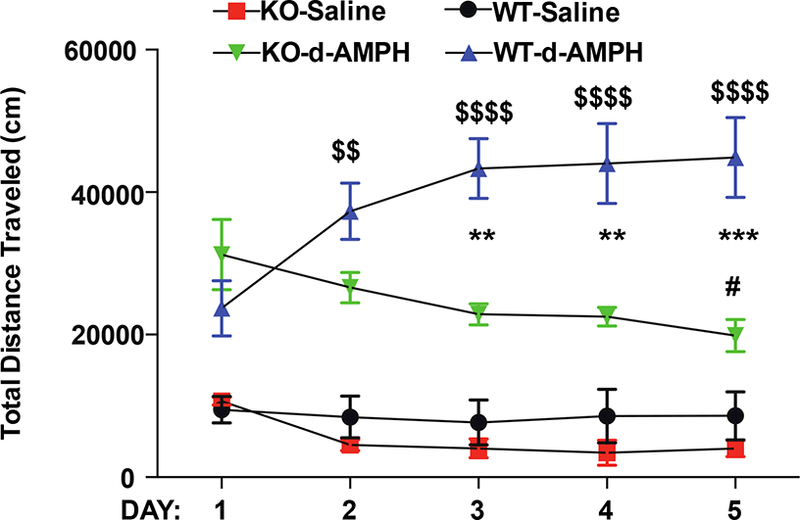
Spinophilin KO or WT control littermates were injected with saline or d-amphetamine daily for 5 days. Immediately following injection, animals were placed in an open-field chamber and total distance traveled over 60 minutes was evaluated. Amphetamine treated WT animals had increasing levels of distance traveled on days 2–5 compared to day 1 ($ symbols). In contrast, spinophilin KO mice had no change in travel on days 2–4 and a decrease in distance traveled on day 5 (# symbol). Moreover, there was a greater distance traveled in the WT compared to KO spinophilin mice on days 3, 4, and 5 (* symbols). (#P<0.05, ** and $$ P< 0.01, *** P<0.001, and $$$$ P<0.0001).
DISCUSSION
Spinophilin-SAPAP3-mGluR5 association
Previous studies have identified group I mGluRs in spinophilin immunoprecipitates (Di Sebastino et al., 2016). Moreover, our previous studies have detected the mGluR-interacting protein, homer, in spinophilin immunoprecipitates isolated from WT, but not spinophilin KO mice (Baucum et al., 2010). In a recent study using proteomics approaches, we found that spinophilin associates with SAPAP3, a regulator of mGluRs (Ade et al., 2016; Chen et al., 2011), and that this association may be attenuated in animal models of PD (Hiday et al., 2017). Our current data demonstrate that spinophilin associates with both SAPAP3 and mGluR5 in a heterologous cell system. Moreover, our data suggests that spinophilin interacts with SAPAP3 via the coiled-coil region of spinophilin (amino acids 665–817) whereas it associates with mGluR5 via the receptor binding domain/PDZ domain (amino acids 151–670). These data are consistent with previous reports of functional and/or biochemical association of spinophilin with G-protein coupled receptors such as: the D2 dopamine receptor via amino acids 100–371 on spinophilin (Smith et al., 1999), the α2-adrenergic receptor via amino acids 169–255 on spinophilin (Richman et al., 2001), the μ-opioid receptor (Charlton et al., 2008), and the muscarinic M3 receptor (Ruiz de Azua et al., 2012). Moreover, our data suggesting that amino acids 151–670 are required for mGluR5 binding, complement a recent study that found that spinophilin associates with the PDZ binding motif of group I mGluRs (Di Sebastino et al., 2016). The PDZ domain on spinophilin contains residues 493–602; however, a fragment containing amino acids 441– 670 of spinophilin was not sufficient to bind mGluR5. Therefore, whereas the PDZ domain on spinophilin is required for binding to mGluR5, it is not sufficient for binding. One possible explanation is that spinophilin binds mGluR via both the spinophilin receptor binding and PDZ domains. This is consistent with observations using rodent spinophilin that suggest spinophilin may associate with multiple domains on group I mGluRs (Di Sebastino et al., 2016).
