Significance
Exosomes, present in most of body fluids, play essential roles in intercellular communication. Here, we found extracellular vesicles, termed “apoptotic exosome-like vesicles” (AEVs), released in the apoptotic cells. Their biogenesis was dependent on the sphingosine-1-phosphate/sphingosine-1-phopshate receptors (S1P/S1PRs) signaling. The S1P/S1PRs complex in the AEVs induced inflammatory mediators such as IL-1α/β, Cox2, Cxcl1, and Ccl2 in macrophages and mice. Therefore, the AEVs in the dying cells are implicated in the pathogenesis of various inflammatory diseases including cancer, autoimmune diseases, chronic allergy, and neurodegenerative diseases. In the future AEVs could be utilized as carriers for delivery of genetic materials or drugs.
Keywords: apoptotic exosome-like vesicles, sphingosine-1-phosphate, sphingosine-1-phosphate receptors, damage-associated molecular patterns, IL-1β
Abstract
Recent research has led to contradictory notions regarding the conventional theory that apoptotic cell death can evoke inflammatory or immunogenic responses orchestrated by released damage-associated patterns (DAMPs). By inducing IL-1β from bone marrow-derived macrophages in an effort to determine the inflammatory mediators released from apoptotic cells, we found that exosomal fractions called “apoptotic exosome-like vesicles” (AEVs) prepared from apoptotic-conditioned medium were the main inflammatory factors. These AEVs showed characteristics of exosomes in their size, density, morphology, and protein expression but had unique marker proteins, sphingosine-1-phosphate receptors 1 and 3 (S1PR1 and 3). Their biogenesis was completely dependent on cellular sphingosine-1-phosphate (S1P)/S1PRs signaling from multiple fine spindles of plasma membrane accompanied by F-actin, S1PR1, S1PR3, and CD63 at the early apoptotic phase and progressing to the maturation of F-actin–guided multivesicular endosomes mediated by Gβγ subunits of S1PRs downstream. S1P-loaded S1PRs on AEVs were critical factors for inducing IL-1β via NF-κB transcriptional factor and p38 MAPK, possibly through the RHOA/NOD2 axis, in differentiating macrophages. The AEVs induced genes of proinflammatory cytokines, chemokines, and mediators in both in vitro and in vivo models. In conclusion, AEVs could be key inflammatory mediators, acting as DAMPs that could explain the pathogeneses of various chronic inflammations, autoimmune diseases, or cancers in the future.
Extracellular vesicles (EVs) are membrane-delineated particles released from cells containing protein, DNA, miRNA, and mRNA that are classified as exosomes, microparticles, and apoptotic bodies. Exosomes are endosome-originating EVs ranging in size from 50–150 nm with specific densities ranging from 1.13–1.19 g/mL. They play important roles in intracellular waste disposal and intercellular communications in normal physiologic processes as well as in cancers. The biogenesis of exosomes involves several stages: invagination of intraluminal vesicles (ILVs) into multivesicular endosomes (MVEs), transport of MVEs to the plasma membrane, fusion of MVEs with the plasma membrane, and subsequent release of ILVs to extracellular space. Upon maturation, the ILVs are sorted into endosomal-sorting complexes required for transport (ESCRT) machinery-dependent or -independent mechanisms (1). The ceramide-dependent process is one of the ESCRT-independent mechanisms (2).
Sphingosine-1-phosphate (S1P), a bioactive lipid, is produced by the phosphorylation of sphingosine, which is processed by the enzymes sphingosine kinase 1 (SPHK1) and SPHK2 and is degraded by S1P lyase (SGPL1). S1P affects a variety of physiologic and pathologic conditions such as proliferation, survival, angiogenesis, immune cell migration, cancers, and inflammatory diseases (3–5). These effects are caused by the inside-out translocalization of S1P and its binding to S1P receptors (S1PR) 1–5. S1PRs are G protein-coupled receptors the signals of which are transduced by activating downstream heterotrimeric G proteins (Gi/o, Gq, and G12/13) (6, 7).
Damage-associated molecular patterns (DAMPs) released from dying cells or injured tissue stimulate surrounding cells or tissues to induce inflammation without pathogenic infection (i.e., sterile inflammation). This inflammation evokes specific immune responses by activating dendritic cells (8). For some time it has been thought that DAMPs are released only from necrotic cells, not from apoptotic cells. However, in specific conditions, especially when massive secondary apoptosis or immunogenic cell death occurs, apoptotic cell death also induces inflammatory or immunogenic responses (9–13). In a previous study we showed that the elements released during apoptotic cell death might induce inflammatory responses (14). In the present study we identified apoptotic exosome-like vesicles (AEVs) as the inflammatory elements released at apoptotic cell death. We then determined the mechanisms of the biogenesis and the inflammatory signal transduction of AEVs, and their biological effects in cellular and animal models.
Results
Conditioned Medium from Apoptotic Cells Activates Macrophages to Induce IL-1β.
To find the inflammatory mediators released from apoptotic cells, we induced apoptosis in Hela cells by staurosporine treatment or amino acid deprivation and primary necrosis by repeated freezing and thawing of Hela cells or treatment of the cells with a high level of H2O2. As a result, the conditioned medium from apoptotic cells contained nucleosomal DNA in laddering patterns, whereas either fully degraded or unbroken genomic DNA was released in primary necrosis (SI Appendix, Fig. S1A). The conditioned medium was then treated with mouse bone marrow-derived macrophages (BMMQs), and BMMQ differentiation was evaluated by measuring mRNAs (15). Interestingly, IL-1β mRNA and protein were markedly increased in the macrophages stimulated with the conditioned media from the apoptotic cells compared with the other macrophages. Also, several genes were expressed more by treatment with the conditioned medium from apoptotic cells than by other treatments (SI Appendix, Fig. S1 B and C and Table S1).
EVs Contribute to the Induction of IL-1β from Macrophages.
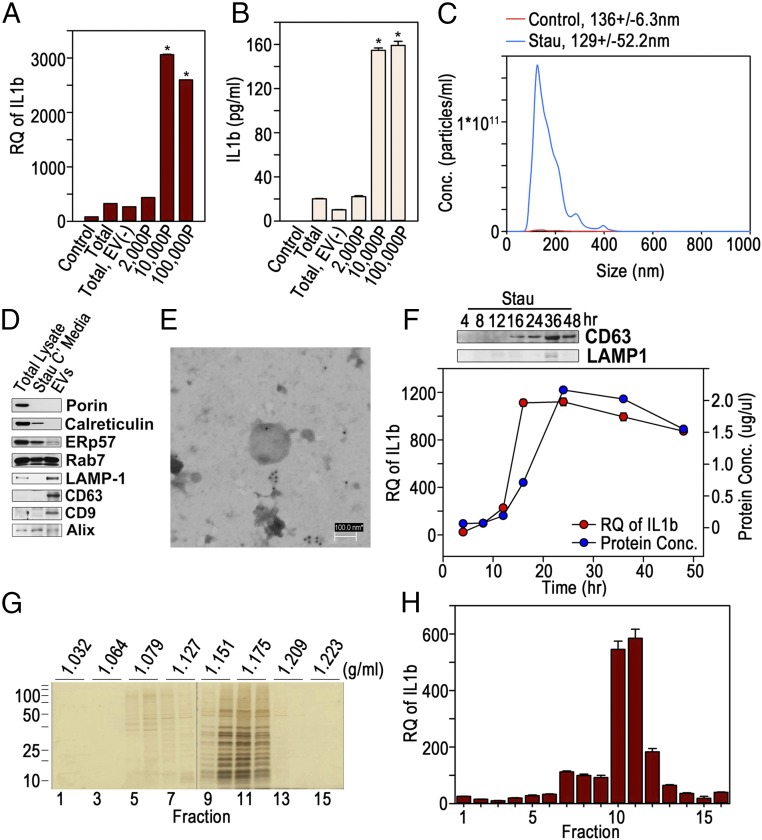
The conditioned medium from apoptotic cells was then sequentially centrifuged, and the EV fractions were collected and used to treat BMMQs. The EV fractions centrifuged at 10,000 × g or 100,000 × g notably induced IL-1β mRNA and protein compared with the conditioned medium (Fig. 1 A and B), suggesting that EVs released from apoptotic cells stimulate macrophages. Therefore, a combined mixture of the 10,000 × g and 100,000 × g fractions was analyzed in the following experiments. The EVs harvested from the conditioned medium of staurosporine-treated HeLa cells were shown to have a mode value of 129.6 ± 52.2 nm in diameter and were released in significantly higher concentrations than from the untreated control cells according to nanoparticle tracking analysis (NTA) (Fig. 1C). They strongly expressed the lysosomal marker LAMP1 and the exosomal markers CD63 and CD9 but showed less expression of the mitochondrial marker Porin, the endoplasmic reticulum (ER) and Golgi markers calreticulin and ERp57, respectively, or the late endosomal markers Rab7 and Alix compared with total lysate or the conditioned medium of staurosporine-treated HeLa cells in Western blots (Fig. 1D). By transmission electron microscopy (TEM), the EVs were found to have a membrane structure composed of a lipid bilayer and to be stained with anti-CD63 Ab (Fig. 1E). Staurosporine-treated HeLa cells released EVs in a time-dependent manner with a peak CD63 level at 36 h by Western blot (Fig. 1F, Upper). The highest release as determined by protein concentrations occurred at 24 h, but IL-1β induction by the EVs showed highest values at 16 h after treatment and then slowly declined (Fig. 1F, Lower). This implies that the activity of a macrophage-stimulating factor within the EVs is likely to have a relatively short half-life. The data showing that EVs incubated at 37 °C for 1 or 2 h lost their activity and failed to stimulate macrophages support this suggestion (SI Appendix, Fig. S2). The EVs loaded onto the OptiPrep density gradient were mainly located between 25% and 30%. The presumed specific density of the vesicles was thus between 1.151 and 1.175 g/mL (Fig. 1G); these fractions were able to activate macrophages to induce IL-1β efficiently (Fig. 1H).
