Significance
Selective autophagy is critical for the regulation of cellular homeostasis and physiology in mammals. Selective autophagy of invading pathogens and damaged mitochondria requires the TBK1 kinase and the autophagy receptors NDP52 and TAX1BP1, but with poorly understood mechanisms. In this study, we present the crystal structures of the SKIP carboxyl homology (SKICH) domains of NDP52 and TAX1BP1 in complex with NAP1, which not only uncover how NDP52 interacts with NAP1 for the recruitment of TBK1 but also provide an atomic picture showing how a SKICH domain functions as a protein–protein interaction module to interact with its binding partners. Furthermore, the determined complex structures also help to evaluate the currently known TBK1-mediated phosphorylation sites in the SKICH domains of NDP52 and TAX1BP1.
Keywords: autophagy receptor, NDP52, TAX1BP1, NAP1, selective autophagy
Abstract
NDP52 and TAX1BP1, two SKIP carboxyl homology (SKICH) domain-containing autophagy receptors, play crucial roles in selective autophagy. The autophagic functions of NDP52 and TAX1BP1 are regulated by TANK-binding kinase 1 (TBK1), which may associate with them through the adaptor NAP1. However, the molecular mechanism governing the interactions of NAP1 with NDP52 and TAX1BP1, as well as the effects induced by TBK1-mediated phosphorylation of NDP52 and TAX1BP1, remains elusive. Here, we report the atomic structures of the SKICH regions of NDP52 and TAX1BP1 in complex with NAP1, which not only uncover the mechanistic bases underpinning the specific interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1 but also reveal the binding mode of a SKICH domain. Moreover, we uncovered that the SKICH domains of NDP52 and TAX1BP1 share a general binding mode to interact with NAP1. Finally, we also evaluated the currently known TBK1-mediated phosphorylation sites in the SKICH domains of NDP52 and TAX1BP1 on the basis of their interactions with NAP1. In all, our findings provide mechanistic insights into the interactions of NAP1 with NDP52 and TAX1BP1, and are valuable for further understanding the functions of these proteins in selective autophagy.
Autophagy is a tightly regulated lysosome-dependent “self-eating” catabolic process to recycle cytoplasmic material in eukaryotic cells, and it plays a critical role in the maintenance of cellular homeostasis and physiology (1–3). Previously, autophagy was regarded as a nonspecific bulk degradation process with little or no selectivity. Recently, increasing evidence reveals that many cytosolic contents, including bulk protein aggregates, dysfunctional organelles, and invading pathogens, are degraded by autophagy in a highly selective manner, which is termed selective autophagy (2, 4–8). The key factors involved in selective autophagy processes are autophagy receptors, such as SQSTM1/p62, NDP52, TAX1BP1, optineurin, NBR1, Nix, FUNDC1, FAM134B, and NCOA4 in mammals, which not only can specifically recognize relevant autophagic cargoes but also can bind to the key autophagic factor, the ATG8 family proteins, thereby serving as bridging adaptors to target specific cargoes to the autophagy machinery for subsequent autophagic degradations (7, 9, 10). In view of crucial roles played by autophagy receptors in selective autophagy, the autophagic functions of autophagy receptors have been tuned well spatially and temporally by other regulatory proteins, particularly protein kinases, such as the casein kinase 2 and the TANK-binding kinase 1 (TBK1) (11–14). For instance, TBK1 was found to phosphorylate the Ser177 residue in optineurin and the Ser403 residue in SQSTM1 to promote their interactions with ATG8 family proteins and ubiquitin, respectively (11, 12). However, until now, many of the detailed molecular mechanism underpinning the specific associations of autophagy receptors with their regulatory proteins, as well as the downsteam effects mediated by these regulatory proteins in selective autophagy processes, have been elusive.
NDP52 and TAX1BP1 are two important ubiquitin-binding and multidomain autophagy receptors in mammals, both of which are demonstrated to participate in selective autophagic degradations of invading infectious pathogens (xenophagy), such as Salmonella enterica Typhimurium, and the depolarized mitochondria (mitophagy) (15–18). In addition, NDP52 was reported to mediate selective autophagic degradations of retrotransposon RNA (19) and specific functional proteins, including DICER and AGO2 in the miRNA pathway and MAVS in immune signaling (20, 21). Notably, genetic mutation of NDP52 is directly linked to Crohn’s disease, a type of inflammatory bowel disease likely caused by a combination of environmental, immune, and bacterial factors (22). Except for the diverse central coiled-coil region, NDP52 and TAX1BP1 share a highly similar domain structure (Fig. 1A), and both contain an N-terminal SKIP carboxyl homology (SKICH) domain, followed by an unconventional LC3-interacting region (LIR) motif that can differentially bind to different LC3/GABARAP orthologs (17, 23), and two C-terminal zinc fingers (Fig. 1A), which participate in the recognition of ubiquitin proteins decorated on autophagic cargoes and the unconventional myosin motor myosin VI (17, 24–26). In contrast to TAX1BP1, NDP52 uniquely contains a galectin 8-interacting region (Fig. 1A), which can specifically interact with the sugar receptor galectin 8 to target vesicle-damaging pathogens (27–29). In addition to NDP52 and TAX1BP1, SKICH domains are found in the CALCOCO1, SKIP, and proline-rich inositol-polyphosphate 5-phosphatase (PIPP) proteins, the SKICH domains of which are important for plasma membrane localization (30). However, due to the lack of systemic characterization, the precise working mode of the SKICH domains is still not well established. Interestingly, the N-terminal SKICH domains of NDP52 and TAX1BP1 are reported to interact with the adaptor proteins, NAP1 and SINTBAD (15), both of which can further directly bind to the TBK1 kinase through their C-terminal TBK1-binding domains (TBDs) (31, 32) (Fig. 1A). Therefore, NDP52 and TAX1BP1 may indirectly associate with TBK1 through NAP1 or SINTBAD. Importantly, recent studies reveal that the recruitment of TBK1, as well as its kinase activity, is required for efficient xenophagy and mitophagy processes (13, 14, 18, 33, 34). Strikingly, TBK1 can directly mediate the phosphorylation of NDP52 and TAX1BP1 at multiple sites, including their SKICH domains (13, 14). However, how TBK1 associates with NDP52 and TAX1BP1 mediated by NAP1 or SINTBAD, as well as the downsteam consequences induced by TBK1-mediated phosphorylation of NDP52 and TAX1BP1, is currently unknown, and the detailed binding mechanism of NDP52 and TAX1BP1 with the TBK1-binding adaptors remains to be elucidated.
