Significance
Light enhances the translation efficiency of thousands of mRNAs during photomorphogenic development in Arabidopsis, but the underlying molecular mechanism remains elusive. Here we show that light activates the auxin-target of rapamycin (TOR)-ribosome protein S6 (RPS6) pathway to enhance translation in deetiolating Arabidopsis. We discovered that CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) represses TOR activity in dark-grown seedlings. The perception of far-red and blue light by photoreceptors inactivates COP1, which leads to the derepression of the auxin-TOR-RPS6 pathway and enhanced de novo protein synthesis. Our study revealed a light-triggered signaling pathway for translational regulation. This sophisticated regulation also functions to ensure that young seedlings have strict skotomorphogenic development in the dark and a timely switch to photomorphogenic development.
Keywords: TOR, RPS6, translation, photomorphogenesis, light
Abstract
Deetiolation is an essential developmental process transforming young plant seedlings into the vegetative phase with photosynthetic activities. Light signals initiate this important developmental process by triggering massive reprogramming of the transcriptome and translatome. Compared with the wealth of knowledge of transcriptional regulation, the molecular mechanism underlying this light-triggered translational enhancement remains unclear. Here we show that light-enhanced translation is orchestrated by a light perception and signaling pathway composed of photoreceptors, CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1), the phytohormone auxin, target of rapamycin (TOR), and ribosomal protein S6 (RPS6). In deetiolating Arabidopsis seedlings, photoreceptors, including phytochrome A and cryptochromes, perceive far-red and blue light to inactivate the negative regulator COP1, which leads to activation of the auxin pathway for TOR-dependent phosphorylation of RPS6. Arabidopsis mutants defective in TOR, RPS6A, or RPS6B exhibited delayed cotyledon opening, a characteristic of the deetiolating process to ensure timely vegetative development of a young seedling. This study provides a mechanistic view of light-triggered translational enhancement in deetiolating Arabidopsis.
Plants are capable of transforming sunlight energy to chemical energy stored in carbohydrates. The presence or absence of light also serves as important environmental cues for plants to deploy appropriate developmental programs for coping with the environment. For example, with growth in the dark, “skotomorphogenesis” represents the morphology of young seedlings with closed cotyledons, an apical hook, and long hypocotyls. However, “photomorphogenesis” represents the light morphology of plants with open cotyledons, development of chloroplasts for photosynthesis, and short hypocotyls (1).
In plants, light signals are perceived by at least four families of photoreceptors, including phytochrome (phy), cryptochrome (CRY), phototropin (phot), and UV B resistance locus 8 (UVR8). CRY and phot families perceive blue and UV A, whereas the phy family perceives red and far-red (FR) light (2). The phy family is found in most plants, and five phy members (phyA–E) were identified in Arabidopsis (3). phyA and phyB are the primary photoreceptors for far-red and red light, respectively (4, 5).
A number of positive signaling components downstream of photoreceptors have been identified in the past few decades (6). Many are transcription factors. For example, Arabidopsis defective in a positive regulator ELONGATED HYPOCOTYL 5 (HY5), a bZIP transcription factor, showed a long hypocotyl phenotype under a broad spectrum of light, which suggests that HY5 acts downstream of all photoreceptors (7, 8). In contrast, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) is a key negative regulator of photomorphogenesis and an E3 ligase responsible for the degradation of photoreceptors (9–11) and various photomorphogenesis-promoting transcription factors such as HY5, HY5 HOMOLOG (HYH), LONG AFTER FAR-RED LIGHT 1 (LAF1) and LONG HYPOCOTYL IN FAR-RED LIGHT 1 (HFR1). COP1 actions could be repressed by various photoreceptors upon light treatments, thus triggering photomorphogenesis (1, 12).
Despite the abundant knowledge of transcriptional and posttranslational regulation in photomorphogenesis, translational control by light signals is much less discussed. We previously showed that light activates the translation of thousands of mRNAs in deetiolating seedlings (13, 14). The selective and dynamic translation of mRNAs encoding components of photosynthetic machinery is also regulated by diurnal signals (15). These findings indicate that translational control constitutes an important regulatory step in light-regulated gene expression. What remains to be revealed is the underlying molecular mechanism responsible for the massive translation triggered by light signals in deetiolating Arabidopsis seedlings.
Target of rapamycin (TOR) has been inferred to regulate ribosome biogenesis, translation initiation, and the elongation process, at least in part by phosphorylating its substrates, such as ribosomal S6 kinase (S6K) in mammals (16). The phosphorylation of S6K is also TOR dependent in plants (17, 18). In plants, TOR is a central regulator for sensing intracellular energy status, nitrogen mobilization, glucose utilization, stresses, and hormone coordination (19, 20). For example, glucose from photosynthesis can activate TOR kinase to regulate root meristem development (21).
Previous studies indicated that the phosphorylation of ribosomal protein S6 (RPS6) primarily depends on the TOR-S6K signaling pathway (22). RPS6 is the substrate of S6K, and its phosphorylation can be stimulated by nutrients and growth factors in plants and mammals (18, 21, 23). In terms of translation control, Arabidopsis with reduced TOR expression has fewer mRNAs under active translation and an early senescence phenotype (24). The protein synthesis rate of the reporter gene GUS was increased in plants overexpressing TOR or RPS6A/B but reduced in tor and rps6 mutants (25). In plants grown under light–dark cycles, the degree of RPS6 phosphorylation was higher in the light period (26). The degree of RPS6 phosphorylation also appeared to be associated with the photosynthetic activities (27). Increased phosphorylated RPS6 was found associated with polysomes in adult plants in the light period (28). This evidence implies that photosynthesis could trigger TOR-RPS6 phosphorelay to regulate translation in adult plants with photosynthetic activities. Whether the TOR-RPS6 pathway is responsible for the globally enhanced translation in young seedlings during their first few hours of light exposure and before the full establishment of photosynthetic capacity is unknown.
