Significance
The spread of Plasmodium falciparum with reduced susceptibility to artemisinin (ART) in Southeast Asia threatens global malaria control. Most failures of ART treatment are attributed to mutations in the pfkelch13 locus acting through an unclear mechanism. The role of pfkelch13 in reducing ART susceptibility was first identified in an in vitro selection experiment. We carried out a similar in vitro selection and discovered mutations in a different gene that reduce susceptibility to ART. The gene encodes PfCoronin, a conserved protein that in other organisms binds with actin to enhance cytoskeletal plasticity or is involved in vesicular transport. PfCoronin and PfKelch13 share structural similarities, and this finding may yield insights into the molecular mechanisms of ART resistance.
Keywords: malaria, artemisinin resistance, evolution, coronin, actin
Abstract
Drug resistance is an obstacle to global malaria control, as evidenced by the recent emergence and rapid spread of delayed artemisinin (ART) clearance by mutant forms of the PfKelch13 protein in Southeast Asia. Identifying genetic determinants of ART resistance in African-derived parasites is important for surveillance and for understanding the mechanism of resistance. In this study, we carried out long-term in vitro selection of two recently isolated West African parasites (from Pikine and Thiès, Senegal) with increasing concentrations of dihydroartemisinin (DHA), the biologically active form of ART, over a 4-y period. We isolated two parasite clones, one from each original isolate, that exhibited enhanced survival to DHA in the ring-stage survival assay. Whole-genome sequence analysis identified 10 mutations in seven different genes. We chose to focus on the gene encoding PfCoronin, a member of the WD40-propeller domain protein family, because mutations in this gene occurred in both independent selections, and the protein shares the β-propeller motif with PfKelch13 protein. For functional validation, when pfcoronin mutations were introduced into the parental parasites by CRISPR/Cas9-mediated gene editing, these mutations were sufficient to reduce ART susceptibility in the parental lines. The discovery of a second gene for ART resistance may yield insights into the molecular mechanisms of resistance. It also suggests that pfcoronin mutants could emerge as a nonkelch13 type of resistance to ART in natural settings.
The World Health Organization (WHO) currently recommends five different artemisinin (ART)-based combination therapies (ACTs) as the treatment for uncomplicated Plasmodium falciparum malaria worldwide (1). Typically, a fast-acting ART-based drug with short half-life is paired with a partner drug that has a longer plasma half-life to reduce the likelihood of resistance that might emerge from monotherapies. The ACTs were adopted worldwide more than a decade ago and have been widely successful in the efforts for malaria control and elimination. However, emerging ACT treatment failure in Southeast Asia is a major threat to malaria control (2–5). Currently, ART resistance is contained within the greater Mekong subregion centered on Cambodia, where slow-clearing ART-treated P. falciparum infections were first identified in 2007 (6). However, chloroquine and pyrimethamine resistance spread to the rest of the world from the same region over the last century. Therefore, given how few new antimalarial drugs are available and how drugs under development take nearly a decade to deploy, it is imperative that we safeguard the efficacy of ACT use worldwide for as long as possible.
The evolution and global spread of ART resistance would result in a major public health disaster, especially in Africa, where ACTs are heavily used in management of the nearly 90% of the cases of P. falciparum malaria occurring in children age <5 y worldwide. Despite the success of ACTs over the past decade, one-half of the world’s population (3.3 billion people) remains at risk for malaria infection. The magnitude of the health and economic threat posed by ART treatment failure has prompted an urgent call to action to develop strategies to circumvent the emergence and spread of ART resistance.
Effective malaria control in public health is aided by the identification of molecular markers of ART resistance for surveillance and an understanding of the underlying biological mechanism of action and resistance. Ariey et al. (7) were the first to report a mutation (M476I) in the pfkelch13 propeller domain that was sufficient to confer reduced ART susceptibility. The mutation was identified in parasites (F32 from Tanzania) selected in vitro by stepwise increases in ART exposure over a 5-y period (7). Ariey et al. then analyzed the pfkelch13 sequence of parasites derived from clinical cases with delayed clearance and in which the phenotype in an in vitro ring-stage survival assay (RSA) was scored as ART resistant. None of these field-isolated parasites carried the M476I mutation discovered by in vitro selection; however, the field-isolated slow-clearing parasites harbored mutations at other sites in the PfKelch13-propeller domain (Y493H, R539T, I543T, and C580Y) that correlated with the ART-resistant phenotype in vitro and in vivo (7–9). The RSA has subsequently become the standard in vitro assay for reduced ART susceptibility.