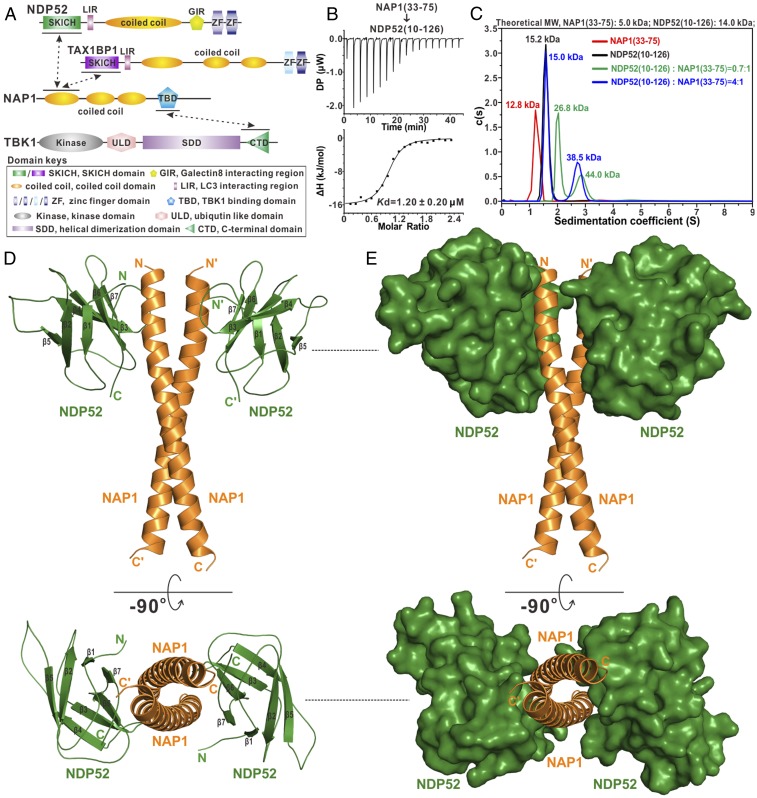
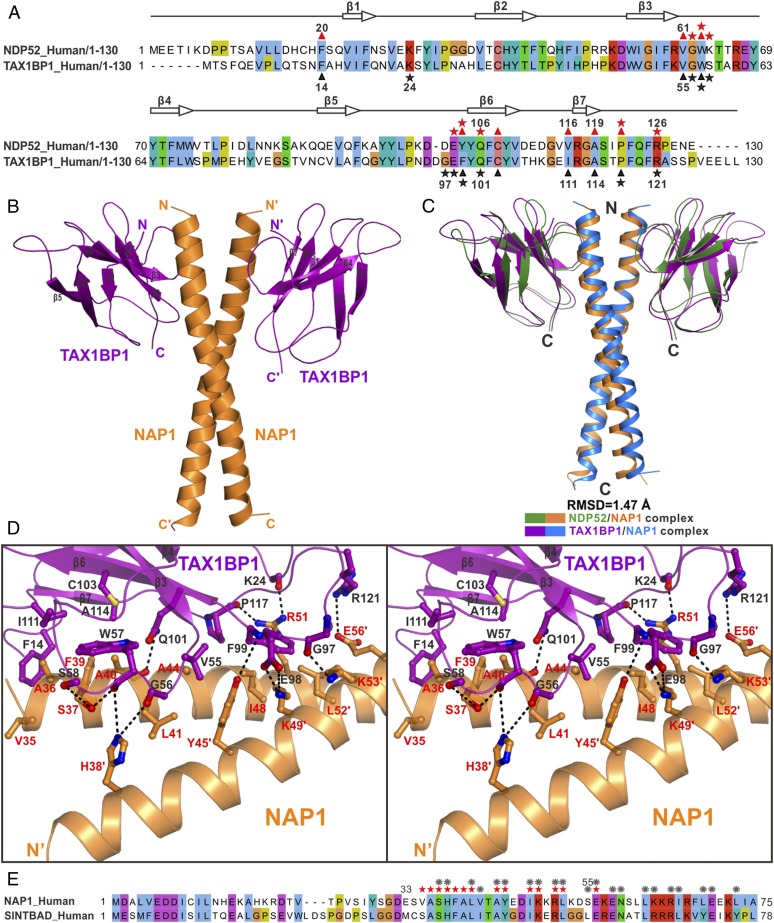
Fig. 1.
Biochemical and structural characterizations of the interaction between NDP52 and NAP1. (A) Schematic diagram showing the domain organizations of NDP52, TAX1BP1, NAP1, and TBK1. In this drawing, the NDP52/NAP1, TAX1BP1/NAP1, and NAP1/TBK1 interactions are further highlighted and indicated by two-way arrows. (B) ITC-based measurement of the binding affinity of NAP1(33–75) with NDP52(10–126). The Kd error is the fitted error obtained from the data analysis software when using the one-site binding model to fit the ITC data. DP, the differential power measured by the ITC machine; ΔH, the heat change measured by the ITC machine. (C) Overlay plot of the sedimentation velocity data of NAP1(33–75), NDP52(10–126), and NAP1(33–75) incubated with a 1:0.7 or 1:4 molar ratio of NDP52(10–126). The results demonstrate that NAP1(33–75) forms a stable dimer and may interact with one or two monomeric NDP52(10–126) receptors to form a 2:1 or 2:2 stoichiometric complex in solution. MW, molecular weight. (D) Ribbon diagram showing the overall structure of NDP52(10–126) in complex with NAP1(33–75). In this drawing, NDP52(10–126) is shown in forest green and NAP1(33–75) is shown in orange. (E) Combined ribbon and surface representation showing the overall architecture of the NDP52/NAP1 complex with the same color scheme as in D.
In this study, we biochemically and structurally characterized the interactions between NAP1 and the SKICH regions of NDP52 and TAX1BP1, and discovered that the N-terminal coiled-coil domain of NAP1 forms a stable dimer and specifically interacts with the SKICH domains of NDP52 and TAX1BP1 to form two unique heterotetramers. The determined structures of the NDP52/NAP1 and TAX1BP1/NAP1 complexes not only uncovered the detailed binding mechanism of NAP1 with the SKICH domains of NDP52 and TAX1BP1 but also unveiled that the SKICH domains of NDP52 and TAX1BP1 share a general binding mode to interact with NAP1 and SINTBAD. Furthermore, we demonstrated that NDP52 and TAX1BP1 adopt different mechanisms to recruit TBK1. Finally, we also evaluated the currently known phosphorylation sites mediated by TBK1 in the SKICH domains of NDP52 and TAX1BP1. In summary, our findings provided mechanistic insights into the interactions of NAP1 with autophagy receptor NDP52 and TAX1BP1, and expanded our understanding of the functions, as well as the working modes, of those proteins in selective autophagy.
Results
Biochemical Characterizations of the Specific Interaction Between NDP52 and NAP1.
