Significance
In the E2 → E1 transition of Ca2+-ATPase, low-affinity Ca2+-binding sites are converted to high affinity by releasing multiple protons bound to some of the Ca2+-coordinating residues. This is one of the transitions comprised of very large and complex structural changes, whose sequence of events remains unknown. Two mutant crystal structures of Glu309, the gating residue to cytoplasmic Ca2+, reveal how such a transition takes place and what is the critical structural component pertinent to the E2 state.
Keywords: ion pump, SERCA, deprotonation, quantum chemical calculation
Abstract
Ca2+-ATPase of sarcoplasmic reticulum (SERCA1a) pumps two Ca2+ per ATP hydrolyzed from the cytoplasm and two or three protons in the opposite direction. In the E2 state, after transferring Ca2+ into the lumen of sarcoplasmic reticulum, all of the acidic residues that coordinate Ca2+ are thought to be protonated, including the gating residue Glu309. Therefore a Glu309Gln substitution is not expected to significantly perturb the structure. Here we report crystal structures of the Glu309Gln and Glu309Ala mutants of SERCA1a under E2 conditions. The Glu309Gln mutant exhibits, unexpectedly, large structural rearrangements in both the cytoplasmic and transmembrane domains, apparently uncoupling them. However, the structure definitely represents E2 and, together with the help of quantum chemical calculations, allows us to postulate a mechanism for the E2 → E1 transition triggered by deprotonation of Glu309.
Ca2+-ATPase of sarcoplasmic reticulum (SERCA1a) translocates two Ca2+ from the cytoplasm per ATP hydrolyzed and thereby establishes a 10,000-fold concentration gradient of Ca2+ across the membrane. This active transport is conventionally explained by the E1/E2 theory. In E1, the transmembrane Ca2+-binding sites have high affinity for Ca2+; in E2, the binding sites have low affinity for Ca2+ and are stabilized by H+ (1, 2). In the E2 → E1 transition, low-affinity transmembrane Ca2+-binding sites release two (or three) protons and become high affinity for Ca2+ (3). This transition occurs spontaneously under physiological conditions but accompanies large and complex structural changes in both the cytoplasmic and transmembrane domains (4).
To explore how these structural changes take place and in what order, mutant structures are helpful. One of the most interesting point mutations is the Gln substitution of Glu309, the gating residue located in the unwound part (“hinge”) connecting the cytoplasmic and luminal halves of the M4 helix (M4C and M4L) (5). This residue caps the second bound Ca2+ (6) and is thought to transmit the Ca2+-binding signal to the phosphorylation site, presumably by changing the path (angle and elevation) of M4C. Substitutions of this residue totally abolish the transport activity (7). In E2, Glu309 is expected to be protonated (1) to stabilize the empty binding site by forming a hydrogen bond with the main chain carbonyl of Val304 at the top of M4L. Thus, under E2 conditions, Glu309Gln substitution is expected to cause little change, but limited proteolysis shows that the mutant under E2 conditions takes an E1-like conformation (5).
Here we describe a crystal structure of this mutant at 2.5-Å resolution, as well as that of Glu309Ala mutant at 3.4 Å, under E2 conditions in the presence of thapsigargin (TG), a very potent inhibitor that fixes the ATPase in E2 (8, 9). As described before (9, 10), there are four prominent features that characterize the E2 structure. They are (i) closed and compact headpiece, (ii) highly curved (“bowed”) M5 helix, (iii) axially lower position of the M4 helix, and (iv) rotation of the unwound part of the M6 helix. This crystal structure of Glu309Gln [designated as Glu309Gln(TG)] is very different from any E2 crystal structures published thus far and, at first glance, closer to that of E1·Mg2+ (4). In this structure, the cytoplasmic domains assume an E1-like arrangement, whereas the transmembrane domain shows a unique but definitely an E2 configuration. Thus, this seemingly insignificant substitution causes drastic rearrangements in both the cytoplasmic and transmembrane domains, apparently uncoupling them. More importantly, the crystal structures, combined with quantum chemical calculations, reveal the critical structural features for establishing the E2 state and give insights into how the E2 → E1 transition proceeds.
Results
Crystal Structures of the Glu309Ala and Glu309Gln Mutants.
The Glu309Ala mutant was crystallized under essentially the same E2(TG) conditions as those for the native enzyme (9), and the crystal structures themselves are almost identical (RMSD = 0.20 Å; SI Appendix, Fig. S1D and Table S1). For instance, the M2 helix is partly unwound as in the native enzyme, consistent with a limited proteolysis study. Even the path of the 308PEGL motif in the hinge connecting M4C and M4L is identical. The only difference is that the density corresponding to the carboxyl to the Cβ atom of the Glu309 side chain is absent (SI Appendix, Fig. S1A). The Val304 carbonyl, which is expected to form a hydrogen bond with protonated Glu309 in the native enzyme, appears to be stabilized in the mutant most likely by an additional water molecule (SI Appendix, Fig. S1B). Thus, it is evident that the side chain of Glu309 is not indispensable in forming the E2 structure.