Since the discovery of mutations in the pfkelch13-propeller domain associated with drug resistance, increased genetic surveillance has identified many additional nonsynonymous SNPs in the pfkelch13-propeller domain in parasites isolated from a wide geographic range (10–15). The role of these pfkelch13 polymorphisms in drug resistance remains to be determined.
Functional validation has confirmed the role of certain SNPs in the pfkelch13-propeller domain in conferring elevated RSA survival. Straimer et al. (8) genetically engineered the four pfkelch13 mutations associated with slow parasite clearance following ART treatment in vivo, as well as the mutation conferring the M476I change identified in vitro into newly culture-adapted Cambodian isolates and other laboratory reference lines of P. falciparum. These authors showed that introduction of the pfkelch13 mutations M476I, Y493H, R539T, I543T, and C580Y was associated with increased RSA survival rate, with I543T providing the greatest RSA survival benefit (40–49%) (8). Significant reductions in the RSA survival rate were observed on reversion of the mutations R539T, I543T, and C580Y back to the wild-type allele. Importantly, the degree of resistance was found to depend not only on the pfkelch13 mutation, but also on the genetic background of the parasite (8). These findings illustrate the power of combining in vitro selection with investigation of extant population diversity to identify and functionally validate key proteins.
Recent work has demonstrated that the resistant RSA survival phenotype can occur in parasites exhibiting a wild-type pfkelch13 sequence, indicating that there may be more than a single pathway to ART resistance (16, 17). In vitro ART selection experiments with P. falciparum and Plasmodium chaubadi have identified several putative targets that do not include pfkelch13 (18–20). This is not surprising given the currently limited understanding of ART’s mechanism of action, with more than 124 protein targets involved in multiple pathways (21, 22), further complicating the biological unraveling of ART resistance.
Our current understanding of ART resistance is based almost exclusively on observations in Southeast Asia. However, the parasite population, the history of ART use, and the selection pressures on the parasite population in Southeast Asia are very different from the situation in Africa. Polymorphisms in the propeller domain of pfkelch13 associated with ART treatment failure have not yet been observed in Africa, although a common polymorphism (K189T) outside the propeller domain of pfkelch13 is present in parasites, especially in West Africa (www.malariagen.net/projects/p-falciparum-community-project).
As history shows, ART pressure in Africa is likely to result in the emergence of resistant parasites, and thus it is important to investigate mechanisms of ART resistance that can arise in the genomic context of African parasites. Toward this objective, we began selecting for ART resistance among culture-adapted parasite isolates from Senegal (23). The objective was to select drug-resistant mutants that could then be interrogated to identify a mechanism of resistance. We began these selection studies before publication of the study by Ariey et al. (7). Our in vitro selection yielded two parasite lines that lack pfkelch13 mutations yet confer reduced ART susceptibility as measured by RSA.
Results
Selection of ART Resistance and Phenotype Characterization.
We hypothesized that we could identify genetic variants in Senegal lines selected under ART drug pressure to find candidate genes that may contribute to ART resistance. We used intermittent in vitro selection with stepwise increasing drug concentrations to derive parasites with reduced ART susceptibility (Fig. 1A), and successfully generated two such lines. As parental lines for the selection, we chose two different culture-adapted Senegal field isolates, one from Pikine (SenP019.04) and the other from Thiès (SenTh032.09). We started with independent selections of both the parental lines, each with ∼109 parasites per experimental group (Fig. 1A). We pulsed the cultures with dihydroartemisinin (DHA), the active metabolite of ART, until parasites were undetectable by microscopy. The treated cultures were then allowed to recover in the absence of drug pressure until ∼1% parasitemia was observed before starting the next drug pulse. Recovery in the absence of drug took between 7 and 60 d (SI Appendix, Fig. S1).
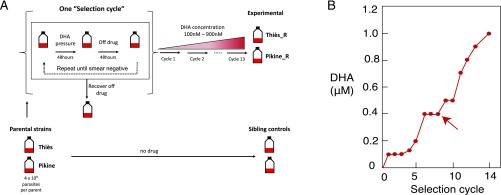
Fig. 1.
(A) Outline of in vitro selection protocol. Parasite lines were pulsed intermittently with (DHA), cycling on and off the drug until the culture was smear-negative, which took 4–12 d. Parasites were allowed to recover to 1% parasitemia in the absence of DHA, a process that took 7–60 d. The complete process of treatment to recovery is considered one selection cycle. Following each selection cycle, cultures were pulsed again with an increased concentration of DHA. (B) Drug concentrations applied to two populations originating from two field isolates from Senegal over 13 drug selection cycles. The arrow indicates the first appearance of Pfcoronin mutations in both selected lines.