To gain molecular insights into the specific interaction of NDP52 with NAP1, we first conducted detailed sequence alignment analyses of the NDP52 SKICH domain and the NAP1 N-terminal coiled-coil regions, which were reported to mediate NDP52/NAP1 complex formation in an earlier study (15). The result showed that these regions of NDP52 and NAP1 are highly conserved during evolution (SI Appendix, Fig. S1), in line with their potential functional roles to interact with each other. Then, based on systematic isothermal titration calorimetry (ITC) and NMR analyses, we further validated and mapped out the precise binding regions of NDP52 and NAP1, which are the NDP52(10–126) and NAP1(33–75) fragments (a detailed account of the construct optimization of NDP52 and NAP1 for mapping their interaction is provided in SI Appendix, SI Results). Specifically, the NDP52(10–126) fragment can directly bind to NAP1(33–75) with a binding affinity (Kd) value of about 1.20 μM (Fig. 1B). Finally, using an analytical ultracentrifugation-based assay, we revealed that NAP1(33–75) forms a stable homodimer and NDP52(10–126) is a monomer in solution (Fig. 1C). Interestingly, although NAP1(33–75) can specifically interact with NDP52(10–126) to form a single complex peak on analytical gel filtration chromatography analysis (SI Appendix, Fig. S3), our sedimentation velocity results revealed that the NAP1(33–75) dimer actually may bind to one or two NDP52(10–126) molecules to form a 1:1 or 1:2 stoichiometric complex under an unsaturated condition (Fig. 1C). However, in the presence of excess amounts of NDP52(10–126) protein, the NAP1(33–75) dimer can simultaneously bind to two NDP52(10–126) molecules to form a heterotetramer (Fig. 1C).
Overall Structures of NDP52(10–126) and Its Complex with NAP1(33–75).
To further uncover the mechanistic basis underlying this unique interaction between NDP52(10–126) and NAP1(33–75), we sought to determine their atomic structures. Although the structure of the NDP52(21–141) fragment has been solved in a previous study (23), the NDP52(10–126) fragment is different from NDP52(21–126) in binding to NAP1 based on our ITC and NMR results (SI Appendix, Fig. S2). Therefore, we first determined the crystal structure of NDP52(10–126) using the molecular replacement method (SI Appendix, Table S1). Notably, in the crystal structure, the extreme N-terminal eight residues of NDP52(10–126) are unsolved due to a lack of electron density, but the critical F20 residue of NDP52 for interacting with NAP1 (more details are provided below) is well defined. The structure of NDP52(10–126) features a β-sandwich, Ig-like architecture assembled by three-stranded, antiparallel β-sheet packing with a four-stranded, antiparallel β-sheet (SI Appendix, Fig. S4A). Unsurprisingly, the overall structure of NDP52(10–126) is highly similar to the previously reported structure of NDP52(21–141), except for the N-terminal and C-terminal loop regions (SI Appendix, Fig. S4B).
Next, we purified the NDP52(10–126)/NAP1(33–75) complex with abundant NDP52(10–126) and successfully obtained good crystals that diffracted to a resolution of 2.02 Å. Using the molecular replacement method with the apo-form structure of NDP52(10–126), we managed to solve the complex structure (SI Appendix, Table S1). In the final complex structural model, each asymmetrical unit contains one NDP52(10–126)/NAP1(33–75) complex molecule, which has 2:2 stoichiometry and forms a symmetrical heterotetramer consisting of two NDP52 molecules and one NAP1 dimer (Fig. 1D), consistent with our analytical ultracentrifugation analysis (Fig. 1C). In the complex structure, the two NAP1 molecules mainly form two continuous α-helices, which pack against each other in a head-to-head manner to form a parallel but crossed coiled-coil homodimer (Fig. 1 D and E), and, intriguingly, through each solvent-exposed side of the four-stranded β-sheet, the two NDP52 molecules symmetrically bind to the N-terminal region of the NAP1 dimer, forming a unique heterotetrameric complex with an overall architecture distinct from any currently known protein structures revealed by a structural similarity search using the program Dali (35) (Fig. 1D). Notably, there is no direct contact between two NDP52 molecules in the complex structure (Fig. 1 D and E). Further structural comparison revealed that the NDP52(10–126) molecules in the complex structure are also missing their N-terminal eight residues owing to the lack of electron density and adopt a similar overall conformation to that of the apo-form of the protein, except for the extreme N-terminal loop, which is directly involved in the NAP1 binding, and the flexible loop linking β4 and β5 (SI Appendix, Fig. S4C).
The Dimerization Interface of NAP1 in the NDP52/NAP1 Complex.
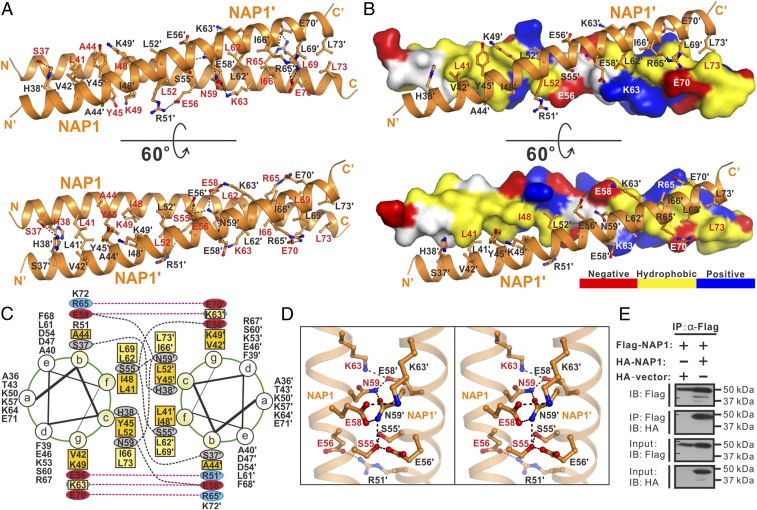
Further structural analysis revealed that in the structure of the NDP52(10–126)/NAP1(33–75) complex, the coiled-coil region (residues 36–73) of NAP1 is composed of five regular heptad repeats and is responsible for the NAP1 dimer formation (Figs. 1D and 2 A–C and SI Appendix, Fig. S4D). The dimerization of NAP1 is mediated by extensive hydrophobic and polar interactions between residues located at the b, c, f, and g positions of the two NAP1 coiled-coil helices (Fig. 2 A–C). In particular, an N-terminal hydrophobic patch formed by L41, V42, A44, Y45, I48, K49, and L52 residues, together with a C-terminal hydrophobic patch assembled by L62, K63, I66, L69, and L73 residues of one NAP1 helix, pack against their corresponding counterparts in the other NAP1 helix to form two separate hydrophobic interfaces of the NAP1 dimer (Fig. 2 A–C). Strikingly, a highly specific polar interaction network composed of unsymmetrical hydrogen bonds and salt bridges was found between the side chains of R51, S55, E56, E58, N59, and K63 residues located in the middle region of the paired NAP1 helixes (Fig. 2 A, B, and D). Moreover, two symmetrical intermolecular hydrogen bonds formed between the side chains of S37 in the b position of one NAP1 helix and H38 in the c position of the other NAP1 helix; in addition, two charge–charge interactions found between the positively charged R65 residue of one NAP1 helix and the negatively charged E70 of the other NAP1 helix further stabilize the NAP1 dimer formation (Fig. 2 A–C). Consistently, further coimmunoprecipitation (co-IP) assays demonstrated that the full-length NAP1 also forms dimers in cells, as the HA-tagged NAP1 could be readily pulled down by the Flag-tagged NAP1 when using anti-Flag antibody–conjugated agarose beads (Fig. 2E).