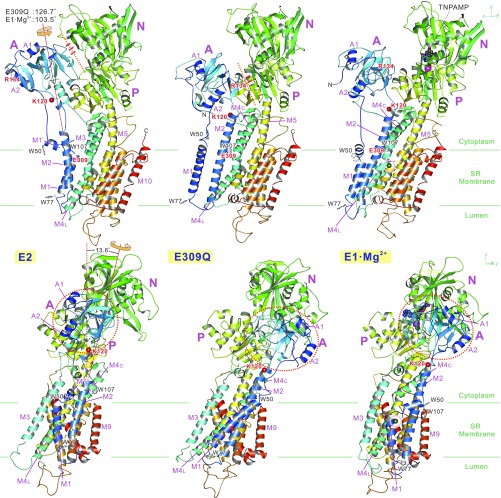
The Glu309Gln mutant formed crystals under E2(TG) conditions only in the presence of decylmaltoside, a detergent, in addition to octaethyleneglycol mono-n-dodecylether (C12E8) used for solubilization. Two molecules of decylmaltoside per ATPase were identified in the crystal structure (SI Appendix, Fig. S2): one in a cleft in the N-domain (11) and the other in the transmembrane region in the cleft between the M2 and M6 helices, normally occupied by a phospholipid molecule (12, 13). More striking differences from the structure of the native enzyme are that the headpiece appears to be open and the M5 helix almost straight (Fig. 1). Furthermore, the A-domain takes an extreme azimuthal position, ∼125° rotated from that in authentic E2 (Fig. 1, SI Appendix, Fig. S3, and Movie S1). This position is similar to that in E1·Mg2+ (4), but even 23° more rotated (Fig. 1 and Movie S2).
Fig. 1.
Ribbon representation of the Glu309Gln(TG) crystal structure compared with those of native ATPase (SERCA1a) in E2(TG) and in E1·Mg2+. Viewed in two orthogonal directions. (Top) Front views. (Bottom) Viewed from the left-hand side. Models are aligned with the M7-M10 helices. Colors change gradually from the amino terminus (blue) to the carboxyl terminus (red). Green horizontal lines indicate the approximate boundaries of the hydrophobic core of the lipid bilayer. The axes of rotation of the A-domain (thin red lines) with respect to the P-domain are common to both Glu309Gln(TG) and E1·Mg2+ and are shown for the transition from E2(TG). K120 (small red balls) is the cleavage site of proteinase K yielding the 95K band (23). Red dotted lines and short red bars indicate the interface between the A- and N-domains and hydrogen bonds between them. Also see Movies S1 and S2 for structural changes.
Cytoplasmic Domains in Glu309Gln(TG).
The compact headpiece has been regarded as one of the hallmarks of the E2 state (9). In the crystal structure of the native enzyme in E2, the A- and N-domains are connected by seven hydrogen bonds, including two salt bridges (9), forming an apparently rigid headpiece (Fig. 1). Indeed, the headpiece appeared always the same if in E2, although overall appearance of the ATPase varied depending on the crystal packing and inhibitors used (14). Soaking of ATP or their derivatives did not destroy the compact headpiece (15, 16). Thus, the energy barrier for opening the headpiece would be high, and a large-scale rearrangement of the transmembrane helices realized in E2 → E1·Mg2+ (4) would be a prerequisite.
In fact, the headpiece in the present structure is not fully opened as in E1·Mg2+. New hydrogen bonds between the A- and N-domains are formed at the very periphery of the A-domain (SI Appendix, Fig. S3B), similar to those in E1·AMPPCP (SI Appendix, Fig. S3C), in addition to those with the P-domain [between Arg139 (A) and the Gly721 carbonyl (P)], stabilizing the extreme azimuthal position (17) (SI Appendix, Fig. S3C). These new hydrogen bonds are reminiscent of the “electrostatic catch” observed in E2·MgF42− on the other side of the A-domain, between an A-domain loop containing Arg198 and residues on the P-domain (18).
Rearrangement of Transmembrane Helices.
The ∼125° rotation of the A-domain requires a large rearrangement of transmembrane helices (Figs. 2 and 3 and SI Appendix, Fig. S4; most accessible in Movie S1), most conspicuous with M1 and M2, connected directly to the A-domain. M1, now a straight helix like that in E1·2Ca2+ (6), is highly inclined (Fig. 1), occupying a unique lateral position (Fig. 3). M2 is now contiguous all of the way from Phe87 to Tyr122, taking an E1-like position parallel to the M4 helix, forming many van der Waals contacts and two hydrogen bonds unique to this mutant structure (Fig. 2). In native ATPase in E2, it is unwound between Asn111 and Ala115 (19) and detached from M4. These differences with M2 will explain the difference in susceptibility to proteinase K attack at Lys120 (5). As the axis of rotation of the A-domain is inclined by 13.6° from the membrane normal (Fig. 1), the 125° rotation causes large downward shifts of the M1 and M2 helices (∼10 Å at the A-domain–M2 and –M1 junctions). Large inclination of M1 and winding of M2 appear to be required for compensating such shifts to prevent extrusion of the transmembrane helices from the bilayer.
