What are the cellular limits that constrain the maximal rate of microbial cell duplication? What are the genetic determinants conferring maximal microbial growth rates? In the case of Cyanobacteria—an ecologically ubiquitous and diverse phylum of prokaryotes capable of oxygenic photosynthesis—these have increasingly become questions of both applied and fundamental significance. In PNAS, in a technical tour de force of reciprocal genome editing by Ungerer et al. (1), two genetically similar cyanobacterial strains with very different growth phenotypes are shown to have their large phenotypic differences controlled by remarkably few alleles. What is perhaps even more remarkable is the nature of the relationship between these alleles and the surprising case of allele synergy that may help illustrate how broad phenotypes such as growth rate and overall metabolic capacity can be controlled.
Cyanobacteria have long proved to be excellent experimental models for the study of photosynthesis and other aspects of autotrophic growth. More recently, cyanobacteria have emerged as important targets for sustainable biotechnology due to their amenability to genetic manipulation and their ability to grow on little more than sunlight, carbon dioxide, and minerals (2). Different species of cyanobacteria exhibit a very wide range of maximal rates of photosynthesis and, correspondingly, a wide range of maximal growth rates (reviewed in ref. 3). This is important since “fast growth” is often an important characteristic in choosing a cyanobacterial strain for engineering.
An especially interesting example of how different strains of cyanobacteria can have very different growth rates are recent comparisons of two closely related strains of the rod-shaped unicellular cyanobacteria, each described as Synechococcus elongatus (4–6). Understanding these strains’ background is important to the whole story. One of the two strains, Synechococcus elongatus PCC 7942 (Pasteur Culture Collection designation) (Synechococcus 7942), was isolated in the early 1970s in California and shown to be the first cyanobacterium that could be reliably transformed using exogenous DNA. Its transformability led to the rapid adoption of Synechococcus 7942 as an experimental model in laboratories across the globe for the development of emerging molecular genetic techniques to study the mechanisms of photosynthesis. The second strain, Synechococcus elongatus UTEX 2973 (Synechococcus 2973), derives from samples obtained from Texas, deposited in the UTEX strain collection, and largely forgotten until inspection of the literature reminded researchers that it grows at nearly three times the rate of its popular cousin (7), Synechococcus 7942. Indeed, it proved to have the very-fast-growth phenotype and a correspondingly high rate of photosynthesis in comparison with Synechococcus 7942 (5). Morphologically, the strains are very similar, but it was unexpected just how closely related the two strains are to one another. Whole-genome sequencing of both strains revealed that they differ at only 55 loci, mostly involving SNPs and other ostensibly minor genotypic variations (1, 7).
Nevertheless, these relatively small genotypic differences underlie dramatic changes in physiology and growth phenotype. Perhaps most significantly, the faster-growing Synechococcus 2973 strain thrives in high light intensities, reflecting an adaptation to a bright, sunlit natural habitat. In fact, under such conditions, it grows as fast as, or faster than, any previously recorded cyanobacterium (∼2 h doubling time). In contrast, the growth of Synechococcus 7942 is inhibited and prone to photodamage under similar high-light conditions. There are other differences, including the fact that Synechococcus 2973 is not readily transformable like its cousin, but the most significant differences relate to an increased metabolic capacity to drive anabolic metabolism, particularly the ability to generate high levels of cellular energy in the form of the energy carriers ATP and NADPH. The ability to use very high light intensities productively is traced to an increased flux capacity for driving electrons through the proton-pumping photosynthetic electron transport chain (5, 6). Compared with Synechococcus 7942, there is an absence of bottlenecks in the electron transport chain, a greater amount of photosystem 1 to drive electrons out of the electron transport chain to ferredoxin, increased energy carriers to receive the enhanced output, and an efficient inorganic nutrient uptake to facilitate the assimilation of carbon dioxide and nitrate, thereby yielding new biomass.
How many genetic differences does it take to convert a relatively slow-growing bacterial strain into the photosynthetically more robust and faster-growing strain? Starting with the observation that Synechococcus 7942 and Synechococcus 2973 have nearly identical genomes yet exhibit dramatically different growth and physiological phenotypes as noted, a systematic genome editing study using CRISPR/Cpf1 was conducted to identify the loci responsible for the large phenotypic differences (1). Using this approach, alleles were exchanged, in both directions, between Synechococcus 7942 and Synechococcus 2973. With 55 differing loci to choose from, the number of responsible loci could, in principle, involve all of the observed DNA differences, yet the actual number of critical differences proved surprisingly small: Only five SNPs in three genes, atpA, ppnK, and rpaA, account for virtually all of the large phenotypic differences between the two strains.
