Significance
It is well known that ribosomal RNA processing is directly impacted by the rate of transcription elongation by RNA polymerase I (Pol I). To understand how these processes are orchestrated, we must carefully define transcription elongation properties in vitro and in living cells. Here, we characterize DNA sequence elements that pause and terminate Pol I transcription in vitro. We also establish methods for analyzing Pol I transcription elongation properties in vivo using native elongating transcript sequencing (NETSeq). Our NETSeq data revealed frequent pausing by Pol I and decreased Pol I occupancy at G residues, suggesting unequal rates of nucleotide incorporation by the enzyme. These findings redefine our understanding of Pol I transcription elongation and its heterogeneity in vivo.
Keywords: transcription elongation, RNA polymerase I, ribosome, transcription pausing, transcription termination
Abstract
DNA sequence motifs that affect RNA polymerase transcription elongation are well studied in prokaryotic organisms and contribute directly to regulation of gene expression. Despite significant work on the regulation of eukaryotic transcription, the effect of DNA template sequence on RNA polymerase I (Pol I) transcription elongation remains unknown. In this study, we examined the effects of DNA sequence motifs on Pol I transcription elongation kinetics in vitro and in vivo. Specifically, we characterized how the spy rho-independent terminator motif from Escherichia coli directly affects Saccharomyces cerevisiae Pol I activity, demonstrating evolutionary conservation of sequence-specific effects on transcription. The insight gained from this analysis led to the identification of a homologous sequence in the ribosomal DNA of S. cerevisiae. We then used native elongating transcript sequencing (NETSeq) to determine whether Pol I encounters pause-inducing sequences in vivo. We found hundreds of positions within the ribosomal DNA (rDNA) that reproducibly induce pausing in vivo. We also observed significantly lower Pol I occupancy at G residues in the rDNA, independent of other sequence context, indicating differential nucleotide incorporation rates for Pol I in vivo. These data demonstrate that DNA template sequence elements directly influence Pol I transcription elongation. Furthermore, we have developed the necessary experimental and analytical methods to investigate these perturbations in living cells going forward.
The production of a single ribosome requires the synthesis, processing, and assembly of more than 80 proteins and four noncoding RNAs known as ribosomal RNAs (rRNAs) (1). Because this process represents the major energetic investment for a rapidly dividing eukaryotic cell, it is subject to tight regulation. In Saccharomyces cerevisiae, the first step in ribosome biogenesis is transcription of the 35S rRNA gene (also referred to as 37S rRNA gene or RDN37) by RNA polymerase I (Pol I) (2). This transcript is co- and posttranscriptionally processed to produce three of the four RNAs required for ribosome biogenesis—18S, 5.8S, and 25S rRNAs (3, 4). Regulation of Pol I activity represents a robust method used by cells to control the rate of ribosome biogenesis. Previous studies concerning the regulation of transcription by Pol I have principally focused on transcription initiation. It is well established that recruitment of Pol I to the ribosomal DNA (rDNA) promoter is a key regulatory target for cellular control of ribosome biosynthesis (5). However, we and others have shown that later steps in the transcription cycle can be influenced by transcription factors (6–9). Furthermore, the efficiency of transcription elongation directly affects processing of the nascent rRNA (10). Thus, transacting factors or template sequence features that influence transcription elongation by Pol I can have substantial consequences on cellular proliferation by affecting the synthesis or processing of rRNA.
While the effect of DNA template sequence on Pol I transcription has not been established, elongation-affecting sequence motifs have been identified as regulatory elements in prokaryotic transcription systems (11, 12). One such motif is the rho-independent terminator motif. This motif is composed of a guanine–cytosine (G–C)-rich region of dyadic symmetry, followed by a thymine-rich tract (T tract). Transcription of the T tract creates active site instability induced by a weak RNA–DNA hybrid (12, 13). This weakened complex is further perturbed by the formation of an RNA hairpin in the upstream G–C-rich tract of the nascent transcript. This combined effect results in efficient termination of transcription by prokaryotic RNA polymerase (RNAP) in a protein factor-independent manner (13). Furthermore, similar motifs that induce termination of eukaryotic RNA polymerase II have also been found in viral and mammalian genes (14–16). Because the rho-independent terminator motif affects the active site of RNAP, which is conserved among all multisubunit RNA polymerases (17), we reasoned that this motif will also affect Pol I.
In this study, we used a fully reconstituted promoter-dependent transcription assay to determine if DNA template sequence can affect Pol I transcription. The sequence motif that we used was the rho-independent terminator motif from the spy gene of Escherichia coli. We demonstrate that this motif induces termination of Pol I transcription in vitro. We then used mutational analysis of this motif to show that both the stem loop and uridine tract (U tract) contribute to its effect on Pol I transcription. Based on these observations, we identified sequence elements present in the native 35S rRNA gene that also influence Pol I transcription elongation kinetics and demonstrated that the effects of both motifs on Pol I transcription are dependent on UTP concentration in our in vitro system.
To determine whether Pol I pauses in vivo and whether the same sequence elements contribute to this pausing, we adapted native elongating transcript sequencing (NETSeq) for use with S. cerevisiae Pol I. This application of NETSeq enables precise mapping of Pol I occupancy of the rDNA gene with single nucleotide resolution. We observed reproducibly heterogeneous occupancy of Pol I on the rDNA in rapidly growing cells. Thorough analysis of the resulting Pol I occupancy data revealed significantly lower occupancy of Pol I at G residues throughout the rDNA. Taken together, these data lead to the following conclusions: (i) The effects of strong rho-independent terminator sequences on RNA polymerase activity are conserved across domains of life; and (ii) Pol I elongation efficiency is sensitive to both the DNA sequence and other factors in vivo and in vitro.
Results
A Rho-Independent Terminator Motif Affects Pol I Transcription Elongation.
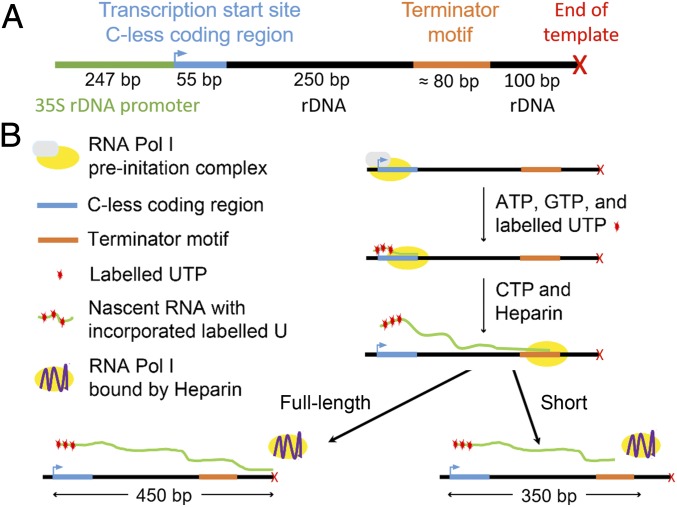
To test whether prokaryotic termination motifs can affect Pol I transcription elongation, we used a fully reconstituted promoter-dependent in vitro transcription assay (18). Individual sequence motifs (e.g., the spy motif) were inserted into native rDNA sequence downstream of the transcription start site (Fig. 1A). This motif was chosen because it was identified as the strongest terminator of E. coli RNAP transcription in a recent survey (19). To this template we added Pol I, Rrn3p, TBP, and core factor (CF) (Rrn6p, Rrn7p, and Rrn11p) to assemble the Pol I preinitiation complex at the promoter. Transcription was initiated by the addition of ATP, GTP, UTP (15 µM of each), and 32P-labeled UTP, resulting in synchronized transcription elongation complexes at the first encoded C (position +56). The synchronized enzymes were then released by addition of 15 µM CTP, and samples were collected as a function of time. Resultant RNA transcripts were resolved via PAGE and visualized by phosphorimage analysis. If a fraction of the Pol I population paused, arrested, or terminated at the spy motif, a product ∼100 nucleotides shorter than the full-length product was observed (Fig. 1B). To quantify the magnitude of the effect of the motif on Pol I transcription elongation, we calculated the ratio of the shortened product to the total RNA signal in each individual lane.
