Significance
Our DNA contains a blueprint for phenotypic traits, which include the immune response to tumors. As cancer immunotherapies continue to show clinical promise, it is important to understand how inherited genetic variation may account for variable immune responses, as reflected by gene expression within the tumor. We systematically identified the germline genetic polymorphisms associated with variable tumor tissue gene expression across 24 human cancer types. We showed that expression of major regulators of immunity, such as ICOSLG and ERAP2, was under strong genetic control. Additionally, we defined germline variants associated with the abundance of immune cells that infiltrated the tumor. These data demonstrate that germline genetics is a major player in shaping the tumor environment and immune response.
Keywords: TCGA, eQTL, eGenes, cancer immunology
Abstract
Cancer immunotherapy has emerged as an effective therapy in a variety of cancers. However, a key challenge in the field is that only a subset of patients who receive immunotherapy exhibit durable response. It has been hypothesized that host genetics influences the inherent immune profiles of patients and may underlie their differential response to immunotherapy. Herein, we systematically determined the association of common germline genetic variants with gene expression and immune cell infiltration of the tumor. We identified 64,094 expression quantitative trait loci (eQTLs) that associated with 18,210 genes (eGenes) across 24 human cancers. Overall, eGenes were enriched for their being involved in immune processes, suggesting that expression of immune genes can be shaped by hereditary genetic variants. We identified the endoplasmic reticulum aminopeptidase 2 (ERAP2) gene as a pan-cancer type eGene whose expression levels stratified overall survival in a subset of patients with bladder cancer receiving anti–PD-L1 (atezolizumab) therapy. Finally, we identified 103 gene signature QTLs (gsQTLs) that were associated with predicted immune cell abundance within the tumor microenvironment. Our findings highlight the impact of germline SNPs on cancer-immune phenotypes and response to therapy; and these analyses provide a resource for integration of germline genetics as a component of personalized cancer immunotherapy.
Cancer can be thought of as a dynamic tug-of-war between the tumor and the host. To thrive, tumor cells acquire characteristics of sustained proliferation, genome instability, and mutation (1). The host immune system counteracts cancer by recognizing, targeting, and eliminating tumor cells, but, in doing so, applies the selective pressure responsible for tumor immune evasion (2). The relative success of the tumor is determined by a myriad of factors, which include genetic susceptibilities that have been mapped to oncogenes or tumor suppressor genes, as well as our lifetime of cumulative environmental exposures. An unexplored set of determinants are the germline genetic and environmental factors that combine to represent the inherent immunological status of a patient, as it plays out at the multiple rate-limiting steps of the cancer-immunity cycle (3). The rapidly emerging field of cancer immunotherapy aims to tip the balance by harnessing the host immune response(s); however, despite significant successes, only a subset of patients benefits from durable responses (3). To explore the role of this underappreciated determinant of heterogeneity among patients with cancer, namely, germline genetic polymorphism, we defined the set of expression/gene signature quantitative trait loci (QTLs) that associated with tumor gene expression and the infiltration of tumors by immune cells.
Inherited genetic variants have been demonstrated to impact baseline and induced host immune responses. Indeed, there is large variation in the abundance and activation state of multiple immune cell types (4–6), molecules (7, 8), and genes (9, 10), which likely translates to distinct immune states. Prior studies have demonstrated that inherited genetic variation affects expression of a substantial number of genes (11–13). In fact, genome-wide association studies (GWASs) have identified hundreds of genetic risk loci for autoimmune and inflammatory diseases (14), underscoring the heritable nature of immune variation. However, such concepts have yet to be systematically applied to cancer immunology.
To assess the association between genetic variation and tumor gene expression, we utilized The Cancer Genome Atlas (TCGA) and systematically performed expression QTL (eQTL) analysis across 24 human cancer types. eQTLs are genomic loci harboring polymorphisms that are associated with gene transcript levels. Because gene expression reflects cellular phenotypes, eQTL studies bridge germline variants and phenotypes via the intermediate trait of mRNA expression. Large-scale eQTL studies, such as the Genotype-Tissue Expression (GTEx) project, have demonstrated widespread genetic control of gene expression across normal tissue types (15–17). To formally test whether similar underlying genetic variants are determinants of gene expression in cancer, we applied a similar approach, establishing the set of common hereditable factors that are associated with cancer-immune phenotypes. Notably, there is considerable overlap between GTEx tissue and TCGA tumor eGenes, indicating that regulation of tumor tissue gene expression shares genetic determinants with healthy tissues. We also identified gene signature QTLs (gsQTLs) that associated with predicted immune cell abundance, thus establishing putative modulators of immune cell infiltration in tumors. This comprehensive eQTL and gsQTL resource helps to define the likelihood that a given person’s tumor will adopt a more inflamed or noninflamed phenotype, effectively defining the person’s immune state, and supporting a path toward personalized immunotherapy.
Results
eQTL Discovery in TCGA Cancer Types.
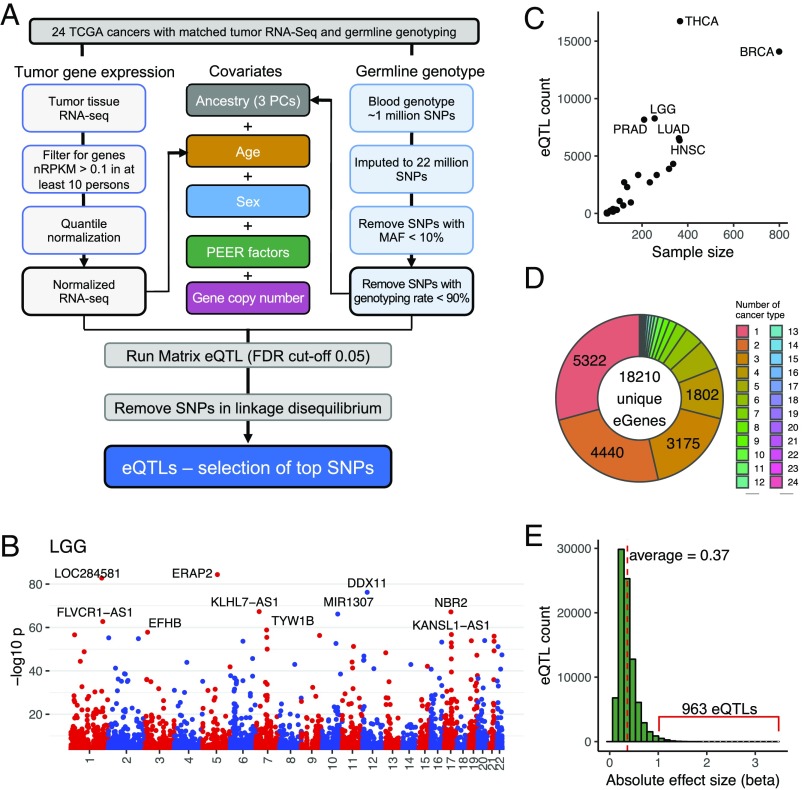
To identify germline genetic variants associated with tumor gene expression, we systematically performed eQTL discovery in each of the 24 cancer types in TCGA that contained matched tumor RNA-sequencing (RNA-seq) and whole-genome genotyping data. Recognizing that tumor gene expression can be impacted by somatic copy number alterations, we modified the eQTL discovery pipeline established by the GTEx Consortium (17) by including gene-level somatic copy number values in the linear models (Fig. 1A). Briefly, genome-wide genotyping data for n = 4,840 patients included in TCGA were derived from either normal blood (for solid tumor cancer types) or normal tissue (for acute myeloid leukemia), imputed to ∼22 million SNPs, and then filtered to a total of ∼4 million common variants with >10% minor allele frequency (MAF) and 90% genotyping call rates. In parallel, TCGA tumor RNA-seq datasets were aligned to GRCh38, filtered to exclude genes with low expression, and then quantile- and normal-transformed to minimize the impact of outliers. Within each cancer type, we performed local eQTL discovery using linear regression, modeling the expression value of each gene as predicted by a nearby SNP (defined by ±1-Mb proximity to the annotated gene boundary), while controlling for ancestry, age, sex, and probabilistic estimation of expression residuals (PEER) factors (18), as in the GTEx workflow. PEER factors were computed from normalized gene expression to capture global expression variability due to hidden factors, such as batch effects. In addition, we included the estimated somatic copy number for the gene in question as a covariate. Matrix eQTL software (19) was modified to enable incorporation of copy number as well as variance factorization to reveal the contribution of all covariates and SNPs to gene expression in serial regression (details are provided in Methods). The numbers of eQTLs and associated eGenes are listed for the 24 cancer types, applying a false discovery rate (FDR) cutoff of 0.05 for significant SNP-gene pairs. To account for linkage disequilibrium (LD), we further trimmed our hits to retain only the most significant eQTL per haplotype block (Fig. 1A and details are provided in Methods). We provide a complete set of eQTL-eGene pairs via https://albertlab.shinyapps.io/tcga_eqtl/.