The complete process of drug treatment to smear-negative culture, followed by recovery of the culture to 1% parasitemia, was considered one “selection cycle.” After 13 selection cycles over a 4-y period, we isolated two laboratory-selected lines with reduced ART susceptibility (Fig. 1B). These lines, designated “Pikine_R” and “Thiès_R,” were derived from the Pikine parent and the Thiès parent, respectively. An accompanying no-drug sibling control for each parental line was cultured in parallel but in the absence of drug pressure for approximately the same number of generations (Fig. 1A).
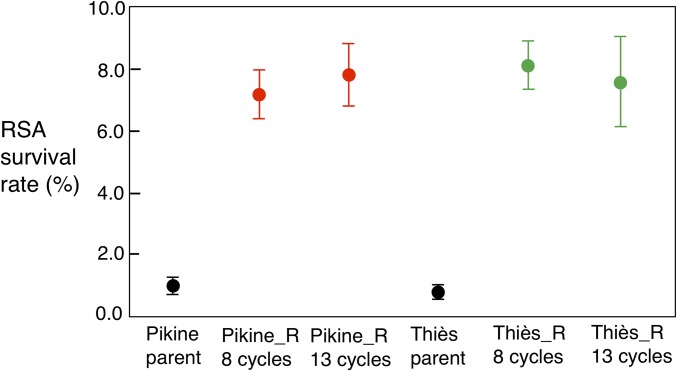
Increased ART resistance was assessed by a variety of phenotypic assays, including RSA and half-maximal effective concentration (EC50) assays, alongside the assessment of copy-number variation (CNV) in the gene pfmdr1, previously associated with antimalarial drug resistance (24–27). Reduced ART susceptibility in vitro is defined as increased survival by RSA following a pulse of DHA at a clinically relevant concentration (700 nM), with a survival percentage >1% considered resistant (7, 9). In our assays, Cambodian isolates with pfkelch13 mutations and delayed clearance exhibited the following RSA survival (%) values (mean ± SEM): C580Y, 18.6 ± 2.49; I543T, 7.51 ± 0.71; R539T, 22.8 ± 4.83; Y493H, 4.57 ± 1.19 (16). These values are in accordance with those reported by other laboratories (7, 8). Fig. 2 shows the RSA phenotypes of the parental lines and those of the selected lines after 8 and 13 cycles of selection under drug pressure. After 13 selection cycles, Pikine_R, and Thiès_R show mean RSA survival percentages of 7.8 ± 1.0% and 7.6 ± 1.5%, respectively, and the parental lines are sensitive (RSA <1%) (Fig. 2 and SI Appendix, Table S1). The RSA values of the selected lines are comparable to those observed for delayed-clearance Cambodian clinical isolates with mutations in pfkelch13 (16) and also to the RSA values of the in vitro selected line described by Ariey et al. (7). Increased RSA values were also observed in bulk cultures of Pikine_R and Thiès_R after eight selection cycles under drug pressure (Fig. 2). In contrast, no changes in EC50 for ART or its derivatives (SI Appendix, Fig. S2) and no changes in pfmdr1 CNV (SI Appendix, Fig. S3) were detected in the selected lines. Importantly, targeted sequencing of the pfkelch13 locus using PCR revealed no genetic changes in this locus compared with the parental strains.
Fig. 2.
RSA survival rates for Senegalese parental lines and their parasite populations that underwent drug selection for 8 or 13 cycles. Survival >1% is considered elevated. Parasites after 8 or 13 drug selection cycles had significantly higher RSA survival rates compared with their parents (P < 0.001). Results from two to three biological replicates with two technical replicates each are included.
Candidate Genes Associated with ART Resistance Identified by Genome Sequencing.
We performed whole-genome sequencing to identify single nucleotide polymorphisms (SNPs) in our ART-resistant selected lines and identified multiple nonsynonymous SNPs that differed between the selected drug-resistant lines relative to the original parental lines and no-drug sibling controls that had been cultured for a similar number of generations in the absence of drug. We eliminated SNPs in the highly variable subtelomeric regions, known to be subject to read mapping error. This narrowed the list of candidate genes to 10 SNPs in seven genes for further follow-up (Table 1). All candidate SNPs were confirmed by PCR amplification and Sanger sequencing.
Table 1.
Candidate genes identified by whole-genome sequencing in the two DHA selected lines
 |
Mutations indicated in red.
Mutations in some candidate genes were unique to a single selected line, while others were shared between the two lines (Table 1). Mutations in PF3D7_1251200 encoding the protein PfCoronin were found in both selected lines. We performed additional whole-genome sequencing of earlier selection steps and found that pfcoronin mutations first appeared after eight selection cycles in both the Pikine_R and Thiès_R lines, with the two mutations R100K and E107V in Pikine_R appearing concurrently (Fig. 1B). Mutations in PF3D7_1433800 were the first to appear, after the sixth and seventh rounds of selection in Thiès_R and Pikine_R, respectively (SI Appendix, Fig. S4).