Fig. 1.
Promoter-dependent in vitro transcription assay for RNA polymerase I. (A) linearized DNA template for promoter-dependent in vitro transcription by RNA polymerase I. (B) Experimental scheme for RNA polymerase I promoter-dependent in vitro transcription assay.
Inclusion of the spy terminator motif in the template resulted in the appearance of a short product (Fig. 2 A and B) not seen in the negative control (Fig. 2B). Since accumulation of this short product is not rescued after an extended incubation time of 20 min, we conclude that these complexes have either terminated transcription or are terminally arrested. These data indicate that the spy terminator motif directly perturbs transcription elongation by Pol I.
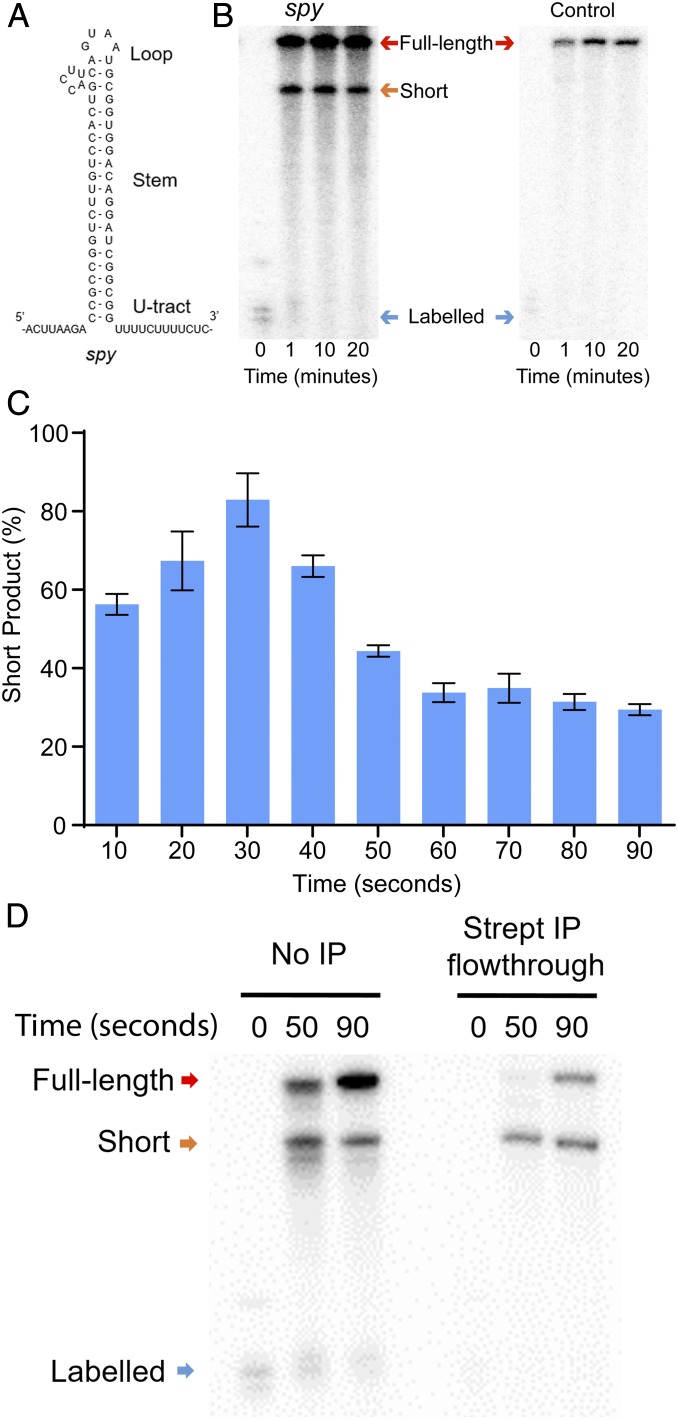
Fig. 2.
The spy rho-independent terminator motif affects RNA polymerase I transcription in vitro. (A) Proposed secondary structure of transcribed spy rho-independent terminator motif. (B) Polyacrylamide gel of long time course in vitro transcription of the spy rho-independent terminator template by RNA polymerase I (Left), compared with the negative control (Right). (C) The short product at each time point of short time course expressed as a percentage of total signal. n = 3, error bars represent SD. (D) Polyacrylamide gel of time course in vitro transcription of the biotinylated spy rho-independent terminator template followed by streptavidin immunoprecipitation (IP) comparing no IP (Left portion) to the IP flowthrough (Right portion).
To define the kinetics of the spy effect on Pol I, we measured reaction progress during shorter time courses. The accumulation of short product reaches a maximum of 83 ± 6.7% of total RNA product at the 30-s time point (Fig. 2C). The accumulation of short product then decreases at each time point before stabilizing at 30 ± 1.6% of total RNA product after 60 s (Fig. 2C). To determine if the observed effect was termination or arrest, the experiment was repeated with a biotinylated template (Fig. 2D). Following immunoprecipitation of the template, the short product was observed in the flowthrough while the labeled product was not. These data demonstrate that the nascent transcript is not associated with the DNA template, indicating that the motif induces termination of Pol I transcription. Based on these data, it is clear that the spy terminator motif induces pausing by the majority of elongation complexes. Most enzymes ultimately escape this pause and reach the end of the template, while a subset undergoes termination. Thus, rho-independent termination motifs can both pause and terminate transcription by eukaryotic Pol I.
Multiple Regions of the spy Motif Contribute to the Effect on Pol I.
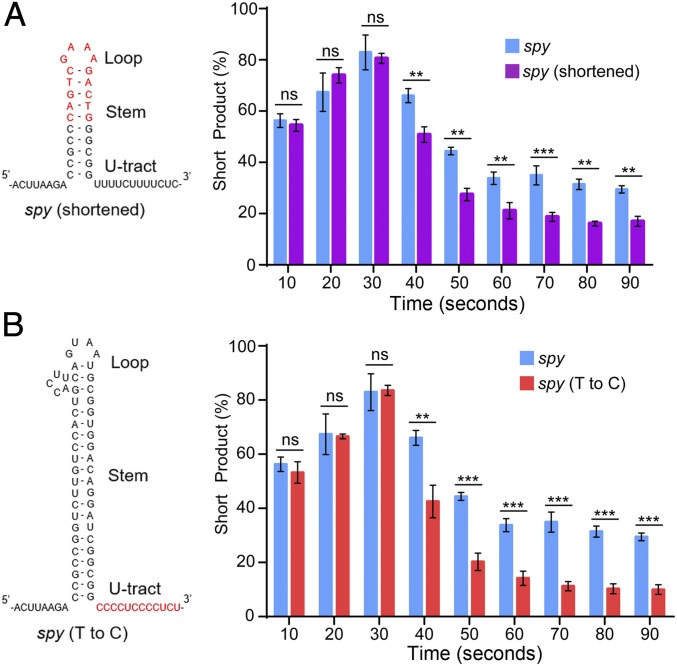
To identify the features of the spy terminator motif that are necessary for its effect on Pol I transcription, we constructed several variants of the motif. We then repeated the transcription assay described above using these variants. In the first variant, we shortened the stem loop to 10 base pairs from 21 (Fig. 3A). Compared with the wild-type spy terminator motif, the shortened stem loop variant accumulates substantially less short product at every time point (Fig. 3A). By 90 s, the percentage of short product is 16 ± 2.0% compared with 30 ± 1.6% for wild type. For the second variant, we mutated the template strand to abolish the U tract in the nascent transcript by exchanging each T residue with a C residue and vice versa (Fig. 3B). The motivation for this mutation strategy is based on the finding that uridine enrichment in the residues directly downstream of the hairpin plays an important role in the effect of the motif on polymerase elongation (20). These mutations stabilize the RNA–DNA complex by replacing the A–U base pairs with stronger G–C base pairs which has been previously shown to reduce pausing by prokaryotic RNAP (13, 19, 21). By 90 s, the percentage of short product is just 10 ± 1.8% compared with 30 ± 1.6% for wild type (Fig. 3B). Taken together, these data demonstrate that both the stem loop length and the U tract are critical for the effect on Pol I transcription. However, abolishing the U tract results in a larger decrease in termination compared with the shortened stem loop variant. These data are consistent with prokaryotic studies showing that both of these features are critical for pausing and terminating RNA polymerases (19). Furthermore, these findings provide guidance for predicting effects of native ribosomal DNA sequence on transcription elongation by Pol I.