In this study, we discovered that the TOR-S6K-RPS6 pathway contributes to the light-triggered translation enhancement in deetiolating Arabidopsis seedlings before they acquire photosynthetic activities. The TOR-S6K-RPS6 pathway can be triggered by far-red and blue light photoreceptors but is repressed by a key negative regulator COP1 in the dark. Our study puts TOR-S6K-RPS6 into the biological context of functioning downstream of COP1 for translation control. An intact TOR-S6K-RPS6 pathway is required for de novo protein synthesis and timely cotyledon opening in deetiolating Arabidopsis. Our study revealed a regulatory cascade involving photoreceptors, COP1, auxin, TOR, and RPS6 as an action mode controlling light-enhanced translation.
Results
phyA and COP1 but Not HY5/HYH Regulate Light-Enhanced Translation.
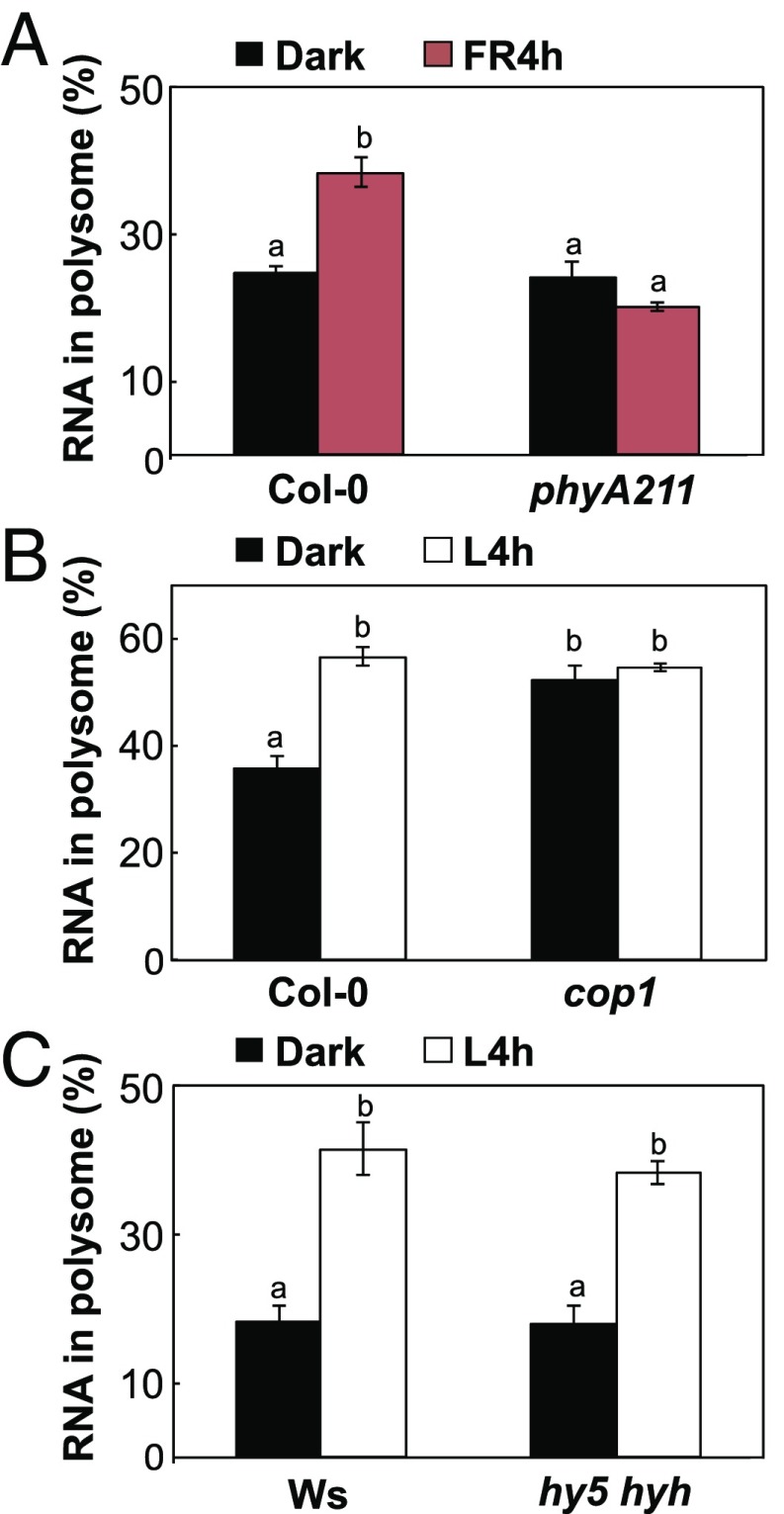
Light could trigger a translational enhancement in deetiolating seedlings, but whether this phenomenon is under the control of photoreceptors or light signaling molecules is unknown. Because phyA is the sole photoreceptor for FR light in plants, we first examined whether FR alone could activate translation. Polysome profiling analysis showed that in the wild type (Col-0), 4-h FR light treatment could trigger a significant shift of RNAs from nonpolysome (NP) to polysome (PL) fractions (Fig. 1A and SI Appendix, Fig. S1A, Upper). The FR-induced translation was compromised in the phyA211 mutant (Fig. 1A and SI Appendix, Fig. S1A, Lower). This result indicates that FR-mediated translational enhancement depends on the FR photoreceptor phyA.