Fig. 1.
Expression of QTLs in tumor tissues. (A) eQTL discovery workflow to identify the association between germline genotype and tumor gene expression in 24 TCGA cancer types. Covariates studied are color-coded. nRPKM, normalized read per kilobase of transcript per million mapped reads. (B) Representative Manhattan plot showing eQTLs in brain lower grade glioma (LGG) along the 22 human chromosomes. Each dot represents a significant eQTL, and the y axis represents −log10 P value. The top 10 most significant eGenes are labeled. Plots for each cancer type can be found in SI Appendix, Fig. S1A. (C) Number of eQTLs in each cancer type correlates with sample size. Definitions of cancer types are provided in Table 1. (D) Donut plot showing the sharing of eGenes across tissues. For example, 5,322 eGenes were discovered in only one cancer type and 4,440 eGenes were discovered in two cancer types. (E) Histogram showing the distribution of the absolute effect size (β) of eQTLs of all cancer types.
eQTLs Associate with Gene Expression of 18,210 Genes in Tumor Tissues.
In total, we identified 76,201 significant SNP-gene pairs, corresponding to 64,094 unique eQTLs and 18,210 unique eGenes across the 24 TCGA cancer types (Fig. 1B). The Manhattan plots highlight the top 10 most significant eGenes per cancer type (Fig. 1B and SI Appendix, Fig. S1A). Across cancer types, 11,406 (17.8%) of SNPs associated with more than one local eGene and 13,286 (73%) eGenes were regulated by two or more independent eQTLs (i.e., not in LD). The number of eQTLs ranged from five in adrenocortical carcinoma to 16,737 in thyroid carcinoma (THCA) (Table 1), which correlated with the sample size (Fig. 1C). Comparison of our findings to results from the GTEx project revealed that 87.4% of tumor eGenes have been cataloged as eGenes based on the study of available healthy tissues. This suggests that germline genetic variants may similarly contribute to the biological processes within tumor tissue.
Table 1.
Number of eQTLs and eGenes per cancer type
| Cancer type | Acronym | Sample size | eQTL count | eGene count |
| Adrenocortical carcinoma | ACC | 49 | 5 | 5 |
| Bladder urothelial carcinoma | BLCA | 151 | 958 | 808 |
| Breast invasive carcinoma | BRCA | 799 | 14,089 | 8,474 |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | CESC | 118 | 698 | 604 |
| Colon adenocarcinoma | COAD | 183 | 3,351 | 2,510 |
| Glioblastoma multiforme | GBM | 135 | 2,295 | 1,776 |
| Head and neck squamous cell carcinoma | HNSC | 364 | 6,361 | 4,314 |
| Kidney renal clear cell carcinoma | KIRC | 61 | 239 | 228 |
| Kidney renal papillary cell carcinoma | KIRP | 102 | 1,085 | 940 |
| Acute myeloid leukemia | LAML | 122 | 2,714 | 2,118 |
| Brain lower grade glioma | LGG | 255 | 8,273 | 5,531 |
| Liver hepatocellular carcinoma | LIHC | 85 | 328 | 298 |
| Lung adenocarcinoma | LUAD | 361 | 6,545 | 4,265 |
| Lung squamous cell carcinoma | LUSC | 264 | 3,349 | 2,313 |
| Ovarian serous cystadenocarcinoma | OV | 318 | 3,882 | 2,656 |
| Pancreatic adenocarcinoma | PAAD | 46 | 74 | 73 |
| Prostate adenocarcinoma | PRAD | 209 | 8,166 | 5,628 |
| Rectum adenocarcinoma | READ | 73 | 391 | 355 |
| Sarcoma | SARC | 90 | 308 | 277 |
| Skin cutaneous melanoma | SKCM | 73 | 155 | 144 |
| Stomach adenocarcinoma | STAD | 234 | 2,710 | 1,933 |
| Thyroid carcinoma | THCA | 366 | 16,737 | 10,153 |
| Uterine corpus endometrial carcinoma | UCEC | 336 | 4,315 | 3,050 |
| Uterine carcinosarcoma | UCS | 46 | 8 | 8 |
To ascertain how sample size influenced the number of eGenes identified, we subsampled each cancer type to a range of sample sizes, where possible, and repeated eGene discovery. As expected, the number of eGenes decreased with down-sampling (SI Appendix, Fig. S1B). Interestingly, several cancer types, including THCA, prostate adenocarcinoma (PRAD), brain lower grade glioma, and acute myeloid leukemia, had a higher number of eGenes than other cancer types when sample size differences were adjusted (SI Appendix, Fig. S1B). For example, in THCA, 26.9–32.2 eGenes were discovered per sample (SI Appendix, Fig. S1C), consistent with previous findings using healthy tissues, which identified thyroid as one of the tissues bearing the greatest number of eGenes (17). For all cancer types, the number of eGenes increased with sample size, with no evidence of saturation (SI Appendix, Fig. S1B), therefore suggesting that the true extent of germline influence on tumor gene expression is likely to be even greater.
To understand how genetic effects vary among cancer types, we next investigated the sharing of eGenes among different cancer types. Notably, 56 eGenes were common to 20 or more cancer types; however, the majority of eGenes (14,739 or 80.9%) were discovered in only four or fewer cancer types (Fig. 1D), with 29.2% (5,322) being identified in only one cancer type (Fig. 1E). Larger datasets will be required to determine if the degree of tumor tissue sharing was limited by statistical power or if these are truly tumor tissue-specific eGenes. The MAFs of eQTLs were evenly distributed from 10–50% (SI Appendix, Fig. S1D). The effect size, defined as the slope (β) of the linear regression, measured the magnitude of normalized gene expression change per additional copy of minor allele, in the units of SDs of gene expression. Across indications, the effect size of all eQTLs had absolute values that ranged from 0.058 to 3.49, with a mean β = 0.37 (Fig. 1E and SI Appendix, Fig. S1E). Nine hundred sixty-three eQTLs had effect sizes larger than 1. As expected, most eQTLs were concentrated proximal to the transcriptional start site of the genes, and these eQTLs were shown to have higher absolute effect sizes (SI Appendix, Fig. S1 F and G).