Fig. 3.
Both the stem loop and U-tract regions of the spy rho-independent terminator contribute to its effect on RNA polymerase I transcription. (A) Predicted secondary structure of the shortened stem loop spy rho-independent terminator motif mutant (Left) and the mutant’s short product compared with wild type (Right). (B) Predicted secondary structure of the T-to-C switch spy rho-independent terminator motif mutant (Left) and the mutant’s short product compared with wild type (Right). For A and B, n = 3, error bars represent SD. Comparison by one-tailed Student’s t test. ns, not significant, *P < 0.05, **P < 0.005, ***P < 0.0005.
Native rDNA Sequence Elements Affect Pol I Transcription Elongation.
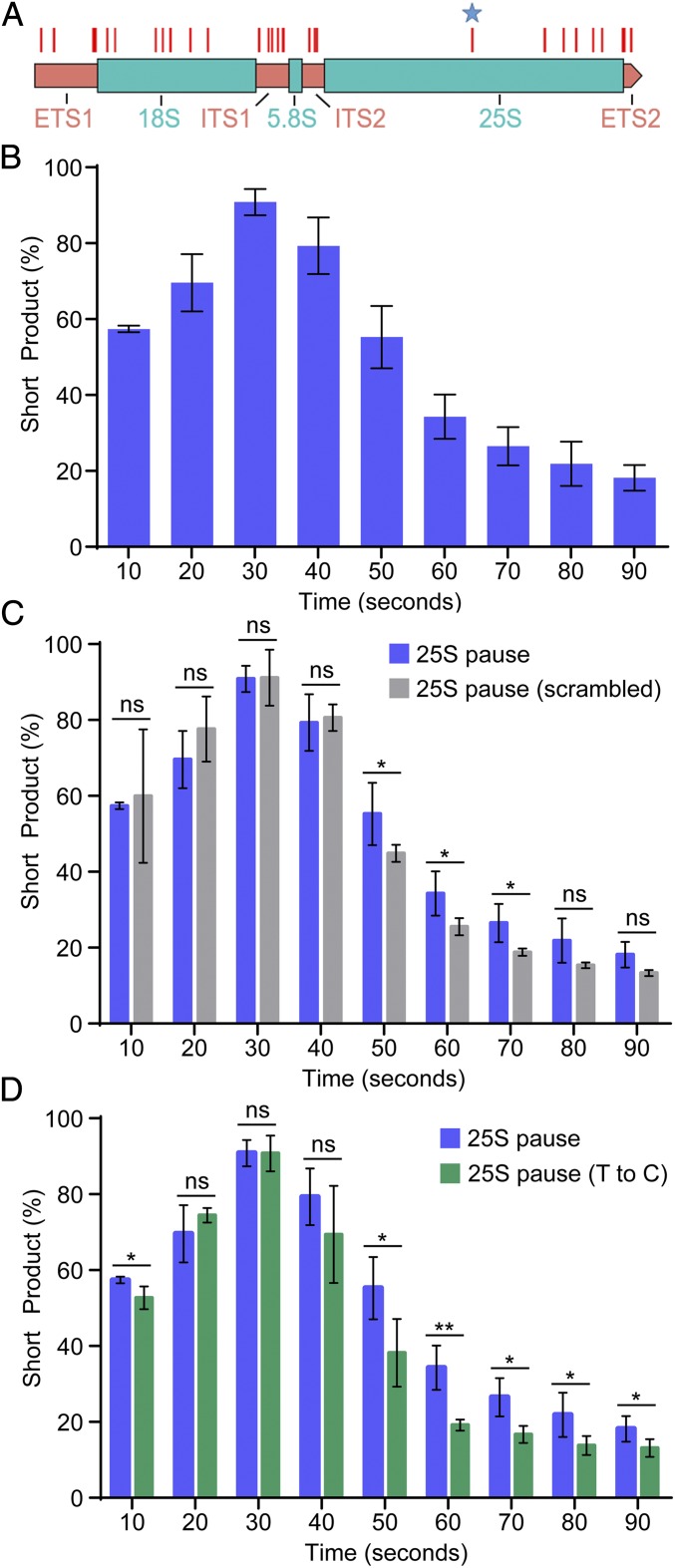
To determine if endogenous elongation-affecting motifs exist, we examined the 35S gene region of the rDNA for T-rich tracts (which encode U tracts in the nascent transcript), which is one of the key elements of the spy terminator motif. We identified 54 tracts that are at least 10 nucleotides long, end with three T residues, and have a T-residue enrichment of 70% or more (Fig. 4A). The longest element observed was in the 25S rRNA coding region of the 35S rDNA gene. We inserted this T-rich tract including 90 nucleotides upstream and 15 nucleotides downstream into our template and transcribed the template in vitro using the system outlined above. We observed a transient pause of a majority of the polymerases at the rDNA sequence 30 s after release. This effect is similar to the effect of the spy terminator motif. Most of these polymerases clear the pause and produce full-length product with the percentage of short product stabilizing at 15 ± 2.2% by 90 s postrelease (Fig. 4B). These data demonstrate that sequences within the rDNA inhibit transcription elongation by Pol I.
Fig. 4.
Endogenous rDNA sequence induces site-specific pausing, with contributions from the T tract and the upstream sequence. (A) T tracts (10 nucleotides or greater in length, greater than 70% T enrichment, ending with three T residue) in the 35S gene (marked with red lines.) The blue star marks the T tract to be analyzed. (B) rDNA sequence wild-type short product at each time point expressed as a percentage of total signal. (C) rDNA sequence upstream scramble short product compared with wild type. (D) rDNA sequence T-to-C switch short product compared with wild type. N = 3, error bars represent SD. Comparison by one-sided Student’s t test. ns, not significant, *P < 0.05, **P < 0.005, ***P < 0.0005.
We then produced two variants to determine which parts of this sequence were necessary for its effect on Pol I transcription. In the first variant, we abolished the possibility of RNA stem loop formation by scrambling the 30 nucleotides upstream of the T tract. This perturbation had relatively little effect on short product accumulation (Fig. 4C). In the second variant, we abolished the T tract by exchanging the T residues for C residues in the DNA template. Abolishing the T tract decreased short product accumulation at almost every point (Fig. 4D). Thus, the T tract is required for the motif’s pausing and termination effects on Pol I transcription. Taken together, these data demonstrate that T tracts within the rDNA can directly influence Pol I activity. This observation is consistent with conclusions drawn using prokaryotic DNA elements.