Fig. 2.
Dimerization interface of the NAP1 dimer in the NDP52/NAP1 complex. (A) Combined ribbon and stick-ball representation showing the detailed interactions between two NAP1 monomers in the NDP52/NAP1 complex. In this drawing, the side chains of the key residues are shown in the stick-ball mode and the hydrogen bonds involved in the binding are shown as dotted lines. (B) Combined surface representation and the ribbon-stick model showing the molecular interface of the NAP1 dimer in the NDP52/NAP1 complex. In this presentation, one NAP1 monomer is shown in the surface model and the other is shown in the ribbon-stick model. The hydrophobic amino acid residues of the NAP1 monomer in the surface model are shown in yellow, the positively charged residues are shown in blue, the negatively charged residues are shown in red, and the uncharged polar residues are shown in gray. (C) Helical wheel presentation showing the detailed dimerization interface formed between the heptad repeats of two NAP1(33–75) monomers. In this presentation, the interhelical hydrogen bonds and salt bridges are depicted by black and pink dashed lines, respectively. Two hydrophobic contacts located at the N-terminal and C-terminal parts of the NAP1 coiled-coil dimer and formed by hydrophobic residues located at the b, c, g, and f positions are further highlighted with orange and yellow boxes, respectively. (D) Stereoview of the ribbon-stick representation showing the unique polar interaction network found in the middle region of the NAP1 dimerization interface. The relevant hydrogen bonds and salt bridges involved in the binding are shown as dotted lines. (E) Co-IP assay showing that the full-length NAP1 may form dimers in cells. IB, immunoblot.
The Binding Interface Between NDP52 and NAP1.
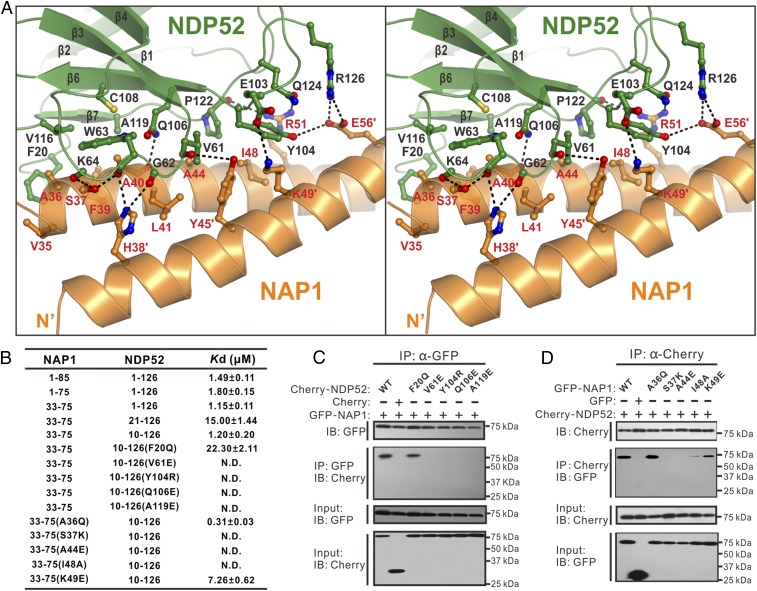
In the complex, the two NDP52 molecules symmetrically bind to two homodimeric interfaces located at the opposite sides of the N-terminal part of the NAP1 dimer, each burying a total surface area of ∼824 Å2 (Fig. 1 D and E). Detailed structural analysis showed that the binding interface between NDP52 and NAP1 is formed by residues from the β6 and β7 regions and loops located at the solvent-exposed face of the four-stranded β-sheet of NDP52, and that it accommodates NAP1 residues located at the N-terminal region of the paired helixes through both hydrophobic and polar interactions (Fig. 3A). In particular, the hydrophobic side chains of V35, A36, F39, and A40 from one chain of the NAP1 dimer pack against a hydrophobic patch formed by the side chains of F20, W63, C108, V116, and A119 from NDP52, and the hydrophobic side chains of V61 and P122 of NDP52 partially occupy a hydrophobic groove formed by the side chains of L41, A44, and I48 from one chain of the NAP1 dimer and the aromatic side chain of Y45 from the other chain of the NAP1 dimer (Fig. 3A). In addition, the backbone carboxyl groups of V61, G62, W63, K64, and P122, together with the polar side chain groups of Y104, Q106, and Q124 from NDP52, interact with the S37, A40, and R51 residues from one chain of the NAP1 dimer and the H38, Y45, and E56 residues from the other chain of the NAP1 dimer to form nine highly specific hydrogen bonds (Fig. 3A). Moreover, an Arg-Glu pair (Arg126NDP52-Glu56NAP1) and a Lys-Glu pair (Lys49NAP1-Glu103NDP52) of salt bridges further strengthen the NDP52 and NAP1 interaction (Fig. 3A). In line with their important structural roles, all of these key residues of NDP52 and NAP1 involved in the binding interface are evolutionarily conserved across different eukaryotic species (SI Appendix, Fig. S1).
Fig. 3.
Molecular details of the NDP52 and NAP1 interaction. (A) Stereoview of the ribbon-stick representation showing the binding interface between NDP52 and NAP1 in the determined NDP52/NAP1 complex structure. The hydrogen bonds and salt bridges involved in the interaction are shown as dotted lines. (B) Measured binding affinities between various forms of NDP52 and NAP1 or their mutants by ITC-based assays. N.D., the Kd value is not detectable. (C and D) Mutagenesis-based co-IP assays to confirm the interactions between NDP52 and NAP1 observed in the determined complex structure. IB, immunoblot.
Using ITC and co-IP analyses, we further verified the specific interactions between NDP52 and NAP1 observed in the complex structure. Consistent with our structural data, the ITC results showed that individual mutations of key interface residues from either NDP52 or NAP1, such as the V61E, Y104R, Q106E, and A119E mutations of NDP52 or the S37K, A44E, I48A, and K49E mutations of NAP1, essentially abolished or largely reduced the specific interaction between NDP52(10–126) and NAP1(33–75) (Fig. 3B and SI Appendix, Fig. S5). Notably, the replacement of the F20 residue located at the N-terminal loop of NDP52 with a Gln residue reduced the binding affinity between NDP52(10–126) and NAP1(33–75) to a value comparable to that of NDP52(21–126) and NAP1(33–75) interaction (Fig. 3B and SI Appendix, Figs. S2E and S5A), further confirming our aforementioned biochemical and structural results (Figs. 1B and 3A and SI Appendix, Fig. S2E). Importantly, in agreement with our in vitro ITC results, further co-IP experiments revealed that point mutations of key interface residues, including the F20Q, V61E, Y104R, Q106E, and A119E mutations of NDP52 and the S37K, A44E, I48A, and K49E mutations of NAP1, all completely disrupt or significantly attenuate the specific interaction between full-length NDP52 and NAP1 in cotransfected cells (Fig. 3 C and D). To our surprise, the A36Q mutation of NAP1 enhances the interaction between NAP1 and NDP52 in vitro and in vivo (Fig. 3D and SI Appendix, Fig. S5F), but the exact reasons still require elucidation. Nevertheless, all these data demonstrated that the specific interaction between NDP52(10–126) and NAP1(33–75) is essential for the NDP52/NAP1 complex formation.