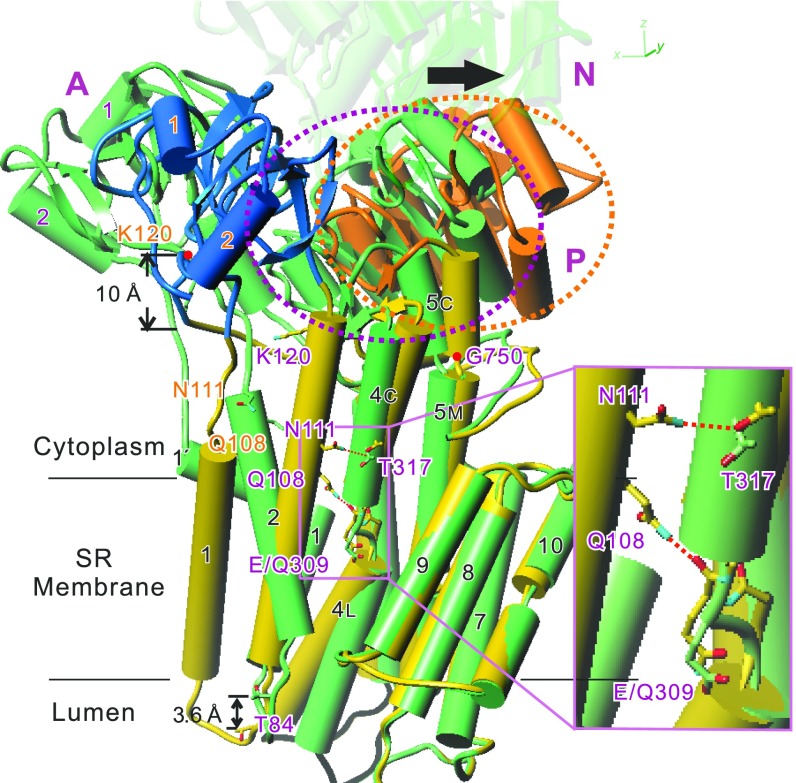
Fig. 2.
Superimposition of atomic models for the Glu309Gln mutant and native SERCA1a in E2(TG). Aligned with the M7-M10 helices. M6 is removed for clarity. The N-domain of Glu309Gln(TG) is removed and that of native ATPase is made transparent. The model for Glu309Gln(TG) appears in yellow with the A-domain in blue, the P-domain in orange, and native SERCA1a in green. Orange and purple dotted circles enclose the P-domain of Glu309Gln(TG) and native ATPase, respectively. M5 is a contiguous helix, but shown with three cylinders (M5C, M5M, and M5L). Note that hydrogen bonds are formed in Glu309Gln(TG) between the side chains of Asn111 and Thr317 and between the side chain of Gln108 and the main chain carbonyl of Gln309 (pink dotted lines). Also note that the downward shift of M2 is large (∼10 Å at Lys120) at the cytoplasmic end but reduced to ∼4 Å (at Thr84) at the other end due to winding of the M2 helix in the mutant. The arrow shows the movement of the P-domain (dotted circles) caused by the Glu309Gln substitution. Inset shows a magnified view of the boxed area.
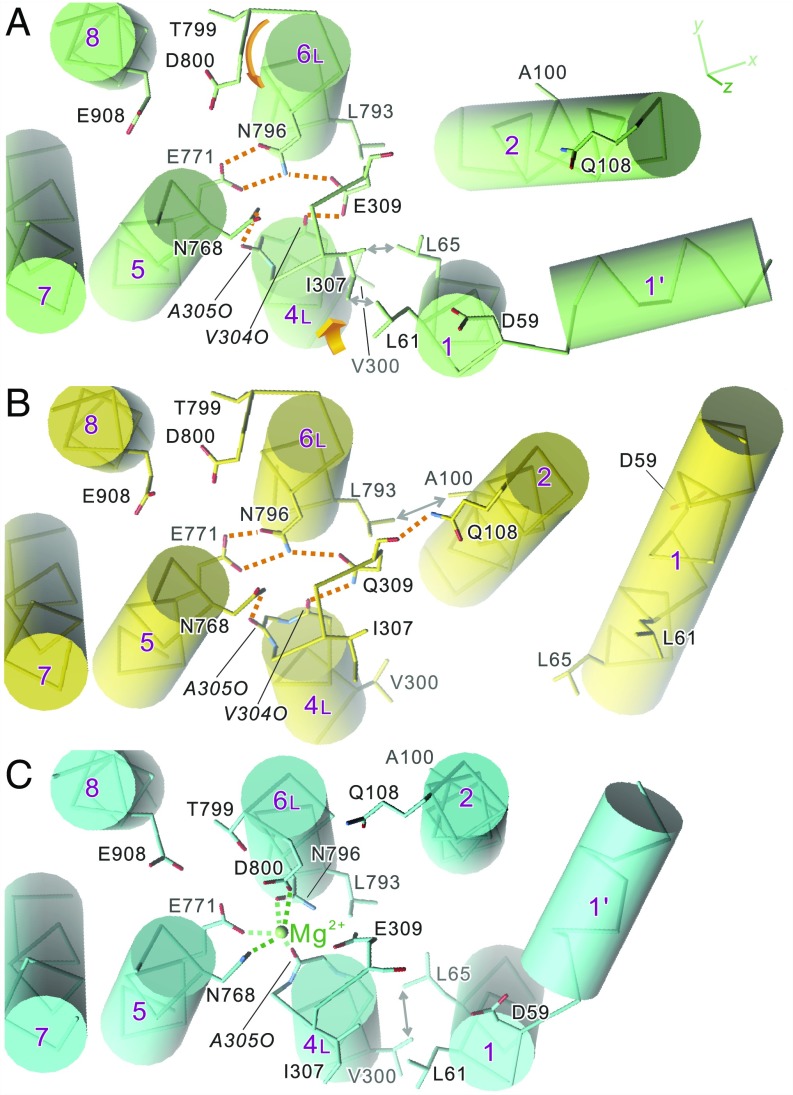
Fig. 3.