Pausing by Pol I at T Tracts Is Sensitive to UTP Concentration.
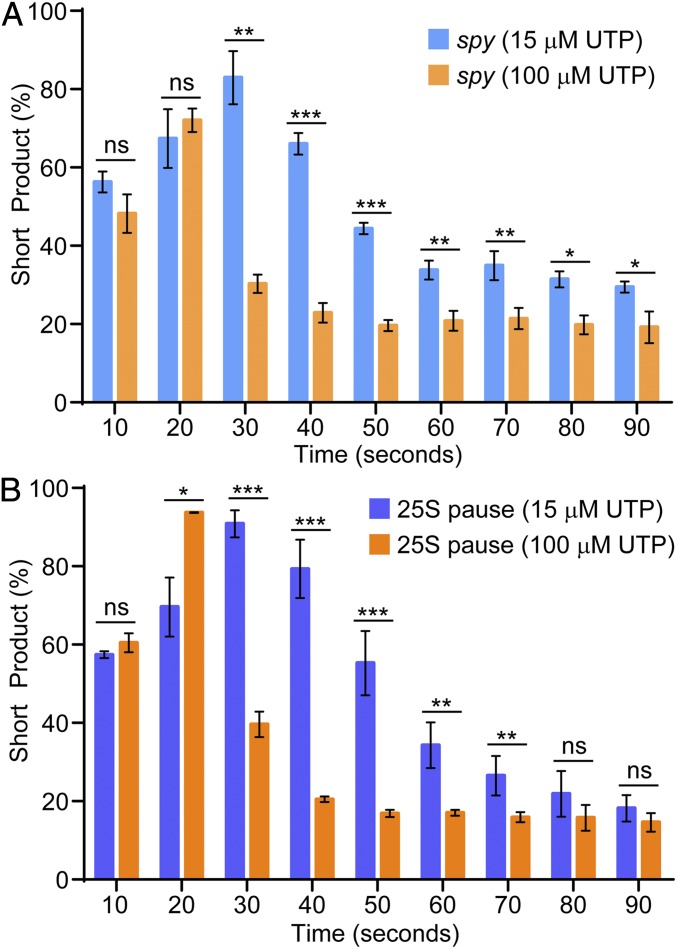
Since we have determined that the T tract is critical for the effects on Pol I transcription, we tested whether the UTP concentration in the reaction alters the kinetics of Pol I elongation. We repeated in vitro transcription assays under two different UTP concentrations: 100 μM UTP and 15 μM UTP. For the spy terminator motif, there is a significant difference between the amount of short product in the 100-μM UTP and 15-μM UTP experiments. This difference is observed starting at 30 s and continues through 90 s after which both values stabilize (Fig. 5A). At 15 μM UTP, short product accounted for 30 ± 1.6% of all RNA by 90 s. At 100 μM UTP, this value decreased to 19 ± 4%. These data demonstrate that elevated UTP concentration suppresses, but does not eliminate, the observed pause and termination effects of the spy terminator motif on Pol I.
Fig. 5.
The effect of both motifs on RNA polymerase I transcription is UTP concentration dependent. (A) Terminator wild-type short product at 15 μM and 100 μM UTP. (B) rDNA sequence wild-type short product at 15 μM and 100 μM UTP. n = 3, error bars represent SD. Comparison by one-sided Student’s t test. ns, not significant, *P < 0.05, **P < 0.005, ***P < 0.0005.
To determine if the eukaryotic sequence also responds to substrate concentration, we repeated these experiments using the T-rich 25S rDNA pause sequence detailed above. We found that, like the spy motif, there is a significant reduction in observed pausing at the T tract in the presence of 100 μM UTP compared with 15 μM UTP (Fig. 5B). Although the percentage of terminated polymerases does not change, the pause dwell time is reduced as UTP concentration increases. All of these in vitro analyses call for analysis of pausing by Pol I in vivo.
NETSeq Reveals Heterogeneous Pol I Occupancy on rDNA.
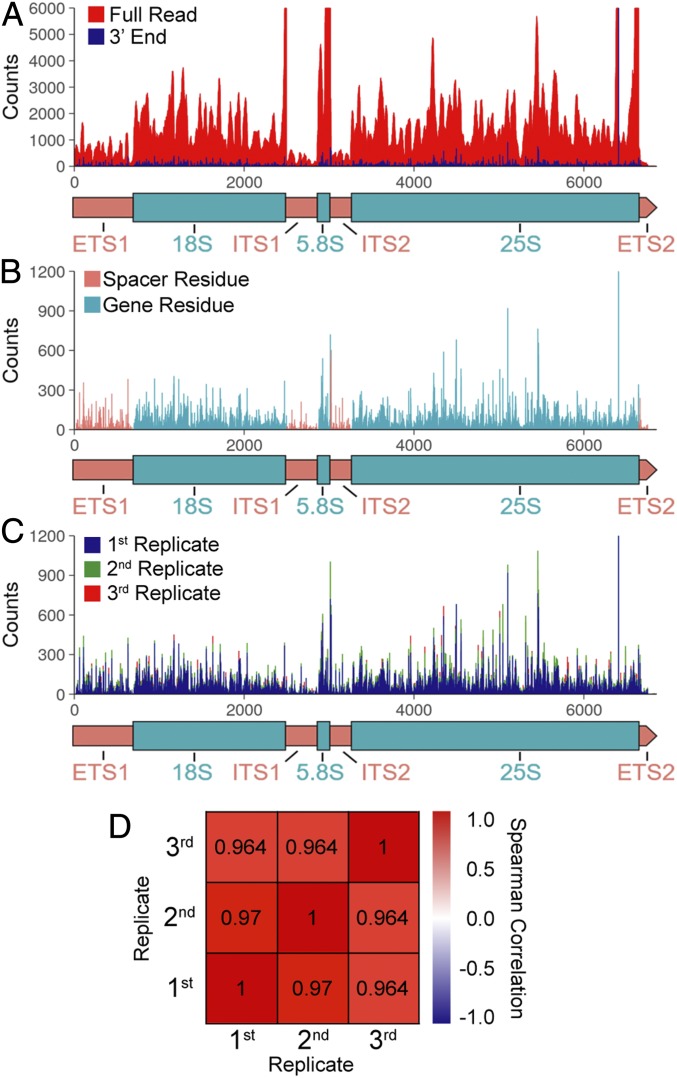
To characterize Pol I pausing in vivo, we performed NETSeq. We adapted our methods from those used previously to characterize RNA polymerase II (SI Appendix) (22). Three biological replicates of NETSeq were performed with a S. cerevisiae strain bearing HA-tagged Pol I, and the resultant reads were mapped to the rDNA gene (Fig. 6A). The 3′-end of each read corresponds to the last phosphodiester bond formed by the polymerase (Fig. 6 A and B). Our data demonstrate obvious heterogeneity in Pol I occupancy throughout the gene. The amplitude of each peak reflects the frequency with which a Pol I complex is observed at that position in vivo. Thus, high peak height can be interpreted as a position that induces pausing by Pol I.
Fig. 6.
RNA polymerase I NETSeq reads map to rDNA and are qualitatively reproducible. (A) NETSeq Pol I full-read density in the 35S gene (blue) compared with 3′-end density (black) with 35S gene diagram below. (B) NETSeq rDNA 3′-end density, with 35S gene diagram below. Spacer residues highlighted in pink, gene residues highlighted in cyan. (C) NETSeq rDNA 3′-end densities from three biological replicates overlaid (blue, green, red) with 35S gene diagram below. (D) Heat map of Spearman’s correlations between replicate position lists ranked by occupancy. All P values <1 × 10−8.