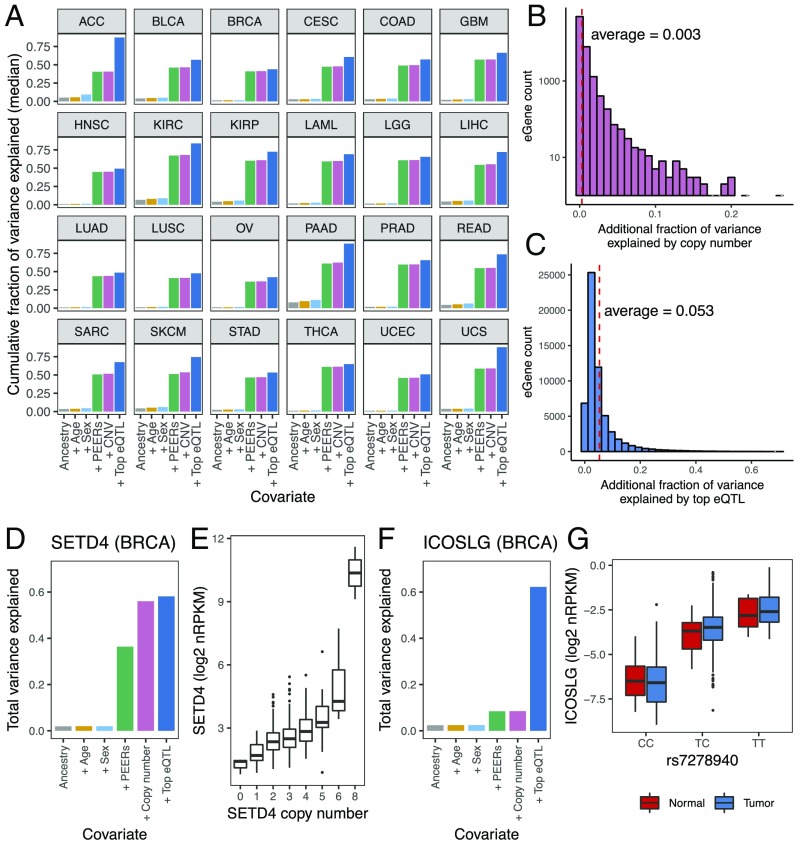
Having determined the set of eQTL-eGene pairs, we integrated the different covariates that contributed to variability in tumor eGene expression. For all genes, we regressed expression on ancestry, age, sex, PEER factors, gene copy number, and the genotype of top associated eQTLs, adding each covariate in sequence and recording the additional fraction of total expression variance explained by each added covariate. The fraction of gene expression variance explained jointly by ancestry, age, and sex was modest, with median values (across all genes) ranging from 3.5% in head and neck squamous cell carcinoma (HNSC) to 28.6% in pancreatic adenocarcinoma (PAAD) (Fig. 2A). By contrast, adding PEER factors to the model increased the explanatory power drastically, with the median cumulative fraction of variance explained ranging from 36.2% in ovarian serous cystadenocarcinoma to 67.3% in kidney renal clear cell carcinoma, suggesting the presence of confounding factors that substantially contributed to the heterogeneity in tumor gene expression (Fig. 2A). We asked if copy number alteration, a common anomaly in cancer, could be one of the unexplained confounding factors. Indeed, we found in many cancer types that select PEER factors correlated with copy number (SI Appendix, Fig. S2A). For example, in colon adenocarcinoma (COAD), PEER factor 2 showed moderate correlation with copy number (Spearman’s correlation coefficient = 0.36 across all COAD eGenes) (SI Appendix, Fig. S2A). This indicated that PEER factors had captured some of the global impact of copy number on gene expression. Nevertheless, to account for the full effect of copy number on the expression level of individual genes, we included the copy number of the local gene in question as an additional covariate, executed after taking into account PEER factors (Fig. 1A). Overall, we found that the additional fraction of expression variance explained by a given gene’s copy number was small (average = 0.3%). It is notable, however, that for 177 eGenes, copy number explained an additional 5–26.4% of expression variance (Fig. 2B).
Fig. 2.
eQTLs explain 5.3% of total tumor gene expression variance. (A) Cumulative fraction of variance in eGene expression was calculated using sequential addition of the following covariates: three ancestry principal components, age, sex, 15 PEER factors, copy number, and the top associated eQTLs. The bars represent the median value for all eGenes across 24 TCGA cancer types. Definitions of cancer types are provided in Table 1. (B) Histogram showing the distribution of additional fraction of variance in gene expression that is explained by copy number, after accounting for ancestry, age, sex and PEER factors, for eGenes aggregated across all cancer types. (C) Histogram showing the distribution of the fraction of variance in gene expression explained by the top associated eQTLs, for eGenes aggregated across all cancer types. (D) Total fraction of variance in SETD4 expression explained by sequential addition of different covariates, in BRCA. (E) Boxplot showing the expression of SETD4 in BRCA, as stratified by SETD4 copy number. (F) Total fraction of variance in ICOSLG expression explained by sequential addition of different covariates, in the BRCA cancer type. (G) Boxplot showing the expression levels of ICOSLG stratified by the genotype of its top associated eQTL, rs7278940, in BRCA. Both normal and tumor samples are shown. nRPKM, normalized read per kilobase of transcript per million mapped reads.
Accounting for aforementioned covariates, we determined that the top eQTLs, on average, explained an additional 5.3% of the total variance in gene expression, showing a wide range at the individual gene level (SD = 6.3%, maximum additional fraction of variance explained = 70.4%) (Fig. 2C). As a majority of the eGenes had more than one associated eQTL present in distinct LD blocks, these results provide a lower limit estimate of the impact of germline variants on tumor gene expression.
To illustrate results from these analyses, we show here two examples in breast invasive carcinoma, where somatic copy number or germline genetics contributed greatly to variable gene expression. In the first example, ancestry, age, sex, and PEERs collectively explained 36.4% of the variance in SETD4 gene expression. Inclusion of copy number markedly increased the total variance explained to 56%, while further inclusion of the top eQTL only marginally improved the total variance explained to 58.1% (Fig. 2D). The impact of copy number on SETD4 expression is illustrated by the boxplot showing a positive correlation between SETD4 copy number and SETD4 mRNA expression levels (Fig. 2E). By contrast, for the inducible T cell costimulator ligand (ICOSLG) gene, ancestry, age, sex, PEERs and copy number collectively explained only 8.5% of the total variance in ICOSLG expression. Upon addition of the top eQTL (rs7278940), the fraction of total variance explained increased to 62.2% (Fig. 2F), suggesting that ICOSLG expression in the tumor was under strong germline control. In fact, the stratification of ICOSLG expression by genotype was observed not only in tumor samples but also in normal breast tissues in TCGA (Fig. 2G). Our online tool provides the ability to query the serial explained variance decomposition for each gene expressed within the dataset of TCGA, highlighting the additional contributions of each factor of interest (https://albertlab.shinyapps.io/tcga_eqtl/).
Immunity-Related Genes Are Under Strong Germline Genetic Control.
