Significance
Nutritional symbionts in sap-feeding insects are characterized by highly degenerate genomes. It is poorly understood how hosts evolve to maintain these symbionts, particularly when hosts rely on more than one symbiont that requires distinct support for basic cell functions. We show that the aster leafhopper (Macrosteles quadrilineatus), which depends on two symbionts with tiny genomes (Sulcia and Nasuia), has differentially reprogramed gene-expression patterns in symbiont-associated cells. The host has acquired novel genetic traits and likely recruited preexisting mitochondrial support mechanisms to meet the specific needs of each symbiont. Broad comparisons across anciently diverged sap-feeding hosts reveal that the evolution of symbiont support mechanisms is largely unique to each host lineage. Important parallels are further observed with organelle evolution.
Keywords: nutritional symbiosis, DNA replication and repair, transcription, translation, eukaryotic genome evolution
Abstract
Plant sap-feeding insects (Hemiptera) rely on bacterial symbionts for nutrition absent in their diets. These bacteria experience extreme genome reduction and require genetic resources from their hosts, particularly for basic cellular processes other than nutrition synthesis. The host-derived mechanisms that complete these processes have remained poorly understood. It is also unclear how hosts meet the distinct needs of multiple bacterial partners with differentially degraded genomes. To address these questions, we investigated the cell-specific gene-expression patterns in the symbiotic organs of the aster leafhopper (ALF), Macrosteles quadrilineatus (Cicadellidae). ALF harbors two intracellular symbionts that have two of the smallest known bacterial genomes: Nasuia (112 kb) and Sulcia (190 kb). Symbionts are segregated into distinct host cell types (bacteriocytes) and vary widely in their basic cellular capabilities. ALF differentially expresses thousands of genes between the bacteriocyte types to meet the functional needs of each symbiont, including the provisioning of metabolites and support of cellular processes. For example, the host highly expresses genes in the bacteriocytes that likely complement gene losses in nucleic acid synthesis, DNA repair mechanisms, transcription, and translation. Such genes are required to function in the bacterial cytosol. Many host genes comprising these support mechanisms are derived from the evolution of novel functional traits via horizontally transferred genes, reassigned mitochondrial support genes, and gene duplications with bacteriocyte-specific expression. Comparison across other hemipteran lineages reveals that hosts generally support the incomplete symbiont cellular processes, but the origins of these support mechanisms are generally specific to the host–symbiont system.
Nutritional symbioses with microorganisms are fundamentally important to the evolutionary success of many insect groups (1, 2). Symbiotic interactions between hosts and particular bacteria can persist for millions of years, underlying the diversification of several of the most diverse insect orders that include the plant sap-feeding species in the Hemiptera (3). Most species in this group depend on obligate symbioses with intracellular bacteria for essential amino acids (EAAs) and vitamins that are deficient in their phloem and xylem diets (4, 5). In exchange, bacteria are maintained in a stable intracellular environment, provided with essential resources and support, and vertically transmitted between host generations (6, 7). Despite clear mutualistic advantages, ancient symbiotic bacteria pose several challenges to their hosts. They experience extreme genome reduction due to streamlining and stochastic gene losses vis-à-vis strong genetic drift (4, 8). As a result, bacteria lose over 90% of their genes, even genes considered essential to the bacterium and the symbiosis (9–11).
Intracellular symbionts of the Hemiptera typically lose the abilities to synthesize critical components of their cellular and metabolic machineries that are considered to be essential in free-living bacteria (4, 9). In particular, it has long been recognized that although bacteria generally retain basic enzymes involved in central cellular information processing (CIP) systems (e.g., nucleic acid and protein synthesis), bacteria with the smallest genomes (<500 kb) are missing many genes involved in DNA and RNA synthesis, DNA repair, transcription, translation, and tRNA aminoacylation (9). Thus, it remains unclear how these organisms can still function, replicate, and express genes despite significant losses from these essential cellular machineries. It has been speculated that some proteins may have expanded catabolic functionalities, or that the host may contribute genetic machineries to its symbionts (12, 13). In at least one case, the obligate symbiont of mealybugs, “Candidatus Tremblaya princeps,” has taken up its own symbiont that can provide missing CIP genes to its bacterial host (14, 15). However, how CIP systems are complemented in all other hemipteran–bacterial symbionts, which generally lack their own symbionts, is largely unknown.
To support and control symbiont functions, hemipteran insects generally maintain bacteria in discrete host organs (bacteriomes) and cells (bacteriocytes) (7, 16). Metabolic exchange between partners occurs across a symbiosomal membrane that provides a locus of control and exchange between hosts and bacteria (17). Several recent studies have demonstrated via comparative tissue-specific transcriptomics that bacteriocytes more highly express host genes that specifically complement those missing from symbiont nutritional metabolisms (15, 18–21). Many support genes appear to be derived from a variety of origins, including the broad overexpression of insect eukaryotic genes (20, 21), de novo duplication of certain gene families in the host genome (22–24), and the horizontal transfer of genes from infecting bacteria to the host nuclear genome (15, 18, 19). To date, examples of these systems are exclusively from the Sternorrhyncha suborder, where hosts generally rely on a single symbiont, including the pea aphid–“Candidatus Buchnera aphidicola,” hackberry psyllid–“Candidatus Carsonella ruddii,” and silverleaf whitefly–“Candidatus Portiera aleyrodidarum” (25–27). In the unusual case of the citrus mealybug, the host relies on two bacterial symbionts (Tremblaya and its intracellular partner, “Candidatus Moranella endobia”), but they essentially function as a fused unit and are housed in a single bacteriocyte type (15, 28). In contrast, symbioses in the Auchenorrhyncha suborder (i.e., leafhoppers, cicadas, spittlebugs, and planthoppers) are often more complex, with host species relying on two or more bacterial partners (29–33). These bacteria are generally housed separately in distinct bacteriocyte types that vary in location, morphology, and nuclei number (16, 29, 32, 34). It remains poorly understood how hosts have evolved to maintain an integrated symbiosis with multiple bacterial partners in distinct organs and that have discrete cellular and metabolic capabilities.
Here, we investigate how the aster leafhopper (ALF), Macrosteles quadrilineatus (Cicadellidae), has evolved to maintain two bacterial symbionts, “Candidatus Sulcia muelleri” (Bacteroidetes) and “Candidatus Nasuia deltocephalinicola” (Betaproteobacteria) (hereafter Sulcia and Nasuia). Both bacteria perfectly complement each other to provide the 10 EAAs: Sulcia synthesizes eight EAAs while Nasuia provides the remaining two (32). Sulcia established in the common ancestor to the Auchenorrhyncha >270 Mya and its descendant lineages are found widely throughout the suborder (35). Nasuia may be equally ancient, nutritionally supporting Sulcia throughout the diversification of the Auchenorrhyncha (29, 36). Both bacteria have two of the smallest known genomes of any insect–symbiont system (Sulcia = 190 kb and Nasuia = 112 kb). Most of the metabolic pathways and cellular functions in these bacteria have been stripped away. Those that are retained are incomplete and appear to require extensive genetic inputs from the host, including synthesis and transport of essential metabolites for nutrition pathways and cellular functions involved in the CIP systems (32).
To investigate how ALF has evolved to support its two symbionts, we developed a bacteriocyte type-specific gene-expression assay and sequenced the host genome. Sulcia and Nasuia are segregated into discrete bacteriocyte types that can be differentiated by cell size and shape, location in the body, and number of nuclei (32, 37, 38). These characteristics provide a target for cell dissociation via microdissections. Our results reveal that ALF employs a range of genetic mechanisms to differentially support Sulcia and Nasuia. Several of these mechanisms are derived from the evolution of novel functional traits via horizontally transferred bacterial genes (HTGs) to the host genome, reassignment of mitochondrial (MT) support genes, and host gene duplications that exhibit bacteriocyte-specific expression. Remarkably, for nonnutritional cellular processes, the host more highly expresses genes that have clear and distinct complementation of incomplete CIP systems in each symbiont. This host complementation pattern likely requires host genes to be expressed as proteins within the bacterial cytosol, obscuring distinctive classification of symbionts and organelles (39). Comparison of the evolution of support mechanisms with other hemipteran bacterial symbiont systems indicates that, although hosts broadly support central bacterial cell functions, the origins of genetic support vary between major host lineages.