Fig. 1.
phyA activates translation in the light and COP1 represses it in the dark. (A) Bar graph shows the ribosome loading efficiency of etiolated 4-d-old Col-0 or phyA221 seedlings grown in the dark or treated with FR4h. (B) Bar graph shows the ribosome loading efficiency of etiolated 2-d-old Col-0 or cop1 seedlings grown in the dark or treated with L4h. (C) Bar graph shows the ribosome loading efficiency of 4-d-old wild-type Ws or hy5 hyh seedlings grown in the dark or L4h. Values are mean percentages ± SE from three biological replicates. Bars with the different letters represent significant difference [Tukey’s honestly significant difference (HSD) P < 0.05].
COP1 is a negative regulator repressing photomorphogenesis in the dark by directing photoreceptors and many positive regulators for degradation via 26S proteasome (12). We next examined whether COP1 also plays a negative role in the translation in dark-grown seedlings. Compared with the wild type (Col-0), the cop1 mutant showed a significant increase in RNAs in the PL fraction in darkness (Fig. 1B and SI Appendix, Fig. S1B). The proportion of RNAs in the PL fraction in the dark-grown cop1 mutant was comparable to that in Col-0 treated with 4-h light (L4h). The light treatment did not further increase the translation in the cop1 mutant (Fig. 1B). Therefore, COP1 represses global translation in the dark.
Previous studies reported that two transcription factors, HY5 and its homolog HYH, are key positive regulators for photomorphogenic development (29). We next investigated whether light-mediated translational enhancement depends on transcriptional activation through these two transcription factors. PL% did not differ between Col-0 and the hy5 hyh double mutant when grown in the dark (Dark) or treated with L4h (Fig. 1C and SI Appendix, Fig. S1C). Therefore, HY5 and HYH function primarily in transcriptomic adjustment but are not required for translational regulation at the early photomorphogenic stage investigated in this study.
Light Activates RPS6 Phosphorylation to Promote Translation and Cotyledon Opening.
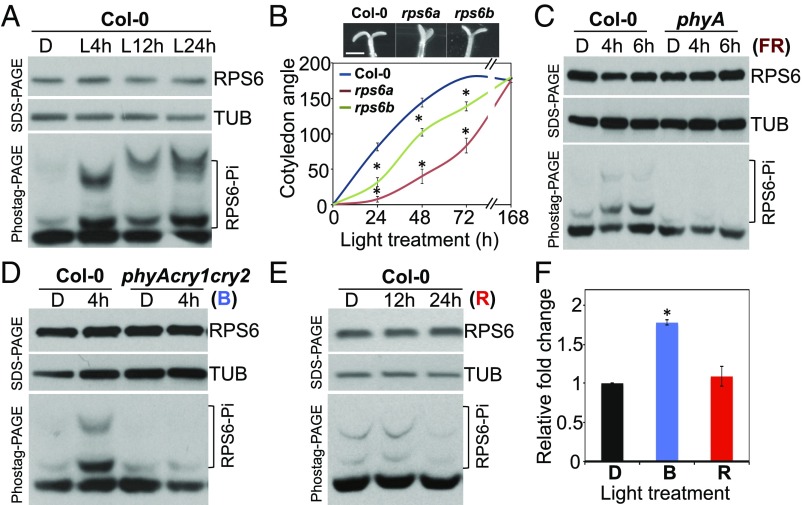
Previous studies indicated that RPS6 phosphorylation increases the binding affinity of RPS6 to 5′-m7-GpppG mRNAs in vitro (30), which suggests a positive role for RPS6 in increasing translation efficiency. To assess whether RPS6 is involved in light-triggered translation enhancement, we first examined whether RPS6 is differentially phosphorylated in Arabidopsis seedlings before and after light treatment. Total proteins from 4-d-old etiolated seedlings with white light treatment for 4 h (L4h), 12 h (L12h), or 24 h (L24h) were isolated and subjected to both SDS/PAGE and Phostag-PAGE analyses; the latter analysis can resolve proteins with differential phosphorylation status. Although the RPS6 protein abundance remained unchanged, the degree of RPS6 phosphorylation increased with time of light treatments (Fig. 2A). We also used phosphatase treatment to confirm that RPS6 proteins with mobility shifts in response to light signals are indeed phosphorylated RPS6 (SI Appendix, Fig. S2).
Fig. 2.