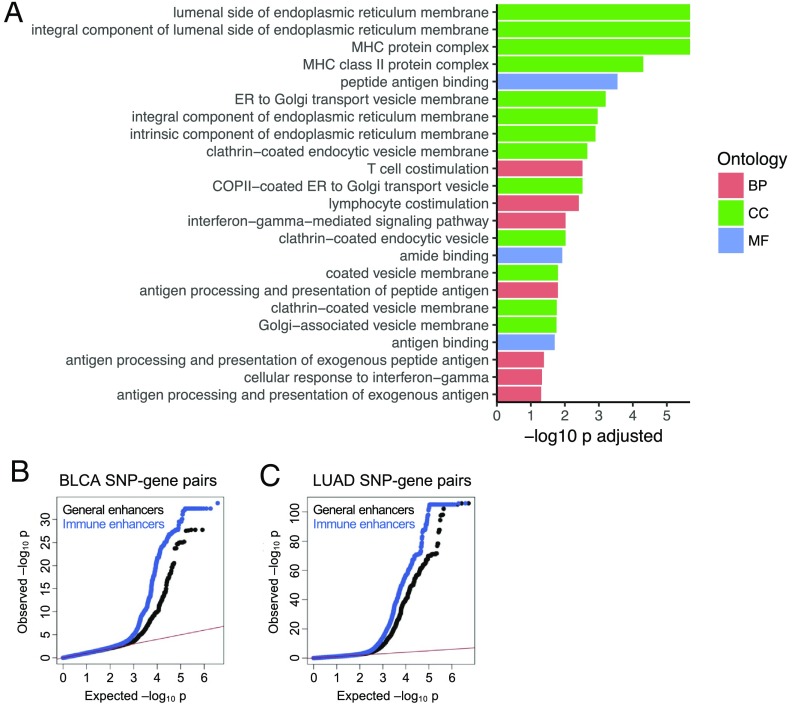
To identify cellular processes under strong influence of germline genetics, we evaluated those gene sets that are under strong influence of germline genetics, as measured by overrepresentation of eGenes. For each cancer type, we performed gene ontology (GO) analysis in three defined categories: biological process, cellular component, and molecular function. GO analysis identified multiple immune-related gene sets that were enriched for eGenes (Dataset S1). For example, in sarcoma, multiple GO terms relevant for T cell immunity were enriched for eGenes (FDR < 0.05): MHC protein complex, peptide antigen binding, and T cell costimulation (Fig. 3A). The enrichment of eGenes in immune-related gene sets was not unique to TCGA tumor tissues, as a similar enrichment was observed in multiple normal tissues in the GTEx project (Dataset S1). These findings are consistent with previous studies, which reported that genetic variants play a major role in shaping an individual’s immune response (8, 20).
Fig. 3.
Immunity-related genes and enhancers are enriched for eQTLs. (A) GO enrichment using eGenes for the sarcoma cancer type. BP, biological process; CC, cellular component; MF, molecular function. (B and C) Quantile-quantile plots showing the deviation of SNP-gene pair association P values from the distribution expected under the null hypothesis (red line). (B) BLCA SNP-gene pair P values for SNPs located within either general (i.e., bladder tissue) (black) or immune (i.e., T cell) enhancer regions (blue). Relative to SNPs located within general enhancers, SNPs located within immune enhancers have more significant gene-SNP pair P values. (C) LUAD SNP-gene pair P values for SNPs located within either general (i.e., lung tissue) (black) or immune (i.e., T cell) enhancer regions (blue).
We further tested whether eQTLs preferentially impacted expression of immune genes relative to other genes. Previous studies have indicated the presence of eQTLs in enhancer regions (15, 17); thus, we asked if immune gene enhancers were more likely to harbor stronger eQTLs than general enhancers. We compared SNP-gene association P values for SNPs that mapped to enhancers active in the tissue of origin (general enhancers) with enhancers active in T cells (immune enhancers). We performed our analyses using SNP-gene pair P values computed from bladder urothelial carcinoma (BLCA) and lung adenocarcinoma (LUAD), where enhancer annotations were available for the corresponding healthy tissue (21). As expected, gene-SNP association P values for SNPs located within the general enhancer regions deviated from the uniform distribution, consistent with the presence of eQTLs in enhancers (Fig. 3 B and C). Interestingly, the global gene-SNP P value distribution for SNPs in immune enhancers was further shifted toward smaller P values (Fig. 3 B and C), suggesting that SNPs in immune enhancer regions harbored more significant gene-SNP associations than SNPs in general enhancers (Wilcoxon rank sum test: P < 2.2 × 10−16 for both BLCA and LUAD). This suggests that expression of immune genes, relative to overall gene expression, was enriched for their being impacted by SNPs.
Endoplasmic Reticulum Aminopeptidase 2 Is a Pan-Cancer eGene That Stratifies Response to Luminal-Subtype Bladder Cancer Immunotherapy.
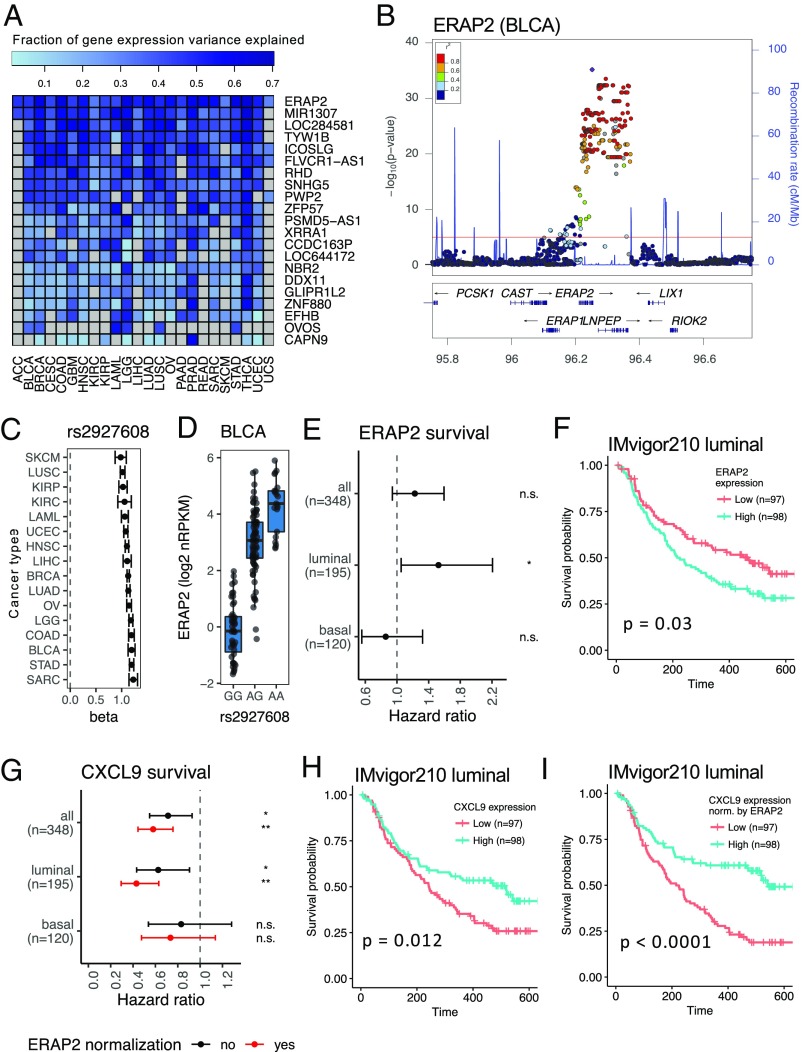
We next aimed to determine if expression of eGenes was associated with response to immunotherapy. We focused on eQTL-eGene pairs for which associated genetic variants most strongly contribute to heterogeneity in tumor gene expression. We filtered the eQTLs by their influence on eGene expression variance (accounting for >50% of the additional variance in gene expression in at least one cancer type). This filtering step resulted in a list of 21 eGenes under strong germline genetic control (Fig. 4A).
Fig. 4.