Genetic deletion of SAPAP3 increases mGluR signaling and leads to motor behavior pathologies such as excessive grooming in rodents, a mouse model of OCD (Ade et al., 2016; Chen et al., 2011; Wan et al., 2014; Welch et al., 2007). While there is a link between group I mGluR signaling and SAPAP3, our data are the first to show that spinophilin association with SAPAP3 is enhanced by mGluR5 expression and/or activity. Moreover, we also found that SAPAP3 increases targeting of spinophilin to mGluR5. Whereas SAPAP3 and mGluR5 bind different domains on spinophilin, we did not directly test if SAPAP3 or mGluR5 enhance in vitro binding of spinophilin to mGluR5 and SAPAP3, respectively. Given that mGluR5 is a membrane protein, proper folding and localization may be important in spinophilin binding and may limit these types of approaches. Using immunofluorescence, we observed areas of co-localization between all 3 proteins in a heterologous cell system. It is important to note that the overlap was not complete and there were many areas of co-localization between two of the three proteins. Moreover, while we used a high-resolution imaging approach (SIM), these data have limitations based on transfection efficiency and imaging resolution/saturation that may limit the ability to detect a direct versus indirect interaction. However, taken with the co-immunoprecipitation data, our data suggest an association between spinophilin, SAPAP3, and mGluR5. While our co-isolation studies denote an interaction, we cannot conclude a direct interaction between the 3 proteins and future studies will need to use direct biochemical analyses in cells (e.g. Forster resonance energy transfer) or in vitro to address this question.
Spinophilin SAPAP3 interaction is regulated by PKC.
The association of spinophilin with SAPAP3 is enhanced by mGluR5 and the interaction between spinophilin and mGluR5 is enhanced by SAPAP3. These data suggest that SAPAP3 and mGluR5 may act as a bridge for spinophilin binding. However, in addition to this bridge function, we wanted to determine if mGluR signaling may regulate the spinophilin/SAPAP3 interaction. Group I mGluRs signal through Gq receptors, activation of which increases PKC activity (Niswender and Conn, 2010). Overexpression and activation of the striatal enriched PKC isoform, PKCβ, (Lein et al., 2007) also enhanced the association between spinophilin and SAPAP3. It is important to note that overexpression and activation of PKCβ enhanced the association between spinophilin and SAPAP3 by ~3.3-fold in HEK293 cells whereas overexpression of mGluR5 enhanced the association of spinophilin with SAPAP3 by ~12-fold. This may suggest both mGluR5-dependent activation of PKCβ and possibly bridging may enhance the binding of spinophilin with SAPAP3. As Gq-coupled receptors also lead to increases in intracellular calcium release, we cannot rule out that calcium is also playing a role in modulating the association between spinophilin and SAPAP3.
In addition to our heterologous cell studies, we found that activation of striatal mGluRs using DHPG showed a trend for an increased interaction between spinophilin and SAPAP3 in a slice pharmacology study. We found that treatment of striatal slices with 50 μΜ DHPG for 30 minutes increased the association of spinophilin with SAPAP3 by −1.5– 3-fold. While this increase was not statistically significant, the fold increase was similar to what was observed in HEK293 cells. Moreover, it is important to note that the main striatal neuron, the GABAergic medium spiny neuron, is separated into direct and indirect pathway classes and there may be differential responses in these two classes. Our KO data suggest specific, but not completely selective, co-immunoprecipitation of SAPAP3 with spinophilin. Therefore, non-specific binding may influence an ability to detect DHPG-dependent changes in the spinophilin/SAPAP3 interaction. Mechanistically, activation of mGluRs by DHPG will increase calcium via both extracellular and intracellular sources as well as enhance production of diacylglycerol (DAG) (Wisniewski and Car, 2002). Conventional PKC isoforms such as PKCβ are activated by both calcium and DAG. However, in addition to mGluR-dependent regulation of calcium and PKC activity in slices, agonist-dependent conformational changes in mGluRs (Niswender and Conn, 2010) may also modulate how spinophilin associates with mGluRs and SAPAP3. Conformational changes in mGluRs have been previously shown to modulate association with proteins such as prion protein (Haas et al., 2014). Moreover, DHPG activation at a higher concentration and time (100 μΜ and 1 hour) has been shown to lead to internalization of mGluRs (Hong et al., 2009) which may differentially regulate the spinophilin/SAPAP3 interaction. Interestingly, when we incubated striatal slices with 50 μΜ DHPG for 1 hour, there was no difference in the interaction between SAPAP3 and spinophilin and the interaction actually trended towards a decrease (24%−32% decrease; data not shown). Therefore, increases in mGluR signaling may be counter-balanced by internalization of the receptor. Taken together, we cannot completely rule out that alterations in mGluR confirmation, in addition to activation of PKC, regulate the spinophilin-SAPAP3 interaction and future studies will need to test if conditions that can increase mGluR signaling without internalizing the receptor enhance the association between spinophilin and SAPAP3.