Fig. 1.
Exosome-like vesicles from apoptotic cells activate BMMQs to induce IL-1β. (A and B) EV fractions were collected from the conditioned medium of staurosporine (1 μM)-treated HeLa cells by a series of centrifugations (2,000P: 2,000 × g for 20 min; 10,000P: 10,000 × g for 20 min; 100,000P: 100,000 × g for 70 min). BMMQs were treated with equal amounts (5 μg/mL) of the conditioned medium (Total), the conditioned medium depleted of EVs [Total EV (−)], or the vesicular fractions, and IL-1β mRNA and secreted IL-1β were detected by real-time PCR (A) and ELISA (B), respectively. *P < 0.001 versus conditioned medium. (C) EVs from staurosporine-treated (Stau) or nontreated (Control) HeLa cells were analyzed by NTA. (D) Equal amounts of total cellular lysate, the conditioned medium (Stau C′ media), and the EVs from staurosporine-treated HeLa cells were Western blotted to detect various proteins. (E) A TEM image of an EV from a staurosporine-treated cell stained with anti-CD63 Ab and secondary Ab tagged with gold particles. (F, Upper) The EVs were prepared from the conditioned medium of HeLa cells treated with 1 μM staurosporine and were Western blotted for CD63. (Lower) Protein concentration (right y axis) and IL-1β mRNA (left y axis) were measured by Bradford assay and real-time PCR, respectively. (G) The EVs were loaded onto OptiPrep density gradient and separated according to specific density. Fractions were collected and silver-stained for protein detection. (H) BMMQs were treated with equal volumes of resuspended vesicles from each fraction, and IL-1β mRNA was measured by real-time PCR. Data in triplicate are presented as mean ± SD in A, B, F, and H; *P < 0.001 versus control in A and B.
MVEs Are Formed Within Apoptotic Cells.
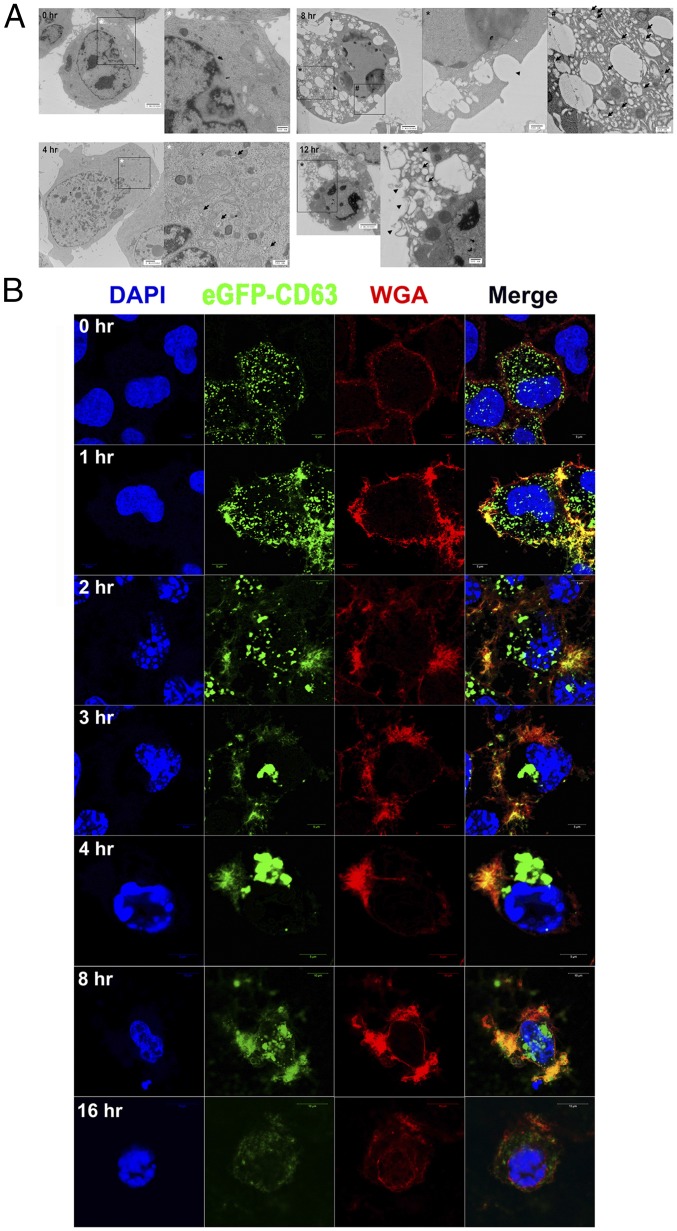
In TEM images of staurosporine-treated HeLa cells, MVEs containing ILVs began to appear at 4 h and increased until 8 h. Vestiges of fusion between the vesicles and plasma membrane were also observed between 8 to 12 h (Fig. 2A). In the confocal microscopic images of HeLa cells expressing eGFP-CD63 and stained with wheat germ agglutinin (WGA) to visualize the plasma membrane, small particles of CD63 were uniformly distributed in both the cytoplasm and plasma membrane at the growing stage. However, numerous fine spindles were formed at the plasma membrane where the eGFP-CD63 particles were colocalized at 1 h after staurosporine treatment. As the apoptotic nuclear changes progressed, the CD63 particles increased in size from the fine spindles and moved to the perinuclear region as MVEs that are clearly shown in the TEM images. The CD63 particles in the perinuclear region then split and moved to the plasma membrane, where they were surrounded by vacuoles. They were released from the vacuoles at 8 h. At the end of the apoptotic process, at 16 h, nearly all the CD63 particles had disappeared from the cells (Fig. 2B). The data suggest that the biogenesis of CD63+ exosome-like vesicles begins with fine spindle-shaped structures at the plasma membrane which subsequently mature into perinuclear MVEs that are released by peripheral movement and fusion with the plasma membrane.
Fig. 2.
MVEs are formed within the apoptotic cells. (A) TEM images of staurosporine-treated HeLa cells at the indicated times. Arrows denote MVEs, and the arrowhead denotes the presumed vestige of fusion between MVEs and the plasma membrane. (B) Confocal microscopic images of staurosporine-treated HeLa cells expressing eGFP-CD63 at the indicated time points and stained with WGA and DAPI.
AEVs Are Released From Various Cells at Various Modes of Apoptotic Cell Death.
The release of EVs was definitely decreased in the cells in which caspase, especially caspase 3 or 9, was knocked down (SI Appendix, Fig. S3A). When treated with macrophages, the EVs of these cells showed decreased induction of IL-1β mRNA compared with EVs from control cells (SI Appendix, Fig. S3B). This implies that the release of EVs is dependent on caspases. The EVs were also able to stimulate differentiated THP-1 cells to generate IL-1β mRNA, as is consistent with BMMQs but not with RAW264.7 cells (SI Appendix, Fig. S4A). The EVs could be released by apoptosis-inducing chemotherapeutic drugs such as cisplatin or VP16, and differentiating BMMQs to produce IL-1β mRNA (SI Appendix, Fig. S4B). Staurosporine treatment could also release EVs to stimulate macrophages in A549, MCF7, and MDA-MB231 cells as well as in HeLa cells (SI Appendix, Fig. S4C).
Taken together, our results revealed that the EVs released during apoptotic cell death showed the characteristics of exosomes with regard to their size, structure, density, and proteins, including CD63, CD9, and LAMP1. The biogenesis of the EVs began with the formation of CD63-containing vesicles close to the fine spindles at the plasma membrane and proceeded through the generation of perinuclear CD63+ MVEs in a caspase-dependent manner. Therefore, the EVs released from apoptotic cells are referred to hereafter as “apoptotic exosome-like vesicles,” AEVs.
Release of AEVs Is Dependent on the S1P/S1PRs Signaling Pathway.