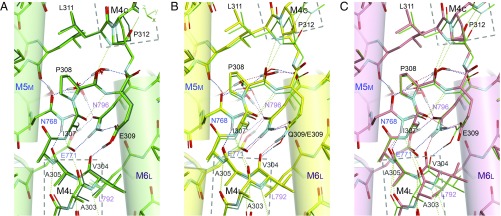
Disposition of the transmembrane helices in crystal structures of SERCA1a: native ATPase in E2(TG) (A), Glu309Gln mutant (B), and native ATPase in E1·Mg2+ (C). Viewed from the cytoplasmic side approximately perpendicular to the membrane. Orange dotted lines represent likely hydrogen bonds, and those in green represent Mg2+ coordination. Orange arrows in A indicate the directions to which the unwound part of M6 rotates and M4L moves in the E2 → E1·Mg2+ transition. Note that dispositions of the side chains of the Ca2+-coordinating residues (Glu309 on M4; Asn768 and Glu771 on M5; Asn796, Thr799, and Asp800 on M6; and Glu908 on M8) in the Glu309Gln mutant (B) are exactly the same as those in native enzyme (A), although the M1 and M2 helices occupy distinctly different positions. Double-headed arrows indicate van der Waals contacts.
The M5 helix appears more or less straight, again at first glance similar to that in E1·Mg2+ (Fig. 1). In the E2 → E1·Mg2+ transition, such straightening of the M5 helix causes a change in inclination of the P-domain and an upward shift of the M4 helix (4) because the cytoplasmic segments of M4 (M4C) and M5 (M5C; Phe740 to Gly750) are both integrated into the P-domain (6, 9) (Fig. 2). A similar event was expected to occur here, but the difference in inclination of the P-domain (∼15°) is substantially smaller than that in the E2 → E1·Mg2+ transition (∼24°), reflecting a small change in inclination of M4C (∼5°). Thus, M4C keeps very much the same axial position, stabilized by the two hydrogen bonds with the residues on M2 (Fig. 2). However, the horizontal shift of the P-domain in the M1-M10 direction exceeds 8 Å at the A-P interface (at Thr725), enough to destroy the compact headpiece. As M5 contains two Gly residues (Gly750 and Gly770) that can work as flexible hinges (9), M5C could follow the movement of M4C, independent of other segments of M5. In fact, the luminal end of M5 (M5L; Gly770-Ala780) is fixed by hydrogen bonds between (protonated) Glu771 (M5) and Asn796 (M6), exactly the same as in authentic E2 (Fig. 3), and the middle part (M5M) changes inclination only 3.8°. This is a critical difference from the situation in E1·Mg2+, where the hydrogen bonds between Glu771 and Asn796 are absent.
The luminal half of the M4 helix (M4L) is inclined by ∼16° toward M3 with an apparent pivot around the Ala305 carbonyl (Fig. 4 and Movie S1). This is because the Ala305 carbonyl is in van der Waals contact with Asn768Cβ and, at the same time, hydrogen-bonded with the Asn768 side chain amide. As Asn768 is located very close to the second hinge (Gly770) connecting M5M and M5L, it cannot move as long as Glu771 forms hydrogen bonds with Asn796 on M6. Conversely, due to the hydrogen bonds between them, the unwound part of M6 takes exactly the same path as that in authentic E2 (Fig. 3 and SI Appendix, Fig. S4).
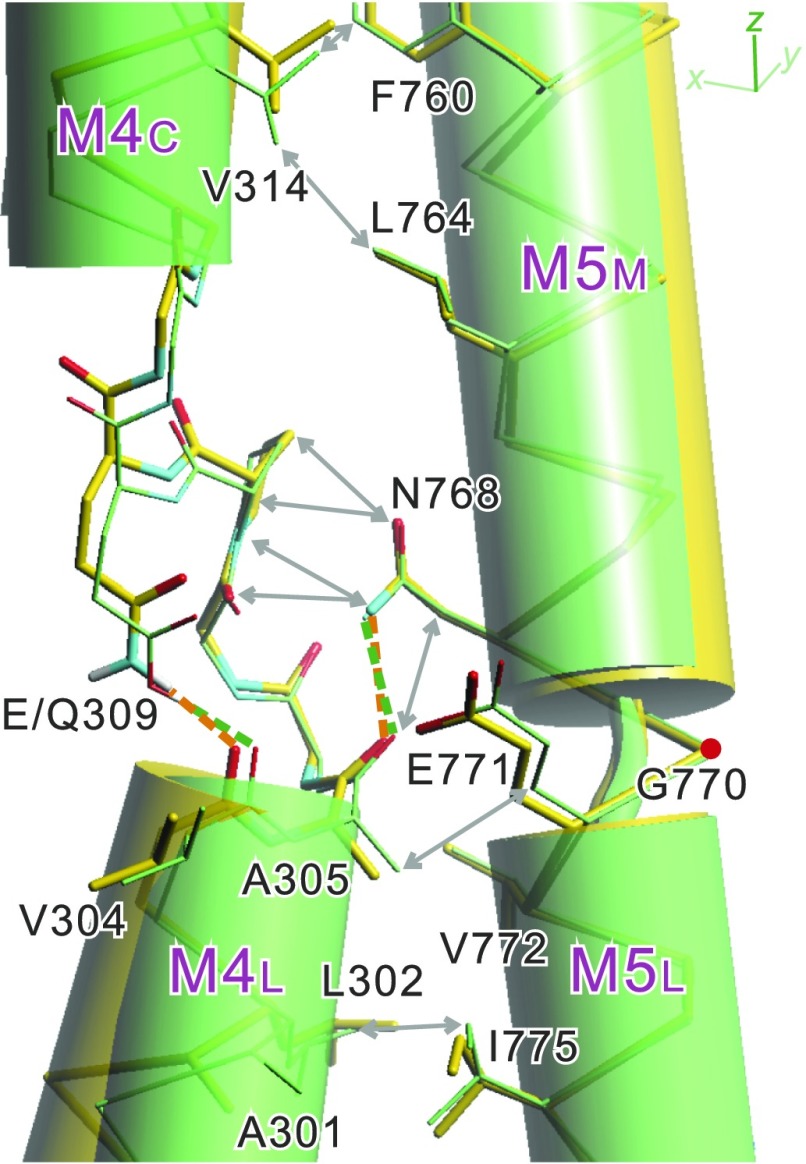
Fig. 4.