Results and Discussion
Differential Expression Patterns of Host Genes Between Bacteriocyte Types.
While both Sulcia and Nasuia exhibit the shared loss of certain metabolisms and cellular functions, they differ widely in their nutritional contributions to the host and in the completeness of their basic cellular capabilities (32, 40). To maintain a stable symbiosis, ALF must provide genetic and cellular support to each bacterium. To determine the mechanisms ALF employs to maintain Sulcia and Nasuia, we sequenced the complete transcriptomes of the two bacteriocyte types and compared them against nonsymbiotic host tissues. Replicated RNA sequencing (RNA-seq; n = 3 biological replicates per tissue) (SI Appendix, Fig. S1A) yielded 13.55–44.91 million paired-end reads per library (SI Appendix, Table S1). Combined de novo assembly of the nine libraries produced total of 155,626 transcripts that represent 79,284 genes. CD-HIT reduced redundant contigs to 121,806 transcripts and 68,533 genes that represent 92% of the core insect genes as identified with BUSCO (41, 42). Global differential expression (DE) analysis revealed that the host more highly expresses over 10,000 genes in bacteriocytes relative to body tissues at a statistical threshold of P ≤ 0.001 and fold-change (FC) ≥ 4× (SI Appendix, Fig. S1B). The total number of more highly expressed genes in bacteriocytes is within the range found in other hemipteran lineages (e.g., ∼6,000 and 11,000 genes differentially expressed in the hackberry psyllid and citrus mealybug symbioses, respectively) (15, 18). FC values discussed below for specific genes and pairwise tissue comparisons are abbreviated as follows: Sulcia bacteriocytes-body (S-B FC) and Nasuia bacteriocytes-body (N-B FC). All differential gene-expression statistics are provided in Dataset S1.
Broadly, differential gene-expression assays reveal that, to meet the needs of Nasuia and Sulcia, ALF employs a range of mechanisms that include HTGs from other infecting bacteria, reassignment of MT support genes, and the duplication of existing genes with bacteriocyte-specific expression (Table 1 and SI Appendix, Table S2). Along with thousands of other eukaryotic host genes, novel gene acquisitions play important roles in supporting the basic cellular processes of both symbionts, including the CIP systems. Below we outline the role of these mechanisms in supporting basic bacterial cell functions.
Table 1.
HTGs in the M. quadrilineatus (ALF) genome
| FPKM (FC) | |||||||
| Trinity ID | Gene | Product | Function | Predicted origin | Body | Nasuia | Sulcia |
| DN64545_c0_g2 | rnc-1 | Ribonuclease III | RNA processing | Wolbachia* | 0 | 503.7 (53,374.4) | 26.5 (2,905.3) |
| DN45236_c0_g1 | rnc-2 | Ribonuclease III | RNA processing | Wolbachia* | 0 | 3.8 (234.3) | 0.3 (26.7) |
| DN55377_c1_g1 | rnc-3 | Ribonuclease III | RNA processing | Wolbachia* | 0 | 5 (667.6) | 92.6 (10,212.5) |
| DN56617_c3_g1 | ileS | Isoleucine-tRNA ligase | Translation | Wolbachia | 0 | 17.4 (315.1) | 294.9 (4,940.9) |
| DN57080_c0_g1 | tmk | dTMP kinase | dTMP synthesis | Vibrio | 0.1 | 51.3 (933.6) | 783.8 (13,679.1) |
| DN33246_c0_g1 | frr | Ribosome recycling factor | Translation | Alphaproteobacteria | 0 | 8.2 (465.7) | 140.5 (7,786.8) |
| DN40971_c0_g1 | alv | Thiol_cytolysin | Cytolysis | Firmicutes | 0.3 | 1 (–) | 5.6 (18.3) |
| DN41609_c0_g1 | ATPase-1 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 0 (–) | 2.2 (96.5) |
| DN58902_c0_g1 | ATPase-2 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 1.3 (46.8) | 19.8 (832.4) |
| DN56403_c1_g4 | ATPase-3 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 5.6 (464.3) | 87 (7,384.9) |
| DN62224_c2_g1 | ATPase-4 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 7.2 (392.5) | 125.6 (7,241) |
| DN62110_c2_g2 | ATPase-5 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 1.9 (174.2) | 24.6 (2,284.6) |
| DN59545_c1_g2 | ATPase-6 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 6.8 (348.2) | 87.9 (4,227.3) |
| DN66588_c0_g3 | ATPase-7 | AAA-ATPase | ATPase activity | Firmicutes* | 0 | 1.6 (28.6) | 48 (756.2) |
| DN33783_c0_g1 | dut | Deoxyuridine triphosphatase | Nucleotide metabolism | Wolbachia* | 0 | 5.2 (40) | 1.3 (9.6) |
| DN47540_c0_g1 | def-1 | Peptide deformylase | Translation factor | Rickettsia | 0 | 13.9 (1,317.7) | 1.5 (149.7) |
| DN48799_c0_g1 | def-2 | Peptide deformylase | Translation factor | Rickettsia | 0 | 23 (2,070.8) | 1.5 (144.4) |
| DN52029_c0_g1 | def-3 | Peptide deformylase | Translation factor | Rickettsia | 0 | 4 (221.9) | 0.2 (–) |
| DN67119_c0_g1 | def-4 | Peptide deformylase | Translation factor | Rickettsia | 0.1 | 619.8 (7,527.4) | 34.6 (387.3) |
| DN50262_c0_g1 | def-5 | Peptide deformylase | Translation factor | Rickettsia | 0 | 0.4 (–) | 1.4 (62.6) |
| DN66033_c3_g1 | ribD | Uracil reductase | Riboflavin synthesis | Wolbachia | 0.1 | 479.3 (13,904.5) | 27 (721.3) |
| DN48507_c0_g1 | rluA | RNA pseudouridine synthase | RNA binding | Gammaproteobacteria | 0 | 72.4 (4,233) | 3.9 (233.3) |
| DN66364_c0_g3 | yebC-1 | Transcriptional regulator | Transcription | Midichloria | 0.2 | 59.8 (348.5) | 818.8 (4,484.5) |
| DN66182_c1_g3 | yebC-2 | Transcriptional regulator | Transcription | Midichloria | 0.1 | 424.9 (7,590.7) | 22.3 (362.2) |
| DN57670_c0_g2 | per | Putative permease | Transport | Rickettsia | 0 | 250.6 (9,311) | 18.1 (694.3) |
| DN64865_c1_g1 | pel | Pectin lyase | Cell wall degradation | Pseudomonas | 0.8 | 3.9 (4.7) | 4.5 (5) |
| DN53310_c0_g1 | gh25-1 | Glycosyl hydrolase family 25 | Lysozyme activity | Pseudomonas | 2.5 | 335.3 (136.3) | 18.6 (6.9) |
| DN45357_c0_g1 | gh25-2 | Glycosyl hydrolase family 25 | Lysozyme activity | Pseudomonas | 472.1 | 0 (2−15) | 0 (2−15) |
| DN54290_c0_g1 | cel-1 | Cellulase | Cell wall degradation | Streptomyces | 9.4 | 0 (2−10) | 0 (2−10) |
| DN43812_c0_g1 | cel-2 | Cellulase | Cell wall degradation | Streptomyces | 644.5 | 5.3 (2−7) | 0.7 (2−10) |
FPKM, fragments per kilobase per million; FC, fold-change in expression of bacteriocytes relative to body tissue; Nasuia, Nasuia bacteriocytes; Sulcia, Sulcia bacteriocytes; FC is represented with “–” if P value is higher than 0.001.
The origin is predicted based on the BLASTP results.
Horizontal Transfer of Bacterial Genes to the ALF Genome.
In some cases, eukaryotic genes may be unable to fill gaps in bacterial cellular functions, necessitating the acquisition of novel functional traits via HTGs from bacteria (15, 18, 19, 43, 44). We found 30 HTGs in the ALF genome; 27 are more highly expressed in bacteriocytes (Table 1). The expression values of eight selected HTGs and one MT support gene (see below) were further verified with additional dissections and qRT-PCR (SI Appendix, Fig. S2). HTGs in the ALF genome are capable of compensating for specific gene losses in the essential cellular functions of each symbiont, including vitamin synthesis (ribD) and CIP systems (ileS, yebC, frr, def, rnc, and rluA). One of the more striking aspects of the ALF HTGs is that several show clear DE patterns between bacteriocytes (e.g., ribD and frr) (Table 1), suggesting that they have been recruited to meet the specific needs of either Sulcia or Nasuia.