Confirmation of the Role of pfcoronin Mutations in Conferring Reduced ART Susceptibility.
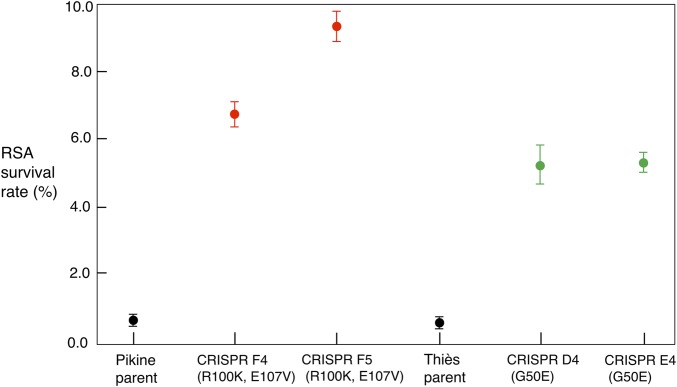
To functionally test the role of these mutations in ART susceptibility, we introduced the pfcoronin mutations into the parental parasite lines using CRISPR/Cas9 genome editing (28, 29) and carried out RSA phenotyping analyses. Successful knockins of pfcoronin mutations identified in Pikine_R (R100K and E107V) or Thiès_R (G50E) parasites into their respective parental backgrounds were verified by Sanger sequencing (SI Appendix, Figs. S5B and S6B). Two independent clones for each background were obtained by limiting dilution of the bulk transfectant cultures and used in further analyses. Pfcoronin mutations were sufficient to confer reduced ART susceptibility based on RSA in CRISPR mutants from both Thiès and Pikine backgrounds (Fig. 3), while the EC50 response to ART and its derivatives remained unchanged (SI Appendix, Fig. S7). CRISPR mutants with PfCoronin R100K and E107V in the Pikine background (clones F4 and F5) had RSA survival of 6.8 ± 0.9% and 9.4 ± 1.1%, respectively, while the mutants with PfCoronin G50E in the Thiès background (clones D4 and E4) showed RSA survivals of 5.2 ± 1.4% and 5.3 ± 0.7%, respectively, compared with their parents (RSA <1%) (SI Appendix, Table S1). These findings were robust and reproducible.
Fig. 3.
RSA survival rates for parental Pikine and Thiès lines and their two CRISPR/Cas9 edited clones with pfcoronin mutation(s). Survival >1% is considered elevated. Pfcoronin mutations identified in Pikine_R (R100K and E107V) and Thiès_R (G50E) were knocked into their respective parents. RSA survival rates were significantly higher for both independent clones in each background, Pikine (CRISPR F4 and CRISPR F5) (P < 0.001) and Thiès (CRISPR D4 and CRISPR E4) (P < 0.01) compared with their respective parental lines. Results from three biological replicates with two technical replicates each are included.
Discussion
After approximately 4 y of intermittent selection in vitro with stepwise increasing concentrations of DHA, we recovered two parasite lines with reduced susceptibility to ART from two recently culture-adapted parental lines from Senegal. Resistant parasites were recovered after approximately 13 selection cycles, with each exposure to drug followed by 7–60 d of recovery (SI Appendix, Fig. S1). The resistant parasites show no change in sensitivity to ART by EC50 measurement and also no change in copy number of pfmdr1, a well-characterized gene known to be amplified in other instances of antimalarial drug resistance (SI Appendix, Figs. S2 and S3) (24, 30).
Notably, no mutations were observed in the propeller domain of the pfkelch13 locus in any of the selected lines. The parental and selected lines of both backgrounds did contain a common West African polymorphism in pfkelch13 K189T, which is outside the propeller domain and does not confer ART resistance. We also found no evidence of SNPs in loci previously associated with ART resistance in Cambodian parasites, for example, mutations in apicoplast ribosomal protein S10 (arps10), multidrug resistance protein 2 (mdr2), ferredoxin (fd), and the chloroquine resistance transporter (pfcrt) (31). The selected ART-resistant lines from Senegal demonstrate that ART resistance not associated with pfkelch13 can evolve in vitro.