Details around the Glu/Gln309 side chain and the difference in inclination of the two segments of the M4 helix. Superimpositions of atomic models for native ATPase in E2(TG) (green) and the Glu309Gln mutant (yellow). Aligned with the M7-M10 helices. Green and orange broken lines represent likely hydrogen bonds. Double-headed arrows indicate van der Waals contacts between the residues on the M4 and M5 helices.
Thus, M4L keeps a low axial position (Fig. 2), and the residues to coordinate Ca2+—namely, Glu/Gln309 on M4, Asn768 and Glu771 on M5, Thr799 and Asp800 on M6, and Glu908 on M8—all occupy the same positions as those in authentic E2 (Fig. 3 and SI Appendix, Fig. S4). Accordingly, the protonation state of the acidic residues must be the same, and hence, by definition, this mutant structure represents an E2 state.
We therefore conclude that establishing the E2 state does not depend on (i) the Glu309 side chain, (ii) a compact headpiece, (iii) bowing of the M5 helix toward M1, (iv) bent conformation of M1, or (v) partial unwinding of the M2 helix. Rather, we see that the path of the unwound part of M6 and the hydrogen bonds between Glu771 and Asn796 are maintained in the mutant structure. These two are related, as the hydrogen bonds between them fix the path of the unwound part of M6.
What Determines the Path of M4 in E2.
Then, we ought to address what has changed the inclination of M4C. The difference must arise from the substitution of Glu309 with Gln, although the Gln substitution would hardly affect the structure, as the Glu309 carboxyl is expected to be protonated in E2 (1). We notice, however, that the distance between the oxygen/nitrogen of Glu/Gln-309 and the carbonyl oxygen of Val304 is different: 2.8 Å in the native ATPase structure and 3.0 Å in that of the mutant (SI Appendix, Table S2). The 2.8-Å distance is too short for (partially) charged oxygen atoms to be juxtaposed, suggesting that Glu309 is indeed protonated, and a hydrogen bond is formed between them.
Then, the question to be addressed is whether there is any geometrical difference between the two types of hydrogen bond: the one between a carbonyl and a protonated carboxyl and the other between a carbonyl and an amide. Quantum chemical calculations using coupled-cluster single-double and perturbative triple [CCSD(T) (20)] showed that the optimal distance for an amide–carbonyl hydrogen bond is 3.16 Å, 0.33 Å longer than that for a protonated carboxyl–carbonyl hydrogen bond (SI Appendix, Fig. S5), corroborating the crystal structures. This difference seems small and easily accommodated by a slight change in side chain conformation of Glu/Gln309.
To go further, we carried out quantum chemical energy minimization of the atomic models derived by X-ray crystallography. The minimizations converged without difficulty and provided us with atomic models around Glu309 with explicit hydrogens and protons. They showed very small deviations from the original models (Fig. 5A and SI Appendix, Figs. S6–S8 and Table S2), if we assume that the Val304 carbonyl is hydrogen-bonded with protonated Glu309 in native E2 and with water in Glu309Ala (SI Appendix, Fig. S1B). The hydrogen bond distances between the Glu/Gln309 side chain and the Val304 carbonyl were very close to those in the original atomic models (2.62 vs. 2.77 Å for the native ATPase and 2.97 vs. 3.02 Å for the mutant; SI Appendix, Table S2).
Fig. 5.
Quantum chemically optimized atomic models of SERCA1a around Glu/Gln309. (A) Superimposition of atomic models derived from the crystal structure of native ATPase in E2(TG), before (thin lines) and after (sticks) energy minimization by DFT-D, assuming a protonated Glu309. (B) Superimposition of the optimized model of Glu309Gln(TG) (sticks) and that of native ATPase in E2(TG) (thin lines). (C) Superimposition of energy-minimized models of native ATPase with deprotonated (sticks) and protonated (thin lines) Glu309. Aligned with the residues on the M5 and M6 helices, the Cα atoms of which are spatially fixed in the optimization. Dotted lines represent hydrogen bonds. Viewed approximately parallel to the membrane. The side chain of Ile307 is removed. Stereo pictures are provided in SI Appendix, Figs. S7A and S9 A and B, respectively.