The overlay of the three biological replicates revealed exceptional reproducibility between cultures (Fig. 6 C and D). In all three replicates, significant sequence coverage was observed only in rDNA genes and the mature ends of snoRNAs, short RNAs associated with nascent rRNA transcripts. In addition to the 3′-nascent RNAs detected throughout the gene, we observed large signal spikes at the position of the mature ends of all four rRNA species (25S, 5.8S, 18S, and 5S genes, SI Appendix, Fig. S3A). It is well established that mature ribosomal RNA is recovered when preparing RNA sequencing libraries. Furthermore, the 5S gene is not transcribed by Pol I, yet it was observed in all of our libraries (SI Appendix, Fig. S4B). Thus, we concluded that these four large peaks represented contaminating mature product, not nascent RNA, and we eliminated these peaks from subsequent analyses and plots.
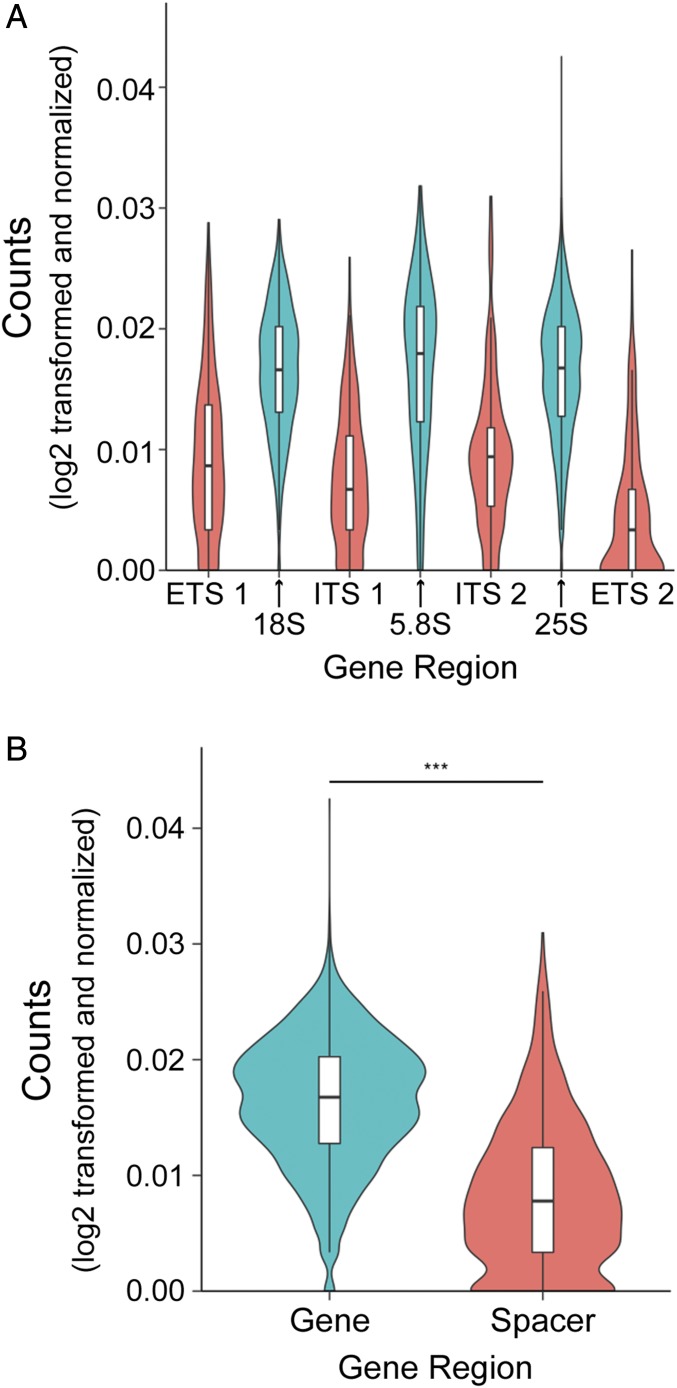
Initial analysis revealed significantly lower Pol I occupancy within the spacer regions of the rDNA compared with the regions that encode mature rRNA (Fig. 7). There are a number of potential explanations for this observation. The simplest interpretation is that Pol I may elongate more quickly through those regions of the template, particularly the spacer between the 18S and 5.8S regions termed “ITS1” (Fig. 7A). It is known that mature rRNA folds into stable secondary and tertiary structures, whereas spacer elements are less well conserved and likely less structured. If rRNA structures begin to form on the nascent RNA, these structures might slow down Pol I. This potential effect would be exacerbated in the sequences that give rise to mature product. It is also possible that there is contamination of our nascent RNA with degradation products of mature rRNA (see Discussion below).
Fig. 7.
RNA polymerase I occupancy is increased in gene regions. (A) Violin plot with incorporated box plots of rDNA position occupancies sorted by region. Counts are log2 transformed. (B) Violin plot with incorporated box plots of all spacer positions and all gene positions. Counts are log2 transformed and normalized. Comparison by Mann–Whitney U test. ***P value <1 × 10−8.
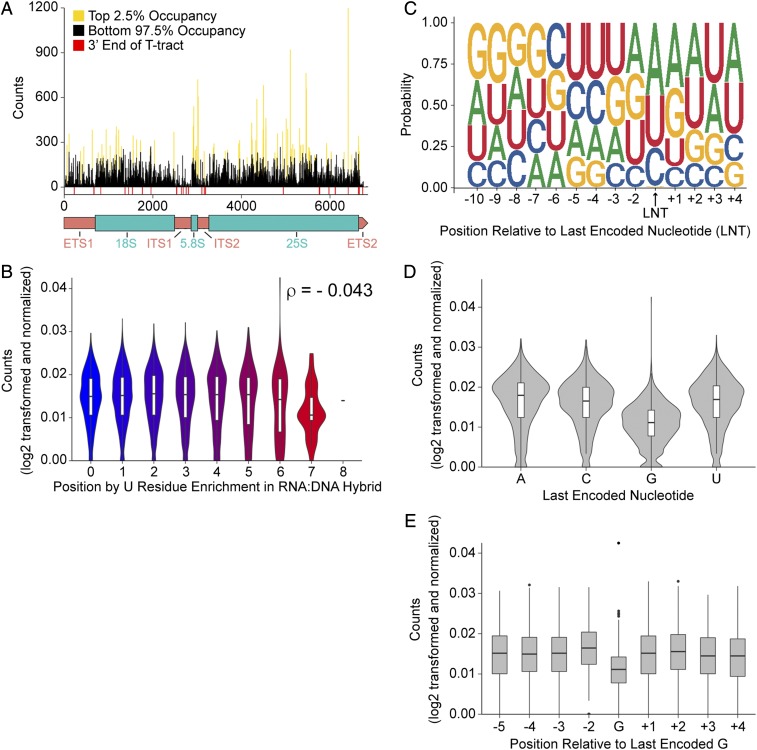
To determine sites of polymerase pausing, we identified the top 2.5% of positions by Pol I occupancy (171 positions) for each replicate as top sites of polymerase enrichment/pausing. A total of 127 positions were found in the top 2.5% by occupancy in all three sets. None of these top positions coincide with the ends of the T tracts identified by our in vitro studies (Figs. 4A and 8A). Broadening our analysis, we examined the sequence content immediately upstream of the polymerase occupancy at each top pause site. We focused on the seven nucleotides upstream of each position, as this would reflect the minimal RNA:DNA hybrid within the polymerase. We found a significant correlation between hybrid U content and Pol I occupancy (Fig. 8B). However, the correlation coefficient was very small [Spearman’s correlation coefficient (ρ) = −0.043, P < 1 × 10−8], indicating that that hybrid U content is not a reliable predictor of Pol I pausing in vivo. Generation of a sequence logo using the top sites yielded the intriguing result that only one of the 127 top sites had a G residue as the last encoded nucleotide (Fig. 8C). We then looked at occupancy across the gene as a factor of the last encoded nucleotide and found a significantly lower median occupancy value for G residues (Fig. 8D). Expanding this analysis, we determined the occupancy at positions proximal to G residues (Fig. 8E). We observed significantly increased occupancy directly upstream of G residues. These data strongly suggest that the incorporation of G nucleotides by Pol I occurs more slowly than the other three, but addition of the next nucleotide after a G residue occurs more rapidly. The relationship between nascent preribosome assembly and Pol I transcription is crucially important, and NETSeq provides a critical tool for characterization of this complex biosynthetic process.