Cellular Colocalization of NDP52 and NAP1 Required Specific Interaction Between NDP52(10–126) and NAP1(33–75).
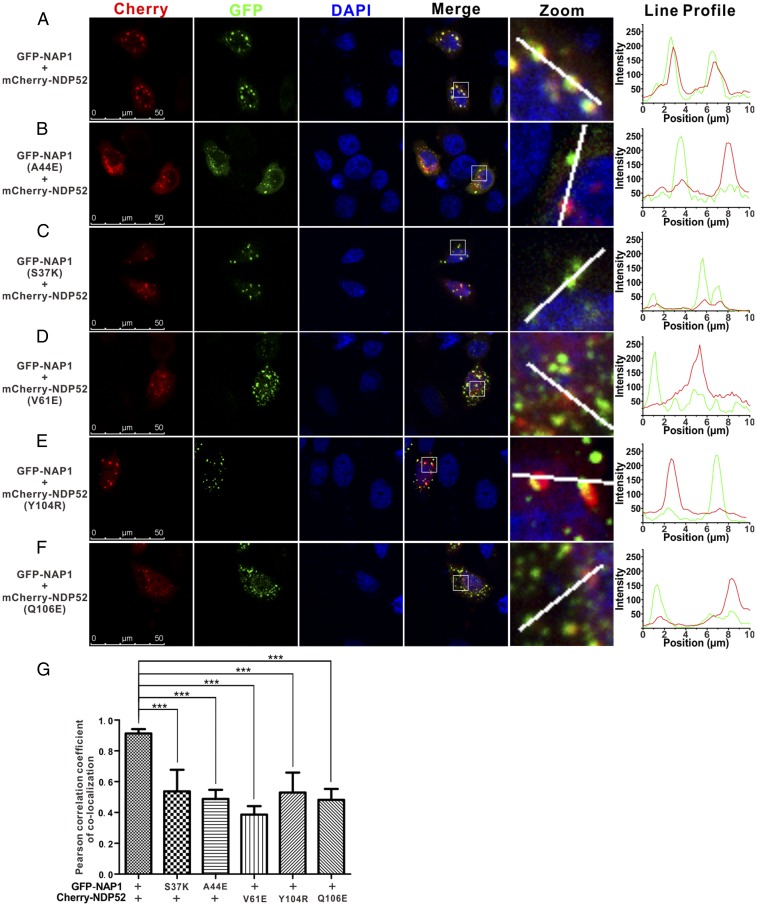
Next, we examined the role of NDP52(10–126)/NAP1(33–75) interaction on the cellular localizations of NDP52 and NAP1 in transfected HeLa cells. When cotransfected, the mCherry-tagged NDP52 mainly displayed a punctate staining pattern, and it colocalized perfectly with the GFP-tagged NAP1 puncta in the cytoplasm of transfected cells (Fig. 4 A and G). As a control, the mCherry-tagged NDP52 and GFP-tagged NAP1 did not colocalize well with the GFP tag or the mCherry tag from the cotransfected empty vector (SI Appendix, Fig. S6). In contrast, when NDP52 was cotransfected with the NAP1 S37K or A44E mutant, each of which was demonstrated to lose its ability to interact with NDP52 (Fig. 3 B and D), the mCherry-tagged NDP52 became more diffused in cells and the colocalization of NDP52 and NAP1 was largely compromised (Fig. 4 B, C, and G). To further evaluate the NDP52(10–126)/NAP1(33–75) interaction in the cellular NAP1/NDP52 colocalization, we also assayed the V61E, Y104R, and Q106E mutants of NDP52, as individual mutations of these three residues of NDP52 would essentially eliminate the NDP52/NAP1 interaction in vitro (Fig. 3 B and C). When the NDP52 V61E, Y104R, or Q106E mutant was coexpressed with the wild-type NAP1, it displayed a diffused localization pattern in the cytosol, and its colocalization with NAP1 puncta was also dramatically reduced (Fig. 4 D–G), in line with our biochemical and structural results (Fig. 3). Taken together, all these data clearly demonstrated that the specific interaction between NDP52(10–126) and NAP1(33–75) is critical for the cellular colocalization of NDP52 and NAP1.
Fig. 4.
Specific interaction between NDP52(10–126) and NAP1(33–75) is required for the cellular colocalization of NDP52 and NAP1 in cotransfected HeLa cells. (A) When coexpressed, NDP52 colocalizes well with the NAP1 puncta. (B–F) Point mutations of key interface residues of NDP52 or NAP1 that disrupted their interaction in vitro largely decrease the colocalization of NDP52 and NAP1. (G) Statistical results related to the colocalizations of different NDP52 and NAP1 variants in cotransfected HeLa cells, shown as Pearson’s correlations. The Pearson’s correlation coefficient analysis was performed using Leica Application Suite X software based on a randomly selected region that roughly contains one cotransfected HeLa cell. The data represent the mean ± SD of >30 analyzed cells. An unpaired Student’s t test analysis was used to define a statistically significant difference, and the asterisks indicate the significant differences between the indicated bars (***P < 0.001).
Biochemical and Structural Characterizations of the Interaction Between the TAX1BP1 SKICH Domain and NAP1.
In addition to autophagy receptor NDP52, NAP1 was implicated in the binding to the SKICH-containing autophagy receptor TAX1BP1 (15). Careful sequence alignment analysis showed that the SKICH region of TAX1BP1 (residues 1–121) is highly conserved during the evolution (SI Appendix, Fig. S7A), and is very similar to that of NDP52 (Fig. 5A). Analytical gel filtration chromatography and ITC-based assays revealed that the SKICH region of TAX1BP1 can directly interact with NAP1(33–75) with a Kd value of ∼2.05 μM (SI Appendix, Fig. S7 B and C), which is close to the Kd value of the NDP52/NAP1 interaction (Fig. 1B). To uncover the detailed binding mode between TAX1BP1 and NAP1, we also determined the high-resolution crystal structure of the TAX1BP1(1–121)/NAP1(33–75) complex (SI Appendix, Table S1). As expected, the TAX1BP1(1–121)/NAP1(33–75) complex structure is composed of a NAP1 coiled-coil dimer and two TAX1BP1 molecules, which pack together to form a symmetrical heterotetramer (Fig. 5B and SI Appendix, Fig. S8A). The overall conformation of the TAX1BP1(1–121)/NAP1(33–75) complex, as well as the NAP1 dimer in the TAX1BP1/NAP1 complex, is very similar to that of the NDP52/NAP1 complex (Fig. 5C and SI Appendix, Fig. S8B). In the complex structure, the TAX1BP1(1–121) molecule misses the 11 residues located at the N-terminal loop owing to the lack of electron density and adopts a similar fold to that of NDP52(10–126) in the NDP52/NAP1 complex (SI Appendix, Fig. S8C).