We noticed that there are many constraints on the side chain conformation of Glu309. First, the other oxygen atom of the carboxyl group of Glu309 is fixed by a hydrogen bond with Asn796 (Fig. 5A). All of the side chain atoms of Glu309 are in van der Waals contacts with surrounding residues (SI Appendix, Fig. S8). A slight change in inclination of M4L, on which Val304 is located, may appear to accommodate the difference. The inclination is indeed changed 16°, but in a direction that shortens the distance, with an apparent pivoting point around the Ala305 carbonyl (Fig. 4). This is because M4L is already in tight contact with M5 (Fig. 4) and cannot move toward M5. As a result, Val304 carbonyl is displaced slightly upward (0.4 Å). Also, two main chain hydrogen bonds (Ala303O-Ile307N and Pro308O-Leu311N) across the unwound part of M4 move M4C and M4L together (Fig. 5).
The difference in hydrogen bond distance is accommodated, in reality, by slightly inclining M4C (∼5°; Fig. 2). This small change is amplified by the M4C helix, enough to move the P- and the N-domains >8 Å away from the A-domain, breaking apart the compact headpiece. The new A-domain position brings M2 toward M4. A new hydrogen bond between Gln108 (M2) and the Gln309 main chain carbonyl (Fig. 2) stabilizes M4C shifted ∼1 Å upward (Fig. 4), at a position possibly higher than that if Gln108 were not there.
Consequence of the Deprotonation of Glu309.
Is Glu309 in E2 really protonated? The answer is yes, as a quantum chemical energy minimization of the atomic model around Glu309 caused little change if Glu309 was protonated but large deviations if not (Fig. 5 and SI Appendix, Figs. S7, S9, and S10 and Table S2). If deprotonated, due to electrostatic repulsion, even the Glu309 main chain detached by 0.4 Å from the Val304 carbonyl. As a result, the entire M4C segment (up to Pro312 in this calculation) is inclined as in the Glu309Gln mutant, partially opening the hinge. This predicted change in inclination is small (2.2°; Fig. 5C), yet it is likely to be enough to open the compact headpiece as the movement at the end of M4C becomes 1.5 Å. Also, the shift of the Glu309 main chain (0.4 Å) compares well with the difference in hydrogen bond distance between a carbonyl and an amide or protonated carboxyl. What is interesting is that Asn796 now forms a hydrogen bond with the Val304 carbonyl, disrupting the one with the Glu771 carboxyl.
This hydrogen-bonding pattern is critically different from that in Glu309Ala, in which the Val304 carbonyl is stabilized without disturbing other residues by, most likely, an additional water molecule (SI Appendix, Fig. S7C). Even when the additional water molecule was excluded in the calculation, the Asn796 amide formed a hydrogen bond with Val304, still keeping the hydrogen bonds between Asn796 and Glu771 intact. In native ATPase, such a compensating water molecule cannot enter the cavity due to the Glu309 side chain.
Discussion
As the E2 state should be denoted, more explicitly, as E2·nH+, protonation states of the acidic residues that coordinate Ca2+ in E1·2Ca2+ must be the most important property characterizing the E2 state. Those Ca2+-coordinating residues in Glu309Gln occupy identical positions to those in authentic E2, indicating that the mutant structure described here represents an E2 state. This arrangement of the acidic residues requires a low axial position of M4 and the unwound part of M6 to take a path that brings the Asn796 side chain into the Ca2+-binding cavity. As the axial position of the M4 helix is directly related to the inclination of the P-domain, we previously thought that a compact headpiece and, accordingly, a highly curved M5 are absolute requirements for E2. We were therefore surprised to see that in the Glu309Gln mutant structure the headpiece is open and the M5 helix more or less straight, yet the E2 state is maintained with only a small change in inclination of M4.
Although not recognized before, the axial position of M4 and the path of the unwound part of M6 are strongly linked. Due to the presence of a Pro residue (Pro803), M6 cannot form a proper α-helix and is partially unwound [or forming a π helix as in E1·2Ca2+ (6)] between Leu797 and Gly801. The unwound part contains two Ca2+-coordinating residues, Thr799 and Asp800, and changes its path and axial position depending on the inclination of M5 (Movie S3). In E1·2Ca2+, in which M5 is even slightly bent toward M10, Thr799 and Asp800 coordinate site I Ca2+ (6). In E2, M5 is deeply inclined toward M1, and the unwound part is rotated clockwise so that Asp800 takes the position of Thr799, bringing in Asn796 to form hydrogen bonds with (protonated) Glu771, and stabilizing the empty Ca2+-binding sites (9).