The functional identity of the critical three genes is as telling as it is interesting. The genes atpA and ppnK are both involved in energy generation, with atpA encoding the alpha subunit of the F1F0 ATP synthase. The ppnK gene encodes NAD+ kinase, which functions to increase the pool of NADP+, thereby enabling a larger store of anabolic reductant. Indeed, it was shown that the Synechococcus 2973 alleles of these enzymes have higher Vmax and affinities, consistent with earlier physiological assays showing increased phosphorylating and reducing power compared with Synechococcus 7942 (4–6). The third critical gene, rpaA, encodes a master transcriptional regulator that has been shown to bind to over 100 promoters and acts to transduce the output of the global circadian clock (8, 9). It has been inferred to exert widespread programmatic changes in gene expression that poise cells for diurnal metabolism based on photosynthesis (day) and respiration (night). Indeed, earlier transcriptional analyses show that large numbers of genes are controlled by RpaA. The present work also shows large numbers of genes whose transcription is affected by exchanging rpaA alleles. As broadly consequential rpaA is to the control of gene expression, and as important as atpA and ppnK are to the generation of ATP and NADP+, no single mutation is sufficient to produce as fast a growth rate as the wild-type Synechococcus 2973. This is because the three gene variants act synergistically (Fig. 1). While each allele of the fast-growing strain increased the rate of growth (and other physiological parameters) when introduced into the slower-growing Synechococcus 7942, the effects were not simply additive. Only when all three were introduced in concert did the growth of the modified Synechococcus 7942 increase greatly so as to closely resemble that of Synechococcus 2973 (1). In fact, the rpaA allele alone induces the smallest growth effect of the three alleles, despite its pervasive effect on transcription (1). It is as if the changes in gene transcription due to the Synechococcus 2973 rpaA allele also require the more active F1F0 ATP synthase and NAD+ kinase to make it an effective growth-changing allele. Perhaps, for example, the rpaA allele promotes the expression of photosynthesis genes, but without the more active NAD+ kinase and F1F0 ATP synthase, the greater photosynthetic capacity is frustrated by bottlenecks in the throughput of NADPH and ATP. If validated, this would provide a nice example of how a small cohort of different gene products cooperate to produce a large phenotypic effect.
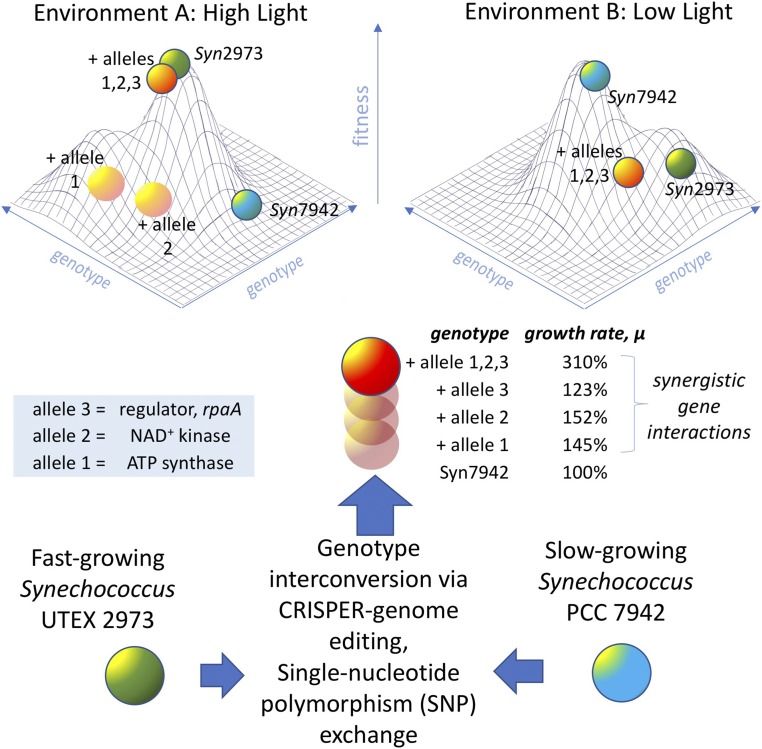
Fig. 1.