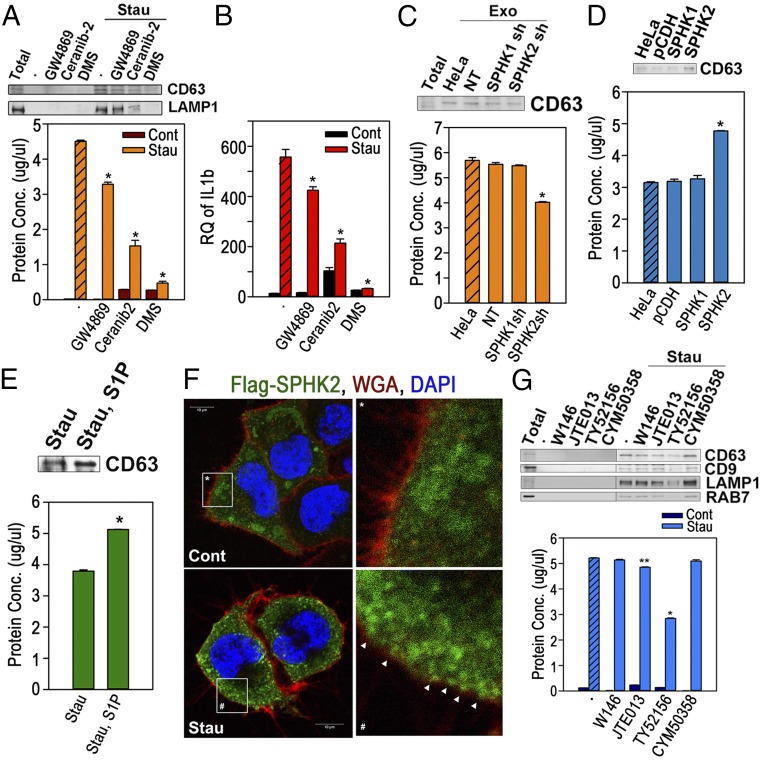
The formation of MVEs is primarily attributed to ESCRT-dependent or -independent mechanisms (16). In addition, RAB27A has been reported to be implicated in the fusion of MVEs with plasma membrane in HeLa cells (17). Therefore, to investigate the role of the ESCRT complex and RAB27A in AEV biogenesis, one of the ESCRT-associated genes, Alix (PDCD6IP) or RAB27A, was knocked out by the CRISPR/Cas9 system (SI Appendix, Fig. S5 A and C). Neither the knockout of Alix nor of RAB27A showed inhibitory effects on the release of AEVs (SI Appendix, Fig. S5 B and D). We then determined whether the sphingolipid-mediated mechanism, one of the ESCRT-independent mechanisms, plays a role in AEV biogenesis (2, 18). The effects of a sphingomyelinase inhibitor, GW4869, a ceramidase inhibitor, Ceranib-2, or a sphingosine kinase inhibitor, dimethyl sphingosine (DMS), on AEV release were determined. Interestingly, DMS cotreatment with staurosporine almost completely blocked the release of AEVs, and the other two inhibitors significantly suppressed the release of AEV as shown by protein concentrations and Western blot (Fig. 3A). The decreased AEVs prepared from cells cotreated with the inhibitors also showed decreased IL-1β induction by macrophages when equal volumes of purified AEVs were treated (Fig. 3B). In addition, the overexpression of SGPL1, an enzyme theat irreversibly degrades S1P, decreased the release of AEVs (SI Appendix, Fig. S6 A–C). Furthermore, the release of AEVs was significantly diminished in the cells in which SPHK2 was knocked down. In contrast, no difference in AEV release was observed in the cells in which SPHK1 was knocked down compared with control cells (Fig. 3C and SI Appendix, Fig. S7 A and B). Also, although the overexpression of SPHK2 significantly increased the release of AEVs, the overexpression of SPHK1 did not (Fig. 3D and SI Appendix, Fig. S8), indicating that the biogenesis of AEVs is undoubtedly related to S1P synthesis in which SPHK2 plays an essential role. Consistent with this, the cotreatment of S1P with staurosporine significantly increased AEV release in HeLa cells (Fig. 3E). In fact, confocal microscopy showed that SPHK2 remained close to the plasma membrane at the early apoptotic phase, in contrast to SPHK1, which moved toward the nucleus (Fig. 3F and SI Appendix, Fig. S9). Collectively, the data show that SPHK2 near the plasma membrane at early apoptosis contributes to the regional increase in S1P, which causes the formation of complex, fine spindle-like structures at the plasma membrane and subsequent maturation of CD63+ vesicles.
Fig. 3.
Release of the AEVs is completely dependent on the S1P/S1PR signaling pathway. (A and B) AEVs were prepared from HeLa cells treated with GW4869 (20 μM), ceranib-2 (20 μM), or DMS (20 μM) in the presence or absence of staurosporine (1 μM) for 16 h. (A, Upper) Equal volumes were Western blotted. (Lower) Protein concentration was measured. (B) BMMQs, treated with equal volumes of the AEVs for 16 h were analyzed for IL-1β mRNA expression. (C) AEVs were prepared from the cells expressing nontargeting shRNA (NT), SPHK1 shRNA, or SPHK2 shRNA. Amounts of the AEVs were measured by Bradford assay (Lower) and Western blots (Upper). Exo, exosome. (D) AEVs prepared from the cells transfected with vector (pCDH), 3×Flag-SPHK1 cDNA, or 3×Flag-SPHK2 cDNA were Western blotted (Upper) and measured for protein concentrations (Lower). (E) AEVs from the cells treated with 5 μM staurosporine in the presence or absence of S1P were Western blotted (Upper), and protein concentrations were measured (Lower). (F) HeLa cells expressing 3×Flag SPHK2 were incubated for 1 h with (Stau) or without (Cont) 1 μM staurosporine and were stained with WGA (red) and anti-Flag Ab (green). The arrowheads indicate SPHK2 spots beneath the plasma membrane. (G) HeLa cells were treated with W146 (20 μM), JTE013 (20 μM), TY52156 (20 μM), or CYM50358 (20 μM) with or without 1 μM staurosporine. (Upper) Equal volumes of the AEVs were Western blotted. (Lower) Protein concentrations were measured. Data in duplicate are presented as mean ± SD; *P < 0.001 and **P < 0.01 vs. hatched bar or control in A–E and G.
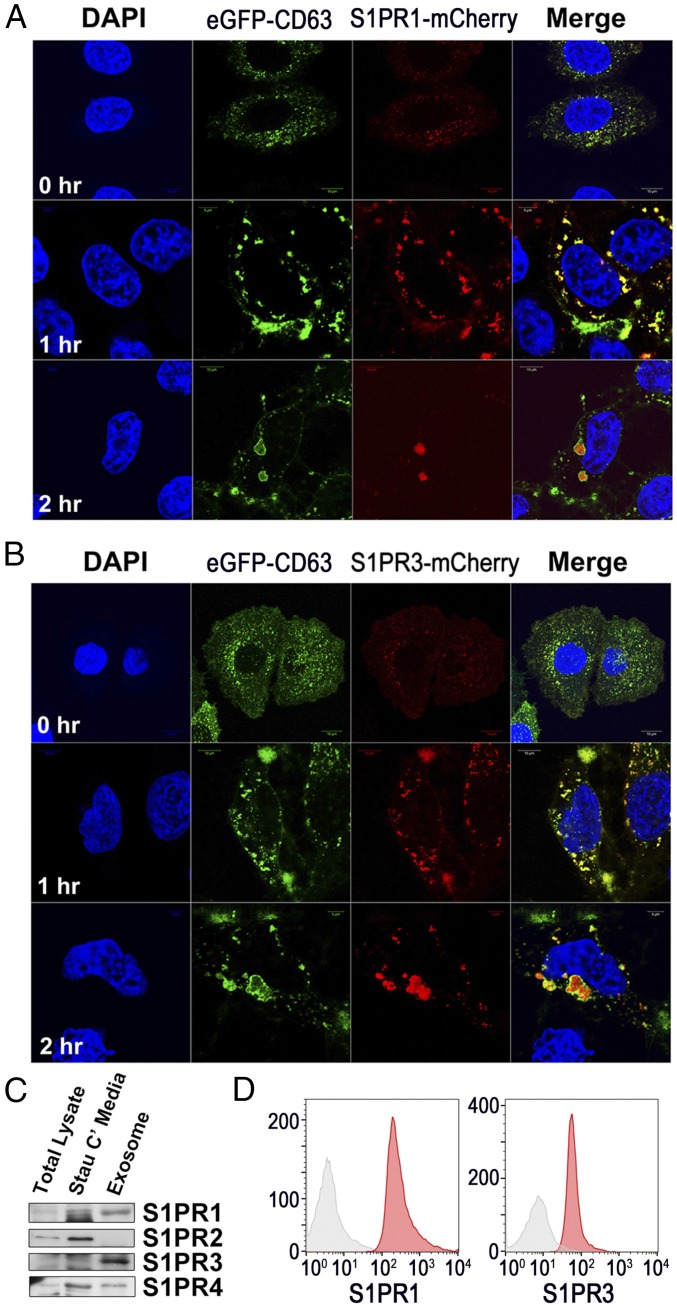
S1P has been shown to be responsible for the formation of ILVs through binding to its receptors S1PR1 and S1PR3 to serve in cargo sorting to exosomes (18). Thus we investigated whether signaling through S1P receptors similarly promoted the biogenesis of AEVs. In the cotreatment experiment with S1PR1–S1PR4 antagonists (W146, JTE013, TY52156, or CYM50258), treatment with the S1PR3 antagonist TY52156 markedly reduced the release of AEVs in apoptotic cells in both Western blots and protein concentrations. The S1PR2 antagonist JTE013 slightly decreased the release of AEVs, but the other antagonists did not reduce the release of AEVs (Fig. 3G). Knockdown of S1PR1 as well as S1PR3 certainly decreased the release of AEVs, and the overexpression of either S1PR1 or S1PR3 significantly increased the amount of released AEVs (SI Appendix, Figs. S7 C and D and S10 A and B). Therefore, low expression of S1PR1 in the parental cells would likely lead to the S1PR1 antagonist W146 having little effect on AEVs’ release. Confocal microscopic images of cells expressing eGFP-CD63 with either S1PR1-mCherry or S1PR3-mCherry showed that S1PR1 and S1PR3 were completely colocalized with CD63 in both the spindle-like structures at the plasma membrane and the cytosolic large vesicles of apoptotic cells 1 h after staurosporine treatment but showed partial overlaps in the growing cells. However, as the size of the perinuclear MVEs increased together with the continued apoptotic process 2 h after staurosporine treatment, CD63 was located mainly at the outer membrane of MVEs, whereas S1PR1 or S1PR3 were localized in the lumen of MVEs (Fig. 4 A and B). These data show that signals through S1PR1 and S1PR3 are likely implicated in the formation of ILVs together with the formation of endosomes originating from the plasma membrane, in accordance with the results reported in a recent publication (18). Furthermore, as shown in Western blots, S1PR1 and S1PR3 were highly concentrated in AEVs compared with total lysate or the conditioned medium of staurosporine-treated HeLa cells and were expressed at the surface of the AEVs in flow-cytometric analysis of the AEV-coated latex beads (Fig. 4 C and D). Collectively, our data imply that the S1P synthetic pathway involving SPHK2 and the subsequent S1P–S1PR1/3 signaling axis induces AEVs biogenesis during apoptotic cell death, proceeding from complex spindle-like structures at the plasma membrane to perinuclear MVEs. In addition, S1PR1 and S1PR3 are highly expressed in AEVs, implying that they can be membrane markers of AEVs.