Phosphorylation and biological functions of RPS6. (A) Four-day-old etiolated Col-0 seedlings (dark, D) were treated with white light (L) for the indicated time. Total protein from each treatment was extracted and subjected to SDS/PAGE and Phostag-PAGE analyses. (B) Four-day-old etiolated Col-0, rps6a, and rps6b seedlings were treated with 15 μE white light to observe cotyledon opening for 7 d. Cotyledon opening angles are shown as mean ± SE *P < 0.001 vs. wild type, Col-0 (n = 16–22). Representative image showed cotyledons from etiolated seedlings treated with 72 h white light. (Scale bar: 1 mm.) (C) Four-day-old etiolated seedlings in the dark (D) of Col-0 and phyA were treated with 1.2 W/m2 FR light for 4 h or 6 h. (D) Four-day-old etiolated seedlings of Col-0 and phyA cry1 cry2 were treated with or without 35 μE blue (B) light for 4 h. (E) Four-day-old etiolated seedlings of Col-0 were treated with 35 μE (R) light for 12 h or 24 h. RPS6 and TUB were detected by RPS6- and α–tubulin-specific antiserum, respectively. The levels of TUB served as internal controls. (F) De novo protein synthesis was determined for 4-d-old etiolated seedlings of Col-0 in the dark (D) or treated with B or R light. Relative fold change was shown by normalizing to the level of newly synthesized proteins in etiolated wild-type seedlings. Values are mean ± SE (Student t test, *P < 0.01, n = 3).
Whether RPS6s are crucial for photomorphogenic development was next investigated. Four-day-old etiolated seedlings of Col-0, rps6a, and rps6b mutants were exposed to white light for the indicated times to observe cotyledon opening kinetics. Cotyledon opening was clearly delayed in rps6 mutants compared with Col-0 (Fig. 2B). Between these two rps6 mutants, rps6a exhibited a more severe phenotype than rps6b, which is consistent with RPS6A playing a more crucial role in plant development (31). Nevertheless, RPS6A and RPS6B are both required for timely cotyledon opening during the deetiolation process.
We next examined whether like white light, monochromatic light, such as FR, blue (B), or red (R) light, could activate RPS6 phosphorylation. Four to 6 h of both FR and B light increased RPS6 phosphorylation (Fig. 2 C and D). The FR- and B-induced RPS6 phosphorylation was diminished in mutants defective in corresponding photoreceptors: phyA and phyA cry1 cry2 triple mutant (Fig. 2 C and D). However, R light could not induce an appreciably increased phosphorylation of RPS6 even by extending the R light treatment to 24 h (Fig. 2E). Also, in contrast to the increased de novo protein synthesis under B light, R light alone failed to trigger de novo protein synthesis (Fig. 2F and SI Appendix, Fig. S3). To exhaust the exploration of the impact of R light on RPS6 phosphorylation, etiolated Arabidopsis seedlings were treated with high-fluence R light (140 μE) for 1 and 2 d. In contrast to the marked RPS6 phosphorylation within hours in B or FR light, RPS6 phosphorylation was observed under 140 μE R light only after 2 d (SI Appendix, Fig. S4). Thus, perception of FR and B light by phyA and CRYs could quickly lead to a coherent phosphorylation of RPS6 and translation enhancement. However, R light works much less efficiently in evoking RPS6 phosphorylation. These observations are consistent with the more apparent cotyledon opening in deetiolating seedlings treated with FR and B rather than R light (32).
Light-Activated RPS6 Phosphorylation Is Independent of Photosynthesis and Exogenous Glucose but Requires Auxin.
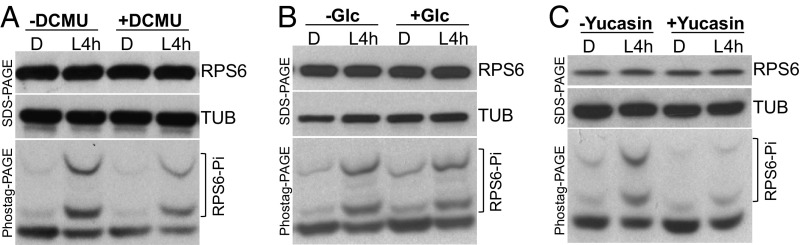
TOR is known to activate S6K for the phosphorylation of RPS6 (21, 33). TOR kinase activity can also be activated by photosynthesis and glucose to regulate the cell cycle and metabolism-related genes for plant growth (21). We next studied whether the phosphorylation of RPS6 in deetiolating Arabidopsis seedlings results from increased photosynthesis or photosynthates (sugar). For this purpose, we treated Col-0 with or without 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), an inhibitor of photosystem II. SI Appendix, Fig. S5 A and B show that 20 μM DCMU could effectively suppress photosystem II activity in deetiolating Arabidopsis seedlings. However, the phosphorylation of RPS6 was still increased after light treatment in the presence of DCMU (Fig. 3A). Supplying exogenous glucose could not effectively activate RPS6 phosphorylation compared with light in etiolated seedlings (Fig. 3B). These findings suggest that the increased phosphorylation of RPS6 can be independent of photosynthesis and its derived glucose. They are also consistent with our observation that FR light, despite its inability to trigger photosynthesis (34), could still effectively enhance RPS6 phosphorylation and translation (Figs. 1A and 2C).
Fig. 3.
Light-activated RPS6 phosphorylation depends on auxin but not photosynthesis or exogenous glucose. Four-day-old seedlings were grown with or without 20 μM DCMU (A), 15 mM glucose (Glc) (B), or 100 μM yucasin (C). Total protein from each treatment was extracted and subjected to SDS/PAGE and Phostag-PAGE analyses.