Fig. 8.
Low RNA polymerase I occupancy correlates strongly with last encoded nucleotide. (A) NETSeq Pol I 3′-end densities (black) with significant positions indicated (wheat) and previously identified U tracts (red) with 35S gene diagram below. (B) Violin plot of 3′-positions sorted by RNA:DNA hybrid U content, with incorporated box plots. Counts are log2 transformed and normalized. Correlation coefficient determined by Spearman’s correlation test, P value <1 × 10−8. (C) Sequence logo of the RNA:DNA hybrid corresponding to top 3′-positions conserved in all three sets. (D) Violin plot with incorporated box plots of rDNA position occupancies sorted by last encoded nucleotide. Comparison by Kruskal–Wallis test produced a P value <1 × 10−8. (E) Box plot of position occupancies proximal to last encoded G residues. Counts are log2 transformed. Comparisons by Mann–Whitney U test. ns, not significant, *P value <5 × 10−6, ***P value <5 × 10−16.
Certain aspects of our data suggest contamination by mature rRNA species or nascent transcripts from different polymerases. We observe no significant reads from Pol II-transcribed genes or from Pol III-transcribed tRNA genes (SI Appendix, Fig. S4 C and D). However, we observe signal at positions throughout the 5S gene, which is transcribed by Pol III (SI Appendix, Fig. S4B). These data suggest that either 5S rRNA is coprecipitated with the nascent 35S rRNA or we are detecting contamination from degrading mature rRNAs and mapping them to the 35S gene. Unlike studies focused on mRNA synthesis, we cannot simply “ribo-deplete” our libraries as we are uniquely focused on the rDNA. To rigorously ensure that our conclusions are reflective of nascent transcription and not artifacts of bulk ribosome decay, we repeated our analysis focusing only on residues in the spacer regions of the 35S gene. These residues are not present in mature ribosomes and are rapidly degraded during pre-rRNA processing. We found that the trends observed through the entire 35S gene were recapitulated in this smaller spacer RNA set (SI Appendix, Fig. S6). These data confirm that contaminating rRNA is not the driver of the observed phenomena.
Our NETSeq data are highly reproducible, revealing sites of increased and decreased Pol I occupancy in vivo. We have also discovered two sequence variables that correlate significantly with low Pol I occupancy. However, these data also suggest that DNA sequence elements alone do not govern Pol I occupancy. This disconnect between the biochemical studies and experiments in live cells is expected, given the relative complexity of the two systems. Importantly, these data suggest that although sequence elements can influence Pol I directly, there may be many additional features that influence and control Pol I in vivo. The search for these factors (transcription elongation factors, nucleosomes, RNA structures, as-yet undiscovered sequence elements, etc.) represents an exciting area of future investigation.
Discussion
Analysis of Pol I pausing in vitro and in vivo reveals important features of the enzyme while raising key questions regarding the cellular factors and conditions that govern pausing. Our data clearly demonstrate that Pol I is prone to pausing. This observation is not surprising, since previous studies have identified or predicted pausing for many RNA polymerases (9, 23–28). Pausing of transcription elongation can have important consequences on gene regulation. Sophisticated regulatory systems, such as attenuation mechanisms in bacteria, rely on programmed pausing of RNAP (25). Furthermore, pausing by Pol II is a recently defined feature of eukaryotic gene regulation that has dramatic impact on cell growth and differentiation (29, 30). In this study we begin to explore the factors that control pausing by Pol I. We not only identify key features that are conserved among RNA polymerases but also raise questions regarding the factors that govern pausing in vivo.
DNA Sequence Effects on Transcription Are Conserved Between Bacteria and Eukaryotes.
Here, we demonstrate that a prokaryotic terminator motif can induce pausing and termination of transcription by a eukaryotic polymerase. The observed pausing and termination of transcription by Pol I can be modulated by modification of the stem loop or U-tract regions of the spy terminator motif. We further demonstrated that the induced pausing and termination were both dependent on UTP concentration. All of these factors are consistent with previous observations in prokaryotic systems (12, 19, 31). Previous studies have suggested that certain DNA sequence elements can directly affect all RNA polymerases (32). Our findings reveal clearly conserved effects of prokaryotic terminator motifs on both Pol I and RNAP. Similar motifs also affect Pol II (15–17). All of these data support the conclusion that critical interactions between the DNA template, the nascent RNA, and the RNA polymerase have been preserved throughout evolution.
Substrate Concentration May Influence rRNA Synthesis Directly.
The data described above provide insight into sequence motifs that affect transcription elongation by Pol I. Our search for similar T tracts in the rDNA yielded 54 sites (Fig. 4A). In vitro transcription of one such site from the 25S region had a similar effect on Pol I transcription as the rho-independent terminator motif. We also determined that the pause, but not the termination effect of this motif, was sensitive to UTP concentration. In the context of gene regulation, this is potentially an important finding. Given the established link between Pol I transcription rate and efficient rRNA processing (4), it is possible that these pause sites mediate RNA secondary structure formation required for rRNA processing, a phenomenon previously observed in prokaryotic expression systems (11). Taking into account the UTP concentration dependence of these T tracts, these pause sites could alternatively serve as cellular sensors of nutrient concentration. Each pause site might be sensitive to UTP concentration around a specific threshold. If pausing by Pol I were responsive to substrate concentration, then rRNA processing might be linked to cellular nutrient conditions. No such factor-independent stress sensor has been identified in eukaryotic rRNA expression. However, substrate NTP concentration plays a pivotal role in the regulation of prokaryotic rRNA synthesis (33, 34). This previously described regulatory mechanism demonstrates that nutrient sensing via the cellular nucleoside triphosphate pool can be exploited for the control of rRNA synthesis.
In Vitro Pausing by Pol I Is Not Predictive of Pause Sites in Vivo.
To characterize Pol I pausing in vivo, we adapted NETSeq methods from those used previously for Pol II (22). We observed obvious heterogeneity in Pol I occupancy. Notably, we saw no significant correlation between identified T tracts and Pol I occupancy (Fig. 8A). In addition, we found that U-residue enrichment in the rDNA hybrid had little effect on Pol I occupancy (Fig. 8B). These results stand in stark contrast to what we observed in vitro. One potential explanation for this discrepancy is that the biochemical studies were performed under conditions of limiting substrate. Our in vitro experiments were performed at a NTP concentration of 15 µM for each nucleotide. Increasing the concentration to 100 µM largely abrogated the effect on Pol I elongation for both motifs (Fig. 5). The K1/2 value for ATP concentration with respect to Pol I elongation in vitro is 170 µM (35), and the nucleolar concentrations of NTP in living cells is expected to be much higher. Thus, our in vitro experiments were performed at subsaturating substrate concentrations. The lack of pausing at these T tracts in vivo may be due to the fact that the nucleolar NTP concentrations during balanced growth were simply too high. Perhaps under conditions when substrates are limited (as discussed above), DNA sequence elements may contribute more substantially to pausing in vivo.