Fig. 4.
S1PR1 and S1PR3 are surface markers on apoptosis-associated exosome-like vesicles. (A and B) An eGFP-CD63 HeLa cell overexpressing S1PR1-mCherry or S1PR3-mCherry was treated with staurosporine for the indicated time periods. Confocal microscopic images were taken. (C) The exosomal fraction was prepared from staurosporine-treated HeLa cells, Western-blotted for S1PR1–S1PR4, and compared with equal amounts of the either total cellular lysate or the conditioned medium. (D) Exosomal fractions were coated onto aldehyde/sulfate beads and stained with S1PR1 or S1PR3 Ab (red) or with negative control Ab (gray) to confirm surface expression.
Comparison Between AEVs and Conventional Exosomes.
We speculated about the difference between AEVs released in apoptotic cells and conventional exosomes (CEs) released in growing cells. In Western blots executed with equal amounts of protein, AEVs showed higher expression of CD63, LAMP1, HSP70, S1PR1, S1PR3, and S1PR4 than CEs. In contrast, CEs showed higher expression of CD9, ALIX, RAB7, and S1PR2 (SI Appendix, Fig. S11A), confirming that AEVs originate not from the ESCRT-dependent pathway but from S1P–S1PR signaling and possibly contain stress-associated proteins such as HSP70 as a result of the conditions under which AEVs are released. Only AEVs were able to activate macrophages to induce IL-1β mRNA (SI Appendix, Fig. S11B). Interestingly, the AEVs also contained a higher amount of S1P than the CEs, as measured by S1P ELISA (SI Appendix, Fig. S11C), implying that the S1P receptor on the surface of AEVs might be occupied by S1P to directly activate the macrophages.
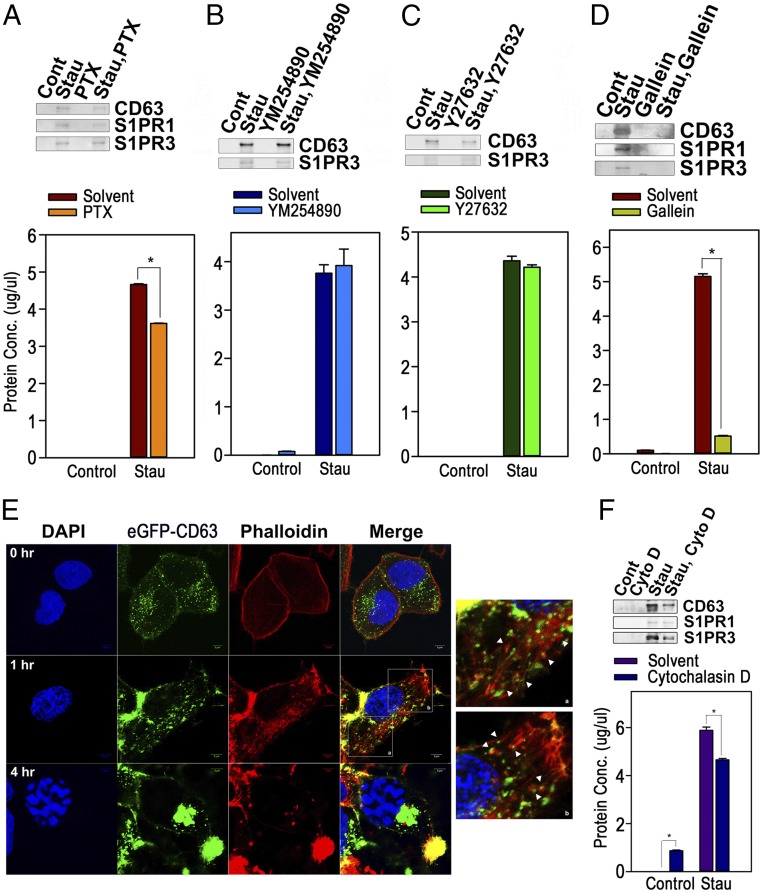
Modulation of the Actin Cytoskeleton Through Signaling of the S1PRs-Coupled Gβγ Subunit Plays a Key Role in the Formation of AEVs.
S1P binding to S1PRs dissociates specific GTP-bound Gα subunits from the Gβγ subunit, resulting in unique down-stream signal transduction, and S1PR1 and S1PR3 are coupled with heterotrimeric complexes of Gi, Gq, or G12/13 (6). Accordingly, we examined the effects of an inhibitor of the Giα subunit [pertussis toxin (PTX)], an inhibitor of the Gqα subunit (YM254890), an inhibitor of ROCK that is a major downstream target of the G12/13α subunit (Y27632), or an inhibitor of the Gβγ subunit (gallein) on the release of AEVs. As shown in Fig. 5 A–D, gallein reduced the release of AEVs most effectively; PTX also partially blocked the release of AEVs, while the others did not. These results suggest that S1P/S1PR signals mediate the biogenesis of AEVs mainly through Gβγ-mediated signaling. In addition, the amount of the exosomal fraction was increased in the serum of LPS-treated mice as compared with control mice in the in vivo mouse model in which a low-dose LPS injection is known to drive systemic apoptosis (19–21). However, pretreatment with gallein blocked the increase in the exosomal fraction (SI Appendix, Fig. S12A). Qualitatively, the exosomal fraction of LPS-treated serum contained more S1PR1+ and S1PR3+ AEVs; AEVs disappeared completely in mice cotreated with gallein and LPS, although treatment with gallein alone could partially increase the AEVs, as shown in Western blots with equal amounts of protein (SI Appendix, Fig. S12B). These results are entirely consistent with the data from the cellular model.
Fig. 5.
Modulation of actin cytoskeleton mediated by Gβγ subunit signaling coupled with S1PRs plays a key role in the formation of exosome-like vesicles. (A–D) HeLa cells pretreated with PTX (200 ng/mL) for 2 h (A), YM254890 (20 μM) for 1 h (B), Y27632 (20 μM) for 1 h (C), or gallein (20 μM) for 1 h (D) were incubated with solvent or staurosporine (1 μM) for 16 h. Exosomal fractions were Western blotted for CD63, S1PR1, or S1PR3 (Upper), and protein concentrations were measured by Bradford assay (Lower). (E) eGFP-CD63 HeLa cells treated with staurosporine were stained with phalloidin. Confocal microscopic images were taken. (Magnification: 400×; Insets: 1,000×.) The arrowheads designate CD63+ endosomes enclosed by F-actin. (F) HeLa cells preincubated with cytochalasin D (20 μM) or solvent were treated with staurosporine for 16 h, and exosomal fractions were prepared. CD63, S1PR1, and S1PR3 were detected by Western blot (Upper), and protein concentrations in the resuspended fractions were measured (Lower). Data in duplicate are presented as mean ± SD; *P < 0.001 versus control in A, D, and F).
Recent research on newly identified roles of the Gβγ subunits has shown that the Gβγ subunit of S1PR can function in the maturation of MVEs for exosomal cargo sorting via the regulation of the actin cytoskeleton through Rho family small GTPase (22). Therefore, we evaluated the changes in the actin cytoskeleton after apoptotic stimuli. Notably, the CD63+ fine, spindle-like structures of plasma membrane observed at 1 h after staurosporine treatment were determined to be exactly coincident with phalloidin-stained F-actin, and the actin filaments extended into the cytoplasm to which CD63+ vesicles were bound or were terminally wrapped. Then, at 4 h after the treatment, CD63+ perinuclear large MVEs were disconnected from the actin filaments (Fig. 5E). Additionally, gallein and DMS cotreatment obliterated the formation of the plasma membrane spindles composed of actin filaments (SI Appendix, Fig. S13), and the release of AEVs was partially prohibited by cotreatment with cytochalasin D, an inhibitor of actin polymerization (Fig. 5F). In the experiments with the overexpression of wild-type, constitutively active, or a dominant negative mutant of Rho family GTPase, the constitutively active mutant of Rac1, Cdc42, or RhoA enhanced the release of AEVs as confirmed by Western blot for CD63, S1PR1, or S1PR3, and by measurement of protein concentrations (SI Appendix, Fig. S14). Collectively, our data show that the S1P/S1PR signaling system stimulates the modulation of the actin cytoskeleton beneath the plasma membrane to prompt endocytosis to form CD63+ and S1PRs+ endosomes and their centripetal movement and maturation to MVEs through the Gβγ-mediated activation of Rho family GTPases.