The rotation is related to the curvature/inclination of M5. When M5 inclines toward M1 (or, more rigorously, into the space between M2 and M6) in the E1·Mg2+ → E2 transition (i.e., backward reaction), the middle section of M5 (M5M) containing Tyr763 and Leu764 pushes the Thr799 and Asp800 side chains to rotate the unwound part of M6 (Movie S3). This rotated configuration appears to be energetically more favorable if Glu771 is protonated and forms two hydrogen bonds with Asn796. In Glu309Gln(TG), the M5 helix appears straighter (M5M 3.8° less inclined), but not as much as that in E1·Mg2+ (12.9° less inclined; Fig. 1). Therefore the path of the M5 helix in Glu309Gln(TG) is actually closer to that in E2 of native ATPase.
When Glu771 becomes deprotonated, Asn796 would detach from Glu771 and the unwound part of M6 change its path so that it protrudes further toward M4C (SI Appendix, Fig. S4). Hydrogen bond formation between Asn796 and the Val304 carbonyl could be a driving force for the rotation of M6. As a result, primarily, Asp800 and Thr805 push Val314 on M4C so that M4C takes a new lateral position detached from M5 and M6 (Movie S3) and more inclined toward M3 near the hinge (Movie S2), displacing Ala305 from direct contact and hydrogen bonding with Asn768 (Fig. 3). Then, an upward movement of M4 is allowed. This is what we see in the E2 → E1·Mg2+ transition (4, 21).
Furthermore, as long as the unwound part of M6 takes the path realized in E2, the A-domain cannot take its position in E1·Mg2+, as residues on M2 would cause steric clashes with those on M6 [in particular, Leu797 (M6) and Asn101 (M2); SI Appendix, Fig. S4]. Thus, we now see important roles of the unwound part of M6 in regulating the E2 → E1 transition.
The point to be emphasized is that, for the unwound part of M6, only the E2 configuration is possible when M5 inclines toward M1 and stable if and only if Glu771 is protonated. As long as the two hydrogen bonds between Asn796 and protonated Glu771 are in place, M4 and M5/M6 are in tight van der Waals contacts, reinforced by a hydrogen bond between the Asn768 amide and the Ala305 carbonyl (Fig. 4), preventing an upward movement of M4. Thus, bowing of M5 appears to be a means to ensure the protonated E2 (i.e., E2·nH+) state and lock the configuration by arranging the cytoplasmic domains to form a compact headpiece.
A charge movement study showed that proton release takes place at substantially higher pH in the Glu309Gln and Glu309Ala mutants than in wild-type ATPase (apparent pKa of 7.8 vs. 6.7) (22). As we showed that the arrangement of the Ca2+-binding residues is identical to the native ATPase in both mutants, the measurements indicate that the acidic residues other than Glu309 [i.e., Glu771 and, presumably, Asp800 (1)] have higher apparent pKa than that of Glu309 and do not release protons at lower than pH 7.2. However, all proton releases take place at once around pH 6.7, as the charge movement curve shows only one rise (22). This observation indicates that, around pH 7, deprotonation of Glu309 is prerequisite to the proton releases from the other acidic residues. In other words, the deprotonation of Glu309 is likely to be the first chemical event in the E2 → E1 transition.
Quantum chemical simulation (Fig. 5C and SI Appendix, Figs. S9B and S10B) showed that deprotonation of Glu309 could indeed cause disruption of one of the two hydrogen bonds between (protonated) Glu771 and Asn796, in addition to the opening of the hinge between M4C and M4L. This is because the new negative charge arising from the deprotonation has to be compensated for by bringing in the Asn796 amide. Resultant structural changes would be much more profound than that by merely opening the hinge because the latch (i.e., the hydrogen bonds between Glu771 and Asn796) that prevents the unwound part of M6 from taking a different path would become unlocked, triggering the transition into E1. As such change did not take place in Glu309Gln, nor in Glu309Ala, introduction of a negative charge is the cause of such events. Conversely, because those critical hydrogen bonds can be realized only when Glu771 is protonated, its protonation is the most critical structural feature in realizing the E2 state. It certainly makes sense, because E2 should actually be denoted as E2·nH+.
We have learned that the overall organization of SERCA1a is very sensitive to the inclination of M4C, which can be changed in two ways. Increasing the length of the hydrogen bond linking the two halves of M4 (as in the Glu309Gln substitution) is one way, and introducing a negative charge (by deprotonation of Glu309) to increase electrostatic repulsion between the two halves is the other way. Only the latter is possible (but inevitably causing other effects from those seen with the mutation) within the reaction cycle of the native pump and is realized in the E2 → E1 transition. Opening of the headpiece per se does not cause the transition, but would certainly increase the likelihood. If the opening happens in E2, the structure of SERCA1a would appear very much the same as that presented here for the Glu309Gln mutant.
Thus, the scenario for the E2 → E1 transition is as follows: (i) Spontaneous deprotonation of Glu309 opens the hinge in M4 by electrostatic repulsion, which in turn opens the headpiece. Now the A-domain rotates horizontally ∼120° and takes an E1-like azimuthal position. M5 straightens somewhat, tending to that in E1. (ii) Deprotonation of Glu309 at the same time pulls the Asn796 side chain toward Val304, destroying one of the two hydrogen bonds between Glu771 (M5) and Asn796 (M6), and the latch that prevents the unwound part of M6 from taking a different path is unlocked. (iii) An anticlockwise rotation of the unwound part brings Thr799 and Asp800 into the Ca2+-binding cavity, pushing M4C, primarily through Val314. (iv) M4C inclines toward M3 near the hinge, detaching from M5 and M6. (v) M5 now straightens completely, bringing the entire M4 helix upward to the E1 position.