To verify that HTGs are integrated into the host genome, we assembled a draft genome of ALF comprising 198,236 scaffolds (1,000–70,047 bp with a BUSCO score of 40%, but see transcriptome BUSCO score above) with an average coverage of 15.6×. National Center for Biotechnology Information (NCBI)-BLASTN found all 30 HTGs among host scaffolds (scaffold size = 2.1–16 kb). Twenty-five HTGs are flanked by known insect genes and 10 genes contain introns with the canonical eukaryotic GT-AG boundary (both evidence for 7 genes). Two genes, rluA and def-2, are present on scaffolds for which neighboring genes could not be identified. However, no bacterial genes were found on these scaffolds, supporting inference of a HTG origin because bacterial genomes are generally densely coding (45).
Phylogenetic analyses and NCBI-BLASTP searches confirm that the HTGs are placed in a wide-range of bacterial groups that contain known environmental bacteria (Table 1; see phylogenies in Dataset S2). We further identified six genes represented by multiple copies (two to seven copies). In general, gene copies are monophyletic (e.g., def, yebC, gh25, and cel) (Dataset S2 H, K, N, and O), indicating that copies are derived from a single origin. However, several duplicated genes form weakly supported clades (e.g., rnc and ATPase) (Dataset S2 A and F) and their origins are unclear. As generally shown in other hemipteran systems, none of the ALF HTGs are derived from Sulcia or Nasuia (15, 18, 19, 44).
HTGs Are Not Shared Between the Major Hemipteran Groups.
To determine whether HTGs in ALF are widespread across the Hemiptera, we compared them against the available genomic resources for insect hosts from the Heteroptera [stink bug (Halyomorpha halys)], Sternorrhyncha [e.g., the pea aphid (Acyrthosiphon pisum), mealybugs (e.g., Planococcus citri), and so forth], and the Auchenorrhyncha [e.g., brown planthopper (Nilaparvata lugens), scrub cicada (Diceroprocta semicincta), and so forth] (see Fig. 1 and SI Appendix, SI Materials and Methods for full taxon sampling) (15, 18, 19, 44, 46–49). ALF does not share HTGs with the Heteroptera species and only shares two genes (ATPase and ribD) with Sternorrhyncha hosts; however, several Sternorrhyncha HTGs are shared within that lineage and may be derived from a common origin (Fig. 1). An ATPase gene is present in ALF, mealybugs, psyllid, spittlebug, and cicada, but its origin and function are unclear. Finally, the ribD gene in mealybugs and ALF appears to be derived from a Wolbachia origin, which is a relatively common source for HTGs (Table 1 and Dataset S2I). Whether ribD shares a common origin is uncertain, as we did not find this gene in any of the other Auchenorrhyncha hosts.
Fig. 1.
Comparison of HTGs involved in nutrition synthesis, CIP, bacterial cell wall synthesis, population regulation, and plant or fungal cell wall degradation in the M. quadrilineatus leafhopper and 10 other hemipteran lineages (three mealybugs harboring all of the HTGs identified in mealybug species are included). Host and symbiont names are given on phylogenetic tips. Phylogenetic relationships between hosts are based on Cryan and Urban (111). HTGs represented by boxes are grouped by their functions. Shaded boxes are genes that are found in each host genome and unshaded ones are not present.
The HTGs encoded in the ALF genome are further distinct from the other Auchenorrhyncha species. We identified three HTGs (excluding the ATPase gene) in ALF that are shared with the spittlebug and cicada. All of the three Auchenorrhyncha hosts share a pel (pectin lyase) gene predicted to be capable of cleaving pectin, which is the main plant cell wall component (50). Plant cell wall-degrading HTGs are commonly found in arthropods and they may play a role in the evolution of herbivory (51). ALF and the cicada further distinctly share two HTGs, yebC (transcriptional regulator) and frr (ribosome recycling factor). These genes may support bacterial symbiont transcription and translation machineries (discussed below). Phylogenetic analyses show that each of the frr and pel genes form monophyletic groups (Dataset S2 D and M), indicating that each of them may be derived from a single transfer to an Auchenorrhyncha ancestor (Table 1), while the yebC gene copies found in different hosts may have independent origins (Dataset S2K). Finally, ALF was found to share no HTGs with the brown planthopper. This result may be explained by the fact that planthoppers are distantly related to leafhoppers (Fig. 1) and that Sulcia and its partner bacterium were more recently replaced by a yeast-like symbiont in Delphacidae planthopper lineages and may have subsequently lost shared HTGs (35, 46).
Reassignment of Eukaryotic MT Support Genes to Bacterial Symbionts.
Animals encode more than 1,000 proteins that are known to support the anciently reduced bacterial genome of the MT (Alphaproteobacteria) (52, 53). In ALF, we identified 211 MT support genes more highly expressed in the bacteriocytes (Dataset S3). We further found that 33 genes have multiple copies with bacteriocyte-type–specific expression patterns (Dataset S3). To verify that the DE of these MT support genes is unlikely to be a function of increased metabolic demands in the bacteriocytes or increased MT abundance, we investigated both the DE levels of MT-encoded genes and the ratio of mtDNA:nuclear DNA between bacteriocytes and body tissues (54, 55). DE analyses of mRNA-enriched and non–mRNA-enriched RNA-seq and further qRT-PCR validation of two genes (cox1 and cox3) reveal that they are not differently expressed between bacteriocytes and body tissues (P ≤ 0.05 and FC ≥ 2×) (SI Appendix, Table S3). The relative copy number of host nuclear and MT genomes is also not significantly different between tissue types (P ≤ 0.05) (SI Appendix, Table S3). Thus, the discrete subset of more highly expressed MT support genes in the bacteriocytes appears to be a distinct symbiont support mechanism, rather than compensation for globally increased MT activity in those cells.
The expression pattern of MT support genes has not been a significant focus of previous studies of insect symbioses. However, in at least one case, it was noted that in the pea aphid-Buchnera symbioses four MT-related transporters are highly expressed in the bacteriocytes (21). The high expression of these genes was predicted to reflect heightened MT activity (21). However, our results identified a much larger set of genes in ALF, suggesting that MT support genes may be broadly retargeted to support nutritional symbionts. Although this pattern has not been reported for other obligate insect symbioses, the retargeting of MT genes to support other ancient symbioses, such as the chloroplast in plants, is well known (56). The dual-targeting proteins are enriched in the essential cellular processes that include cell-cycle control, DNA synthesis, and protein synthesis (57). For example, 15 of the 24 MT aminoacyl-tRNA synthetases (aaRSs) are dual-targeted to chloroplasts in Arabidopsis thaliana (58). The dual-targeting proteins harbor twin or ambiguous signal peptides (SPs) that target both organelle types (56). In our study, only a small number of the more highly expressed MT support genes in the bacteriocytes were predicted to possess targeting signals, which may reflect the long evolutionary history of sequential gene losses and the variable evolutionary mechanisms employed to support those losses (see below).
Remarkably, several MT support genes in ALF show distinct expression between bacteriocytes that may complement unique gene losses in either Nasuia or Sulcia. For example, the translation initiation factor IF-2 (infB) gene is uniquely missing from Nasuia and the MT IF2 gene is more highly expressed in Nasuia bacteriocytes (SI Appendix, Table S2). In contrast, the MT single-stranded DNA-binding protein (SSBP) that may complement the missing ssb gene is only overexpressed in Sulcia bacteriocytes (SI Appendix, Table S2). Reassigned MT support genes also appear to prop-up Sulcia’s and Nasuia’s CIP systems, including DNA replication and repair, and translation (discussed below). Finally, MT support genes are likely to be an important source of transporters. ALF bacteriocytes highly express MT amino acid transporters (AATs) and ADP/ATP translocase that may play a role in the transport of nutritional metabolites and cellular energy in the ALF symbionts (SI Appendix, Table S2).