AEV-Mediated IL-1β Induction Is Dependent on Both NF-κB and p38 MAPKs.
To examine the molecular mechanisms of the AEVs’ effects on macrophages, macrophages were pretreated with various inhibitors of inflammatory signaling (the NF-κB inhibitors JSH23 and Bay 11-7082; an MEK-ERK1/2 inhibitor, PD98059; a JNK inhibitor, SP600125; a p38 inhibitor, SB203580; a PI3K inhibitor, LY294002; and a STAT3 inhibitor, cucurbitacin I) before the addition of AEVs to macrophages. Pretreatment with JSH23, Bay 11-7082, or SB203580 completely prohibited but pretreatment with PD98059, SP600125, LY294002, or cucurbitacin I enhanced IL-1β induction from macrophages (SI Appendix, Fig. S15A). In Western blots executed with control and AEVs-treated macrophages, increases in pro–IL-1β and IL-1β were observed simultaneously with an increase in the processed form of pro-caspase 1 (SI Appendix, Fig. S15B, first and second panels from the top). AEV-treated macrophages also showed increased lactate dehydrogenase release compared with control cells, implying that pyroptic changes in the cells were caused by AEVs (SI Appendix, Fig. S16). Therefore, it is suggested that AEVs might have a factor that stimulates the formation of inflammasomes to degrade pro-caspase 1 to caspase 1, subsequently producing mature IL-1β from pro–IL-1β released through the pores formed by pyroptosis (23, 24). Phosphorylation of MAPKs (ERK 1/2, JNK 1/2, and p38) and processing of the p100 NF-κB subunit to p52 were also increased. However, the degradation of IκBα or an increase of phosphor-IκBα was not observed (SI Appendix, Fig. S15B, panels 3–12 from the top).Therefore, the data indicate that the NF-κB and p38 MAP kinase play a central role in AEV-mediated IL-1β induction, although other MAPKs can be activated also. The activation of NF-κB seems to be caused by mechanisms other than the classic canonical pathway, because neither degradation nor phosphorylation of IκBα was observed. In the ChIP assay, immunoprecipitates with anti-p65, anti-IκBβ, and anti–c-Rel Abs could amplify the NF-κB–responsive sequences of the mouse IL-1β promoter after AEV treatment (SI Appendix, Fig. S15C), which was consistent with the findings that nuclear localization of p65, c-Rel, and IκBβ increased at relatively earlier times than p50 and p52 (SI Appendix, Fig. S15D) and implying that p65, c-Rel, and IκBβ are crucial NF-κB and IκB subunits for IL-1β induction.
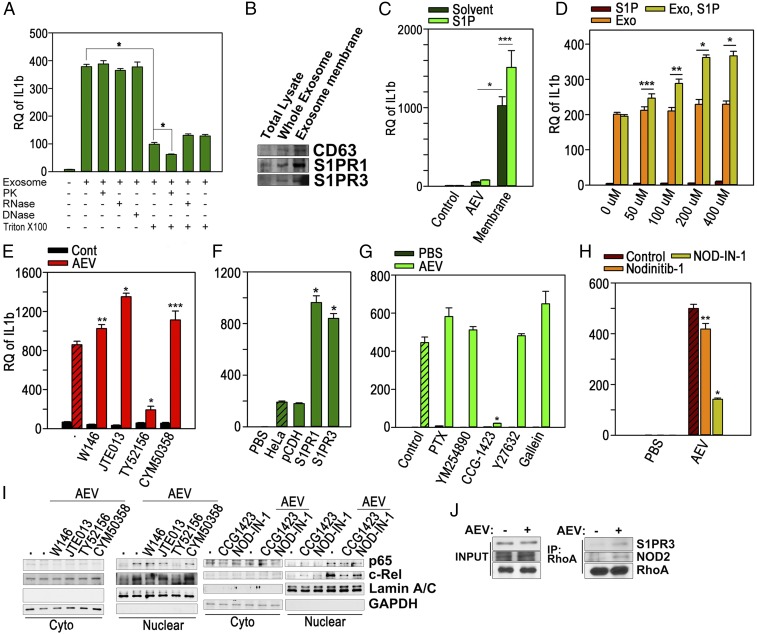
S1P-Loaded S1PR of AEVs Activates BMMQs to Induce IL-1β.
We then determined the component of AEVs that stimulated BMMQs, resulting in IL-1β induction. The AEVs expressing eGFP-CD63 were internalized to the BMMQs within 2 h after the treatment (SI Appendix, Fig. S17), suggesting that any component of the AEVs might stimulate the BMMQs. Pretreatment with C34, an inhibitor of TLR4, or with CPT22, an inhibitor of TLR1/2, showed no effect on AEV-mediated IL-1β induction (SI Appendix, Fig. S18), indicating that TLRs are not implicated in macrophage activation. Therefore, to find the molecular entities of the factor that activates macrophages, AEVs were preincubated with proteinase K, RNase A, DNase I, and/or Triton X-100 before the macrophages were stimulated. As shown in Fig. 6A, IL-1β induction was notably blocked by the preincubation with Triton X-100 but was not influenced by proteinase K, RNase A, or DNase I. However, proteinase K in combination with Triton X-100 resulted in a significant decrease in AEV-mediated IL-1β induction. Additionally, the membrane fraction prepared from the AEVs showed a markedly greater effect on macrophages than the intact AEVs (Fig. 6 B and C). These data imply that either a membrane lipid or a membrane protein of AEVs could be an inflammatory factor. Indeed, when comparing the effects of lysophosphatidic acid, sphingosylphosphorylcholine, or S1P, which is reported to be capable of activating NF-κB as a bioactive lipid (25–27), S1P could induce IL-1β only at a very high concentration (50 μM) (SI Appendix, Fig. S19). We then determined whether S1P-loaded S1PR1 or S1PR3 activated BMMQs to induce IL-1β. A higher IL-1β level was induced in a dose-dependent manner when complete or membrane fractions of AEVs were preincubated with S1P at the final concentration at which macrophages could not be stimulated, compared with the nontreated control (Fig. 6 C and D). Also, cotreatment with the S1PR3 antagonist TY52156 and AEVs almost completely blocked IL-1β induction, but the other antagonists significantly enhanced the IL-1β induction (Fig. 6E). The AEVs prepared from the cells overexpressing S1PR1 or S1PR3 were found to be more potent inducers of IL-1β than the parental or vector-transfected cells (Fig. 6F and SI Appendix, Fig. S20). The cotreatment with TY52156 also inhibited the AEV-mediated nuclear translocation of NF-κB subunits p65 and c-Rel (Fig. 6I, two left panels). Collectively, these data indicate that S1P-loaded S1PR3 directly activates macrophages to induce IL-1β via the NF-κB pathway, and S1P-loaded S1PR1 can probably perform similar roles.
Fig. 6.
S1P-loaded S1PRs on AEVs stimulate macrophages to induce IL-1β. (A) BMMQs were treated for 16 h with AEVs (40 μg) preincubated with proteinase K (PK) (0.2 mg/mL), RNase (10 μg/mL), DNase (10 μg/mL), or Triton X-100 (1%) for 30 min. IL-1β mRNA was observed. (B) Equal amounts of total cellular lysate, AEVs, or membrane fractions of AEVs were Western blotted. (C) BMMQs were treated for 16 h with equal amounts of AEVs or membrane fractions that had been preincubated with S1P (200 μM) at 37 °C for 1 h. IL-1β mRNA was measured. (D) BMMQs were treated with 100 μL AEVs (200 μg) or PBS with/without S1P preincubation for 1 h at 37 °C. IL-1β mRNA was measured. (E) BMMQs were treated with AEVs (5 μg/mL) or PBS with/without W146 (20 μM), JTE013 (20 μM), TY52165 (50 μM), or CYM50358 (5 μM). IL-1β mRNA was measured. (F) BMMQs were treated for 16 h with AEVs prepared from parental HeLa cells or cells transfected with vector, S1PR1-mCherry, or S1PR3-mCherry. IL-1β mRNA was measured. (G) BMMQs were incubated with AEVs (10 μg/mL) or PBS after 1-h pretreatment with PTX (200 ng/mL), YM254890 (20 μM), CCG-1432 (50 μM), Y27632 (20 μM), or gallein (20 μM). IL-1β mRNA was measured. (H) BMMQs, pretreated with DMSO, Nodinitib-1 (20 μM), or NOD-IN-1 (50 μM), for 1 h, were incubated with AEVs or PBS for 16 h. IL-1β mRNAs were measured. (I) BMMQs were treated for 1 h with AEVs (10 μg/mL) in 10% exosome-free FBS MEM in the presence of W146 (20 μM), JTE013 (20 μM), TY52165 (50 μM), CYM50358 (5 μM), CCG-1432 (50 μM), or NOD-IN-1 (50 μM). The nuclear and cytosolic fractions were prepared, and Western blotting was performed. (J) BMMQs treated for 1 h with PBS or AEVs were, immunoprecipitated with anti-RhoA Ab. The lysates (INPUT) and the immunoprecipitates (IP) were Western blotted. Experiments were performed in triplicate. Data are presented as mean ± SD in A and C–H. *P < 0.001; **P < 0.01; ***P < 0.05.