Adaptive landscape for the cyanobacterium Synechococcus 2973 (Syn2973, green spheres) and the nearly genetically identical, widely used experimental model Synechococcus 7942 (Syn7942, blue spheres). Syn2973 exhibits high fitness in Environment A, characterized by high light, but is not as well adapted to Environment B, characterized by low light. Syn7942 has the opposite fitness profile. Environmental factors other than light are likely involved in the fitness profiles of both. With only 55 single-nucleotide differences between the two genomes, a systematic genome editing study using CRISPR/Cpf1 was conducted to identify the loci by exchanging alleles in both directions. The number of critical differences was surprisingly small: Only five SNPs in three genes, atpA, ppnK, and rpaA, account for the large phenotypic differences. Interestingly, the introduction of the fast genes to Syn7942 + alleles 1,2,3 (orange spheres) showed considerable synergy shown in terms of the relative rate constants for exponential growth, μ (obtained from the doubling times in ref. 1), and this combination apparently renders the converted strain highly fit in Environment A.
There is an important twist to the story: When the amino acid sequences were analyzed for the three critical alleles from Synechococcus 2973 conferring fast growth in Synechococcus 7942, it was observed that the “fast alleles” are typical in many cyanobacteria and, perplexingly, also found in very-slow-growing cyanobacteria. Ungerer et al. (1) offer a reasonable scenario in which the transfer of fast-growth alleles from Synechococcus 2973 to Synechococcus 7942 is actually a restoration of the original alleles found in a common ancestor, which is more like Synechococcus 2973. In this scenario, the fast alleles were the original alleles, but these recently mutated and became fixed as an adaptation to a different environment with lower light intensities, leading to the slower-growing strain. This could have occurred in nature before the collection of the two strains, or even during the early period of laboratory domestication of Synechococcus 7942. Unfortunately, the strain history, at least to our knowledge, is not decisive on this point, although the authors’ general explanation is reasonable.
This twist clearly indicates that the three identified fast alleles are not necessarily conferring faster growth in all genetic contexts. Although the transcriptional information is useful, the molecular mechanisms for this unique growth phenotype remain a mystery. Nevertheless, the larger point here is that a handful of mutations can lead to profound phenotypic alterations, which has important implications for the interpretation of sequence diversity in natural environments and the evolvability of microbes in changing environments. Despite exceedingly high sequence similarity—identical rRNA in this case—these two strains could, in principle, occupy very different niches (Fig. 1). One could imagine the accumulation of just several mutations leading to the surprisingly facile occupation of a new environmental niche as the initial stage of speciation, followed by further genetic diversification (neutral genetic drift as well as adaptive refinements). At least the initial stages of niche occupation would not be discernible by sequenced-based rRNA classification.
Acknowledgments
This research was funded by the US Department of Energy (DOE), Office of Science, Basic Energy Sciences, Grant DE-FG02-08ER15968.
Footnotes
The authors declare no conflict of interest.
See companion article on page E11761.
References
- 1.Ungerer J, Wendt KE, Hendry JI, Maranas CD, Pakrasi HB. Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973. Proc Natl Acad Sci USA. 2018;115:E11761–E11770. doi: 10.1073/pnas.1814912115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berla BM, et al. Synthetic biology of cyanobacteria: Unique challenges and opportunities. Front Microbiol. 2013;4:246. doi: 10.3389/fmicb.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnap RL. Systems and photosystems: Cellular limits of autotrophic productivity in cyanobacteria. Front Bioeng Biotechnol. 2015;3:1. doi: 10.3389/fbioe.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller TJ, Ungerer JL, Pakrasi HB, Maranas CD. Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973. Sci Rep. 2017;7:41569. doi: 10.1038/srep41569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abernathy MH, et al. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol Biofuels. 2017;10:273. doi: 10.1186/s13068-017-0958-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ungerer J, Lin P-C, Chen H-Y, Pakrasi HB. Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. MBio. 2018;9:e02327-17. doi: 10.1128/mBio.02327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markson JS, Piechura JR, Puszynska AM, O’Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond S, Jun D, Rubin BE, Golden SS. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci USA. 2015;112:E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]