Another potential explanation for the discrepancy between pause site selection in vitro versus in vivo is the relative complexity of the two experimental systems. The biochemical assays that we deploy are fully reconstituted. The DNA template is free of histones, and all of the proteins other than Pol I are expressed and purified from E. coli. Thus, these assays are designed to characterize the minimal set of factors that influence RNA synthesis. Our data demonstrate that under these purified conditions, DNA sequence elements can have robust effects on transcription by distantly related RNA polymerases. However, in vivo these potential effects may be secondary to effects by other factors (DNA binding proteins or elongation assisting factors). Based on this model, there exists a collection of factors that directly influence the rate of Pol I transcription throughout the rDNA gene.
One candidate factor is the Spt4/5 complex. SPT5 is conserved throughout eukarya (36). NusG, the homolog of Spt5 in bacteria, has been shown to increase the elongation rate and general processivity of the E. coli RNA polymerase in vitro (37). The Spt4/5 complex has been shown to affect the processivity of Pol II (38). We demonstrated previously that Spt4/5 interacts directly with Pol I (39, 40). Furthermore, deletion of SPT4 or mutation of SPT5 resulted in defective rRNA synthesis and processing (40, 41). The interaction of the Spt4/5 complex with elongating Pol I may increase its transcription elongation rate or processivity, allowing it to read through sequences which might otherwise cause Pol I to pause or arrest. Defining the factors that perturb Pol I activity in vivo will impact our understanding of how cells orchestrate the complex process of ribosome assembly.
Pol I Occupancy Is Reduced at G Residues.
Analysis of the sequence context of the most robust sites of pausing revealed an antipreference for G as the last encoded nucleotide (Fig. 8C). Expanded analysis revealed significantly lower occupancy at G residues throughout the rDNA, indicating that this is a gene-wide trend instead of being specific to high-occupancy sites (Fig. 8D). Analysis of occupancy at positions proximal to G residues also revealed a significant increase in occupancy directly upstream. This G antipreference has not been described in any previous NETSeq studies, suggesting that the phenomenon is not simply an artifact of library generation. While a similar phenomenon has been observed in vivo for RNAP (42), there are no previous examples of differential nucleotide incorporation by Pol I in vivo. Interestingly, Pol I occupancy at A residues was not similarly reduced compared with G residues, suggesting that the difference is not due simply to purine or pyrimidine base identity. The simplest explanation for these findings is that incorporation of G nucleotides is slow, but incorporation by Pol I after addition of a G to the nascent RNA is fast. We do not see any significant effect of the identity of the next encoded nucleotide (Fig. 8C). Thus, this effect is apparently due to unique features of the deoxyC:riboG hybrid in the active site of the polymerase. The structural basis for this observation is not yet clear. Perhaps the deoxyC:riboG hybrid provides a particularly efficient substrate for isomerization/translocation of the polymerase or a favorable substrate for the next nucleotide addition. Mutational analyses and biochemical characterization will reveal the nature of this unexpected effect of rDNA sequence.
Methods
Generation of DNA Template.
We synthesized linear DNA fragments containing a NotI restriction site, the S. cerevisiae RNA polymerase I distal and core promoter sequences, the first 55 transcribed residues of the 35S gene mutated such that no C residues are encoded, 250 residues of rDNA from the 25S gene, the DNA sequence to be assayed, another 100 residues of rDNA from the 25S gene, an SfoI restriction site, and an XhoI restriction site, as diagrammed in Fig. 1A. These fragments served as templates for all in vitro assays. For the study of the effect of rho-independent terminator motifs on Pol I, the sequence to be assayed was the rho-independent terminator motif from the E. coli gene spy, using the sequence described in the SI Appendix from Chen et al. (19). The Terminator T-to-C mutant template was prepared similarly, except residues 5′-TTTTCTTTTCTCTTCT-3′ in the terminator motif were replaced with 5′-CCCCTCCCCTCTCCTC-3′. For the Terminator short stem loop mutant template, the residues 5′-TCTTGTCCACTACCTTGCAGTAATGCGGTGGACAGGATC-3′ in the terminator motif were replaced with 5′-CAGTCGAAAGACTG-3′. For the rDNA sequence analysis, the sequence to be assayed consisted of residues 1591–1710 of the S. cerevisiae 25S gene. The rDNA sequence T-to-C switch mutant template was prepared similarly, except residues 5′-TTATCTTTTCTTCTT-3′ from the rDNA sequence were replaced with 5′-CCACTCCCCTCCTCC-3′. In the rDNA sequence upstream scramble mutant template the residues 5′-TGGAGACGTCGGCGCGAGCCCTGGGAGGAG-3′ were replaced with 5′-CCGTAGTGCTAAGGTAACCTACAACGTGCT-3′. For the negative control, the sequence to be assayed is the 52 residue scramble sequence 5′-CGTGGCGCGGACTCGAGGAACGGACGCGAAGCTAACGGAATAGTGACTCTTC-3′. The full sequences for all linear DNA fragments are available in SI Appendix. The fragment was ligated into the pBluescript vector plasmid using the NotI and XhoI restriction sites and linearized using the SfoI restriction site. For the bioitinylated template, the linear DNA fragment was amplified by PCR with a 5′-biotinylated primer. DNA concentrations were determined using a Nanodrop 2000c spectrophotometer.
Purification of Proteins.
S. cerevisiae Pol I, CF (Rrn6p, Rrn7p, Rrn11p), and Tata binding protein (TBP) were purified as previously described (43).
S. cerevisiae Rrn3p was purified from E. coli bearing pDAS903. This plasmid consists of the pSUMO vector plasmid with the RRN3 gene cloned into it such that the sequence MHHHHHH followed by the 101-aa SUMO fusion protein was added to the N terminus of Rrn3p. Protein expression was induced by growth in phosphate-buffered TB supplemented with 0.25% vol/vol glycerol, 0.025 wt/vol glucose, and 0.1% wt/vol galactose. Cells from a 1-L culture with Abs600 = 0.6 were pelleted at 4,400 × g for 30 min. The pellet was resuspended in 50 mL breakage buffer (50 mM Tris⋅Cl, 500 mM KCl, 10 mM imidazole, 1% vol/vol glycerol pH, 7.8). The cells were lysed via a French pressure cell at 15,000 psi. The lysate was then cleared at 36,000 × g for 30 min. The supernatant was incubated with a 50% vol/vol GE Fast-Flow nickel resin slurry in breakage buffer for 2 h at 4 °C. The nickel resin was pelleted via centrifugation at 500 × g for 3 min and washed with 10 mL wash buffer (50 mM Tris⋅Cl, 200 mM KCl, 10 mM imidazole, 1% vol/vol glycerol, pH 7.8). The protein was eluted with 7.5 mL elution buffer (50 mM Tris⋅Cl, 200 mM KCl, 250 mM imidazole, 1% vol/vol glycerol, pH 7.8.) The nickel elution fraction was then supplemented with 1 mM DTT and purified SUMO protease and incubated overnight to cleave the 6xHis-SUMO tag off of Rrn3p. The nickel eluent containing now untagged Rrn3p was then loaded onto a GE Mono Q 5/50 GL anion exchange column and washed with 10 mL buffer A (50 mM Tris⋅Cl, 200 mM KCl, 10% vol/vol glycerol, pH 7.8.) The bound protein was eluted using a gradient of 0–100% buffer B (50 mM Tris⋅Cl, 1 M KCl, 10% vol/vol glycerol, pH 7.8.) All protein concentrations were determined via dual-beam spectrophotometry.
Promoter-Dependent in Vitro Transcription.