Next, we tried to inspect the downstream signals of S1PRs leading to NF-κB activation; therefore we determined the effects of inhibitors of G proteins on AEVs-mediated IL-1β induction. Among the inhibitors, CCG-1423, an inhibitor of RhoA transcriptional activity, completely prohibited IL-1β induction in the AEV-treated macrophages (Fig. 6G). RhoA is known to be an effector of S1PR-coupled Gα12/13 (28), and RhoA in S1PR signals has been reported to mediate the activation of NF-κB, but its exact mechanisms are largely unknown (29–33). Furthermore, recent reports have shown that the Rho GTPase–nucleotide-binding oligomerization domain-containing protein 1/2 (NOD1/2) axis is implicated in NF-κB activation and in the subsequent expression of inflammatory genes in the cellular responses to DAMPs (34). Therefore, based on these reports, we determined the effects of NOD inhibitors. As shown in Fig. 6H, NOD-IN-1, an inhibitor of NOD1/2, significantly blocked IL-1β induction, but Nodinitib-1, a NOD1-specific inhibitor, showed a little effect. In accord with these data, CCG-1423 and NOD-IN-1 inhibited nuclear localization of p65 and c-Rel in AEV-treated macrophages (Fig. 6I, two right panels). Additionally, the interaction of RhoA with S1PR3 or NOD2 was increased after AEV treatment, as proven by immunoprecipitation (Fig. 6J). Taken together, our data suggest that AEV-mediated NF-κB activation might involve the interplay of SIPRs-Gα12/13-RhoA-NOD2. In this regard, the discrepancy between responses to AEVs in Raw264.7 and PMA-differentiated THP-1 cells (SI Appendix, Fig. S4A) appears to come from the low expression of Nod1/2 and a critical protein kinase of NOD signals, Rip2, in Raw264.7 cells (35), because LPS-primed Raw264.7 cells were able to induce IL-1β although Raw264.7 cells did not respond to the AEVs (SI Appendix, Fig. S21).
AEVs Have DAMPs.
To investigate the way in which AEVs differentiate macrophages, the transcriptome from AEV-stimulated BMMQs was compared with that of nontreated controls. The results showed that 431 genes were up-regulated and 179 genes were down-regulated with a cutoff value of 4 for fold-change of gene expression and P value < 0.05 in AEV-treated BMMQs compared with the controls according to the analysis of differentially expressed genes after RNA sequencing (SI Appendix, Figs. S22A and S24A). In the evaluation of gene ontology (GO) annotation for the 431 up-regulated genes, the largest set of genes was associated with innate immune response or inflammatory responses in a biological process, located in the cytosol, intracellular vesicles, ER, plasma membrane, or extracellular regions, and assorted as cytokines, chemokines, 2′5′-oligoadenylate synthases, binding protein to dsDNA, dsRNA, or protein in molecular function. However, no critical finding was observed in the GO annotation of 79 down-regulated genes (cutoff value = 4; P < 0.01) (SI Appendix, Figs. S23 and S24B). When 431 up-regulated genes were classified according to their governing transcriptional factors in an upstream examination using Ingenuity Pathway Analysis (Qiagen), 201 genes could be controlled by NF-κB, IRF1, IRF3, IRF5, IRF6, IRF7, MAPK, and AP1 or by CREBB1 and CREBBP, which are known to be activated by TLR signals, which are the best known pattern-recognition receptor (PRR) signals (36). NF-κB was the single transcriptional factor affecting the largest set of genes, consistent with our data proving the NF-κB dependency of IL-1β induction (SI Appendix, Figs. S15 and S22B). In addition to PRR signals, a smaller population was controlled by STAT1, STAT2, STAT4, and STAT5a/b, SMARC4A and SMARCB1, CEBPA/B/D, or TP53 (SI Appendix, Fig. S22C). These data clearly imply that the NF-κB signals through the S1P-loaded S1PRs of AEVs play major roles in the differentiation of macrophages to evoke innate immune responses and inflammation, similar to the roles of TLR-mediated signals.
Therefore we confirmed the possibility that the AEVs function as DAMPs in an in vivo mice model. Various proinflammatory genes such as Il1b, Nos2, Cxcl1, Tnfa, Il6, and Cox2 (Ptgs2) were significantly up-regulated in the liver and spleen of the mice i.v. injected with the AEVs, although Il1b was less increased in the mice than in the macrophages (SI Appendix, Fig. S25). Coinjection of TY52156 partially blocked the increase of all genes except Tnfa and Il6; however, W146 showed little blockade or even positive influences on the expression of the genes (SI Appendix, Fig. S26). Proinflammatory genes were expressed at higher values in the mice i.p. injected with the AEVs than in the i.v.-injected mice, and TY52156 inhibited the increased expression of the genes with a statistically significant effect (SI Appendix, Fig. S27). Serum Il1b was markedly increased in the i.p.-injected mice, and this increase was blocked by coinjection with TY52156 (SI Appendix, Fig. S28). These data from in vivo animal studies were largely analogous to the in vitro data from the macrophages and imply that, as functional DAMPs, AEVs are likely to be associated with various pathologic or physiologic processes accompanying cell death and sterile inflammation in the human body.
Discussion
The schematic illustrations in SI Appendix, Fig. S29 summarize our results together with previous findings. At the early phase of apoptosis, SPHK2 remains close to the plasma membrane where it catalyzes the phosphorylation of sphingosine to S1Ps. The S1Ps are then translocalized outside the membrane, where they bind with S1PR1 or S1PR3, resulting in activation of the receptors. The activated receptors transduce signals through the Gβγ subunits of the downstream G proteins and provoke actin polymerization mediated by Rho family GTPases. Next, the modulated actin cytoskeleton enhances the formation of endosomes, which mature to simultaneously form MVEs and ILVs containing both S1PRs and CD63 with possible interplay by Gβγ- and Giα-mediated F-actin modulation. The MVE fuses with the plasma membrane and releases AEVs. The released AEVs enter the macrophages by endocytosis and fuse with the endosomal membrane or fuse directly with the plasma membrane of the macrophages; by this process, the S1P-loaded S1PRs are expressed at the membrane of the macrophages and expose their cytosolic parts to be ready for signal transduction. S1P-loaded S1PRs and their coupled Gα12/13 provide the platform for the NF- κB–activation complex including RhoA/NOD2/RIP2/IKK, which transduces the signals for translocating the NF-κB complex composed of IkBβ, p65, and c-Rel to the nucleus, finally enhancing mRNA transcription of IL-1β as well as various inflammatory cytokines and chemokines in macrophages.
Our data show that in early apoptosis the plasma membrane forms fine, spindle-like structures composed of filamentous actin where S1PR1 and S1PR3 as well as CD63 are colocalized and the maturation of endosomes begins. However, the inhibition of Gβγ subunits by gallein treatment or of sphingosine kinase by DMS almost completely obliterated the formation of the spindle-like structures and subsequently block the release of AEVs (SI Appendix, Fig. S12). This implies that the formation of the plasma membrane fine, spindle-like structures and successive endosome maturation are undoubtedly dependent on S1P/S1PRs signaling. Interestingly, these spindle-like structures closely resemble PANX1-inhibitable apoptopodia, which are known to consist of filamentous actin and to contribute to the release of apoptotic bodies (37). If plasma membrane spindles and apoptopodia are closely analogous structures, this might suggest that S1P/S1PRs’ activation and the following actin polymerization are closely associated with apoptopodia formation in addition to the biogenesis of AEVs.
Nakamura et al. published a series of results that partially concur with our results. They demonstrated novel mechanisms of ILV formation that are dependent on the SPHK2–S1P–S1PRs axis during endosome maturation, contributing to exosomal cargo sorting (18, 22). Although the signaling systems of S1P-S1PR–dependent ILV maturation are likely to be quite similar to those for AEV formation, our data also reveal their differences. First, we show that AEV maturation by the S1P/S1PR axis begins with endosome formation at the plasma membrane in the early apoptotic phase, so that the vesicles are already fated to be AEVs at the stage of endosome formation. Secondly, S1P/S1PR signals regulate not only a specific type of EV but also the total amount of released AEVs. Therefore, the total amount of AEVs could be changed by modulation of the SPHK2–S1P–S1PRs axis. Finally, S1PR1 and S1PR3 are specific markers of AEVs. Therefore, exosomes from S1P/S1PRs signals could also have S1PR1 and S1PR3 in their membrane. However, the relatively small proportions of S1P/S1PR signal-mediated exosomes in the total exosomes released from the growing cells might make detecting the S1PRs difficult. In this context, the AEVs and S1P/S1PRs-mediated exosomes are assumed to be identical entities, but more vesicles are released in the apoptotic situation than in nonstressful conditions.