Fig. 5.
SKICH domains of TAX1BP1 and NDP52 share a general binding mode to interact with NAP1 and SINTBAD. (A) Structure-based sequence alignment analysis of the SKICH regions of NDP52 and TAX1BP1. In this alignment, the conserved residues are highlighted in colors, using Jalview2.8.1 software (www.jalview.org/). The key residues of NDP52 that interact with NAP1 are highlighted with red stars (polar interaction) or red triangles (hydrophobic interaction), and those of TAX1BP1 are highlighted in black. (B) Ribbon diagram showing the overall structure of TAX1BP1(1–121) in complex with NAP1(33–75). (C) Structural comparison of the overall structures of the TAX1BP1(1–121)/NAP1(33–75) complex and the NDP52(10–126)/NAP1(33–75) complex. In this drawing, the TAX1BP1(1–121) and NAP1(33–75) molecules in the TAX1BP1(1–121)/NAP1(33–75) complex are shown in purple and blue, respectively, while the NDP52(10–126) and NAP1(33–75) in the NDP52(10–126)/NAP1(33–75) complex are drawn in green and orange, respectively. (D) Stereoview of the ribbon-stick representation showing the detailed binding interface of the TAX1BP1/NAP1 complex. The related hydrogen bonds and salt bridges involved in the TAX1BP1 and NAP1 interaction are shown as dotted lines. (E) Sequence alignment analysis of the NDP52-binding regions in NAP1 and SINTBAD showing that most key interface residues of NAP1 that are crucial for dimerization or interacting with NDP52 are also conserved in SINTBAD. The interface residues of NAP1, which are critical for the interactions with NDP52 and/or TAX1BP1, are highlighted with red stars, while the NAP1 residues that are involved in the dimerization of NAP1 are labeled with gray gears.
Further structural analysis revealed that the association between TAX1BP1(1–121) and NAP1(33–75) is driven by extensive hydrophobic contacts and polar interactions (Fig. 5D and SI Appendix, Fig. S8 D and E). Specifically, the hydrophobic side chains of V35, A36, F39, and A40 from one chain of the NAP1 dimer contact extensively with the hydrophobic side chains of F14, W57, C103, I111, and A114 from TAX1BP1 (Fig. 5D and SI Appendix, Fig. S8D), and the hydrophobic side chains of V55, F99, and P117 from TAX1BP1 fit into a hydrophobic pocket assembled by the side chains of L41, A44, and I48 residues from one chain of the NAP1 dimer and Y45, K49, and L52 from the other chain (Fig. 5D). Interestingly, similar to the corresponding interaction found in the NDP52/NAP1 complex (Fig. 3A), a unique polar Q101 residue of TAX1BP1 is buried in the hydrophobic core of the interface by forming a strong hydrogen bond with the backbone carboxyl group of the NAP1 A40 residue (Fig. 5D). In addition, the backbone groups of K24, G56, W57, S58, G97, F99, and P117 residues of TAX1BP1 form eight hydrogen bonds with the side chains of S37, H38, Y45, R51, and K53 from the paired NAP1 dimer (Fig. 5D). Furthermore, two charge–charge interactions formed between the side chains of E98 and R121 of TAX1BP1 and K49 and E56 of NAP1 further contribute to the NAP1/TAX1BP1 complex formation (Fig. 5D).
The SKICH Domains of NDP52 and TAX1BP1 Share a General Binding Mode to Interact with NAP1 and SINTBAD.
Detailed structural and sequence comparison analyses of the NAP1/NDP52 complex and the NAP1/TAX1BP1 complex revealed that the NAP1 dimer employs almost the same residues to interact with NDP52 and TAX1BP1, except for the L52 and K53 residues, which are only important for binding to TAX1BP1 (Figs. 3A and 5D and SI Appendix, Fig. S1A), and the corresponding residues involved in the interactions with NAP1 of NDP52 and TAX1BP1 are also highly similar (Figs. 3A and 5 A and D). Interestingly, like NAP1, another TBK1-binding adaptor, SINTBAD, was also reported to associate with NDP52 through its N-terminal coiled-coil region (15, 31). Careful sequence alignment analysis of the NDP52-binding regions in these two adaptor proteins revealed that most key residues of NAP1 that are critical for homodimer formation or interaction with NDP52 can be also found in SINTBAD (Fig. 5E). Accordingly, based on these structural and sequence observations, we concluded that the SKICH domains of NDP52 and TAX1BP1 share a similar binding model to interact with NAP1 and SINTBAD, thereby forming distinct complexes.
Evaluations of the Potential TBK1-Mediated Phosphorylation Sites in the SKICH Domains of NDP52 and TAX1BP1.
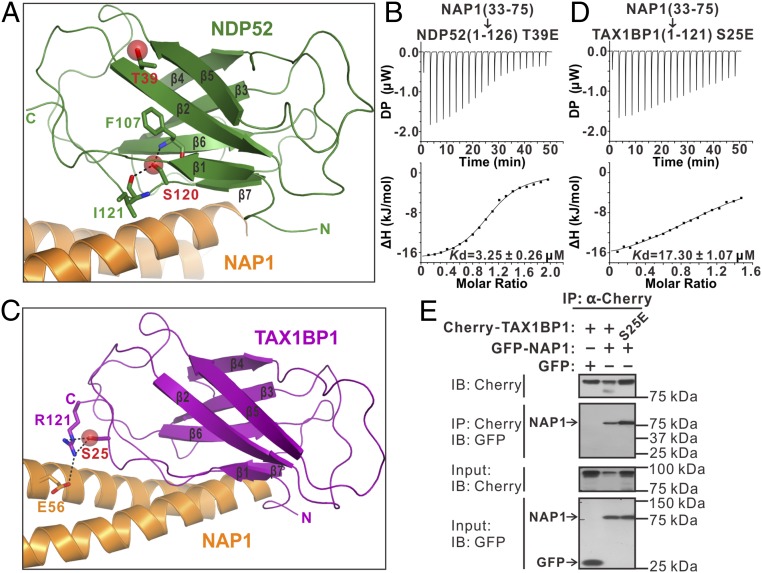
Recent studies reported that the TBK1 kinase can directly or indirectly phosphorylate NDP52 and TAX1BP1; in particular, it can mediate the phosphorylation of the T39 and S120 residues of NDP52 as well as the S25 residue of TAX1BP1, all of which are within the SKICH regions of these two proteins (13, 14). In our determined NDP52/NAP1 complex structure, the T39 residue of NDP52 is located in the β2 strand of the SKICH domain and is far away from the NAP1-binding site (Fig. 6A), while the side chain of the NDP52 S120 residue is buried in the structural core of the SKICH domain by forming two strong hydrogen bonds with the backbone amide of F107 and the backbone carboxyl group of I121, respectively (Fig. 6A). Consistent with these structural observations, we were unable to obtain soluble recombinant protein for the phosphomimetic S120E mutant of NDP52(1–126), and further ITC-based measurement revealed that the phosphomimetic T39E mutant of NDP52(1–126) has a similar binding ability to NAP1(33–75) as that of the wild-type protein (Fig. 6B and SI Appendix, Fig. S2C). Therefore, the phosphorylation of the NDP52 T39 residue is unable to affect the interaction between NDP52 and NAP1 but, instead, may alter the ability of NDP52 to interact with other unknown partners, and the S120 residue of NDP52 is unlikely to be phosphorylated as it is tightly packed in the structural core of the SKICH domain. Interestingly, the TAX1BP1 S25 residue is located in the loop connecting the β1 and β2 strands, and is adjacent to the NAP1-binding interface in the TAX1BP1/NAP1 complex (Fig. 6C). Importantly, the side chain of S25 forms two hydrogen bonds with the side chain of the TAX1BP1 R121 residue, which is further coupled with the negatively charged E56 residue of NAP1 through a charge–charge interaction (Fig. 6C). Thus, once the S25 residue of TAX1BP1 was phosphorylated, the negatively charged phosphate group might disturb the interaction between TAX1BP1 and NAP1. In line with our structural analysis, further ITC-based measurement revealed that the phosphomimetic S25E mutant of TAX1BP1(1–121) showed a much weaker binding to NAP1(33–75) than that of the wild-type TAX1BP1 protein (Fig. 6D and SI Appendix, Fig. S7C). However, further co-IP analysis showed that the phosphomimetic S25E mutant of full-length TAX1BP1 was still able to interact with NAP1 in cells (Fig. 6E). Unfortunately, due to technological limitations, we failed to further characterize the downstream functional effects induced by this TBK1-mediated S25 phosphorylation of TAX1BP1 in vivo. Nevertheless, additional functional studies are required to elucidate the precise downstream effects induced by TBK1-mediated phosphorylation of NDP52 and TAX1BP1 in selective autophagy in the future.