Materials and Methods
Details of all methods used in this study are described in SI Appendix, Materials and Methods, including (i) construction, expression, and purification of SERCA1a mutants; (ii) crystallization; (iii) data collection and structure determination; (iv) quantum chemical refinement of atomic models; and (v) quantum chemical calculation of the energy profile of the hydrogen bond.
Supplementary Material
Acknowledgments
We thank Drs. K. Hasegawa and H. Okumura (Japan Synchrotron Radiation Research Institute), R. Kanai, and Y. Kabashima for their help in data collection at BL41XU of SPring-8. We are indebted to Dr. D.B. McIntosh for his help in improving the manuscript. This work is a part of long-term projects (2016A0133 and 2018A0144) at SPring-8 and was supported in part by Japan Society for the Promotion of Science KAKENHI Grant JP13311708 and JP16804623 (to C.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 5ZMV (Glu309Ala[TG]) and 5ZMW (Glu309Gln[TG])].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815472115/-/DCSupplemental.
References
- 1.Obara K, et al. Structural role of countertransport revealed in Ca(2+) pump crystal structure in the absence of Ca(2+) Proc Natl Acad Sci USA. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiesi M, Inesi G. Adenosine 5′-triphosphate dependent fluxes of manganese and hydrogen ions in sarcoplasmic reticulum vesicles. Biochemistry. 1980;19:2912–2918. doi: 10.1021/bi00554a015. [DOI] [PubMed] [Google Scholar]
- 3.Tadini Buoninsegni F, Bartolommei G, Moncelli MR, Inesi G, Guidelli R. Time-resolved charge translocation by sarcoplasmic reticulum Ca-ATPase measured on a solid supported membrane. Biophys J. 2004;86:3671–3686. doi: 10.1529/biophysj.103.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyoshima C, et al. Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state. Nature. 2013;495:260–264. doi: 10.1038/nature11899. [DOI] [PubMed] [Google Scholar]
- 5.Inesi G, Lewis D, Toyoshima C, Hirata A, de Meis L. Conformational fluctuations of the Ca2+-ATPase in the native membrane environment. Effects of pH, temperature, catalytic substrates, and thapsigargin. J Biol Chem. 2008;283:1189–1196. doi: 10.1074/jbc.M707189200. [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 7.Andersen JP. Dissection of the functional domains of the sarcoplasmic reticulum Ca(2+)-ATPase by site-directed mutagenesis. Biosci Rep. 1995;15:243–261. doi: 10.1007/BF01788358. [DOI] [PubMed] [Google Scholar]
- 8.Sagara Y, Inesi G. Inhibition of the sarcoplasmic reticulum Ca2+ transport ATPase by thapsigargin at subnanomolar concentrations. J Biol Chem. 1991;266:13503–13506. [PubMed] [Google Scholar]
- 9.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 10.Toyoshima C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim Biophys Acta. 2009;1793:941–946. doi: 10.1016/j.bbamcr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Akin BL, Hurley TD, Chen Z, Jones LR. The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum. J Biol Chem. 2013;288:30181–30191. doi: 10.1074/jbc.M113.501585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drachmann ND, et al. Comparing crystal structures of Ca(2+) -ATPase in the presence of different lipids. FEBS J. 2014;281:4249–4262. doi: 10.1111/febs.12957. [DOI] [PubMed] [Google Scholar]
- 13.Norimatsu Y, Hasegawa K, Shimizu N, Toyoshima C. Protein-phospholipid interplay revealed with crystals of a calcium pump. Nature. 2017;545:193–198. doi: 10.1038/nature22357. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi M, Kondou Y, Toyoshima C. Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors. Proc Natl Acad Sci USA. 2007;104:5800–5805. doi: 10.1073/pnas.0700979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyoshima C, Yonekura S, Tsueda J, Iwasawa S. Trinitrophenyl derivatives bind differently from parent adenine nucleotides to Ca2+-ATPase in the absence of Ca2+ Proc Natl Acad Sci USA. 2011;108:1833–1838. doi: 10.1073/pnas.1017659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen AM, Sørensen TL, Olesen C, Møller JV, Nissen P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyoshima C, Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430:529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 18.Toyoshima C, Nomura H, Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- 19.Toyoshima C, Norimatsu Y, Iwasawa S, Tsuda T, Ogawa H. How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump. Proc Natl Acad Sci USA. 2007;104:19831–19836. doi: 10.1073/pnas.0709978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pople JA, Head-Gordon M, Raghavachari K. Quadratic configuration interaction. A general technique for determining electron correlation energies. J Chem Phys. 1987;87:5968–5975. [Google Scholar]
- 21.Winther AM, et al. The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature. 2013;495:265–269. doi: 10.1038/nature11900. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, et al. High-yield heterologous expression of wild type and mutant Ca(2+) ATPase: Characterization of Ca(2+) binding sites by charge transfer. J Mol Biol. 2009;391:858–871. doi: 10.1016/j.jmb.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juul B, et al. Do transmembrane segments in proteolyzed sarcoplasmic reticulum Ca(2+)-ATPase retain their functional Ca2+ binding properties after removal of cytoplasmic fragments by proteinase K? J Biol Chem. 1995;270:20123–20134. doi: 10.1074/jbc.270.34.20123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.