As shown in SI Appendix, Fig. S12, the formation in the plasma membrane of spindles composed of filamentous actin was controlled by the Gβγ subunits of heterotrimeric G proteins downstream of S1PRs. Indeed, Gβγ subunits were reported to control the Rho family GTPases Rac, Cdc42, and Rho by activating Rho family-specific guanine nucleotide exchange factors (GEFs) such as P-Rex1, PLEKHG2, and ARHGEF18 (38–41). Hence, since the overexpression of constitutively active mutants of Rac1, Cdc42, or RhoA partly enhanced the biogenesis AEVs (SI Appendix, Fig. S13), the activation of Gβγ subunits transfers signals to Rho GTPase through Rho family-specific GEFs, resulting in the modulation of actin cytoskeleton.
In this study we have shown that the release of AEVs relies on caspases 3 and 9 (SI Appendix, Fig. S3). However, it should be noted that the biogenesis of AEVs was initiated from fine, spindle-like structures of the plasma membrane before caspases 3 and 9 were activated. Thus, caspases are assumed to have a role in the relatively late phase of the AEV biogenesis. One possible hypothesis for the role of caspases is that a pore-forming protein, DFNA5, cleaved by caspase 3 (42, 43), contributes to the release of the AEVs from MVEs. Further studies are needed to elucidate this possibility.
Our data demonstrated that S1Ps were already loaded on the S1PRs of AEVs at the time the AEVs activate the BMMQs. First, the AEVs had a much greater amount of S1P compared with exosomes. Second, an S1P/SIPRs signaling system was required for the biogenesis of AEVs; consequently, S1P needed to be loaded onto S1PRs (Fig. 3). Third, despite the time-dependent increase in AEVs after apoptotic stress, the relatively early-phase AEVs could stimulate macrophages more efficiently than late-phase AEVs (Fig. 1F), consistent with the data in SI Appendix, Fig. S2 indicating that the S1P–S1PRs interaction was disrupted with long-term incubation.
It has recently been suggested that activation of Rho GTPases and subsequent cytoskeletal changes provoked by DAMPs might stimulate the intracellular PRRs NOD1/2, resulting in their recruitment to the plasma membrane and their interaction with RIP2 to activate TAK1 and the IKK complex (34). The IKK complex then phosphorylates members of the IκB family (IκBα, IκBβ, and IκBε) (44). In this respect, it is probable that S1P/S1PRs in the AEVs mobilize cytoskeleton through their coupled G protein and downstream Rho GTPases and that NOD 1/2 is then recruited to the membrane of the AEVs within the macrophages. These possibilities agree with our data (Fig. 6 H and I).
The phosphorylation and degradation of IκBα were not observed in the AEV-mediated activation of macrophages despite the NF-κB dependency of IL-1β induction (SI Appendix, Fig. S15A). However, for several reasons these results cannot rule out the possibility that phosphorylation and degradation of IκBα occurred in our model. The AEV-mediated nuclear localization of NF-κB subunits (p65, p50, c-Rel, and RelB) and subsequent IL-1β transcription in FBS-containing medium progressed very slowly (SI Appendix, Fig. S15 B and D), whereas the degradation of phosphorylated IκBα has been known to be an instantaneous process and to be rapidly restored (45). Probably, the competition of the AEVs with exosomes contained in FBS during the process of the cellular entry would result in slow and sequential cellular signal transduction, disturbing the observation of rapid and partial phosphorylation and degradation of IκBα in our system.
According to our data, IκBβ complexed with p65/c-Rel bound to IL-1β promotors (SI Appendix, Fig. S15C). These results are consistent with the previous reports showing that IκBβ complexed with c-Rel plays an essential role in the induction of subsets of inflammatory genes, including IL-1β, in macrophages and in a mouse model (46–48). A newly synthesized, nonphosphorylated form of IκBβ complexed with NF-κB dimers can bind to promotors in the inflammatory genes, helping stabilize the interaction between NF-κB and DNA, because IκBβ has no signaling sequences for nuclear export (48, 49).
Dying cells or tissues release DAMPs that have been known to function as inflammatory mediators similar to pathogen-associated molecular patterns (PAMPs) originating from external pathogens (8), but the exact molecular mechanisms that induce inflammation and the intensities of the inflammatory responses via DAMPs have not been completely understood until now (50). Moreover, it is argued that the IL-1 family cytokines could be genuine DAMPs, functionally reminiscent of PAMPs (51). On the other hand, apoptosis has long been believed to be noninflammatory or even antiinflammatory. However, in some situations apoptosis can be an inducer of inflammation or an adaptive immune response (9–13). The S1P-loaded SIPRs on the AEVs might provide answers to these questions, since they can efficiently induce IL-1α/β, and thus the inflammatory response induced by AEVs would be amplified, eventually reaching the levels of inflammation brought about by PAMPs. The inflammatory response evoked by S1P-loaded S1PRs in the AEVs would be exactly proportional to the total apoptotic load, because the release of the AEVs increases in proportion to the increase in apoptotic cell deaths, and the S1P-loaded S1PRs are not ligands dependent on the cellular receptors for the action but are ligand–receptor complexes independent of the receptors. This explains why massive apoptosis is inflammatory. Nonetheless, the signaling mechanisms from the S1P/S1PR complexes to inflammation should be investigated further, especially to elucidate the presence of other downstream molecules for the signal transduction within the AEVs and the signal mechanisms connecting S1P/S1PR to NF-κB.
Materials and Methods
Animal Experiments.
All animal experiments were performed under protocols and guidelines approved by the review board of Laboratory Animal Research Center of Ajou University Medical Center, Suwon, South Korea.
Preparation of Exosomal Fractions and OptiPrep Density Gradient Centrifugation.
Exosomal fractions from culture supernatants of cells incubated in 10% exosome-free FBS Eagle’s minimum essential medium for 48 h or in conditioned medium from HeLa cells treated with 1 μM staurosporine were purified as described previously (52). For the preparation of AEVs, the conditioned medium from apoptotic cells was centrifuged for 10 min at 200 × g and twice for 20 min at 2,000 × g to remove cellular debris and apoptotic bodies. The pellets were collected and washed twice by ultracentrifugation at 100,000 × g for 70 min. The protein concentration of pellets resuspended with PBS was quantified using the Bradford assay. The vesicles resuspended in sucrose buffer (0.25 M sucrose, 1 mM EDTA, 10 mM Tris⋅HCl, pH 7.4) were loaded onto the discontinuous OptiPrep (Sigma Aldrich) density gradient (5%, 10%, 20%, 25%, 30%, 35%, and 40%) and were ultracentrifuged for 15 h at 100,000 × g, from which fractions were collected. An equal volume of the fraction was electrophoresed in SDS/PAGE gel and silver-stained. For fractionation of vesicular membranes, the exosomes were incubated on ice with 100 mM Na2CO3 (pH 11) for 1 h, washed once and resuspended in PBS.
Confocal Microscopy.
Cells stably expressing CD63-eGFP, S1PR1-mCherry2, S1PR3-mCherry2, 3XFlag-SPHK1, and/or 3×Flag-SPHK2 grown on Lab-Tek four-well glass chamber slides (NUNC A/S) were incubated in medium or medium containing 1 μM staurosporine for the indicated times. In some experiments, the cells were incubated with WGA (2.5 μg/mL) for 10 min at 37 °C and were washed twice with HBSS or anti-Flag M2 Ab for 1 h at 37 °C and were washed five times with PBS. The cells were fixed with 4% paraformaldehyde and were mounted with DAPI-containing mounting medium (Vector Laboratories Ltd). For visualization of F-actin, the fixed cells were incubated with phalloidin (6.6 μM) for 20 min at room temperature and were washed twice with PBS. Images were collected using an LSM710 laser-scanning confocal microscope (Carl Zeiss) equipped with argon (488-nm) and krypton (568-nm) lasers, using a 40× water-immersion objective. Images were processed with ZEN 2009 light edition software (Carl Zeiss).
Real-Time PCR.
Total RNA was isolated using an RNeasy kit (Qiagen). A PrimeScript Reverse Transcriptase reagent Kit (TaKaRa) was used to reverse-transcribe mRNA into cDNA. PCR was then performed on an ABI PRISM 7000 machine (Applied Biosystems) using SYBR Premix Ex Taq II (TaKaRa). The sequences of primers for BMMQ differentiation are given in SI Appendix, Table S2. Analysis of each sample in triplicate was performed more than twice for each experiment, and data in the figures are reported as relative quantification (RQ): average values of 2−ΔΔCT ± SD.
Supplementary Material
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by Ministry of Education Grants NRF-2018R1D1A1B07048257, NRF-2017R1D1A1B03034312, NRF-2016R1D1A1B03934488, and NRF-2013R1A2A2A01008507 and by Grant HI16C0992from the Korea Health Technology Research and Development Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare of the Republic of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra/SRP151316).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811432115/-/DCSupplemental.