Whole-genome sequencing identified multiple candidate SNPs, none of which has been reported previously (Table 1). The prime candidate was pfcoronin, which appeared in both independent resistant lines. All three SNPs identified in PfCoronin (G50E, R100K, and E107V) were in positions that are conserved in Plasmodium gaboni and another apicomplexan parasite, Toxoplasma gondii (SI Appendix, Fig. S8). G50 and R100 residues were also conserved in other Plasmodium species (SI Appendix, Fig. S8). The Senegalese parental lines also had a common pfcoronin African polymorphism, S183G, in the WD40 domain (www.malariagen.net/projects/p-falciparum-community-project), which by itself was not sufficient for elevated RSA survival. Some of the other candidate genes had more than one background mutation in the Senegalese parental lines.
Successful CRISPR/Cas9 editing to knockin pfcoronin mutations in both Senegalese parental lines resulted in an RSA-resistant phenotype (Fig. 3), confirming that pfcoronin mutations are sufficient for increased RSA survival. Although RSA phenotype levels are similar in Pikine_R and Pikine pfcoronin CRISPR mutants, in the Thiès background, RSA phenotype is slightly lower in the pfcoronin CRISPR mutants compared with the Thiès_R parasites. Thus, other mutations could be contributing to the reduced ART susceptibility in the Thiès background, which had only one pfcoronin mutation in the selected Thiès_R parasites.
SNPs in other candidate genes (Table 1) could also be contributing to parasite fitness, and their roles in facilitating or modulating ART resistance warrant further investigation. Of particular interest is PF3D7_1433800, a conserved protein of unknown function with no annotated domains on PlasmoDB (32) (Table 1). Mutation in this locus was one of the earliest to appear in both independently selected lines, suggesting a role in the acquisition of resistance or fitness observed in vitro.
PfCoronin has a seven-bladed propeller domain composed of WD40 repeats and β-propeller folds in its N terminus, which is structurally similar to the six-bladed propeller domain found in the C terminus of PfKelch13 (4, 16). Although Kelch and Coronin proteins have distinct functions in eukaryotes, some Kelch domain-containing proteins are also actin-binding proteins, a function typical of Coronin proteins (33). The genomes of unicellular pathogens, including P. falciparum, encode only one single-copy gene for coronin. The protein that it encodes generally binds F-actin and has functions in proliferation, locomotion, and phagocytosis in eukaryotes (34). In the apicomplexan parasite T. gondii, Coronin is involved in F-actin organization as well as endocytosis and membrane recycling, processes crucial in the endoplasmic reticulum stress response (35). In Plasmodium berghei, Coronin knockouts have motility defects in the sporozoite stage (36), although their biological function outside of F-actin organization remains unexplored. PfCoronin is also involved in F-actin organization via its N-terminal propeller domain and localizes to the parasite membrane (37, 38). Its disruption using the piggyBac transposon insertion system identifies PfCoronin as dispensable to parasite growth (39).
Biological elucidation of ART resistance is complicated by the limited understanding of its mechanism of action and its promiscuity. Although ART activity is dependent on hemoglobin digestion (40), it touches players in multiple pathways (21, 22). Transcriptomics analyses of slow-clearing parasites from patients in Southeast Asia showed an up-regulation of stress-response pathways including the unfolded protein response (UPR) (41). Recent in vitro studies using pfkelch13 mutants have corroborated the involvement of UPR protein synthesis (42, 43) while also implicating the phosphatidylinositol-3-kinase (PI3K) pathway (44) and global attenuation of protein synthesis via eiF2α phosphorylation (45). An up-regulation of genes involved in UPR has been reported even in in vitro-selected 3D7 parasites without PfKelch13 mutations (19). The same study implicated two loci, pftrx1 (PF3D7_1457200) and pfspp (PF3D7_1457000), involved in antioxidant defense and endoplasmic reticulum-associated degradation, respectively, in the decreased ART sensitivity (19). DHA selection studies have also been performed in Dd2 parasites, implicating pfmdr1 copy number and increased antioxidant activity (24). Another locus connected to reduced ART sensitivity independent of pfkelch13 includes pfap2-mu, which codes for a subunit of clathrin-associated adaptor protein 2, suggesting involvement of the vesicular trafficking network (20, 46).
The resistance mutations of PfCoronin identified in the present study are all in the N-terminal WD40 domain, which by itself has been confirmed to be involved in actin-organization (37). Therefore, given what is known about T. gondii Coronin and the mechanism of ART resistance in general, an attempt to unravel the resistance mechanism of PfCoronin mutations will require an exploration of the protein’s function in vesicular trafficking in addition to its functions in actin binding and stress response.
Our results demonstrate that reduced ART susceptibility can be mediated by genes other than pfkelch13. The WD40 domain of PfCoronin is polymorphic in natural populations of P. falciparum; however, the variants identified in this study have not been observed in natural populations (www.malariagen.net/projects/p-falciparum-community-project). It may be that mutants selected under laboratory conditions in vitro may be so deleterious in nature that they cannot spread in a population. Given these considerations, whether pfcoronin mutants will become a significant contributor to clinical ART resistance in Africa remains an open question.