ERAP2 expression stratifies overall survival in patients with bladder cancer. (A) Heat map showing the fraction of variance in gene expression explained by eQTLs for 21 genes associated with high-impact variants (accounting for >50% of variance in gene expression in at least one cancer type). (B) LocusZoom plot displaying a 1-Mb region around the ERAP2 gene. The dots represent SNPs; the height of the dots represents the −log10 P value of the association between the SNPs and ERAP2 expression in BLCA. rs2927608 is the most significantly associated SNP with ERAP2 expression. The color of the dots represents LD (r2) of a particular SNP with rs2927608. cMMb, centimorgan per megabase. (C) Effect size (β) of ERAP2 eQTL rs2927608 across TCGA cancer types. The error bars represent SEs. Definitions of cancer types are provided in Table 1. (D) Boxplot showing BLCA ERAP2 expression in log2 normalized read per kilobase of transcript per million mapped reads (nRPKM) for the three genotype groups of the eQTL rs2927608. Each dot represents ERAP2 expression in a patient tumor sample. (E) Hazard ratio of overall survival, as stratified by median ERAP2 expression level, of patients with bladder cancer in the IMvigor210 phase 2 PD-L1 trial, for all molecular subtypes or only the luminal or basal subtype. The error bars represent the 95% confidence intervals (*P < 0.05). n.s., not significant. (F) Overall survival of patients with bladder cancer with tumors of the luminal subtype in the IMvigor210 trial, stratified by median ERAP2 expression level. (G) Hazard ratio of overall survival, as stratified by median CXCL9 expression level, of patients with bladder cancer in the IMvigor210 trial, for all molecular subtypes or only the luminal or basal subtype. Black bars represent the hazard ratio calculated using CXCL9 expression alone; red bars represent the hazard ratio calculated using ERAP2 normalized CXCL9 expression level. The error bars represent the 95% confidence intervals (*P < 0.05; **P < 0.005). Overall survival of patients with the luminal subtype as stratified by median CXCL9 level, without (H) or with (I) ERAP2 normalization, is shown.
Given our interest in cancer immunology and immunotherapy, we focused our analysis on the endoplasmic reticulum aminopeptidase 2 (ERAP2) and ICOSLG genes (Fig. 4A). In all 24 cancer types, the associated ERAP2 SNPs fell into a large (∼150 kb) haplotype block that spans the ERAP2 gene and the neighboring LNPEP gene (also known as IRAP) (Fig. 4B). For the top eQTL, rs2927608, the effect sizes (β) on ERAP2 gene expression ranged from 1.02 in lung squamous cell carcinoma (LUSC) to 0.98 in skin cutaneous melanoma (SKCM) (Fig. 4C). For example, in BLCA, the genotype of rs2927608 was associated with expression levels of ERAP2 (β = 1.19, P = 6.69 × 10−36) (Fig. 4D). The effect of eQTLs on ERAP2 was not unique to tumor samples, as ERAP2 was also a significant eGene in all healthy tissues in the GTEx catalog (17) (SI Appendix, Fig. S4A). Interestingly, the haplotype associated with low ERAP2 expression, tagged by the rs2927608-G allele, contains an SNP that has been previously reported to alter a splice donor site, and thus to result in intronic read-through and the introduction of a stop codon, which, in turn, leads to nonsense-mediated decay (NMD) of the ERAP2 mRNA (22) (haplotype B in SI Appendix, Fig. S4C). ICOSLG was an eGene in 21 TCGA cancer types. The most significant eQTL for ICOSLG, rs7278940, mapped to a haplotype with a known GWAS hit for celiac disease (23). The rs7278940 eQTL showed an effect size on ICOSLG gene expression that ranged from 0.85 in stomach adenocarcinoma (STAD) to 1.2 in LUSC (SI Appendix, Fig. S4F).
In the context of immune regulation, ERAP2 and ICOSLG play important roles. ERAP2 participates in the processing and presentation of antigen, acting as a heterodimer with a closely related aminopeptidase, ERAP1. Together, ERAP1 and ERAP2 trim precursor peptides to mature epitopes that can be loaded onto HLA class I molecules and presented on the surface of both antigen-presenting cells and tumor cells to elicit and engage a cytotoxic T cell response (24). Interestingly, we found that ERAP1 was also an eGene in 16 cancer types. For example, an SNP located within ERAP1, rs28119, was an eQTL for ERAP1, with effect sizes ranging from 0.49 in PRAD to 0.74 in glioblastoma multiforme (SI Appendix, Fig. S4 D and E). While ERAPs have been established as important for the generation of the peptide repertoire, paradoxically, attenuation of the mouse ortholog, ERAAP, resulted in an enhanced antitumor T cell response (25). This enhanced immunogenicity was thought to be a consequence of presentation of an altered repertoire that included novel immunogenic peptides in the absence of ERAAP (26). ICOSLG acts at a distinct step, regulating adaptive immunity by acting as a costimulatory molecule expressed by antigen-presenting cells and engaging ICOS on T cells, inducing activation and differentiation (27). ICOSLG signaling in dendritic cells also serves to amplify pattern recognition-induced cytokine stimulation (28). Additionally, activating ICOS has been shown to enhance tumor immunity in mice (29), and agonist antibodies are being developed for cancer immunotherapy in humans (27).
Given the role of ERAP2 and ICOSLG in adaptive immunity and the preclinical data supporting their respective roles in antitumor immunity in mice, we asked if variable ERAP2 or ICOSLG expression in humans was associated with different survival outcome in patients with cancer. Using RNA-seq data from the anti–PD-L1 (atezolizumab) urothelial bladder cancer phase 2 clinical trial [IMvigor210 (30)], we stratified patients into two groups, split by the median expression of ERAP2 or ICOSLG (of note, genetic data are not available for this trial). While ICOSLG did not serve as a prognostic marker in any bladder cancer subtype (SI Appendix, Fig. S4H), ERAP2 levels were significantly associated with overall survival in patients with the luminal subtype of bladder cancer (Fig. 4E). Consistent with mouse studies, patients with low ERAP2 expression had better overall survival relative to those with higher ERAP2 expression (P = 0.03) (Fig. 4F). One potential caveat concerns ERAP2 expression being induced by IFN-γ (31), which suggested that ERAP2 could simply be a secondary effect of differential IFN-γ stimulation in responders and nonresponders. Indeed, IFN-γ–induced genes, including CXCL9, have been shown to be associated with both enhanced CD8+ T cell infiltration in tumors and response to anti–PD-L1 (30). We interpreted that it was unlikely for ERAP2-associated survival benefit to be a consequence of the IFN-γ response, as enhanced survival was associated with low, rather than high, ERAP2 expression. Nonetheless, to determine whether variable IFN-γ response confounded the association between ERAP2 and patient survival, we included CXCL9 expression (shown to be more robust than IFN-γ expression) into the same model and found that the association between ERAP2 and survival significantly enhanced the predictive value of CXCL9 (likelihood ratio test: P < 2.2 × 10−16) (Fig. 4 G–I). These data suggest that ERAP2 is an independent prognostic predictor of survival in patients with luminal subtype bladder cancer receiving anti–PD-L1 therapy, and, in fact, low ERAP2 expression can be used along with the IFN-γ response to establish a further improved prognostic biomarker signature. We do note that ERAP2 expression was not prognostic for the basal subtype of bladder cancer (Fig. 4E), and may reflect possible interactions with HLA molecules or other unknown factors.
gsQTLs Impact Immune Cell Infiltration in Tumors.