Spinophilin SAPAP3 interaction is regulated by PP1
Overexpression of the catalytic subunit of PP1α decreased the association of spinophilin with mGluR5 and SAPAP3. PP1 binding to spinophilin occurs in a centrally-located PP1 binding domain containing a key residue, Phe451 (Ragusa et al., 2011; Ragusa et al., 2010). Spinophilin targets PP1 to modulate synaptic protein phosphorylation, either by promoting dephosphorylation or by inhibiting PP1 activity to increase phosphorylation (Bielas et al., 2007; Hu et al., 2015; Ragusa et al., 2010). Overexpression of PP1α may compete with binding of spinophilin and associated proteins and/or it may regulate spinophilin and associated protein phosphorylation in a heterologous cell. SAPAP3 binds to the coiled-coil region, upstream of the PP1 binding domain, suggesting that PPla is not displacing SAPAP3 from spinophilin. Moreover, mutation of the PP1-binding domain on spinophilin (F451A) did not modulate the association of spinophilin with mGluR5 or SAPAP3. In addition to SAPAP3, overexpression of PP1 significantly decreased spinophilin binding to mGluR5. As such, modulation of the spinophilin/mGluR5 interaction by PP1 may be part of the reason why the SAPAP3/spinophilin interaction is decreased. Taken together, these data suggest PP1α-dependent decreases in the association of spinophilin with mGluR5 and SAPAP3 may be due to alterations in phosphorylation of any of these proteins and future studies will need to delineate specific phosphorylation sites that regulate this interaction.
Spinophilin/SAPAP3/mGluR5 in physiology and OCD
Spinophilin and mGluR5 both regulate AMPAR function. Specifically, loss of SAPAP3 increases mGluR5 activity and decreases AMPAR surface expression and activity (Wan et al., 2011). Moreover, spinophilin KO or disruption of the spinophilin/PP1 interaction decreases AMPAR current rundown (Allen et al., 2006; Yan et al., 1999). In addition to AMPAR function, activation of striatal mGluRs induces corticostriatal and hippocampal LTD (Sung et al., 2001; Wu et al., 2015). Moreover, loss of SAPAP3 induces a group I mGluR-dependent synaptic depression (Chen et al., 2011) and genetic deletion of spinophilin abrogates mGluR-dependent LTD (Allen et al., 2006; Di Sebastino et al., 2016). The above suggest a synaptic physiology link between spinophilin, SAPAP3, and mGluR5. Interestingly, we observed behavioral changes in spinophilin KO animals that are also associated with behavioral changes in OCD. On the first day, there were no deficits in locomotion between the WT and Spinophilin KO animals in either the saline-treated or amphetamine treated mice. This is consistent with a previous study showing no difference in open field time following a single, 10-minute recording in the open field in young adult mice (Wu et al., 2017). However, we found that loss of spinophilin attenuates amphetamine-induced locomotor sensitization, a behavior associated with OCD, in young adult (~3–6 months of age) mice. Interestingly, the previous study by Wang and colleagues found that older (11–13 months) spinophilin KO animals have greater locomotion in the open field compared to WT animals (Wu et al., 2017). Moreover, we have previously found that striatal spinophilin interactions are altered in adult mice compared to weanlings (postnatal day 21) (Baucum et al., 2012). Future studies will need to determine if loss of spinophilin attenuates more specific OCD-like behaviors (e.g. quinpirole-induced checking) and if loss of spinophilin can ameliorate the excessive grooming observed in a SAPAP3 KO model of OCD (Welch et al., 2007). In addition, the age-dependence of these changes needs to be evaluated. However, our data suggest a functional role of spinophilin in modulating amphetamine-induced striatal locomotor learning pathologies.
Conclusion
Taken together, our data are the first to describe mechanisms modulating the interaction between spinophilin and SAPAP3 and have potential implications in normal striatal MSN physiology and biochemical pathologies in striatal MSNs associated with PD and OCD. Future studies will need to delineate how modulation of the spinophilin/mGluR5/SAPAP3 complex regulates MSN function and striatal disease pathology.
Supplementary Material
Highlights.
SAPAP3 and mGluR5 co-immunoprecipitate with spinophilin
Spinophilin association with SAPAP3 is enhanced by mGluR5
Spinophilin association with SAPAP3 is enhanced by PKC activity
Spinophilin association with mGluR5 is enhanced by SAPAP3
Loss of spinophilin attenuates amphetamine-induced locomotor sensitization
Acknowledgements and Funding.