Evolution of Symbiont Support via Host Gene Duplications.
Gene duplications are an important source of functional novelty in organismal adaptation (59, 60). Broadly, paralog analysis of the ALF transcriptome identified ∼1,600 genes (identity: 40–90% and sequence length coverage: >80%) that are derived from duplication events in the host genome and that are more highly expressed in one or both bacteriocytes. The copy number of these genes ranges from 2 to 98. Similar to other insect–symbiont systems, gene duplications in ALF were widely observed in AAT and sugar transporter families (22–24, 61). For the AATs, we identified 25 more highly expressed genes in the bacteriocytes and 17 of them are derived from gene-duplication events (SI Appendix, Table S2). Phylogenetic analyses reveal that many ALF AAT paralogs do not form monophyletic clades with other hemipteran taxa, suggesting that gene-duplication events occurred multiple times throughout the diversification of the leafhopper lineage and possibly the suborder (Dataset S2 P–R). We further identified 15 DE genes encoding sugar transporters in the bacteriocytes (SI Appendix, Table S2). Thirteen genes are paralogs of the insect bidirectional facilitated trehalose transporter Tret1. Trehalose is the principle sugar in insect hemolymph and is involved in osmoregulation, cell energy, and membrane and protein stability (62, 63). It is likely the main source of energy to power Sulcia and Nasuia metabolisms.
The ALF host appears to complement a wide range of other nonnutrition bacterial cell functions via gene duplications. For example, duplicated eukaryotic genes [MT elongation factor Ts (TSFM), MT translation factor GUF1 homolog (GUF), and MT arginine-tRNA ligase (ARGS)] and HTGs (def, rnc, and yebC) are predicted to support informational transcription and translation (see below) (Table 1 and SI Appendix, Table S2). Similarly, duplicated gene copies are likely to be involved in the regulation of symbiont populations [e.g., peptidoglycan-recognition proteins (PGRPs) and glycosyl hydrolase family 25 (gh25; HTG)] (Table 1 and SI Appendix, Table S2). PGRP genes play important roles in insect–symbiont homeostasis through antibacterial activities, regulation of immune signaling pathways, or by triggering autophagy (64–69). In ALF, the gh25 gene has a bacterial origin where it operates as a peptidoglycan-degrading lysozyme that cleaves the bacterial cell wall (70). This gene was further duplicated in the ALF genome; the gh25-1 paralog is more highly expressed in both Sulcia and Nasuia bacteriocytes, while gh25-2 shows no expression in the bacteriocytes (Table 1). Because PGRP and gh25 are predicted to target the bacterial cell wall (70, 71), their roles in ALF are uncertain as Sulcia and Nasuia are incapable of synthesizing a cell envelope.
Host Support of CIP Systems Predicts Expression of Eukaryotic Genes in Bacterial Symbiont Cells.
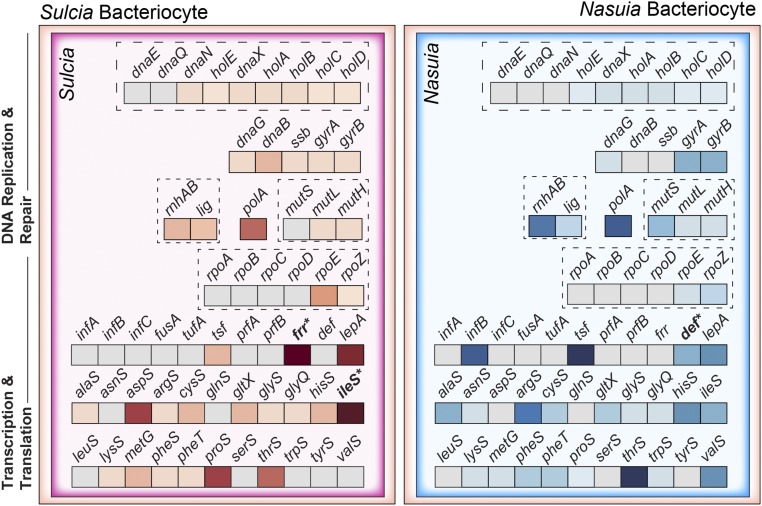
Although nutritional symbionts have undergone extreme genome reduction, they tend to retain some genes involved in the core cellular processes of DNA replication and repair, transcription, and translation. This feature has been used as an argument for their classification as independent cellular entities (4, 9). However, as more bacterial symbiont genomes have become available, it is now apparent that these mechanisms are widely incomplete across nearly all tiny-genome bacterial symbionts of insects (15, 26, 27, 33, 72). It has remained uncertain as to how—or if—these cellular processes function independently of the host. Results from our study suggest that ALF plays a large role in supporting all CIP systems (Fig. 2). While both Sulcia and Nasuia are missing the same genes involved in DNA replication and repair, transcription, and translation, they are also differentially missing distinct genes in each of these cellular machineries. ALF differentially expresses genes that appear to complement bacterial genes missing from these cellular functions in the bacteriocytes where they are required.
Fig. 2.
Host compensation of missing CIP genes in Sulcia and Nasuia. Symbiont genes are colored in gray. HTGs are shown in bold and labeled with an asterisk. More highly expressed host genes in bacteriocytes relative to body tissues are colored based on the log2 FC ratio (see inset legend in Fig. 3). Genes grouped by dashed boxes are predicted to interact directly within the bacterial cytosol.
In order for host genes to complement incomplete symbiont CIP systems, they must be expressed in the bacterial cytosol. This complementary pattern mirrors the host protein translocation typical of eukaryotic nuclear–organelle interactions. Protein-targeting signals (N-terminal sequence) and translocation apparatuses are defining features of the MT and chloroplast organelles (73). These systems have evolved distinct protein import machineries (Tim/Tom complex and Tic/Toc complex, respectively) for recognizing the organelle-specific protein-targeting signals to ensure their proper localization (74, 75). Subcellular localization analysis of HTGs and identified MT support genes involved in CIP (Table 1 and SI Appendix, Table S2) shows that four MT support genes were predicted to be targeted to MT with weak scores [MT targeting peptide (mTP) score < 0.75]. Five genes (three MT support genes and two HTGs) were further predicted to have secretory SPs, and the rest of HTGs and MT support genes were assigned to other cellular localizations (e.g., frr) (SI Appendix, Table S4). The prediction of three mTPs and all SPs were further confirmed with SignalP (76). MTPs and SPs consist of 17- to 31- and 16- to 37-aa residues, respectively, and no sequence conservation was found (SI Appendix, Table S4). These results suggest that gene products targeting Sulcia and Nasuia may not solely depend on targeting peptides at the N terminus and could rely on other secretory pathways or a mixture of mechanisms (77–79). It is also possible that novel mechanisms have evolved to traffic these genes to the bacteria as was recently found to have happened in the amoeba, Paulinella chromatophora, and its intracellular phototrophic cyanobacterial symbiont (80).
The difficulty in identifying a conserved protein import mechanism in the ALF may be due to challenges in current targeting-signal prediction methods and the relative evolutionary complexity of symbioses in the Auchenorrhyncha. Current targeting-signal prediction methods perform poorly with dual-targeting proteins (56, 81) and symbiont-targeting proteins (80). Moreover, symbioses in the Auchenorrhyncha have an ancient and complex history of gene losses and compensatory evolution. First, host proteins expressed in either symbiont must transit two membranes: the symbiosomal membrane and the bacterial cell membrane (82). Second, Sulcia and Nasuia have separate origins and occur in exclusive bacteriocyte types that support highly specific host–symbiont interactions. Therefore, it is plausible that multiple mechanisms have evolved to distinguish between proteins that interact with symbionts.
ALF Uses Multiple Evolutionary Mechanisms to Support Sulcia’s and Nasuia’s CIP Systems.