References
- 1.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 3.Kunkel GT, Maceyka M, Milstien S, Spiegel S. Targeting the sphingosine-1-phosphate axis in cancer, inflammation and beyond. Nat Rev Drug Discov. 2013;12:688–702. doi: 10.1038/nrd4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyne NJ, et al. Role of sphingosine 1-phosphate receptors, sphingosine kinases and sphingosine in cancer and inflammation. Adv Biol Regul. 2016;60:151–159. doi: 10.1016/j.jbior.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Spiegel S, Milstien S. Sphingosine-1-phosphate: An enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 6.Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S. Sphingosine 1-phosphate receptor signaling. Annu Rev Biochem. 2009;78:743–768. doi: 10.1146/annurev.biochem.78.072407.103733. [DOI] [PubMed] [Google Scholar]
- 7.Takabe K, Paugh SW, Milstien S, Spiegel S. “Inside-out” signaling of sphingosine-1-phosphate: Therapeutic targets. Pharmacol Rev. 2008;60:181–195. doi: 10.1124/pr.107.07113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rock KL, Kono H. The inflammatory response to cell death. Annu Rev Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morillas P, et al. Inflammation and apoptosis in hypertension. Relevance of the extent of target organ damage. Rev Esp Cardiol (Engl Ed) 2012;65:819–825. doi: 10.1016/j.recesp.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Joshi VD, Kalvakolanu DV, Cross AS. Simultaneous activation of apoptosis and inflammation in pathogenesis of septic shock: A hypothesis. FEBS Lett. 2003;555:180–184. doi: 10.1016/s0014-5793(03)01271-7. [DOI] [PubMed] [Google Scholar]
- 11.Kuwano K, Hara N. Signal transduction pathways of apoptosis and inflammation induced by the tumor necrosis factor receptor family. Am J Respir Cell Mol Biol. 2000;22:147–149. doi: 10.1165/ajrcmb.22.2.f178. [DOI] [PubMed] [Google Scholar]
- 12.Faouzi S, et al. Anti-Fas induces hepatic chemokines and promotes inflammation by an NF-kappa B-independent, caspase-3-dependent pathway. J Biol Chem. 2001;276:49077–49082. doi: 10.1074/jbc.M109791200. [DOI] [PubMed] [Google Scholar]
- 13.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 14.Yoon S, et al. Caspase-dependent cell death-associated release of nucleosome and damage-associated molecular patterns. Cell Death Dis. 2014;5:e1494. doi: 10.1038/cddis.2014.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray PJ, et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 17.Ostrowski M, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30; suppl pp 11–13. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 18.Kajimoto T, Okada T, Miya S, Zhang L, Nakamura S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat Commun. 2013;4:2712. doi: 10.1038/ncomms3712. [DOI] [PubMed] [Google Scholar]
- 19.Hamesch K, Borkham-Kamphorst E, Strnad P, Weiskirchen R. Lipopolysaccharide-induced inflammatory liver injury in mice. Lab Anim. 2015;49(Suppl 1):37–46. doi: 10.1177/0023677215570087. [DOI] [PubMed] [Google Scholar]
- 20.Zhong J, Deaciuc IV, Burikhanov R, de Villiers WJ. Lipopolysaccharide-induced liver apoptosis is increased in interleukin-10 knockout mice. Biochim Biophys Acta. 2006;1762:468–477. doi: 10.1016/j.bbadis.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 21.Munshi N, Fernandis AZ, Cherla RP, Park IW, Ganju RK. Lipopolysaccharide-induced apoptosis of endothelial cells and its inhibition by vascular endothelial growth factor. J Immunol. 2002;168:5860–5866. doi: 10.4049/jimmunol.168.11.5860. [DOI] [PubMed] [Google Scholar]
- 22.Kajimoto T, et al. Involvement of Gβγ subunits of Gi protein coupled with S1P receptor on multivesicular endosomes in F-actin formation and cargo sorting into exosomes. J Biol Chem. 2018;293:245–253. doi: 10.1074/jbc.M117.808733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H, Callaway JB, Ting JP. Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma D, Kanneganti TD. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J Cell Biol. 2016;213:617–629. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirrig C, Hunter I, Mathieson FA, Nixon GF. Sphingosylphosphorylcholine is a proinflammatory mediator in cerebral arteries. J Cereb Blood Flow Metab. 2011;31:212–221. doi: 10.1038/jcbfm.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun W, Yang J. Molecular basis of lysophosphatidic acid-induced NF-κB activation. Cell Signal. 2010;22:1799–1803. doi: 10.1016/j.cellsig.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siehler S, Wang Y, Fan X, Windh RT, Manning DR. Sphingosine 1-phosphate activates nuclear factor-kappa B through Edg receptors. Activation through Edg-3 and Edg-5, but not Edg-1, in human embryonic kidney 293 cells. J Biol Chem. 2001;276:48733–48739. doi: 10.1074/jbc.M011072200. [DOI] [PubMed] [Google Scholar]
- 28.Donati C, Bruni P. Sphingosine 1-phosphate regulates cytoskeleton dynamics: Implications in its biological response. Biochim Biophys Acta. 2006;1758:2037–2048. doi: 10.1016/j.bbamem.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 29.Tong L, Tergaonkar V. Rho protein GTPases and their interactions with NFκB: Crossroads of inflammation and matrix biology. Biosci Rep. 2014;34:e00115. doi: 10.1042/BSR20140021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu OM, Brown JH. G protein-coupled receptor and RhoA-stimulated transcriptional responses: Links to inflammation, differentiation, and cell proliferation. Mol Pharmacol. 2015;88:171–180. doi: 10.1124/mol.115.097857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dusaban SS, Chun J, Rosen H, Purcell NH, Brown JH. Sphingosine 1-phosphate receptor 3 and RhoA signaling mediate inflammatory gene expression in astrocytes. J Neuroinflammation. 2017;14:111. doi: 10.1186/s12974-017-0882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G, et al. Critical role of sphingosine-1-phosphate receptor 2 (S1PR2) in acute vascular inflammation. Blood. 2013;122:443–455. doi: 10.1182/blood-2012-11-467191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, et al. Sphingosine-1-phosphate receptor-2 deficiency leads to inhibition of macrophage proinflammatory activities and atherosclerosis in apoE-deficient mice. J Clin Invest. 2010;120:3979–3995. doi: 10.1172/JCI42315. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Keestra-Gounder AM, Tsolis RM. NOD1 and NOD2: Beyond peptidoglycan sensing. Trends Immunol. 2017;38:758–767. doi: 10.1016/j.it.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KH, Biswas A, Liu YJ, Kobayashi KS. Proteasomal degradation of Nod2 protein mediates tolerance to bacterial cell wall components. J Biol Chem. 2012;287:39800–39811. doi: 10.1074/jbc.M112.410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill LA, Golenbock D, Bowie AG. The history of Toll-like receptors–Redefining innate immunity. Nat Rev Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 37.Atkin-Smith GK, et al. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat Commun. 2015;6:7439. doi: 10.1038/ncomms8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayeenuddin LH, McIntire WE, Garrison JC. Differential sensitivity of P-Rex1 to isoforms of G protein betagamma dimers. J Biol Chem. 2006;281:1913–1920. doi: 10.1074/jbc.M506034200. [DOI] [PubMed] [Google Scholar]
- 39.Barber MA, et al. Membrane translocation of P-Rex1 is mediated by G protein betagamma subunits and phosphoinositide 3-kinase. J Biol Chem. 2007;282:29967–29976. doi: 10.1074/jbc.M701877200. [DOI] [PubMed] [Google Scholar]
- 40.Ueda H, et al. Heterotrimeric G protein betagamma subunits stimulate FLJ00018, a guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. 2008;283:1946–1953. doi: 10.1074/jbc.M707037200. [DOI] [PubMed] [Google Scholar]
- 41.Niu J, Profirovic J, Pan H, Vaiskunaite R, Voyno-Yasenetskaya T. G protein betagamma subunits stimulate p114RhoGEF, a guanine nucleotide exchange factor for RhoA and Rac1: Regulation of cell shape and reactive oxygen species production. Circ Res. 2003;93:848–856. doi: 10.1161/01.RES.0000097607.14733.0C. [DOI] [PubMed] [Google Scholar]
- 42.Rogers C, et al. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 44.Clohisy JC, Hirayama T, Frazier E, Han SK, Abu-Amer Y. NF-kB signaling blockade abolishes implant particle-induced osteoclastogenesis. J Orthop Res. 2004;22:13–20. doi: 10.1016/S0736-0266(03)00156-6. [DOI] [PubMed] [Google Scholar]
- 45.Xiao G, Fu J. NF-κB and cancer: A paradigm of Yin-Yang. Am J Cancer Res. 2011;1:192–221. [PMC free article] [PubMed] [Google Scholar]
- 46.Scheibel M, et al. IkappaBbeta is an essential co-activator for LPS-induced IL-1beta transcription in vivo. J Exp Med. 2010;207:2621–2630. doi: 10.1084/jem.20100864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao P, et al. IkappaBbeta acts to inhibit and activate gene expression during the inflammatory response. Nature. 2010;466:1115–1119. doi: 10.1038/nature09283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sen R, Smale ST. Selectivity of the NF-kappaB response. Cold Spring Harb Perspect Biol. 2010;2:a000257. doi: 10.1101/cshperspect.a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam WF, Sen R. IkappaB family members function by different mechanisms. J Biol Chem. 2001;276:7701–7704. doi: 10.1074/jbc.C000916200. [DOI] [PubMed] [Google Scholar]
- 50.Davidovich P, Kearney CJ, Martin SJ. Inflammatory outcomes of apoptosis, necrosis and necroptosis. Biol Chem. 2014;395:1163–1171. doi: 10.1515/hsz-2014-0164. [DOI] [PubMed] [Google Scholar]
- 51.Martin SJ. Cell death and inflammation: The case for IL-1 family cytokines as the canonical DAMPs of the immune system. FEBS J. 2016;283:2599–2615. doi: 10.1111/febs.13775. [DOI] [PubMed] [Google Scholar]
- 52.Thery C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3.22. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.