Support for these studies was from NIH (K01NS073700, R21/R33DA041876 to AJB), Department of Biology Start-up funds and Bridge Funding (to AJB), and a summer Undergraduate Research Opportunity Program (UROP) fellowship (to CWM). We thank the Calakos lab (Duke University School of Medicine) for providing SAPAP3 KO and WT brains and the Mastracci lab (Indiana Biosciences Research Institute) for use of the Zeiss Elyra Microscope. The authors have no competing interests to declare.
1Abbreviations
- SAPAP3
SAP90/PSD-95 associated protein 3
- mGluR
Metabotropic glutamate receptor
- PKC
Protein kinase C
- PP1
Protein phosphatase 1
- AMPAR
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor
- OCD
Obsessive Compulsive Disorder
- PD
Parkinson disease
- HEK293
Human Embryonic Kidney 293 cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Ade KK, Wan Y, Hamann HC, O’Hare JK, Guo W, Quian A, Kumar S, Bhagat S, Rodriguiz RM, Wetsel WC, Conn PJ, Dzirasa K, Huber KM, Calakos N, 2016. Increased Metabotropic Glutamate Receptor 5 Signaling Underlies Obsessive-Compulsive Disorder-like Behavioral and Striatal Circuit Abnormalities in Mice. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen PB, Zachariou V, Svenningsson P, Lepore AC, Centonze D, Costa C, Rossi S, Bender G, Chen G, Feng J, Snyder GL, Bernardi G, Nestler EJ, Yan Z, Calabresi P, Greengard P, 2006. Distinct roles for spinophilin and neurabin in dopamine-mediated plasticity. Neuroscience 140, 897–911. [DOI] [PubMed] [Google Scholar]
- Baucum AJ 2nd, Jalan-Sakrikar N, Jiao Y, Gustin RM, Carmody LC, Tabb DL, Ham AJ, Colbran RJ, 2010. Identification and validation of novel spinophilin-associated proteins in rodent striatum using an enhanced ex vivo shotgun proteomics approach. Mol Cell Proteomics 9, 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucum AJ 2nd, Strack S, Colbran RJ, 2012. Age-dependent targeting of protein phosphatase 1 to Ca2+/calmodulin-dependent protein kinase II by spinophilin in mouse striatum. PLoS One 7, e31554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, Ellisman MH, Gleeson JG, 2007. Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell 129, 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody LC, Baucum AJ 2nd, Bass MA, Colbran RJ, 2008. Selective targeting of the gamma1 isoform of protein phosphatase 1 to F-actin in intact cells requires multiple domains in spinophilin and neurabin. Faseb J 22, 1660–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton JJ, Allen PB, Psifogeorgou K, Chakravarty S, Gomes I, Neve RL, Devi LA, Greengard P, Nestler EJ, Zachariou V, 2008. Multiple actions of spinophilin regulate mu opioid receptor function. Neuron 58, 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Wan Y, Ade K, Ting J, Feng G, Calakos N, 2011. Sapap3 deletion anomalously activates short-term endocannabinoid-mediated synaptic plasticity. J Neurosci 31, 9563–9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbran RJ, Bass MA, McNeill RB, Bollen M, Zhao S, Wadzinski BE, Strack S, 1997. Association of brain protein phosphatase 1 with cytoskeletal targeting/regulatory subunits. J Neurochem 69, 920–929. [DOI] [PubMed] [Google Scholar]
- Di Sebastino AR, Fahim S, Dunn HA, Walther C, Ribeiro FM, Cregan SP, Angers S, Schmid S, Ferguson SS, 2016. Role of Spinophilin in Group I Metabotropic Glutamate Receptor Endocytosis, Signaling and Synaptic Plasticity. The Journal of biological chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edler MC, Salek AB, Watkins DS, Kaur H, Morris CW, Yamamoto BK, Baucum AJ 2nd, 2018. Mechanisms Regulating the Association of Protein Phosphatase 1 with Spinophilin and Neurabin. ACS Chem Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MJ, Arnold JC, Cain ME, 2012. Impact of mGluR5 during amphetamine-induced hyperactivity and conditioned hyperactivity in differentially reared rats. Psychopharmacology (Berl) 221, 227–237. [DOI] [PubMed] [Google Scholar]
- Haas LT, Kostylev MA, Strittmatter SM, 2014. Therapeutic molecules and endogenous ligands regulate the interaction between brain cellular prion protein (PrPC) and metabotropic glutamate receptor 5 (mGluR5). The Journal of biological chemistry 289, 28460–28477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiday AC, Edler MC, Salek AB, Morris CW, Thang M, Rentz TJ, Rose KL, Jones LM, Baucum AJ 2nd, 2017. Mechanisms and Consequences of Dopamine Depletion-Induced Attenuation of the Spinophilin/Neurofilament Medium Interaction. Neural Plast 2017, 4153076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YH, Kim JY, Lee JH, Chae HG, Jang SS, Jeon JH, Kim CH, Kim J, Kim SJ, 2009. Agonist-induced internalization of mGluR1alpha is mediated by caveolin. J Neurochem 111, 61–71. [DOI] [PubMed] [Google Scholar]