A recent report indicated that light could stimulate auxin biosynthesis to activate TOR in leaf primordia for regulating true leaf development in Arabidopsis (35). Therefore, we tested whether auxin is also involved in regulating light-induced phosphorylation of RPS6 in the deetiolating stage. The light-induced RPS6 phosphorylation was abolished on pretreatment with yucasin, a chemical inhibitor of YUCCAs, to block auxin biosynthesis (Fig. 3C). Together, these results indicate that light signals, instead of sugars derived from photosynthetic activities, activate RPS6 phosphorylation in an auxin-dependent manner.
COP1 Represses the Auxin Pathway and RPS6 Phosphorylation in Darkness.
Results in Fig. 1B indicated that COP1 plays a negative role in repression of translation in dark-grown seedlings. We then examined whether repression of translation by COP1 was associated with phosphorylation of RPS6. The level of RPS6 phosphorylation in etiolated cop1 seedlings was comparable to that in Col-0 seedlings treated with 4 h of light (Fig. 4A). Four hours of light treatment did not further increase the phosphorylation of RPS6 in cop1 (Fig. 4A), which was similar to the comparable PL% observed for both the dark- and light-grown cop1 mutant (Fig. 1B).
Fig. 4.
COP1 represses RPS6 phosphorylation and auxin pathway. (A) Two-day-old etiolated (dark, D) seedlings of Col-0 and cop1 were treated with L4h. Total protein from each treatment was extracted and subjected to SDS/PAGE and Phostag-PAGE analyses for RPS6 and RPS6 phosphorylation. (B) Two-day-old etiolated seedlings of Col-0 and cop1 were treated with (+) or without (−) yucasin. Representative photographs of seedlings are shown. (Scale bar: 2.5 mm.) (C) Total proteins isolated from 4-d-old etiolated seedlings treated with 0.1% DMSO, 100 nM naphthalene acetic acid (NAA), or 100 nM NAA plus 15 mM glucose (NAA+Glc) underwent SDS/PAGE and Phostag-PAGE analyses.
Yucasin treatment compromised the constitutively open cotyledon observed in the dark-grown cop1 mutant (Fig. 4B) and also delayed the light-induced cotyledon opening in wild-type Col-0 (SI Appendix, Fig. S6). These results indicate that auxin is required for the cotyledon opening phenotype seen in both the dark-grown cop1 mutant and light-grown Col-0. This finding prompted us to examine whether the treatment with auxin or auxin combined with glucose could bypass the inhibitory role of COP1 on RPS6 phosphorylation in dark-grown wild-type seedlings. Neither auxin alone nor auxin plus glucose was sufficient to trigger RPS6 phosphorylation in etiolated seedlings (Fig. 4C). Thus, COP1 may function to repress factors downstream of auxin biogenesis. However, our study cannot exclude the possibility that COP1 also represses additional factors to regulate cotyledon opening during etiolation.
Taken together, in dark-grown wild-type seedlings, COP1 functions to repress the auxin pathway to keep translation in check until light derepresses the actions of COP1. Also, although auxin could substitute the light treatment in activating cell proliferation activity in the shoot apex (35), auxin could only trigger RPS6 phosphorylation in the presence of light signals in deetiolating young seedlings.
Light Activates TOR-Dependent RPS6 Phosphorylation for Enhanced Translation and Cotyledon Opening.
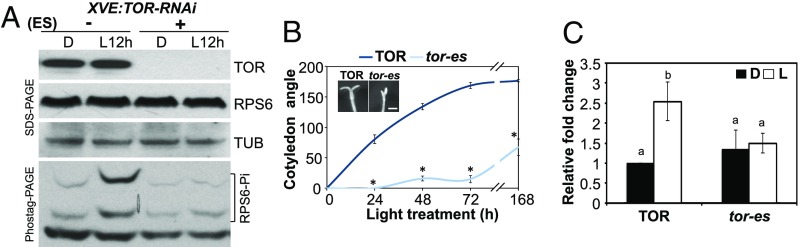
RPS6 phosphorylation mainly depends on the TOR-S6K pathway (21, 33), a pathway inferred to function in translation initiation and the elongation stage in mammals (16). We next examined whether the TOR-S6K pathway is responsible for the light-induced RPS6 phosphorylation by monitoring the phosphorylation of RPS6 in seedlings with reduced expression of TOR. TOR proteins were confirmed to be undetectable in an inducible TOR-RNAi line (XVE:TOR-RNAi) (17) upon the addition of 20 μM estradiol in both etiolated and deetiolating seedlings (Fig. 5A, Upper). Without TOR protein, light treatment could not induce RPS6 phosphorylation (Fig. 5A). These results indicate that light-induced RPS6 phosphorylation is TOR dependent.
Fig. 5.
Light activates TOR-dependent RPS6 phosphorylation to enhance translation and cotyledon opening. (A) Four-day-old XVE:TOR-RNAi seedlings (dark, D) were grown with (+) or without (−) 20 μM estradiol in L12h. Total protein from each treatment was extracted and subjected to SDS/PAGE and Phostag-PAGE analyses. (B) Four-day-old etiolated seedlings of the inducible XVE:TOR-RNAi line grown with (tor-es) or without (TOR) estradiol were treated with 15 μE white light to observe cotyledon opening for 7 d. Cotyledon opening angles are shown as mean ± SE *P < 0.001 vs. wild type (TOR) (n = 13–20). Representative image showed cotyledons from 7-d-old seedlings. (Scale bar: 1 mm.) (C) De novo protein synthesis was determined for TOR or tor seedlings. Four-day-old etiolated seedlings were collected in the dark (D) or treated with 11-h white light (L). Bars with the different letters represent significant difference (Tukey’s HSD P < 0.05).