Materials and Methods
In Vitro Resistance Selection.
Culture-adapted field isolates Thiès (SenTh032.09) and Pikine (SenP019.04) were maintained in culture in RPMI 1640 medium (Gibco) supplemented with 10% O+ serum. Long-term in vitro resistance selection followed an intermittent and stepwise selection protocol, whereby separate flasks of ∼109 parasites were pulsed with increasing concentrations of DHA, beginning at 0.1 μM and increasing in 0.1-μM increments to 1.0 μM at the final selection step (Fig. 1A). Parasites were exposed to DHA for 48 h and then taken off DHA for 48 h. This treatment was repeated until the culture was smear-negative by Giemsa-stained thin smear microscopy (usually 4–12 d). Parasites were then allowed to recover off DHA. Recovery took between 7 and 60 d, with recovery time for cultures defined as the time until smear-negative culture reached 1% parasitemia. The complete process of drug treatment to a smear-negative culture followed by recovery time is considered one drug selection cycle. Following successful recovery, parasites were pulsed again with the same or a higher dose of DHA.
RSA.
The 0–3 h RSA was performed as described by Witkowski et al. (7, 9). In brief, highly synchronized 0–3 h postinvasion rings were exposed to 700 nM DHA for 6 h, followed by removal of the drug. At 72 h after initial drug treatment, parasitemia was assessed by Giemsa-stained smear microscopy of thin smears. The ring-stage survival percentage was calculated as the fraction of surviving DHA-treated parasites over the DMSO-treated control of the same parasite line. An RSA survival percentage >1% is considered to indicate resistance. An RSA for each parasite line was conducted at least twice with two technical replicates per biological replicate. Smears were blinded and counted twice, with at least 10,000 RBCs examined per smear count.
Drug Sensitivity Assays with SYBR Green I.
Drug sensitivity assays were performed as described previously (47). In brief, parasites were grown to 0.8–1% parasitemia in 2% hematocrit in 100 μL total volume in 96-well plates. Parasite viability was determined by SYBR Green I staining of parasite DNA following 72 h or 96 h in culture. Dose–response curves for standard antimalarial drugs were generated from a 12-point dilution series of drugs, carried out in triplicate, centered on expected EC50 values reported in the literature, with three to four biological replicates performed for each drug. A SpectraMax M5 plate reader (Molecular Devices) was used to measure fluorescence, and data were analyzed with GraphPad Prism version 6. EC50 was calculated using nonlinear regression with the log(inhibitor) vs. response with a four-parameter variable slope curve-fitting equation.
Pfmdr1 Copy Number Variation.
Pfmdr1 copy number was determined by quantitative RT-PCR, following the method described by Ribacke et al. (48).
Genome Sequencing.
Parasite DNA was extracted from in vitro cultures using QIAamp DNA blood kits (Qiagen). DNA samples were sequenced on the Illumina HiSEq 2500 for an average read coverage of 60–100×. Reads were aligned to P. falciparum v3 3D7 reference (MRA-102, NIH) genome assembly using BWA (49), and SNPs were called using GATK UnifiedGenotyper (50, 51).
PCR Amplification and Sanger Sequencing.
Parasite DNA was extracted from in vitro cultures using QIAamp DNA blood kits (Qiagen), in accordance with the manufacturer’s instructions. Primers were designed and ordered from Integrated DNA technologies to amplify candidate gene sequences in 500- to 800-bp overlapping amplicons, three to four amplicons per gene. For the verification of transfectants, primer combinations within and outside of the homology region were used to avoid amplifying the residual plasmid from transfection. PCR amplification was conducted as follows: 5 min at 95 °C; followed by 40 cycles of three-step amplification of 95 °C for 30 s, 55–58 °C for 45 s, and 68 °C for 30 s; and followed by a final extension at 72 °C for 7 min. DNA was purified using a ZymoKit DNA Purification Kit according to the manufacturer’s instructions. Sanger sequencing was performed by Macrogen USA. The primers used for PCR are listed in SI Appendix, Table S2.
Gene Editing of pfcoronin Locus.