Both Sulcia and Nasuia maintain some of the basic enzymatic machinery to replicate, repair, and transcribe their DNA. However, they have experienced extensive gene losses in enzymes considered to be essential in other bacteria, such as Escherichia coli (4, 11). Our results demonstrate that the host likely provides extensive and distinct support of these cellular functions in both bacteria. Both Sulcia and Nasuia have highly reduced DNA replication and repair systems that appear to be supplemented by the host (Fig. 2 and SI Appendix, Table S2). For example, although both symbionts retain only three and two genes in the DNA polymerase III holoenzyme, respectively, the host highly expresses genes encoding DNA polymerase subunits in both bacteriocytes that are likely to replace those that are missing (Fig. 2 and SI Appendix, Table S2). Furthermore, Sulcia and Nasuia have differentially lost DNA replication and repair genes that include mutS in Nasuia, and dnaB and ssb in Sulcia. ALF more highly expresses genes that may fill these gaps in their respective bacteriocytes: eukaryotic DNA repair protein mutS homolog 4 in Nasuia bacteriocytes (MUTS: N-B FC = 55.9×), and ATP-dependent DNA helicase (DNA2: S-B FC = 7.7×) and SSBP (S-B FC = 2.4×) in Sulcia bacteriocytes.
Sulcia and Nasuia have also lost essential genes involved in the transcription system, including those required in the RNA polymerase holoenzyme (rpoEZ). We identified DE host genes encoding RNA polymerase subunits in the bacteriocytes that may complement the bacterial RNA polymerase (Fig. 2 and SI Appendix, Table S2). In addition, Sulcia and Nasuia are jointly missing accessory transcriptional enzymes that appear to be complemented by more highly expressed HTGs in the bacteriocytes, including rluA (RNA pseudouridine synthase), rnc (ribonuclease III), and yebC (transcriptional regulator). Remarkably, rnc and yebC have multiple copies that are differentially expressed in each bacteriocyte type. The rnc-1, rnc-2, and yebC-2 copies are more highly expressed in Nasuia bacteriocytes, while rnc-3 and yebC-1 are more highly expressed in Sulcia bacteriocytes (Table 1).
Sulcia and Nasuia have lost a number of essential genes involved in translation, including many aaRSs required to charge tRNAs (15 in Sulcia and 17 in Nasuia) and an array of translation factors (32). The ALF host more highly expresses genes that may differentially fill the gaps of missing genes in the respective bacteriocytes (Fig. 2 and SI Appendix, Table S2). For aaRS genes (14 genes) missing from both symbiont genomes, genes capable of filling these gaps are highly expressed in both bacteriocytes [e.g., MT cysteine-tRNA ligase (CYSS): S-B FC = 11.2× and N-B FC = 11.2×]. Both symbionts have differentially lost sets of aaRSs that appear to require distinct host support. For those lost, the host more highly expresses genes that may replace them in their respective bacteriocytes [e.g., aspartate-tRNA ligase (ASPS): S-B FC = 16.5× and N-B FC = 269.7×, and valine-tRNA ligase (VALS): S-B FC = 113.4× and N-B FC = 6.8×]. NCBI-BLASTP searches show that six of these aaRSs originate from MT support genes.
We found two cases where host genes of different origins appear to complement shared missing aaRS genes in Sulcia and Nasuia. For the jointly lost bacterial proline-tRNA ligase (proS), the host gene encoding MT proline-tRNA ligase (PROS) is more highly expressed in Nasuia bacteriocytes (N-B FC = 4.4×), while the gene for glutamate/proline-tRNA ligase (EPRS) is more highly expressed in Sulcia bacteriocytes (S-B FC = 218.7×). Similarly, isoleucine-tRNA ligase (ileS) has been lost from both Nasuia and Sulcia. In the Sulcia bacteriocytes, an ileS gene horizontally transferred from Wolbachia is more highly expressed (Table 1). Nasuia bacteriocytes, however, more highly express an MT copy of the ILES gene (N-B FC = 28.3×) (SI Appendix, Table S2). This pattern of convergent bacterial gene loss, but differential support by the host, suggests that the bacteria lost these aaRS genes at different times during the evolution of the symbioses. The host likely evolved bacteriocyte-specific mechanisms to support these losses.
Finally, Sulcia and Nasuia are missing additional essential translation-associated enzymes. Each missing gene is likely supplemented by a corresponding host gene that is more highly expressed in the bacteriocytes (Fig. 2). For example, two genes—ribosome recycling factor (frr) and peptide deformylase (def)—were identified as HTGs from different Alphaproteobacteria lineages (Table 1 and Dataset S2 D and H). The frr gene is more highly expressed in Sulcia bacteriocytes, where it is uniquely lost. It is also notable that the def gene has been duplicated into five copies; four of which are more highly expressed in Nasuia bacteriocytes, where they likely replace its missing ortholog. Finally, several genes that may compensate for missing translation factors are likely derived from MT support systems. In Nasuia bacteriocytes, the host IF2 gene may replace bacterial infB (N-B FC = 727×) (SI Appendix, Table S2). The DE genes, TSFM and GUF, likely fill the gaps of the missing genes tsf and lepA in both Sulcia and Nasuia. Strikingly, each of the TSFM and GUF genes has two copies that are differentially expressed between the two types of bacteriocytes (SI Appendix, Table S2). Phylogenetic analyses show the copies of each gene group together, indicating that the duplication took place independently in leafhoppers (Dataset S2 S and T).
Host Support of CIP Systems in Other Hemipteran Symbioses.
To detect if the commonly degenerate CIP systems of other hemipteran symbionts are also supported by their hosts, we compared gene-expression patterns across previously completed studies in the pea aphid–Buchnera, citrus mealybug–Moranella–Tremblaya, hackberry psyllid–Carsonella, silverleaf whitefly–Portiera, and scrub cicada–Sulcia–Hodgkinia symbiont systems (15, 18–20, 47). Overall, results from this analysis reveal that hemipteran hosts do indeed differentially express genes capable of filling missing genetic gaps of CIP systems in their symbionts. The CIP systems of the Carsonella (psyllid), Portiera (whitefly), and Hodgkinia and Sulcia (cicada) are broadly complemented by host genes (Dataset S4). In the whitefly and psyllid symbioses, we were able to identify genes that could fill most gaps of the missing genes in Portiera’s and Carsonella’s CIP systems. However, in the latter case, we were unable to find genes that could complement DNA polymerase and mismatch repair systems, raising questions about how or if these functions are supported in the psyllid symbiosis. De novo assembly for the cicada transcriptomes revealed considerable variation in expression levels, likely due to the fact that samples were acquired from the field, lacking experimental controls on age and environment (e.g., ∼1,700 genes were identified as differentially expressed at a less stringent P ≤ 0.05) (83). Nevertheless, similar to ALF and other hemipterans, the cicada also differentially expresses eukaryotic genes capable of filling gaps in the Hodgkinia and Sulcia CIP systems.
In contrast to symbionts with clear gaps in their CIP systems, we identified few to no highly DE host genes involved in CIP support in the pea aphid or citrus mealybug. The Buchnera genome in the pea aphid has relatively complete CIP machineries and no highly expressed host genes involved in CIP systems were found in the bacteriocytes (20). We similarly found few highly expressed CIP genes in the citrus mealybug bacteriocytes, despite the fact that the Tremblaya genome has lost a large number of genes underlying CIP (e.g., all aaRSs) (14, 15). These missing genes are likely complemented by Tremblaya’s own intracellular symbiont, Moranella (15).
We further investigated whether the origins of CIP support genes are similar across hemipteran systems. As discussed above, the HTGs employed by ALF to fill genetic gaps in the CIP machineries are not found in any Sternorrhyncha hosts. However, ALF and the cicada do share two HTGs, frr and yebC. In contrast, the reassignment of MT support genes to support CIP systems appears to be a common evolutionary mechanism across hemipteran lineages. Several overexpressed MT aaRS genes in the bacteriocytes were identified across hemipteran systems [e.g., MT asparagine-tRNA ligase (ASNS) in the psyllid and whitefly]. Previous studies have shown that the MT aaRSs can have relaxed discrimination capable of aminoacylating bacterial tRNAs (84–86). Thus, it is plausible that the MT aaRS genes in hemipteran lineages are flexible to fill the gaps of symbiont homologs. Finally, several CIP support genes are also derived from duplications with bacteriocyte-specific expression in other hemipteran hosts. For example, we detected two DNA polymerase III POLC genes in the whitefly, with both of them more highly expressed in bacteriocytes (19).
Distinct Host–Symbiont Collaboration in the Synthesis of Essential Nutrition.