Whether the activation of the TOR-RPS6 pathway affects the deetiolating process remains to be evaluated. The defective cotyledon opening phenotype observed in rps6a and rps6b mutants (Fig. 2B) prompted us to examine this phenotype in the wild type and TOR-RNAi line. Estradiol-treated TOR-RNAi seedlings (tor-es) showed significantly delayed cotyledon opening even after 7 d of white light treatment (Fig. 5B). The role of TOR in light-regulated translational enhancement was also evaluated by examining the light-induced de novo protein synthesis in the wild type and XVE:TOR-RNAi line. The de novo protein synthesis was clearly enhanced by light in wild-type seedlings, but such increase was compromised in XVE:TOR-RNAi seedlings in the presence of estradiol (tor-es) (Fig. 5C and SI Appendix, Fig. S7). These results together demonstrated the essential role of TOR in light-enhanced RPS6 phosphorylation and de novo protein synthesis.
COP1 Represses TOR Activity During Skotomorphogenic Development.
We next addressed whether COP1 has negative roles in translation by regulating TOR protein level or activity in dark-grown seedlings. TOR protein abundance was comparable in etiolated and deetiolating Col-0, cop1, and phyA mutants (Fig. 6A and SI Appendix, Fig. S8). However, compared with etiolated Col-0 seedlings, the dark-grown cop1 mutant showed increased S6K phosphorylation level (Fig. 6B). In addition, enhanced RPS6 phosphorylation in the dark-grown cop1 mutant could be repressed by torin2, a specific inhibitor for the TOR ATP-binding site (Fig. 6C). These data demonstrate that COP1 represses TOR activity rather than TOR protein level in etiolated seedlings.
Fig. 6.
A functional TOR is required for photomorphogenic development of dark-grown cop1 mutant. (A) Two-day-old etiolated seedlings of Col-0 and cop1 were treated with white light for 0 h (dark, D), 4 h (L4h), or 12 h (L12h). (B) Two-day-old etiolated seedlings of Col-0 and cop1 mutant were treated with L4h. TOR activity was measured by detection of S6K phosphorylation with anti–phospho-p70 S6 kinase (Thr389) antiserum. Levels of TOR (A) and S6K-Pi (B) were normalized to tubulin in etiolated Col-0 from three biological replicates and presented as mean ± SE (C) RPS6 phosphorylation patterns in 2-d-old etiolated seedlings of cop1 treated with or without 10 μM Torin2. (D) cop1, XVE:TOR-RNAi and cop1 XVE:TOR-RNAi were grown with DMSO (mock, ES−) or treated with estradiol (ES+) for 2 d in the dark. Photographs of representative seedlings are shown. (Scale bar: 2.5 mm.) (E) Cotyledon opening angle and (F) hypocotyl length of 2-d-old dark-grown seedlings shown in D were measured. Values are mean percentage ± SE. Bars with different letters represent data groups with significant difference (Tukey’s HSD P < 0.05, n = 28–56). Not detected (ND) samples remained closed cotyledons. (G) RPS6 phosphorylation patterns in 2-d-old etiolated seedlings of cop1 XVE:TOR-RNAi in the absence (−) or presence (+) of estradiol (ES). (H) An action model illustrated based on this study.
To further establish the regulatory relation between COP1 and TOR, we generated a cop1 XVE:TOR-RNAi line by crossing the cop1 mutant with the TOR-RNAi line. In the absence of estradiol (ES−), cop1 showed a constitutive photomorphogenic phenotype with an open apical hook and cotyledons as previously reported (36), whereas XVE:TOR-RNAi had typical skotomorphogenic phenotypes with an apical hook and closed cotyledons like wild-type seedlings (Fig. 6D, ES−). When estradiol was added in the growth media (ES+), cop1 exhibited similar phenotypes as with ES−, but XVE:TOR-RNAi showed stunted growth with shorter hypocotyl length (Fig. 6D, ES+). However, cop1 XVE:TOR-RNAi showed a cop1 phenotype under ES− but a tor-like phenotype under ES+ (Fig. 6D). Quantitative data for both cotyledon opening and hypocotyl length are shown in Fig. 6 E and F. Compared with cop1, the slightly compromised cotyledon opening angles in cop1 XVE:TOR-RNAi in the absence of estradiol (ES−) (Fig. 6E) likely reflected the leakiness of the XVE system used, as described previously (37). Interestingly, in contrast to the opposite roles of COP1 and TOR in cotyledon opening, both COP1 and TOR are required for full hypocotyl elongation in etiolated seedlings (Fig. 6F).
The above genetic study indicated that, for cotyledon opening, TOR is epistatic to COP1 (i.e., COP1 functions upstream of TOR). Consistent with this notion, the phosphorylation of RPS6 in dark-grown cop1 was compromised when TOR was silenced in the cop1 XVE:TOR-RNAi line (Fig. 6G). These results indicate that both the phosphorylation of RPS6 and the constitutive cotyledon opening of cop1 rely on the presence of TOR.