Immune cell infiltration has been shown to be another important variable that may positively or negatively shape tumor growth, progression, or clinical outcome (32). It has yet to be evaluated if germline genetics influences immune cell infiltration in tumors. Therefore, we next aimed to examine SNPs associated with the abundance of immune cells in tumors, taking advantage of predictive methods that permit the estimation of cellular composition based on gene expression data.
Specifically, we inferred the abundance of immune cells in TCGA tumor samples using a cellularity deconvolution method based on the aggregate expression of marker genes, recently established by Aran et al. (33). These gene signature scores estimate the abundance of 34 lymphoid and myeloid cell types (SI Appendix, Fig. S5A). We next modified our analytical method to determine gsQTLs associated with predicted immune cell abundance. We focused on SNPs that were within 50 kb of annotated genes to reduce the burden of multiple testing. As with the eQTL discovery workflow, we initially controlled for covariates by including three ancestry principal components, 15 PEER factors, sex, and age. Concerned that PEER factors would capture variable gene expression resulting from the heterogeneous presence of immune cells in tumors, we asked if PEER factors correlated with the immune cell gene signatures. Indeed, PEER factors highly correlated with some of the gene signatures (SI Appendix, Fig. S5 B–D and Dataset S1), indicating that cellular composition is a major contributor to the variance in gene expression. To mitigate the potential loss of signal to PEER, while still accounting for unexplained variance, we computed, for each gene, residuals from regressing its expression on gene signature scores. The resulting residual matrix thus reflected global variation in expression that is not correlated with immune cell composition but is due to other factors, such as copy number and batch effects. We then used this residual matrix to calculate a new set of PEER factors. With this improved pipeline (SI Appendix, Fig. S5A), we performed gsQTL discovery using Matrix eQTL software, utilizing the P value threshold of 2.1 × 10−8 for significant hits (details are provided in Methods). Finally, we pruned the hits to generate a final list of independent gsQTLs (i.e., not in LD) (SI Appendix, Fig. S5A).
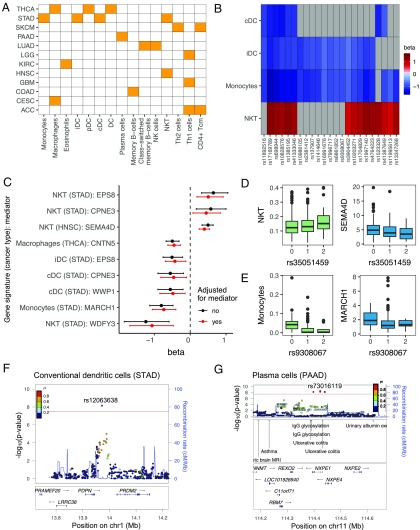
We discovered a total of 103 gsQTLs that were significantly associated with the immune gene signatures (31 lymphoid and 72 myeloid signatures) (Dataset S1). Cancer types with at least one significant gsQTL, along with the associated gene signatures, are shown in Fig. 5A. The significant immune gene signatures included signatures for T cells, B cells, natural killer (NK) cells, dendritic cells, eosinophils, macrophages, and monocytes, suggesting the widespread impact of germline genetics on immune cellularity within the tumor microenvironment (Fig. 5A). Interestingly, 23 gsQTLs were significantly associated with multiple gene signatures, all of which were in the STAD cancer type, a possible reflection of their being a common causal infectious agent responsible for most gastric cancer (Fig. 5B). These gsQTLs were associated with two or more of the following gene signatures: conventional dendritic cells (cDCs), immature dendritic cells (iDCs), monocytes, and NK T (NKT) cells, suggesting the presence of common modulators for these signatures. Alternatively, the gsQTL is impacting a common progenitor, which, in turn, is regulating multiple cell types [e.g., monocytes and iDCs are part of a common hematopoietic lineage (34)]. The gsQTLs associated with cDCs, iDCs, and monocytes had opposite direction effect sizes compared with the gsQTLs associated with the NKT cell signature (Fig. 5B). In agreement, the cDC, iDC, and monocyte signatures were positively correlated, and they negatively correlated with the NKT cell signature (SI Appendix, Fig. S5E).
Fig. 5.
TCGA tumor immune cellularity gsQTLs. (A) Heat map showing gene signatures (x axis) with one or more significant gsQTLs and the cancer type(s) (y axis) in which they were shown to be statistically significant (P < 2.1 × 10−8). Definitions of cancer types are provided in Table 1. Cancer types without any significant gsQTLs were not shown. (B) Heat map showing gsQTLs associated with two or more of the following gene signatures: cDCs, iDCs, monocytes, and NKT cells (in STAD). The color represents the effect size (β) of the association. Gray boxes are nonsignificant associations. (C) Forest plot showing significant mediator genes for the gsQTL-gene signature associations. The dots represent the effect sizes (β) of the gsQTL-gene signature associations before (black) and after (red) adjusting for the expression of the mediator genes. The error bars represent confidence intervals. (D, Left) Boxplot showing NKT cell gene signature scores stratified by genotypes of its associated gsQTL, rs35051459. (D, Right) Boxplot showing the expression of the mediator gene SEMA4D [log2 normalized read per kilobase of transcript per million mapped reads (nRPKM)] stratified by genotypes of the same gsQTL. (E, Left) Boxplot showing the monocyte gene signature scores stratified by genotypes of its associated gsQTL, rs9308067. (E, Right) Boxplot showing the expression of the mediator gene MARCH1 (log2 nRPKM) stratified by genotypes of the same gsQTL. (F) LocusZoom plot for the cDC gene signature in STAD. The gsQTL rs12063638 is located downstream of the PDPN gene. cMMb, centimorgan per megabase; chr, chromosome. (G) LocusZoom plot for the plasma cell gene signature in PAAD. The gsQTL rs73016119 is located in the same LD block as GWAS risk loci for ulcerative colitis.
A possible mechanism by which the gsQTLs influence immune gene signatures was by acting as eQTLs for “mediator” genes that are upstream regulators of immune cell infiltration in tumors. Thus, we asked if any of the gsQTLs were also eQTLs, determined in tumor gene expression (in this study) or in healthy tissues as reported in previous studies (17, 35). Indeed, 19 gsQTLs were also eQTLs with at least one associated eGene. For gsQTLs that were not known eQTLs, we assigned the nearest gene(s) to gsQTLs, thereby generating a list of candidate mediator genes (Dataset S1). A mediator analysis (Methods) was performed to determine if the gsQTL-gene signature associations were likely regulated by expression of the respective candidate mediator genes. Interestingly, nine gsQTL-gene signature associations were significantly attenuated, as reflected by decreased absolute effect size (β), after adjusting for expression of the mediator genes (500 simulations; P < 0.05) (Fig. 5C). However, while attenuated, the gsQTL-gene signature associations were not completely abolished, indicating that the mediation effects were partial. For example, the gsQTL rs35051459 was associated with the NKT cell gene signature in HNSC (β = 0.44, P = 1.79 × 10−10) (Fig. 5D, Left). This association was partially mediated by the SEMA4D gene (Fig. 5D, Right), which was previously shown to be an eGene for rs35051459 (17). Interestingly, expression of SEMA4D has been shown to influence the infiltration and distribution of leukocytes in the tumor microenvironment, and the inhibition of SEMA4D promoted immune infiltration into the tumor (36). In agreement, the rs35051459 genotype associated with lower SEMA4D expression was associated with a higher NKT cell gene signature (Fig. 5D). As a second example in STAD, the association between the gsQTL rs9308067 and the monocyte gene signature (β = −0.95, P = 6.25 × 10−9) was partially mediated through the MARCH1 gene (Fig. 5E), which was previously shown to regulate innate immune responses by modulating monocyte functions (37).