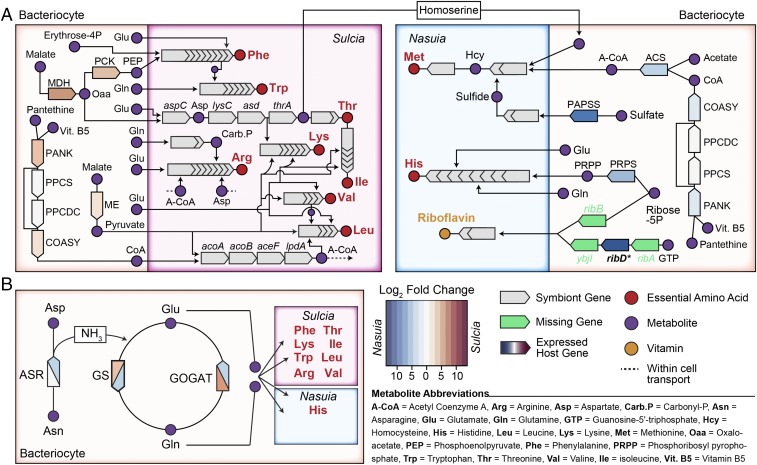
The primary role of bacterial symbioses in the Hemiptera is to provide essential nutrition to the host (2). In contrast to the Sternorrhyncha symbioses, provisioning of the 10 EAAs in ALF is partitioned between each bacterial partner. These EAA pathways are also relatively complete and do not require host complementation, which is distinct from the Sternorrhyncha lineages (18, 19). However, both Sulcia and Nasuia in ALF do require distinct sets of essential metabolites from the host to initiate the synthesis of each EAA (Fig. 3 and SI Appendix, Table S2). Both symbionts require acetyl-CoA (A-CoA), glutamine (Gln), and glutamate (Glu) (Fig. 3A). As has been widely found in other hemipteran symbiont systems, both Glu and Gln are likely supplied via the glutamine synthetase/glutamate synthase (GS/GOGAT) cycle, which is more highly expressed in both of the ALF bacteriocytes (Fig. 3B) (15, 18, 20, 87). The CoA metabolite is likely supplied by two eukaryotic genes, pantothenate kinase (PANK) and bifunctional coenzyme A synthase (COASY) (Fig. 3A) (88, 89). However, unlike Sulcia, Nasuia cannot synthesize A-CoA directly. Remarkably, ALF appears to supplement this need by more highly expressing A-CoA synthase (ACS), specifically in the Nasuia bacteriocytes (Fig. 3A).
Fig. 3.
The inferred metabolism in Sulcia and Nasuia bacteriocytes. (A) Integrated nutrition pathways in Sulcia and Nasuia bacteriocytes. Relevant genes underlying precursor metabolites synthesized by the bacteria and host are shown. (B) The GS/GOGAT cycle for recycling NH3. More highly expressed host genes in bacteriocytes relative to body tissues are colored based on the genome they occur in and the log2 FC ratio. See inset legend for additional details and explanation of metabolite abbreviations. See text for full names of gene products. HTGs are shown in bold and labeled with an asterisk.
Because Sulcia and Nasuia provision different EAAs, they require discrete metabolites from ALF. To meet these needs, the host appears to more highly express specific genes that complement each symbiont’s metabolite requirements (Fig. 3 and SI Appendix, Table S2). For example, Nasuia’s histidine pathway requires phosphoribosyl pyrophosphate (PRPP), which is likely complemented by the more highly expressed host gene, ribose-phosphate pyrophosphokinase (PRPS) (Fig. 3A) (90). Nasuia further requires homoserine and sulfide for methionine synthesis (31, 32). While homoserine has been previously proposed to be provided by Sulcia (13, 91), the host more highly expresses 3′-phosphoadenosine 5′-phosphosulfate synthase (PAPSS) in Nasuia bacteriocytes that may complement Nasuia’s missing cysNC genes in the sulfide pathway (Fig. 3A). In contrast, Sulcia is responsible for most of the EAAs required by the symbioses; additionally, its metabolites appear to be discretely met by the host. To produce aspartate for threonine, isoleucine, lysine, and arginine synthesis, Sulcia requires oxaloacetate that is likely supplied by the host-encoded malate dehydrogenase (MDH) (Fig. 3A). Sulcia also requires pyruvate for isoleucine, valine, leucine, and lysine synthesis that is likely supplied by host-encoded NADP-dependent malic enzyme (ME) (Fig. 3A). Finally, to synthesize tryptophan and phenylalanine the host appears to contribute phosphoenolpyruvate (PEP) via PEP carboxykinase (PCK) (Fig. 3A).
Concluding Remarks
Symbionts of the Hemiptera have tiny genomes that require distinct support mechanisms from their insect hosts (92). In the Auchenorrhyncha, where insect species generally rely on more than one microbial symbiont, a stable symbiosis requires extensive host evolution to integrate multiple beneficial partners that can differ dramatically in their metabolic contributions and basic cellular capabilities. The two obligate symbionts of ALF have two of the most degraded bacterial genomes known from any animal symbiosis, requiring extensive and distinct genetic and cellular support (32). To meet the shared and distinct needs of Sulcia and Nasuia, the host has reprogrammed the expression of a large number of genes in each bacteriocyte type. Many of the genes recruited to support these symbionts are derived from HTGs from other infecting bacteria, reassignment of MT support genes, and endogenous gene duplications. Results from our study reveal that these evolutionary processes are fundamental symbiont support mechanisms in ALF and other hemipteran lineages. Remarkably, these processes operate discretely to jigsaw together support of degenerate symbiont genomes not only among the major hemipteran lineages, but also among the multiple bacterial symbionts within a single host.
The evolutionary success of eukaryotes is due to a complex history of symbioses and horizontal transfer of microbial genes to the nuclear genome to support those symbionts (93, 94). These processes have continued to shape eukaryote diversity and complexity, particularly in the later diversification of plant-specialized insects that use HTGs to maintain obligate nutritional symbioses (95). Although the evolutionary mechanism of horizontally acquiring novel genes to support bacterial symbionts is known from a wide-range of host lineages across the Hemiptera (15, 18, 19, 44), the recruitment of specific genes appears to be relatively lineage-specific. Only a few HTGs that likely support bacterial symbionts are shared across the major lineages of the Hemiptera, particularly between the Sternorrhyncha and Auchenorrhyncha. Furthermore, it is surprising that we were able to recover only four symbiont-support HTGs shared between Auchenorrhyncha host lineages that all harbor Sulcia, although this symbiont sometimes pairs with different coprimary symbionts (e.g., Hodgkinia and Nasuia) (31). The sampled host–symbiont lineages in this study are separated by tens to hundreds of millions of years of evolution, having experienced extensive independent gene losses. Thus, at least at this evolutionary time scale, HTG appears to be a tailored response by particular host–symbiont systems and possibly down to the species level (49).
The reassignment of MT support genes also appears to be a widespread evolutionary mechanism for maintaining obligate symbionts (96). Our study indicates that a wide-range of genes targeting the MT (e.g., MT aaRSs and translation factors) have been recruited to support intracellular nutritional symbionts. This dual-targeting strategy has also occurred during the evolution of chloroplasts in plants, which similarly have highly reduced genomes (107–218 kb) (97) and require extensive host nuclear-encoded support (98). One mechanism to establish essential symbioses with chloroplasts was the co-option of a large number of nuclear-encoded proteins targeting the MT to support chloroplast cellular functions, including cell-cycle control, DNA synthesis, and protein synthesis (56, 57). It is possible that the dual-targeting of these genes facilitated the loss of most cyanobacterial genes as it established as an organelle (99). In the insect ALF host, a similar pattern emerges where dual-targeting of MT support mechanisms may support many of the degenerate symbiont functions in both Nasuia and Sulcia. Comparative analyses with the other Hemiptera (see above) suggest that this is a widespread pattern (18, 19, 47). The use of MT support genes to maintain symbioses with bacteria that have tiny genomes is a logical evolutionary step to shore-up the rapid and widespread loss of symbiont genes.
Our results further provide explicit predictions that—like MT symbioses across eukaryotes—a large set of proteins are likely imported into the cellular matrix of both Sulcia and Nasuia in ALF. In the CIP systems alone, host genes that specifically complement incomplete cell functions not only exhibit compensatory expression in the bacteriocytes, but they cannot support these systems unless their protein products are imported into bacterial cells. Given the dramatic gene losses and apparent incomplete cellular functions across bacterial symbionts in the Hemiptera, intracellular support of symbionts by host-encoded proteins is likely to be a widespread phenomenon (100).