Discussion
In addition to transcriptomic reprogramming, light also enhances the translation of thousands of transcripts (13, 14). This study uncovered a mechanistic perspective underlying light-enhanced translation. Our data support a signaling cascade starting from the well-established light perception by photoreceptors for the repression of COP1 activity. One key finding in this study was the inhibitory role of COP1 in the TOR-S6K-RPS6 phosphorelay that depends on the auxin pathway, for control of de novo protein synthesis and cotyledon opening in deetiolating seedlings (Fig. 6H). Many studies have concluded that COP1 negatively regulates photoreceptors and multiple transcription factors by targeting them for degradation in the dark to promote skotomorphogenesis (12). Along with results from this study, COP1 is a master switch repressing transcription and translation in darkness to ensure that photomorphogenic development remains dormant until young seedlings perceive light signals. Phytochrome interacting factors (PIFs) function to repress the expression of light-inducible genes in dark-grown seedlings (38). Consistent with their negative roles in photomorphogenesis, the pifq/pif1345 mutant exhibited a cop1-like phenotype (39). PIF1 could also enhance the substrate recruitment and E3 activity of COP1 (40). Whether the reduced COP1 activity in pifq leads to partial derepression of the light-triggered translation in pifq as in cop1 remains to be studied.
We previously reported that light enhances massive translation (13, 14) and photoreceptors are involved in transmitting the light signals (this study). Of note, the light-activated phytochrome B has been shown to negatively regulate the translation of protochlorophyllide reductase (PORA) mRNA in cytosol (41). Additional mRNAs whose translation is negatively affected by light await to be identified.
The translational enhancement by light was independent of key transcriptional regulators HY5 and HYH in early photomorphogenic development (Fig. 1C and SI Appendix, Fig. S1C). Thus, target mRNAs of light-enhanced translation differ from those induced by HY5/HYH at early stages of photomorphogenic development. Indeed, we previously found that light enhances the translation of thousands of transcripts preexisting in dark-grown seedlings (13). This observation supports the idea that light targets largely nonoverlapping genes for transcriptional or translational control.
During photomorphogenesis, light has distinct morphological affects on hypocotyls and cotyledons. The light-induced inhibition of hypocotyl elongation could be achieved by repressing auxin signaling, a process dependent on photoreceptors, CRY1 and phyB, and the transcriptional regulator HY5 (42, 43). However, our current study indicated a positive role of auxin pathway in light-induced cotyledon opening (SI Appendix, Fig. S6A). Our data agree with light-triggered auxin signaling in shoot and root apexes (35, 44). The different roles of auxin signaling between hypocotyls and cotyledons might reflect the light-triggered reduction of auxin level in hypocotyls but increased auxin level in cotyledons (45).
TOR acts as a central coordinator in many organisms to respond to stresses or varying availability of nutrients and energy. In plants, TOR integrates light, photosynthesis, metabolites (glucose/sucrose), or phytohormone (auxin) signaling pathways in the seedling stage (21, 35, 44, 46). In this study, phosphorylation of RPS6 depended on light and auxin pathway but was independent of photosynthetic activities, which differed from a previous study finding that TOR-dependent cell proliferation in shoot apex depended on light, auxin, and photosynthesis (35). However, light and sugar worked additively to activate TOR for the increased expression of WUSHEL (WUS) for shoot apical meristem activation (46). The above studies were conducted in different tissues and developmental stages. The molecular and physiological readouts of TOR signaling also varied. The different requirements of light, metabolites, and auxin for TOR activation in these studies suggest complex and sophisticated tissue- and/or stage-dependent modes of TOR activation poised for distinct physiological responses.
Our study indicated that during the first few hours of young seedlings transitioning from the dark to light environment, TOR could be quickly activated by light signals to trigger RPS6 phosphorylation for timely enhancement of translation to establish the photosynthetic apparatus and the opening of the apical hook and cotyledons. This immediate photomorphogenic development may next be followed by the coordinated activation of TOR by light, auxin, and sugar in the shoot apical meristem and leaf primordia to initiate the vegetative developmental program (35).
The degree of RPS6 phosphorylation is positively associated with the time of white light treatment, as resolved by Phostag-PAGE (Fig. 2A). Candidate phosphorylation sites of RPS6 proteins include multiple serines at C-terminal regions. The RPS6 C terminus appears to be evolutionally conserved in multiple eukaryotes, including yeast, plants, invertebrates, and vertebrates (47–49). Whether light-enhanced translation is fine tuned by RPS6s carrying different phosphorylation codes awaits future characterization.
Materials and Methods
Detailed description of plant materials, plant growth conditions, and methods for the isolation of total, nonpolysomal, and polysomal RNAs, protein analyses, cotyledon opening kinetic assay, and the analyses of de novo protein synthesis can be found in SI Appendix, SI Material and Method.
Supplementary Material
Acknowledgments
This research was supported by research grants from the Ministry of Science and Technology, Taiwan (MOST103-2321-B-001-060-, MOST104-2321-B-001-027-, MOST105-2321-B-001-017-, MOST106-2321-B-001-011-, and MOST107-2321-B-001-007-) and an Academia Sinica Investigator Award (to S.-H.W.). G.-H.C. received a postdoctoral fellowship from the Ministry of Science and Technology, Taiwan. J.S. was supported by National Institutes of Health Grant R01GM60493.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809526115/-/DCSupplemental.