Fig. 6.
Structural and biochemical analyses of the currently known TBK1-mediated phosphorylation sites in the SKICH domains of NDP52 and TAX1BP1. (A) Combined ribbon and stick-sphere representations showing the detailed structural roles of the NDP52 T39 and S120 residues in the NDP52/NAP1 complex, both of which are the TBK1-mediated phosphorylation sites found in the SKICH domain of NDP52. In this drawing, the side chains of T39 and S120 are highlighted in the stick-sphere mode and the F107 and I121 residues of NDP52, which are coupled with S120, are shown in the stick mode. The related hydrogen bonds are indicated by black dashed lines. (B) ITC-based measurement of the binding affinity of the phosphomimetic T39E mutant of NDP52(10–126) with NAP1(33–75). The Kd error is the fitted error obtained from the data analysis software when using the one-site binding model to fit the ITC data. DP, the differential power measured by the ITC machine.; ΔH, the heat change measured by the ITC machine. (C) Combined ribbon and stick-sphere representations showing the detailed role of the TAX1BP1 S25 residue, which is the reported phosphorylation site mediated by TBK1 in the SKICH domain of TAX1BP1, in the TAX1BP1(1–121) and NAP1(33–75) interaction. In this drawing, the side chain of S25 is shown in the stick-sphere mode and the related NDP52 R121 and NAP1 E56 residues are displayed in the stick mode. The relevant hydrogen bonds are indicated by black dashed lines. (D) ITC-based measurement of the binding affinity of the phosphomimetic S25E mutant of TAX1BP1(1–121) with NAP1(33–75). The Kd error is the fitted error obtained from the data analysis software when using the one-site binding model to fit the ITC data. (E) Co-IP assay showing that the phosphomimetic S25E mutant of full-length TAX1BP1 can still bind well to NAP1 in cells. IB, immunoblot.
Discussion
In this study, we revealed that the N-terminal coiled-coil region of NAP1 forms a homodimer and uses a similar binding mode and almost the identical key residues to associate with the SKICH regions of NDP52 and TAX1BP1, forming two unique heterotetramer complexes. Interestingly, although our analytical ultracentrifugation assay showed that the NAP1(33–75) dimer could form two stoichiometric types of complexes with the monomeric NDP52(10–126) fragment under an unsaturated condition in solution (Fig. 1C), further analyses showed that the dimeric form of NDP52, such as the NDP52(1–316) fragment, which includes the central coiled-coil region of NDP52, only forms a stable heterotetramer with the NAP1(33–75) dimer (SI Appendix, Fig. S9). Therefore, the full-length NDP52 and TAX1BP1, both of which were reported to form a dimer via their middle coiled-coil regions (24, 36, 37), are predicated to form heterotetrameric complexes when binding to NAP1. In addition, based on structural and sequence analyses, we inferred that the two TBK1-binding adaptors, NAP1 and SINTBAD, likely share a similar binding mode to interact with NDP52. In the future, it will be interesting to know the detailed relationship between NAP1 and SINTBAD in binding to NDP52 and TAX1BP1 during different selective autophagy processes.
So far, the SKICH domain has been identified in many different proteins, including NDP52, TAX1BP1, CALCOCO1, SKIP, and PIPP. However, due to a lack of detailed structure characterization, the precise working mode of the SKICH domain is still elusive. Therefore, the structures of the NDP52/NAP1 and TAX1BP1/NAP1 complexes determined in this work provide an atomic picture showing how a SKICH domain functions as a protein–protein interaction module to interact with its binding partners. Interestingly, although the SKICH domains of NDP52 and TAX1BP1 can both bind to NAP1, detailed sequence and structural analyses revealed that the NAP1-binding residues are not conserved in the SKICH domains of CALCOCO1, SKIP, and PIPP (SI Appendix, Fig. S10 A and B). Therefore, rather than binding to NAP1, the SKICH domains of CALCOCO1, SKIP, and PIPP are likely to interact with other unknown proteins. More work is definitely required to clarify the functions of those SKICH domains.
Previous studies indicated that TBK1, together with NDP52, TAX1BP1, and optineurin, was recruited to the ubiquitin-decorated damaged mitochondria in the depolarization-dependent mitophagy as well as the invading pathogen in xenophagy, and cooperated with those autophagy receptors in selective autophagy (13–15, 18, 34). Specifically, TBK1 can directly bind to and phosphorylate optineurin to regulate its interactions with Atg8 family proteins and ubiquitin proteins (12–14, 38). In contrast to optineurin, our co-IP assay showed that there is no direct interaction between NDP52 and TBK1, and that TBK1 may indirectly associate with NDP52 through the adaptor protein NAP1 (SI Appendix, Fig. S11A), in line with a previous report (15). Importantly, mutations of key interface residues of NAP1, such as the S37K and A44E mutations, which were proved to abolish the specific interaction between NDP52 and NAP1 in this study (Fig. 3 B and D), also essentially disrupted the association of NDP52 with TBK1 mediated by NAP1 in cells (SI Appendix, Fig. S11A). Unexpectedly, TAX1BP1 somehow can directly associate with TBK1, and the NAP1 S37K and A44E mutations that were demonstrated to abolish the specific interaction between the SKICH domain of TAX1BP1 and NAP1(33–75) in vitro (SI Appendix, Fig. S11 B and C) and to reduce the binding of full-length TAX1BP1 with NAP1 in cells (SI Appendix, Fig. S11D) had little effect on the association of TAX1BP1 and TBK1 in cells (SI Appendix, Fig. S11E), suggesting that in addition to the SKICH domain, other regions of TAX1BP1 may bind to NAP1 and, unlike NDP52, TAX1BP1 may adopt a different mechanism to recruit TBK1. Therefore, further studies are required to elucidate the detailed interaction mechanisms of TAX1BP1 with NAP1 and TBK1.