The mediator analysis, while useful, only captured genes that mediated the gsQTL-gene signature associations by acting as eGenes. Besides influencing mRNA expression, gsQTLs may also affect associated genes by influencing mRNA conformation, stability, and localization or by resulting in changes in the encoded amino acids (38). This, in turn, may impact the functional role of the associated genes in regulating immune cell infiltration in the tumor. For example, the gsQTL rs12063638 was associated with the cDC gene signature in STAD (β = 0.44, P = 1.79 × 10−10) (Dataset S1). Although rs12063638 has not been shown to be an eQTL, it is located downstream of the gene that encodes for glycoprotein podoplanin (PDPN) (Fig. 5F), a key modulator of dendritic cell trafficking (39). We also highlight the gsQTL rs73016119, which was associated with the plasma cell gene signature in PAAD (β = 1.39, P = 3.39 × 10−9). Notably, rs73016119 is in LD with rs561722 (R2 = 0.74), which has been shown through GWASs to be associated with ulcerative colitis (Fig. 5G), an inflammatory condition characterized, in part, by infiltration of plasma cells in the gut (40). An understanding of how the gsQTL impacts plasma cells may shed light upon the pathogenesis of ulcerative colitis.
Together, the presence of gsQTLs for immune gene signatures suggests that germline genetic variants may influence the abundance, infiltration, and composition of immune cells in tumors. Moreover, gsQTLs, along with the candidate genes, provide a resource for functional studies of immune infiltration in the tumor microenvironment.
Discussion
Large-scale efforts by the GTEx Consortium have demonstrated the impact of eQTLs on gene expression in healthy tissues (17). Previous studies that aimed to characterize eQTLs in tumors were limited in scope as they were performed in only a small number of cancer types (41, 42). A recent pan-cancer analysis identified eQTLs across TCGA cancer types (43); however, their assessment applied the GTEx-eQTL matrix without consideration for copy number alterations present in tumor genomes; moreover, they did not provide detailed evaluation of the impact of genetic variation on tumor phenotypes. To fully account for heterogeneity of gene expression in tumor samples, we incorporated copy number in our eQTL discovery pipeline. Moreover, we modified the Matrix eQTL software to reveal the contribution of all covariates and SNPs to the variability in eGene expression. This allowed us to assign with confidence the impact of germline variants on the heterogeneous genes in patient tumors. While it will be interesting to assess if these variants similarly impact gene expression in healthy tissues, the limited sample size of TCGA healthy tissues precluded eQTL discovery. However, we note that the majority (87.4%) of tumor eGenes were also eGenes in healthy tissues as cataloged by the GTEx Consortium, suggesting that germline genetic control of gene expression is not lost upon tumorigenesis. Direct comparison of the dataset of TCGA and the GTEx dataset, however, was challenging due to different histological compositions of the tumor compared with the underlying normal tissue. For example, melanoma constitutes the expansion of a cell type, the melanocyte, which is relatively rare in healthy skin and contributes modestly to total mRNA expression. As the field moves toward single-cell-type eQTL studies, such comparisons may be enabled. Additionally, tissue acquisition in TCGA and the GTEx project was distinct, hence introducing batch effects and precluding direct effect size comparisons.
We demonstrated that, in the context of tumors, the expression of immunity-related genes was most highly enriched for their being associated with germline variants. This supported the concept that the strength of a person’s antitumor immune response is an intrinsic characteristic, controlled, in part, by one’s inherited genome. Of the immune genes most highly enriched for their being under genetic control, ERAP2 is a striking example, where a defined haplotype block contributed up to 65% of total variability in tumor gene expression. As discussed, haplotype B is known to generate an RNA transcript that is susceptible to NMD, providing a likely explanation for the low level of ERAP2 expression that is associated with the rs2927608-G allele. The other major ERAP2 haplotype, tagged by the rs2927608-A allele, contains multiple risk alleles for autoimmune and inflammatory diseases, as identified in prior GWASs (44–50) (SI Appendix, Fig. S4C). Given the high MAF, ERAP2 eQTLs are impactful in terms of both the magnitude of their effect on ERAP2 expression and their prevalence in the population. It is precarious to draw inferences from mouse models due to the presence of two ERAP genes in humans compared with one gene in mice (24). Nonetheless, experimental studies have demonstrated that down-regulation of ERAAP results in enhanced immunogenicity, tumor growth arrest, and improved survival (25, 26). One study established that the observed tumor immunity was dependent on CD8+ T cells (25); however, it remains to be established if the mechanism of tumor killing was a result of conventional αβT cells reacting to the enhanced presentation of immunogenic tumor peptides. While seemingly paradoxical, ERAAP deficiency does not necessarily reduce the number of epitopes presented; instead, it alters the peptide repertoire (51), which may confer enhanced T cell responses in some individuals or mouse models. Alternatively, ERAAP deficiency in the tumor may unleash killing by nonclassical CD8+ T cells (52). Our results are also supported by a recent in vivo CRISPR screen, which demonstrated that depletion of ERAAP in mouse tumors increased the efficacy of anti–PD-1 immunotherapy (53). The presence of both ERAP1 and ERAP2 in humans, as opposed to a single ERAAP in mice, makes it challenging to disentangle the aminopeptidases’ contributions in modulating the peptide repertoire for T cell recognition. However, consistent with the mouse studies, we show that low ERAP2 levels were associated with improved response to anti–PD-L1 in patients with the luminal subtype of bladder cancer (Fig. 4E). Further studies will be necessary to evaluate whether the prognostic effect of low ERAP2 expression can be generalized to other cancer types or other clinical trial datasets. Other caveats will also need to be addressed, including potential interactions between ERAP2 and patient HLA haplotypes, which work in concert to define the set of immunogenic peptides available in the tumor. Studies of autoimmune diseases, including ankylosing spondylitis and birdshot chorioretinitis, have already demonstrated strong interactions between ERAP2 polymorphisms and HLA haplotypes (44, 54).
We also investigated the ICOSLG variants identified in the eQTL analysis, querying whether expression level impacts clinical outcome in anti–PD-L1 immunotherapy trials. It has been shown previously that ICOSLG regional polymorphisms were associated not only with decreased ICOSLG expression but also with decreased signaling in response to pattern recognition receptor-induced cytokine secretion in dendritic cells (28). While we did not observe stratification in our sample of patients with bladder cancer receiving anti–PD-L1, the strong underlying role for germline genetics may be an important consideration for active clinical development programs that are attempting to extend preclinical observations from mice. In addition to ERAP2 and ICOSLG, it will be interesting to evaluate how other eGenes, individually and collectively, regulate the antitumor response.
The presence of immune cell infiltrations is considered to be an important biomarker for stratifying human tumors. Notably, inflamed tumors are more sensitive to anti-PD1/anti–PD-L1 therapies compared with those with large stromal infiltrations or those tumors showing an absence of T cells (55, 56). We show that germline-encoded gsQTLs influence the abundance of immune cells within the tumor microenvironment. For example, the gsQTL associated with the cDC gene signature was located near the PDPN gene. PDPN activates the C-type lectin receptor (CLEC-2) to rearrange the actin cytoskeleton in dendritic cells to promote efficient motility along stromal surfaces (39). Functional experiments will be necessary to determine if and how gsQTL affects PDPN function and modulates dendritic cell migration. In fact, clear understanding of the exact mechanisms by which gsQTLs regulate the immune gene signatures will be valuable to the identification of drivers of immune cell infiltration in tumors and likely other inflammatory conditions.