Based on a previously described in vitro transcription assay (18), standard experiments were performed at 25 °C with a volume of 20 μL. Experiments contained standard reaction buffer (17.5 mM Tris acetate pH 7.9, 87.5 mM potassium glutamate, pH 7.95, 7 mM magnesium acetate, 1.75 mM DTT, 0.035 units/μL RNase inhibitor, 0.175 mg/mL BSA, 2.6% glycerol) 169 nM CF, 169 nM TBP, 169 nM Rrn3p, 169 nM RNA polymerase I, 2.3 nM linearized DNA template, 15 μM ATP, CTP, GTP, UTP, ∼300 nM [α-32P] NTP (UTP for the rho-independent terminator studies and GTP for the 25S pause region studies), and 0.025 mg/mL heparin. Rrn3p and RNA polymerase I were incubated at 25 °C for 1 h before the transcription experiment.
Linearized DNA template was added to a 2.5-fold concentrated mixture of standard reaction buffer, followed by CF, TBP, and Rrn3P/RNA polymerase I complex. ATP, GTP, UTP, and radiolabeled nucleotide were then added to the mixture to allow for transcription of the 55 nucleotide C-less coding region for 3 min. CTP and heparin were then added to allow the polymerases to transcribe past the C-less coding region and prevent binding of unbound polymerases to the template, respectively. After the indicated amount of time, reactions were quenched with 250 μL 1 M ammonium acetate in 95% ethanol and cooled overnight at −20 °C. The nascent RNA was pelleted by centrifugation at 16,800 × g for 15 min. The pellet was resuspended in 20 μL RNA loading dye (90% formamide, 25 mM EDTA, 0.025 mg/mL bromophenol blue, pH 8.54). After 10 min of incubation, 10 μL of each sample was loaded into an 8% polyacrylamide gel, which was run for 65 min at 700 V in 1× TBE. The gel was then dried for 2 h and exposed to a phosphor screen overnight.
The in vitro transcription experiment with the biotinylated template was performed as above, except the reactions were quenched with two volumes of 38 mM EDTA in standard reaction buffer. Half of each reaction was incubated with streptavidin-coated magnetic beads for 25 min at 25 °C. The beads were then sequestered by magnet and the flowthrough was isolated. Both fractions for each time point were then combined with 250 μL 1 M ammonium acetate in 95% ethanol and resolved by polyacrylamide gel as above.
Gel Imaging and Data Analysis.
Gels were imaged using the GE Typhoon scanner. All analysis was performed using the ImageQuant TL software package. The volume of the full-length and truncated product bands was determined. The volume for each band was then normalized to radioactive nucleotide incorporation. To produce the fractional product value at each time point, the normalized volume of the truncated product band in that lane was divided by the sum of the normalized truncated product volume and the normalized full-length product volume, and multiplied by 100.
Native Elongating Transcript Sequencing for Pol I.
NETSeq was adapted for use with Pol I, based in large part on previous studies targeting Pol II (22). Detailed descriptions of the cell growth, harvest, RNA isolation, library preparation, and data analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Drs. Mike Crowley and David Crossman (Heflin Center for Genomic Sciences, University of Alabama at Birmingham) for their advice and technical assistance throughout this study; Dr. Tom Broker for providing the cell disruptor that made NETSeq possible; Dr. Mikhail Kashlev for informal conversations that led us to explore the effects of prokaryotic terminators on RNA polymerase I; and members of the D.A.S. laboratory for critical evaluation of this project. This study was funded by NIH Grants GM084946 (to D.A.S.) and AI134693 (to C.M.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809421115/-/DCSupplemental.
References
- 1.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Woolford JL, Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hein N, Hannan KM, George AJ, Sanij E, Hannan RD. The nucleolus: An emerging target for cancer therapy. Trends Mol Med. 2013;19:643–654. doi: 10.1016/j.molmed.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Koš M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grummt I. Life on a planet of its own: Regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 6.Stefanovsky VY, Langlois F, Bazett-Jones D, Pelletier G, Moss T. ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF. Biochemistry. 2006;45:3626–3634. doi: 10.1021/bi051782h. [DOI] [PubMed] [Google Scholar]
- 7.Panov KI, Friedrich JK, Russell J, Zomerdijk JC. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006;25:3310–3322. doi: 10.1038/sj.emboj.7601221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, French SL, Beyer AL, Schneider DA. The transcription factor THO promotes transcription initiation and elongation by RNA polymerase I. J Biol Chem. 2016;291:3010–3018. doi: 10.1074/jbc.M115.673442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci USA. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider DA, et al. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iost I, Dreyfus M. The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J. 1995;14:3252–3261. doi: 10.1002/j.1460-2075.1995.tb07328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: The role of the RNA hairpin. Proc Natl Acad Sci USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 14.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 15.Resnekov O, Aloni Y. RNA polymerase II is capable of pausing and prematurely terminating transcription at a precise location in vivo and in vitro. Proc Natl Acad Sci USA. 1989;86:12–16. doi: 10.1073/pnas.86.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seiberg M, Kessler M, Levine AJ, Aloni Y. Human RNA polymerase II can prematurely terminate transcription of the adenovirus type 2 late transcription unit at a precise site that resembles a prokaryotic termination signal. Virus Genes. 1987;1:97–116. doi: 10.1007/BF00125689. [DOI] [PubMed] [Google Scholar]
- 17.Reines D, Wells D, Chamberlin MJ, Kane CM. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987;196:299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- 18.Schneider DA. Quantitative analysis of transcription elongation by RNA polymerase I in vitro. Methods Mol Biol. 2012;809:579–591. doi: 10.1007/978-1-61779-376-9_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YJ, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds R, Bermudez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 21.Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of bacterial transcription termination: All good things must end. Annu Rev Biochem. 2016;85:319–347. doi: 10.1146/annurev-biochem-060815-014844. [DOI] [PubMed] [Google Scholar]
- 22.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yakhnin AV, Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc Natl Acad Sci USA. 2002;99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larson MH, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 29.Tastemel M, et al. Transcription pausing regulates mouse embryonic stem cell differentiation. Stem Cell Res (Amst) 2017;25:250–255. doi: 10.1016/j.scr.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson KS, von Hippel PH. Stability of Escherichia coli transcription complexes near an intrinsic terminator. J Mol Biol. 1994;244:36–51. doi: 10.1006/jmbi.1994.1702. [DOI] [PubMed] [Google Scholar]
- 32.Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 2012;31:630–639. doi: 10.1038/emboj.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider DA, Gaal T, Gourse RL. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc Natl Acad Sci USA. 2002;99:8602–8607. doi: 10.1073/pnas.132285199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 35.Appling FD, Lucius AL, Schneider DA. Transient-state kinetic analysis of the RNA polymerase I nucleotide incorporation mechanism. Biophys J. 2015;109:2382–2393. doi: 10.1016/j.bpj.2015.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner F. A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. doi: 10.1016/j.jmb.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbert KM, et al. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viktorovskaya OV, Appling FD, Schneider DA. Yeast transcription elongation factor Spt5 associates with RNA polymerase I and RNA polymerase II directly. J Biol Chem. 2011;286:18825–18833. doi: 10.1074/jbc.M110.202119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider DA, et al. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci USA. 2006;103:12707–12712. doi: 10.1073/pnas.0605686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson SJ, et al. The transcription elongation factor Spt5 influences transcription by RNA polymerase I positively and negatively. J Biol Chem. 2011;286:18816–18824. doi: 10.1074/jbc.M110.202101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imashimizu M, et al. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015;16:98. doi: 10.1186/s13059-015-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bedwell GJ, Appling FD, Anderson SJ, Schneider DA. Efficient transcription by RNA polymerase I using recombinant core factor. Gene. 2012;492:94–99. doi: 10.1016/j.gene.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.