In addition to NAP1, a previous study showed that NDP52 can specifically bind to LC3C through its unconventional LIR motif, which is C-terminally adjacent to the SKICH domain (23). Our biochemical analyses using purified proteins demonstrated that NDP52 can simultaneously bind to NAP1(33–75) and LC3C to form a ternary complex in vitro (SI Appendix, Fig. S12 A and B). To further elucidate the relationship between NAP1 and LC3C in binding to NDP52, we also carried out a co-IP assay. The results showed that NDP52 can simultaneously associate with NAP1 and LC3C and that NAP1 is unable to directly interact with LC3C in cells (SI Appendix, Fig. S12C). In summary, we proposed a model depicting the recruitment of TBK1 to NDP52 mediated by NAP1, as well as the potential regulation of NDP52 by TBK1 in mitophagy and/or xenophagy (SI Appendix, Fig. S13). In this model, NDP52 formed dimers through its coiled-coil region and associated with the NAP1 dimer mediated by the unique interaction between its SKICH domain and the N-terminal coiled-coil domain of NAP1 (SI Appendix, Fig. S13). Meanwhile, the C-terminal TBDs of the NAP1 dimer could further interact with the C-terminal domain of the TBK1 dimer, thereby forming a heterohexamer (SI Appendix, Fig. S13). Then, the NDP52/NAP1/TBK1 heterohexameric complex was recruited to the ubiquitin-decorated mitochondria and/or pathogen through the C-terminal ubiquitin-binding ZF2 domain of NDP52 (SI Appendix, Fig. S13). Finally, after activation of TBK1, the activated TBK1 molecules could directly or indirectly phosphorylate NDP52 to regulate its binding abilities to other unknown binding partners.
Materials and Methods
All proteins used in this study were expressed in BL21 (DE3) Escherichia coli cells and purified by affinity chromatography, followed by size-exclusion chromatography or ion-exchange chromatography. Crystals were obtained by the sitting drop vapor diffusion method at 16 °C. An extended description of the materials and methods used for protein preparation, NMR spectroscopy, crystallography, and biochemical and cellular assays is included in SI Appendix.
Supplementary Material
Acknowledgments
We thank Shanghai Synchrotron Radiation Facility BL19U1 and BL17U1 for X-ray beam time, Dr. Jianchao Li for help with X-ray diffraction data collection, Prof. Jiahuai Han for the full-length NDP52 and TAX1BP1 cDNA, Dr. Chengjiang Gao for the NAP1 plasmid, and Prof. Mingjie Zhang for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (Grants 31470749, 21621002, 91753113, and 21822705), the National Key R&D Program of China (Grant 2016YFA0501903), the Science and Technology Commission of Shanghai Municipality (Grant 15JC1400400), and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB20000000); by a “Thousand Talents Program” young investigator award; and by start-up funds from State Key Laboratory of Bioorganic and Natural Products Chemistry and Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors of the NDP52(10–126), NDP52(10–126)/NAP1(33–75) complex, and TAX1BP1(1–121)/NAP1(33–75) complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5Z7A, 5Z7L, and 5Z7G, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811421115/-/DCSupplemental.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randow F, Youle RJ. Self and nonself: How autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 8.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 12.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter B, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- 16.Newman AC, et al. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS One. 2012;7:e50672. doi: 10.1371/journal.pone.0050672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumbarello DA, et al. The autophagy receptor TAX1BP1 and the molecular motor myosin VI are required for clearance of Salmonella typhimurium by autophagy. PLoS Pathog. 2015;11:e1005174. doi: 10.1371/journal.ppat.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun. 2014;5:5276. doi: 10.1038/ncomms6276. [DOI] [PubMed] [Google Scholar]
- 20.Gibbings D, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin S, et al. Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52-mediated selective autophagic degradation in human cells. Mol Cell. 2017;68:308–322.e4. doi: 10.1016/j.molcel.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Ellinghaus D, et al. Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology. 2013;145:339–347. doi: 10.1053/j.gastro.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Muhlinen N, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie X, et al. Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2. Autophagy. 2015;11:1775–1789. doi: 10.1080/15548627.2015.1082025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tumbarello DA, et al. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu S, et al. Mechanistic insights into recognitions of ubiquitin and myosin VI by autophagy receptor TAX1BP1. J Mol Biol. 2018;430:3283–3296. doi: 10.1016/j.jmb.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BW, Hong SB, Kim JH, Kwon DH, Song HK. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat Commun. 2013;4:1613. doi: 10.1038/ncomms2606. [DOI] [PubMed] [Google Scholar]
- 29.Li S, et al. Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Sci Signal. 2013;6:ra9. doi: 10.1126/scisignal.2003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurung R, et al. Identification of a novel domain in two mammalian inositol-polyphosphate 5-phosphatases that mediates membrane ruffle localization. The inositol 5-phosphatase skip localizes to the endoplasmic reticulum and translocates to membrane ruffles following epidermal growth factor stimulation. J Biol Chem. 2003;278:11376–11385. doi: 10.1074/jbc.M209991200. [DOI] [PubMed] [Google Scholar]
- 31.Ryzhakov G, Randow F. SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK. EMBO J. 2007;26:3180–3190. doi: 10.1038/sj.emboj.7601743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, et al. Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat Commun. 2016;7:12708. doi: 10.1038/ncomms12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore AS, Holzbaur EL. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci USA. 2016;113:E3349–E3358. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thurston TL, et al. Recruitment of TBK1 to cytosol-invading Salmonella induces WIPI2-dependent antibacterial autophagy. EMBO J. 2016;35:1779–1792. doi: 10.15252/embj.201694491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling L, Goeddel DV. T6BP, a TRAF6-interacting protein involved in IL-1 signaling. Proc Natl Acad Sci USA. 2000;97:9567–9572. doi: 10.1073/pnas.170279097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sternsdorf T, Jensen K, Züchner D, Will H. Cellular localization, expression, and structure of the nuclear dot protein 52. J Cell Biol. 1997;138:435–448. doi: 10.1083/jcb.138.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F, et al. Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation. Autophagy. 2018;14:66–79. doi: 10.1080/15548627.2017.1391970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.