References
- 1.Wu SH. Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu Rev Plant Biol. 2014;65:311–333. doi: 10.1146/annurev-arplant-050213-040337. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 3.Mathews S. Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell. 2010;22:4–16. doi: 10.1105/tpc.109.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mccormac AC, et al. Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome B-encoding cDNAs: Evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. Plant J. 1993;4:19–27. [Google Scholar]
- 5.Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chory J. Light signal transduction: An infinite spectrum of possibilities. Plant J. 2010;61:982–991. doi: 10.1111/j.1365-313X.2009.04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalitin D, et al. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 11.Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370–2383. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Liu MJ, Wu SH, Chen HM, Wu SH. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol. 2012;8:566. doi: 10.1038/msb.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu MJ, et al. Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 2013;25:3699–3710. doi: 10.1105/tpc.113.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juntawong P, Bailey-Serres J. Dynamic light regulation of translation status in Arabidopsis thaliana. Front Plant Sci. 2012;3:66. doi: 10.3389/fpls.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonseca BD, et al. The ever-evolving role of mTOR in translation. Semin Cell Dev Biol. 2014;36:102–112. doi: 10.1016/j.semcdb.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012;287:2836–2842. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong F, et al. Brassinosteriod insensitive 2 (BIN2) acts as a downstream effector of the target of rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 2017;213:233–249. doi: 10.1111/nph.14118. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Y, Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014;164:499–512. doi: 10.1104/pp.113.229948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobrenel T, et al. TOR signaling and nutrient sensing. Annu Rev Plant Biol. 2016;67:261–285. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Y, et al. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyuhas O. Ribosomal protein S6 phosphorylation: Four decades of research. In: Kwang WJ, editor. International Review of Cell and Molecular Biology. Vol 320. Academic; New York: 2015. pp. 41–73. [DOI] [PubMed] [Google Scholar]
- 23.Meyuhas O. Physiological roles of ribosomal protein S6: One of its kind. In: Kwang WJ, editor. International Review of Cell and Molecular Biology. Vol 268. Academic; New York: 2008. pp. 1–37. [DOI] [PubMed] [Google Scholar]
- 24.Deprost D, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren M, et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell. 2012;24:4850–4874. doi: 10.1105/tpc.112.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turkina MV, Klang Årstrand H, Vener AV. Differential phosphorylation of ribosomal proteins in Arabidopsis thaliana plants during day and night. PLoS One. 2011;6:e29307. doi: 10.1371/journal.pone.0029307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boex-Fontvieille E, et al. Photosynthetic control of Arabidopsis leaf cytoplasmic translation initiation by protein phosphorylation. PLoS One. 2013;8:e70692. doi: 10.1371/journal.pone.0070692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enganti R, et al. Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Front Plant Sci. 2018;8:2210. doi: 10.3389/fpls.2017.02210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson JA, Shanware NP, Chang H, Tibbetts RS. Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J Biol Chem. 2011;286:8688–8696. doi: 10.1074/jbc.M110.141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creff A, Sormani R, Desnos T. The two Arabidopsis RPS6 genes, encoding for cytoplasmic ribosomal proteins S6, are functionally equivalent. Plant Mol Biol. 2010;73:533–546. doi: 10.1007/s11103-010-9639-y. [DOI] [PubMed] [Google Scholar]
- 32.Boccalandro HE, Rossi MC, Saijo Y, Deng X-W, Casal JJ. Promotion of photomorphogenesis by COP1. Plant Mol Biol. 2004;56:905–915. doi: 10.1007/s11103-004-5919-8. [DOI] [PubMed] [Google Scholar]
- 33.Pende M, et al. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCree KJ. The action spectrum, absorptance and quantum yield of photosynthesis in crop plants. Agric Meteorol. 1971;9:191–216. [Google Scholar]
- 35.Li X, et al. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA. 2017;114:2765–2770. doi: 10.1073/pnas.1618782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNellis TW, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Degenhardt RF, Bonham-Smith PC. Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol. 2008;147:128–142. doi: 10.1104/pp.107.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, et al. PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell. 2014;26:1992–2006. doi: 10.1105/tpc.114.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paik I, Yang S, Choi G. Phytochrome regulates translation of mRNA in the cytosol. Proc Natl Acad Sci USA. 2012;109:1335–1340. doi: 10.1073/pnas.1109683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu F, et al. Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant. 2018;11:523–541. doi: 10.1016/j.molp.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- 44.Cai W, et al. COP1 integrates light signals to ROP2 for cell cycle activation. Plant Signal Behav. 2017;12:e1363946. doi: 10.1080/15592324.2017.1363946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun N, et al. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA. 2016;113:6071–6076. doi: 10.1073/pnas.1604782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeiffer A, et al. Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife. 2016;5:e17023. doi: 10.7554/eLife.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wettenhall RE, Erikson E, Maller JL. Ordered multisite phosphorylation of Xenopus ribosomal protein S6 by S6 kinase II. J Biol Chem. 1992;267:9021–9027. [PubMed] [Google Scholar]
- 48.Bandi HR, Ferrari S, Krieg J, Meyer HE, Thomas G. Identification of 40 S ribosomal protein S6 phosphorylation sites in Swiss mouse 3T3 fibroblasts stimulated with serum. J Biol Chem. 1993;268:4530–4533. [PubMed] [Google Scholar]
- 49.Radimerski T, et al. Identification of insulin-induced sites of ribosomal protein S6 phosphorylation in Drosophila melanogaster. Biochemistry. 2000;39:5766–5774. doi: 10.1021/bi9927484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.