The evolution of hemipteran endosymbioses shows important parallels with MT and chloroplast organelles, including extreme gene loss and reliance on the host to complete even the most basic metabolic and cellular functions (96). These parallels raise the fundamental question of what characteristics differentiate obligate bacterial endosymbionts in insects from eukaryotic organelles with symbiotic origins. Currently, a key defining criteria has been the transfer of most metabolic and cellular functions to the host nucleus with compensatory evolution of protein import systems that permit host-encoded gene products to enter the organellar matrix (101, 102). Mounting evidence from a number of systems now strongly challenges the evolutionary specificity of this definition. Protein import of more than 200 genes has been identified from the single-celled eukaryote P. chromatophora (80, 100). Protein translocation to the cellular matrix of bacterial symbiont was similarly demonstrated in the trypanosomatid Angomonas deanei (103). Furthermore, in the insect symbiosis between the pea aphid and Buchnera, it was also shown that a host encoded protein (rlpA4) is imported into the bacterial symbiont’s cytosol (104). Currently, the mechanisms of protein import into hemipteran endosymbionts are completely unknown and offer an area of exciting future work. Investigation of these systems will undoubtedly yield valuable insights into the origin and integration of symbioses throughout eukaryotic evolution.
Materials and Methods
M. quadrilineatus (ALF) specimens were collected from laboratory-reared lines established previously (32). Bacteriocytes and body tissues were dissected from 30 pooled females in three biological replicates. Total RNA was extracted from each pooled sample. Illumina library construction and sequencing were conducted at the University of Texas at Austin Genomic Sequencing and Analysis Facility. De novo transcriptome assembly was performed with Trinity v2.1.1 (105). ORFs (open reading frames) were predicted for each transcript using EMBOSS’s getorf (106) and identified with NCBI-BLASTP v2.2.30+ searches. Differential gene-expression analysis was conducted using edgeR (107). For the ALF draft genome, DNA was extracted from the head and thorax of 45 individuals and sequenced using PacBio approach (Yale Center for Genome Analysis). A hybrid assembly of PacBio and Miseq reads (32) was performed with Spades v3.6.2 (108). HTGs were processed with MEGAN v6.4.0 (109) and individually inspected to remove contaminants. All HTGs were searched against the draft ALF genome using NCBI-BLASTN to verify their location relative to eukaryotic genes. The origins of HTGs were inferred with maximum-likelihood phylogenetic methods in RAxML v8.2.10 (110). The MT support genes and gene duplications were detected based on the NCBI-BLAST results. The location of duplicated genes listed in SI Appendix, Table S2 on the ALF genome was verified with the ALF genome. Phylogenetic reconstructions were performed for duplicated amino acid transporter genes and MT support genes listed in SI Appendix, Table S2. To further verify the expression results of RNA-seq analyses, qRT-PCR was conducted for eight HTGs, one MT support gene, and two MT-encoded genes with the primers listed in SI Appendix, Table S5. The relative abundance of MT in bacteriocytes vs. body tissues was measured with qPCR. Additional information on the experimental and computational methods are available in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank three anonymous reviewers and Dr. John McCutcheon (University of Montana) for their helpful comments and suggestions; Dr. Nancy Moran (The University of Texas at Austin) for her help, laboratory resources, and guidance on the development and implementation of this work; Dr. McCutcheon and Dr. James Van Leuven (University of Montana) for providing the transcriptome data for the citrus mealybug and scrub cicada; Kim Hammond (The University of Texas at Austin) for rearing insects; and The University of Texas at Austin Genomics and Analysis Facility and Yale Center for Genome Analysis for assistance with sequencing. This work was supported by National Science Foundation Award IOS1347116 and start-up support from the University of California, Merced.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw transcriptome and genome reads have been deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession nos. SRP135830 and SRP135903).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811932115/-/DCSupplemental.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 3.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 4.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. The B vitamin nutrition of insects: The contributions of diet, microbiome and horizontally acquired genes. Curr Opin Insect Sci. 2017;23:65–69. doi: 10.1016/j.cois.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Szklarzewicz T, Michalik A. 2017. Transovarial transmission of symbionts in insects. Oocytes: Maternal information and functions, Results and Problems in Cell Differentiation, ed Kloc M (Springer, Cham, Switzerland), Vol 63, pp 43–67.
- 7.Koga R, Meng X-Y, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA. 2012;109:E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wernegreen JJ. Endosymbiont evolution: Predictions from theory and surprises from genomes. Ann N Y Acad Sci. 2015;1360:16–35. doi: 10.1111/nyas.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 10.Dale C, Moran NA. Molecular interactions between bacterial symbionts and their hosts. Cell. 2006;126:453–465. doi: 10.1016/j.cell.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 11.Bennett GM, Moran NA. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCutcheon JP. The bacterial essence of tiny symbiont genomes. Curr Opin Microbiol. 2010;13:73–78. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husnik F, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 16.Buchner P. Endosymbiosis of Animals with Plant Microorganims. Interscience; New York: 1965. [Google Scholar]
- 17.Charles H, et al. A genomic reappraisal of symbiotic function in the aphid/Buchnera symbiosis: Reduced transporter sets and variable membrane organisations. PLoS One. 2011;6:e29096. doi: 10.1371/journal.pone.0029096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloan DB, et al. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 2014;31:857–871. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan J-B, et al. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol Evol. 2015;7:2635–2647. doi: 10.1093/gbe/evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price DR, et al. Sugar transporters of the major facilitator superfamily in aphids; from gene prediction to functional characterization. Insect Mol Biol. 2010;19:97–112. doi: 10.1111/j.1365-2583.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- 23.Price DR, Duncan RP, Shigenobu S, Wilson AC. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Mol Biol Evol. 2011;28:3113–3126. doi: 10.1093/molbev/msr140. [DOI] [PubMed] [Google Scholar]
- 24.Duncan RP, et al. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol Ecol. 2014;23:1608–1623. doi: 10.1111/mec.12627. [DOI] [PubMed] [Google Scholar]
- 25.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 26.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 27.Sloan DB, Moran NA. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett. 2012;8:986–989. doi: 10.1098/rsbl.2012.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Dohlen CD, Kohler S, Alsop ST, McManus WR. Mealybug β-proteobacterial endosymbionts contain γ-proteobacterial symbionts. Nature. 2001;412:433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- 29.Koga R, Bennett GM, Cryan JR, Moran NA. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 2013;15:2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 30.Urban JM, Cryan JR. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea) BMC Evol Biol. 2012;12:87. doi: 10.1186/1471-2148-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett GM, Moran NA. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 2013;5:1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douglas A. Experimental studies on the mycetome symbiosis in the leafhopper Euscelis incisus. J Insect Physiol. 1988;34:1043–1053. [Google Scholar]
- 35.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol. 2005;71:8802–8810. doi: 10.1128/AEM.71.12.8802-8810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett GM, Mao M. Comparative genomics of a quadripartite symbiosis in a planthopper host reveals the origins and rearranged nutritional responsibilities of anciently diverged bacterial lineages. Environ Microbiol. July 25, 2018 doi: 10.1111/1462-2920.14367. [DOI] [PubMed] [Google Scholar]
- 37.Ishii Y, Matsuura Y, Kakizawa S, Nikoh N, Fukatsu T. Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl Environ Microbiol. 2013;79:5013–5022. doi: 10.1128/AEM.01527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noda H, et al. Bacteriome-associated endosymbionts of the green rice leafhopper Nephotettix cincticeps (Hemiptera: Cicadellidae) Appl Entomol Zool. 2012;47:217–225. [Google Scholar]
- 39.McCutcheon JP, Keeling PJ. Endosymbiosis: Protein targeting further erodes the organelle/symbiont distinction. Curr Biol. 2014;24:R654–R655. doi: 10.1016/j.cub.2014.05.073. [DOI] [PubMed] [Google Scholar]
- 40.Bennett GM, Abbà S, Kube M, Marzachì C. Complete genome sequences of the obligate symbionts “Candidatus Sulcia muelleri” and “Ca. Nasuia deltocephalinicola” from the pestiferous leafhopper Macrosteles quadripunctulatus (Hemiptera: Cicadellidae) Genome Announc. 2016;4:e01604–e01615. doi: 10.1128/genomeA.01604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 42.Waterhouse RM, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2017;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikoh N, et al. Bacterial genes in the aphid genome: Absence of functional gene transfer from Buchnera to its host. PLoS Genet. 2010;6:e1000827. doi: 10.1371/journal.pgen.1000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 46.Xue J, et al. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014;15:521. doi: 10.1186/s13059-014-0521-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Leuven JT, Mao M, Bennett GM, McCutcheon JP. July 11, 2018. Cicada endosymbionts have tRNAs that are correctly processed despite having genomes that do not encode all of the tRNA processing machinery. bioRxiv, 10.1101/365791.