- Hu XD, Liu YN, Zhang ZY, Ma ZA, Suo ZW, Yang X, 2015. Spinophilin-Targeted Protein Phosphatase-1 Alleviated Inflammatory Pain by Negative Control of MEK/ERK Signaling in Spinal Cord Dorsal Horn of Rats. J Neurosci 35, 13989–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR, 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Lu R, Chen Y, Cottingham C, Peng N, Jiao K, Limbird LE, Wyss JM, Wang Q, 2010. Enhanced hypotensive, bradycardic, and hypnotic responses to alpha2-adrenergic agonists in spinophilin-null mice are accompanied by increased G protein coupling to the alpha2A-adrenergic receptor. Mol Pharmacol 78, 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludowyke RI, Elgundi Z, Kranenburg T, Stehn JR, Schmitz-Peiffer C, Hughes WE, Biden TJ, 2006. Phosphorylation of nonmuscle myosin heavy chain IIA on Ser1917 is mediated by protein kinase C beta II and coincides with the onset of stimulated degranulation of RBL-2H3 mast cells. J Immunol 177, 1492–1499. [DOI] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ, 2010. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol 50, 295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Allaire M, Nairn AC, Page R, Peti W, 2011. Flexibility in the PP1:spinophilin holoenzyme. FEBS Lett 585, 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W, 2010. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol 17, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman JG, Brady AE, Wang Q, Hensel JL, Colbran RJ, Limbird LE, 2001. Agonist-regulated Interaction between alpha2-adrenergic receptors and spinophilin. The Journal of biological chemistry 276, 15003–15008. [DOI] [PubMed] [Google Scholar]
- Ruiz de Azua I, Nakajima K, Rossi M, Cui Y, Jou W, Gavrilova O, Wess J, 2012. Spinophilin as a novel regulator of M3 muscarinic receptor-mediated insulin release in vitro and in vivo. FASEB J 26, 4275–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FD, Oxford GS, Milgram SL, 1999. Association of the D2 dopamine receptor third cytoplasmic loop with spinophilin, a protein phosphatase-1-interacting protein. The Journal of biological chemistry 274, 19894–19900. [DOI] [PubMed] [Google Scholar]
- Sung KW, Choi S, Lovinger DM, 2001. Activation of group I mGluRs is necessary for induction of long-term depression at striatal synapses. J Neurophysiol 86, 2405–2412. [DOI] [PubMed] [Google Scholar]
- Wan Y, Ade KK, Caffall Z, Ilcim Ozlu M, Eroglu C, Feng G, Calakos N, 2014. Circuit-selective striatal synaptic dysfunction in the Sapap3 knockout mouse model of obsessive-compulsive disorder. Biol Psychiatry 75, 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Feng G, Calakos N, 2011. Sapap3 deletion causes mGluR5-dependent silencing of AMPAR synapses. J Neurosci 31, 16685–16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, Feliciano C, Chen M, Adams JP, Luo J, Dudek SM, Weinberg RJ, Calakos N, Wetsel WC, Feng G, 2007. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 448, 894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski K, Car H, 2002. (S)-3,5-DHPG: a review. CNS Drug Rev 8, 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J, LaPalombara Z, Ahmari SE, 2018. Monoamine abnormalities in the SAPAP3 knockout model of obsessive-compulsive disorder-related behaviour. Philos Trans R Soc Lond B Biol Sci 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Cottingham C, Chen L, Wang H, Che P, Liu K, Wang Q, 2017. Age-dependent differential regulation of anxiety- and depression-related behaviors by neurabin and spinophilin. PLoS One 12, e0180638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YW, Kim JI, Tawfik VL, Lalchandani RR, Scherrer G, Ding JB, 2015. Input- and cell-type-specific endocannabinoid-dependent LTD in the striatum. Cell Rep 10, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hsieh-Wilson L, Feng J, Tomizawa K, Allen PB, Fienberg AA, Nairn AC, Greengard P, 1999. Protein phosphatase 1 modulation of neostriatal AMPA channels: regulation by DARPP-32 and spinophilin. Nat Neurosci 2, 13–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