For the knockin of pfcoronin mutations into the parental background, guides targeting pfcoronin were designed using two separate online tools: Benchling (https://benchling.com/) and CHOPCHOP (chopchop.cbu.uib.no). Two guides were designed per SNP knockin SI Appendix, Figs. S5 and S6. All guides were individually annealed and ligated into the BbsI-digested pDC2-Cas9-U6-hDHFR plasmid, generously provided by Marcus Lee, Wellcome Sanger Institute, Hinxton, UK, which contains the U6 snRNA polymerase III promoter and regions for expression of the Cas9 enzyme and human DHFR drug selection cassette, as described previously (52, 53). Homology regions (SI Appendix, Figs. S5 and S6) were amplified from the resistant parasite gDNA using the primer sets listed in SI Appendix, Table S2 and cloned into Zeroblunt TOPO vector (Thermo Fisher Scientific). Site-directed mutagenesis was performed using the QuickChange II Site-Directed Mutagenesis Kit (Agilent) to scramble guide targeting sequences in the homology region.
Transfection was performed on 6–8% sorbitol synchronized rings from SenTh032.09 (Thiès parent) or SenP019.04 (Pikine parent) with 50 μg each of two Cas9 plasmids containing two different guides and 50 μg of circular TOPO vector containing the target homology region using a Bio-Rad Gene Pulser at 0.31 kV and 960 µF, as described previously (54). After transfection, RBCs were plated at 5% hematocrit in complete medium. Transfected parasites were allowed to recover overnight before the addition of 5 nM WR99210. Drug selection was continued for 72 h. Parasite recovery in drug-free media was monitored by microscopy twice a week. Parasites started to appear in the transfection cultures after approximately 15–25 d posttransfection. Once parasitemia was >1%, DNA was isolated from bulk transfectants and verified by Sanger sequencing as described above (SI Appendix, Table S2). After confirming successful transfection via PCR screening of the bulk transfectant gDNA, dilutional cloning was performed in 96-well plates to obtain at least two confirmed CRISPR-edited parasite clones.
Supplementary Material
Acknowledgments
We thank Ngayo Sy, Younouss Diedhiou, Lamine Ndiaye, and Amadou Moctar Mbaye for their help collecting the original samples from Thiès and Pikine, Senegal used in this study. We also thank Cory L. Schlesener and Courtney Edison for technical support. Funding for this project was provided by National Institutes of Health Grant R01AI099105 (to D.F.W. and D.L.H.) and Grant OPP1156051 from the Bill and Melinda Gates Foundation. A.R.D. was partly supported by National Science Foundation GRFP DGE1144152.
Footnotes
The authors declare no conflict of interest.
Data deposition: Whole-genome sequencing data have been deposited to the National Center for Biotechnology Information’s Sequence Read Archive BioProject (accession no. PRJNA494861).
See Commentary on page 12556.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812317115/-/DCSupplemental.
References
- 1.World Health Organization 2017. World Malaria Report 2017 (World Health Organization, Geneva)
- 2.Leang R, et al. Evidence of Plasmodium falciparum malaria multidrug resistance to artemisinin and piperaquine in western Cambodia: Dihydroartemisinin-piperaquine open-label multicenter clinical assessment. Antimicrob Agents Chemother. 2015;59:4719–4726. doi: 10.1128/AAC.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spring MD, et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: An observational cohort study. Lancet Infect Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 4.Ashley EA, et al. Tracking Resistance to Artemisinin Collaboration (TRAC) Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imwong M, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: A molecular epidemiology observational study. Lancet Infect Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noedl H, et al. Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 7.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straimer J, et al. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witkowski B, et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: In-vitro and ex-vivo drug-response studies. Lancet Infect Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torrentino-Madamet M, et al. K13-propeller polymorphisms in Plasmodium falciparum isolates from patients in Mayotte in 2013 and 2014. Antimicrob Agents Chemother. 2015;59:7878–7881. doi: 10.1128/AAC.01251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor SM, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: A molecular epidemiologic study. J Infect Dis. 2015;211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boussaroque A, et al. Emergence of mutations in the K13 propeller gene of Plasmodium falciparum isolates from Dakar, Senegal, in 2013-2014. Antimicrob Agents Chemother. 2015;60:624–627. doi: 10.1128/AAC.01346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayih AG, et al. A unique Plasmodium falciparum K13 gene mutation in northwest Ethiopia. Am J Trop Med Hyg. 2016;94:132–135. doi: 10.4269/ajtmh.15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mvumbi DM, et al. Molecular surveillance of Plasmodium falciparum resistance to artemisinin-based combination therapies in the Democratic Republic of Congo. PLoS One. 2017;12:e0179142. doi: 10.1371/journal.pone.0179142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang C, et al. Polymorphisms of Plasmodium falciparum k13-propeller gene among migrant workers returning to Henan Province, China from Africa. BMC Infect Dis. 2017;17:560. doi: 10.1186/s12879-017-2634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee A, et al. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar J. 2017;16:195. doi: 10.1186/s12936-017-1845-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutherland CJ, et al. Pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria given artemether-lumefantrine in the UK. Antimicrob Agents Chemother. 