In conclusion, we show that expression of more than half of the genes in tumor tissues was associated with germline genetics, highlighting an underappreciated determinant of variable tumor gene expression and immune cell infiltration. These findings provide insight into stratification of patients receiving anti–PD-L1 therapy, and our analyses serve as a resource for integrating human genetics into the development of novel therapeutics.
Methods
Within each cancer type, we performed local eQTL discovery with Matrix eQTL software (19). PEER factors were computed with the R package PEER (18), with the aim of removing effects of confounding variables (“batch” effects) from the gene expression matrix. We modified Matrix eQTL so that copy number for the gene in question was regressed after regression of PEER factors and other covariates. We applied an FDR cutoff of 0.05 for significant SNP-gene pairs. Using Plink’s clump command, we trimmed the list of eQTLs to retain only the most significant eQTL per haplotype block (r2 = 0.2, clump distance = 500 kb). Full details are provided in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Richard Bourgon for helpful discussions and critical feedback on the manuscript.
Footnotes
Conflict of interest statement: All authors are employees of Genentech, Inc.
This article is a PNAS Direct Submission.
Data deposition: A complete set of eQTL-eGene pairs is available at https://albertlab.shinyapps.io/tcga_eqtl/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804506115/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Vesely MD, Schreiber RD. Cancer immunoediting: Antigens, mechanisms, and implications to cancer immunotherapy. Ann N Y Acad Sci. 2013;1284:1–5. doi: 10.1111/nyas.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 4.Orrù V, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roederer M, et al. The genetic architecture of the human immune system: A bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161:387–403. doi: 10.1016/j.cell.2015.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patin E, et al. Milieu Intérieur Consortium Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat Immunol. 2018;19:302–314, and erratum (2018) 19:645. doi: 10.1038/s41590-018-0049-7. [DOI] [PubMed] [Google Scholar]
- 7.Duffy D, et al. Milieu Intérieur Consortium Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity. 2014;40:436–450. doi: 10.1016/j.immuni.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, et al. A functional genomics approach to understand variation in cytokine production in humans. Cell. 2016;167:1099–1110.e14. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Urrutia A, et al. Milieu Intérieur Consortium Standardized whole-blood transcriptional profiling enables the deconvolution of complex induced immune responses. Cell Rep. 2016;16:2777–2791. doi: 10.1016/j.celrep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Ben-Ali M, et al. Functional characterization of naturally occurring genetic variants in the human TLR1-2-6 gene family. Hum Mutat. 2011;32:643–652. doi: 10.1002/humu.21486. [DOI] [PubMed] [Google Scholar]
- 11.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 13.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 15.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonsdale J, et al. GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battle A, Brown CD, Engelhardt BE, Montgomery SB. GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) Groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration & Visualization—EBI; Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead Analysts; Laboratory, Data Analysis & Coordinating Center (LDACC) NIH Program Management; Biospecimen Collection; Pathology; eQTL Manuscript Working Group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 18.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shabalin AA. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quach H, et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrés AM, et al. NISC Comparative Sequencing Program Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois PCA, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saveanu L, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 25.James E, Bailey I, Sugiyarto G, Elliott T. Induction of protective antitumor immunity through attenuation of ERAAP function. J Immunol. 2013;190:5839–5846. doi: 10.4049/jimmunol.1300220. [DOI] [PubMed] [Google Scholar]
- 26.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2007;8:101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 27.Burugu S, Dancsok AR, Nielsen TO. Emerging targets in cancer immunotherapy. Semin Cancer Biol. 2018;52:39–52. doi: 10.1016/j.semcancer.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Hedl M, Lahiri A, Ning K, Cho JH, Abraham C. Pattern recognition receptor signaling in human dendritic cells is enhanced by ICOS ligand and modulated by the Crohn’s disease ICOSLG risk allele. Immunity. 2014;40:734–746. doi: 10.1016/j.immuni.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan X, Quezada SA, Sepulveda MA, Sharma P, Allison JP. Engagement of the ICOS pathway markedly enhances efficacy of CTLA-4 blockade in cancer immunotherapy. J Exp Med. 2014;211:715–725. doi: 10.1084/jem.20130590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg JE, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanioka T, et al. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 32.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–360. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 33.Aran D, Hu Z, Butte AJ. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans EE, et al. Antibody blockade of semaphorin 4D promotes immune infiltration into tumor and enhances response to other immunomodulatory therapies. Cancer Immunol Res. 2015;3:689–701. doi: 10.1158/2326-6066.CIR-14-0171. [DOI] [PubMed] [Google Scholar]
- 37.Galbas T, et al. MARCH1 E3 ubiquitin ligase dampens the innate inflammatory response by modulating monocyte functions in mice. J Immunol. 2017;198:852–861. doi: 10.4049/jimmunol.1601168. [DOI] [PubMed] [Google Scholar]
- 38.Komar AA, editor. Single Nucleotide Polymorphisms. Humana Press; Totowa, NJ: 2009. [Google Scholar]
- 39.Acton SE, et al. Podoplanin-rich stromal networks induce dendritic cell motility via activation of the C-type lectin receptor CLEC-2. Immunity. 2012;37:276–289. doi: 10.1016/j.immuni.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cupi ML, et al. Plasma cells in the mucosa of patients with inflammatory bowel disease produce granzyme B and possess cytotoxic activities. J Immunol. 2014;192:6083–6091. doi: 10.4049/jimmunol.1302238. [DOI] [PubMed] [Google Scholar]
- 41.Li Q, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, et al. Expression QTL-based analyses reveal candidate causal genes and loci across five tumor types. Hum Mol Genet. 2014;23:5294–5302. doi: 10.1093/hmg/ddu228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong J, et al. PancanQTL: Systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 2018;46:D971–D976. doi: 10.1093/nar/gkx861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuiper JJW, et al. A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum Mol Genet. 2014;23:6081–6087. doi: 10.1093/hmg/ddu307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu JZ, et al. International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jostins L, et al. International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li YR, et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat Med. 2015;21:1018–1027. doi: 10.1038/nm.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cortes A, et al. International Genetics of Ankylosing Spondylitis Consortium (IGAS); Australo-Anglo-American Spondyloarthritis Consortium (TASC); Groupe Française d’Etude Génétique des Spondylarthrites (GFEGS); Nord-Trøndelag Health Study (HUNT); Spondyloarthritis Research Consortium of Canada (SPARCC); Wellcome Trust Case Control Consortium 2 (WTCCC2) Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. 2013;45:730–738. doi: 10.1038/ng.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagarajan NA, et al. ERAAP shapes the peptidome associated with classical and nonclassical MHC class I molecules. J Immunol. 2016;197:1035–1043. doi: 10.4049/jimmunol.1500654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagarajan NA, Gonzalez F, Shastri N. Nonclassical MHC class Ib-restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nat Immunol. 2012;13:579–586. doi: 10.1038/ni.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson PC, et al. ERAP2 is associated with ankylosing spondylitis in HLA-B27-positive and HLA-B27-negative patients. Ann Rheum Dis. 2015;74:1627–1629. doi: 10.1136/annrheumdis-2015-207416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gajewski TF, et al. Cancer immunotherapy strategies based on overcoming barriers within the tumor microenvironment. Curr Opin Immunol. 2013;25:268–276. doi: 10.1016/j.coi.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Hegde PS, Karanikas V, Evers S. The where, the when, and the how of immune monitoring for cancer immunotherapies in the era of checkpoint inhibition. Clin Cancer Res. 2016;22:1865–1874. doi: 10.1158/1078-0432.CCR-15-1507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.