- 48.Ioannidis P, et al. Rapid transcriptome sequencing of an invasive pest, the brown marmorated stink bug Halyomorpha halys. BMC Genomics. 2014;15:738. doi: 10.1186/1471-2164-15-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Husnik F, McCutcheon JP. Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci USA. 2016;113:E5416–E5424. doi: 10.1073/pnas.1603910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanholme B, et al. Molecular characterization and functional importance of pectate lyase secreted by the cyst nematode Heterodera schachtii. Mol Plant Pathol. 2007;8:267–278. doi: 10.1111/j.1364-3703.2007.00392.x. [DOI] [PubMed] [Google Scholar]
- 51.Wybouw N, Pauchet Y, Heckel DG, Van Leeuwen T. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol Evol. 2016;8:1785–1801. doi: 10.1093/gbe/evw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray MW. Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci USA. 2015;112:10133–10138. doi: 10.1073/pnas.1421379112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 54.Nicklas JA, Brooks EM, Hunter TC, Single R, Branda RF. Development of a quantitative PCR (TaqMan) assay for relative mitochondrial DNA copy number and the common mitochondrial DNA deletion in the rat. Environ Mol Mutagen. 2004;44:313–320. doi: 10.1002/em.20050. [DOI] [PubMed] [Google Scholar]
- 55.Hunter SE, Jung D, Di Giulio RT, Meyer JN. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010;51:444–451. doi: 10.1016/j.ymeth.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peeters N, Small I. Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta. 2001;1541:54–63. doi: 10.1016/s0167-4889(01)00146-x. [DOI] [PubMed] [Google Scholar]
- 57.Carrie C, Small I. A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:253–259. doi: 10.1016/j.bbamcr.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 58.Duchêne A-M, et al. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Treangen TJ, Rocha EP. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Innan H, Kondrashov F. The evolution of gene duplications: Classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 61.Price DR, et al. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. 2014;111:320–325. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson SN. Trehalose—The insect ‘blood’sugar. Adv Insect Physiol. 2003;31:205–285. [Google Scholar]
- 63.Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: A multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 64.Ratzka C, Gross R, Feldhaar H. Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus. J Insect Physiol. 2013;59:611–623. doi: 10.1016/j.jinsphys.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA. 2009;106:12133–12138. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anselme C, et al. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 2008;6:43. doi: 10.1186/1741-7007-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anselme C, Vallier A, Balmand S, Fauvarque M-O, Heddi A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microbiol. 2006;72:6766–6772. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yano T, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voronin D, Cook DA, Steven A, Taylor MJ. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc Natl Acad Sci USA. 2012;109:E1638–E1646. doi: 10.1073/pnas.1203519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Metcalf JA, Funkhouser-Jones LJ, Brileya K, Reysenbach A-L, Bordenstein SR. Antibacterial gene transfer across the tree of life. eLife. 2014;3:e04266. doi: 10.7554/eLife.04266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: Modulators of the microbiome and inflammation. Nat Rev Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 72.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: From endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 74.Soll J, Schleiff E. Protein import into chloroplasts. Nat Rev Mol Cell Biol. 2004;5:198–208. doi: 10.1038/nrm1333. [DOI] [PubMed] [Google Scholar]
- 75.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 76.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 77.Villarejo A, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- 78.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee CM, Sedman J, Neupert W, Stuart RA. The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J Biol Chem. 1999;274:20937–20942. doi: 10.1074/jbc.274.30.20937. [DOI] [PubMed] [Google Scholar]
- 80.Singer A, et al. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Curr Biol. 2017;27:2763–2773.e5. doi: 10.1016/j.cub.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 81.Millar AH, Whelan J, Small I. Recent surprises in protein targeting to mitochondria and plastids. Curr Opin Plant Biol. 2006;9:610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 82.Baumann P, et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 83.Williams KS, Simon C. The ecology, behavior, and evolution of periodical cicadas. Annu Rev Entomol. 1995;40:269–295. [Google Scholar]
- 84.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 85.Salinas-Giegé T, Giegé R, Giegé P. tRNA biology in mitochondria. Int J Mol Sci. 2015;16:4518–4559. doi: 10.3390/ijms16034518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kumazawa Y, Himeno H, Miura K, Watanabe K. Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J Biochem. 1991;109:421–427. doi: 10.1093/oxfordjournals.jbchem.a123397. [DOI] [PubMed] [Google Scholar]
- 87.Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. The central role of the host cell in symbiotic nitrogen metabolism. Proc Biol Sci. 2012;279:2965–2973. doi: 10.1098/rspb.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rana A, et al. Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc Natl Acad Sci USA. 2010;107:6988–6993. doi: 10.1073/pnas.0912105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balibar CJ, Hollis-Symynkywicz MF, Tao J. Pantethine rescues phosphopantothenoylcysteine synthetase and phosphopantothenoylcysteine decarboxylase deficiency in Escherichia coli but not in Pseudomonas aeruginosa. J Bacteriol. 2011;193:3304–3312. doi: 10.1128/JB.00334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hove-Jensen B, Harlow KW, King CJ, Switzer RL. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem. 1986;261:6765–6771. [PubMed] [Google Scholar]
- 91.Douglas AE. How multi-partner endosymbioses function. Nat Rev Microbiol. 2016;14:731–743. doi: 10.1038/nrmicro.2016.151. [DOI] [PubMed] [Google Scholar]
- 92.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 93.Keeling PJ, Koonin EV. The Origin and Evolution of Eukaryotes: A Subject Collection from Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor Lab Press; New York: 2014. [Google Scholar]
- 94.Ku C, et al. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 95.Husnik F, McCutcheon JP. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol. 2018;16:67–79. doi: 10.1038/nrmicro.2017.137. [DOI] [PubMed] [Google Scholar]
- 96.McCutcheon JP. From microbiology to cell biology: When an intracellular bacterium becomes part of its host cell. Curr Opin Cell Biol. 2016;41:132–136. doi: 10.1016/j.ceb.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schleiff E, Soll J. Travelling of proteins through membranes: Translocation into chloroplasts. Planta. 2000;211:449–456. doi: 10.1007/s004250000357. [DOI] [PubMed] [Google Scholar]
- 99.Carrie C, Giraud E, Whelan J. Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276:1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- 100.Nowack EC, Grossman AR. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc Natl Acad Sci USA. 2012;109:5340–5345. doi: 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Theissen U, Martin W. The difference between organelles and endosymbionts. Curr Biol. 2006;16:R1016–R1017, author reply R1017–R1018. doi: 10.1016/j.cub.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 102.Keeling PJ, Archibald JM. Organelle evolution: What’s in a name? Curr Biol. 2008;18:R345–R347. doi: 10.1016/j.cub.2008.02.065. [DOI] [PubMed] [Google Scholar]
- 103.Morales J, et al. Development of a toolbox to dissect host-endosymbiont interactions and protein trafficking in the trypanosomatid Angomonas deanei. BMC Evol Biol. 2016;16:247. doi: 10.1186/s12862-016-0820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima SY. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr Biol. 2014;24:R640–R641. doi: 10.1016/j.cub.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 105.Grabherr MG, et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 107.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huson DH, Mitra S, Ruscheweyh H-J, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cryan JR, Urban JM. Higher‐level phylogeny of the insect order Hemiptera: Is Auchenorrhyncha really paraphyletic? Syst Entomol. 2012;37:7–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.