2017;61:e02382-16. doi: 10.1128/AAC.02382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt P, et al. Gene encoding a deubiquitinating enzyme is mutated in artesunate- and chloroquine-resistant rodent malaria parasites. Mol Microbiol. 2007;65:27–40. doi: 10.1111/j.1365-2958.2007.05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocamora F, et al. Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLoS Pathog. 2018;14:e1006930. doi: 10.1371/journal.ppat.1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriques G, et al. Artemisinin resistance in rodent malaria: Mutation in the AP2 adaptor μ-chain suggests involvement of endocytosis and membrane protein trafficking. Malar J. 2013;12:118. doi: 10.1186/1475-2875-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Lin Q. Chemical proteomics approach reveals the direct targets and the heme-dependent activation mechanism of artemisinin in Plasmodium falciparum using an artemisinin-based activity probe. Microb Cell. 2016;3:230–231. doi: 10.15698/mic2016.05.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat Commun. 2015;6:10111. doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park DJ, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc Natl Acad Sci USA. 2012;109:13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui L, et al. Mechanisms of in vitro resistance to dihydroartemisinin in Plasmodium falciparum. Mol Microbiol. 2012;86:111–128. doi: 10.1111/j.1365-2958.2012.08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duraisingh MT, et al. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 26.Foote SJ, et al. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 27.Okombo J, et al. The polymorphic linker domain of pfmdr1 is associated with resistance-conferring mutations in Plasmodium falciparum populations from East and West Africa. Antimicrob Agents Chemother. 2013;57:4595–4598. doi: 10.1128/AAC.00455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghorbal M, et al. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 29.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. Efficient CRISPR-Cas9-mediated genome editing in Plasmodium falciparum. Nat Methods. 2014;11:915–918. doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price RN, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miotto O, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47:226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahl A, et al. PlasmoDB: The Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 34.Xavier CP, Eichinger L, Fernandez MP, Morgan RO, Clemen CS. Evolutionary and functional diversity of coronin proteins. Subcell Biochem. 2008;48:98–109. doi: 10.1007/978-0-387-09595-0_9. [DOI] [PubMed] [Google Scholar]
- 35.Salamun J, Kallio JP, Daher W, Soldati-Favre D, Kursula I. Structure of Toxoplasma gondii coronin, an actin-binding protein that relocalizes to the posterior pole of invasive parasites and contributes to invasion and egress. FASEB J. 2014;28:4729–4747. doi: 10.1096/fj.14-252569. [DOI] [PubMed] [Google Scholar]
- 36.Bane KS, et al. The actin filament-binding protein coronin regulates motility in Plasmodium sporozoites. PLoS Pathog. 2016;12:e1005710. doi: 10.1371/journal.ppat.1005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olshina MA, et al. Plasmodium falciparum coronin organizes arrays of parallel actin filaments potentially guiding directional motility in invasive malaria parasites. Malar J. 2015;14:280. doi: 10.1186/s12936-015-0801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardieux I, et al. A Plasmodium falciparum novel gene encoding a coronin-like protein which associates with actin filaments. FEBS Lett. 1998;441:251–256. doi: 10.1016/s0014-5793(98)01557-9. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M, et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science. 2018;360:eaap7847. doi: 10.1126/science.aap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klonis N, et al. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc Natl Acad Sci USA. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mok S, et al. Drug resistance: Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dogovski C, et al. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 2015;13:e1002132. doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharjee S, et al. Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood. 2018;131:1234–1247. doi: 10.1182/blood-2017-11-814665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mbengue A, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M, et al. Inhibiting the Plasmodium eIF2alpha kinase PK4 prevents artemisinin-induced latency. Cell Host Microbe. 2017;22:766–776.e4. doi: 10.1016/j.chom.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henriques G, et al. The Mu subunit of Plasmodium falciparum clathrin-associated adaptor protein 2 modulates in vitro parasite response to artemisinin and quinine. Antimicrob Agents Chemother. 2015;59:2540–2547. doi: 10.1128/AAC.04067-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JD, et al. Assessment and continued validation of the malaria SYBR Green I-based fluorescence assay for use in malaria drug screening. Antimicrob Agents Chemother. 2007;51:1926–1933. doi: 10.1128/AAC.01607-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribacke U, et al. Genome-wide gene amplifications and deletions in Plasmodium falciparum. Mol Biochem Parasitol. 2007;155:33–44. doi: 10.1016/j.molbiopara.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng CL, et al. CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol Microbiol. 2016;101:381–393. doi: 10.1111/mmi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim MY, et al. UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug-resistance genes. Nat Microbiol. 2016;19:16166. doi: 10.1038/nmicrobiol.2016.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.