Significance
Cell chirality, or handedness of the cell, is a newly discovered, fundamental property of the cell, so far studied in cell culture only with micropatterning or graded biomaterial-based approaches. The relevance of intrinsic cell chirality on organ laterality is yet to be established. Cardiac looping is the first organ-specific left–right asymmetry evident during embryogenesis. Despite extensive insights into the molecular signals regulating cardiac left–right asymmetry, the biophysical mechanism is still unknown. Our findings establish intrinsic cell chirality as a regulator of cardiac laterality. This study combines an in vitro chirality assay with embryonic left–right asymmetry in vivo and will significantly impact the understanding and future studies of embryonic left–right asymmetry and congenital heart diseases.
Keywords: cardiac looping, cell chirality, protein kinase C, left–right asymmetry, tissue morphogenesis
Abstract
The vertebrate body plan is overall symmetrical but left–right (LR) asymmetric in the shape and positioning of internal organs. Although several theories have been proposed, the biophysical mechanisms underlying LR asymmetry are still unclear, especially the role of cell chirality, the LR asymmetry at the cellular level, on organ asymmetry. Here with developing chicken embryos, we examine whether intrinsic cell chirality or handedness regulates cardiac C looping. Using a recently established biomaterial-based 3D culture platform, we demonstrate that chick cardiac cells before and during C looping are intrinsically chiral and exhibit dominant clockwise rotation in vitro. We further show that cells in the developing myocardium are chiral as evident by a rightward bias of cell alignment and a rightward polarization of the Golgi complex, correlating with the direction of cardiac tube rotation. In addition, there is an LR polarized distribution of N-cadherin and myosin II in the myocardium before the onset of cardiac looping. More interestingly, the reversal of cell chirality via activation of the protein kinase C signaling pathway reverses the directionality of cardiac looping, accompanied by a reversal in cellular biases on the cardiac tube. Our results suggest that myocardial cell chirality regulates cellular LR symmetry breaking in the heart tube and the resultant directionality of cardiac looping. Our study provides evidence of an intrinsic cellular chiral bias leading to LR symmetry breaking during directional tissue rotation in vertebrate development.
The embryonic heart is the first evident organ to undergo left–right (LR) asymmetric morphogenesis during development. The heart starts as a straight tube along the midline of the embryo. Subsequently, it rotates and bends toward the right, forming a C-shaped loop, with its convexity pointing toward the right of the embryo. Correct handedness of cardiac looping is of crucial importance for proper cardiac morphogenesis during embryonic and postnatal development (1–5). Reversed cardiac looping (leftward rotation) leads to dextrocardia, a congenital heart disorder with severe consequences, characterized by the apex of the heart pointing toward the right side and often associated with other cardiac defects such as double-outlet right ventricle (6). Therefore, it is clinically important to understand the mechanisms associated with asymmetric cardiac looping.
Despite extensive knowledge obtained in the past decades on biochemical and molecular signals for cardiac development, little is known about the biophysical mechanism of cardiac LR asymmetry at the cellular level. No apparent LR asymmetric cellular proliferation, growth, or cell death has been observed between the right and left sides of the heart tube (7–13). Cardiac directional looping is widely believed to be intrinsic to the heart, is independent of the cardiac jelly and cardiac contractions (2, 9), and may arise from a tissue-intrinsic actomyosin-dependent mechanism (11, 14). Recent evidence suggests that chirality at a cellular level, or cell chirality, regulates chiral morphogenesis at a tissue level during development (15–20), as found in directional rotation of Drosophila hindgut and genitalia (18, 19, 21). Cell chirality is a fundamental property of the cell arising from the chiral nature of intracellular macromolecules such as the cytoskeleton and is often observed as biased cell alignment, migration, and rotation as well as intracellular organelle positioning and cytoskeleton dynamics (19, 20, 22–29). We wondered whether cell chirality controls chiral morphogenesis of the heart during vertebrate development.
In this study, we first demonstrate that chick cardiac cells isolated from embryonic hearts before and during C looping are intrinsically chiral with an in vitro cell chirality assay. Then we show that cells in the developing myocardium exhibit overt chirality as evident by a rightward bias of cell alignment and a rightward polarization of the Golgi complex. Concomitantly, N-cadherin and myosin II are enriched on cell boundaries with a right bias before cardiac looping. Furthermore, we demonstrate that the reversal of cell chirality via activation of the protein kinase C (PKC) signaling pathway reverses the directionality of cardiac looping. Our study, therefore, provides evidence of a tissue-intrinsic cellular chiral bias leading to LR symmetry breaking during directional cardiac looping.
Results
Chick Cardiac Cells Isolated from Hearts Before and During C Looping Exhibit Clockwise Chiral Rotation in Vitro.
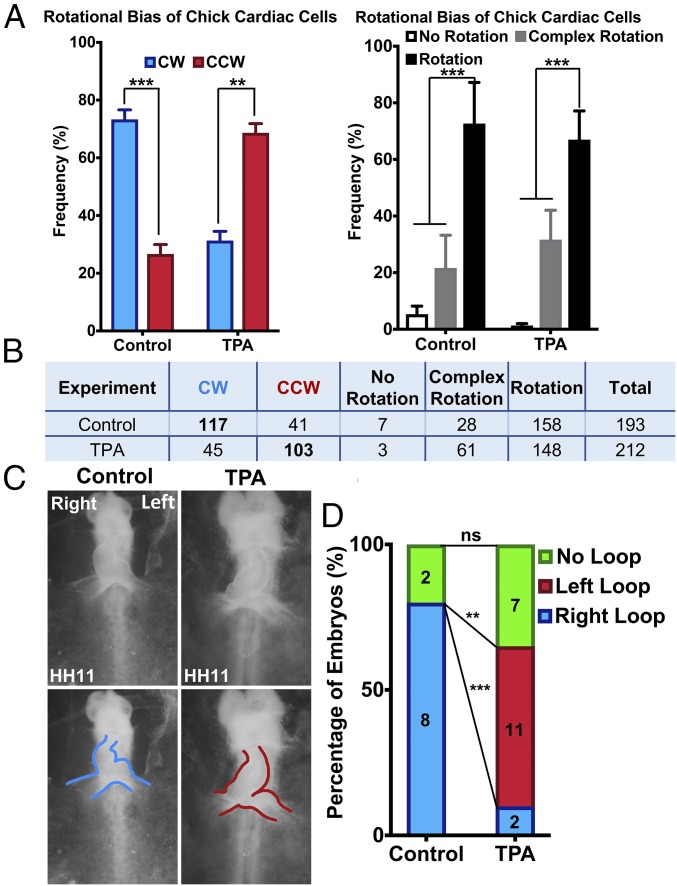
During early embryonic development, the bilateral splanchnic mesoderm merges and folds in a cranial to caudal direction, forming a relatively straight heart tube at Hamburger–Hamilton stage 9 (HH9), which is open along its dorsal side (Fig. 1A and SI Appendix, Fig. S1). Around HH10, the straight heart tube undergoes 90° asymmetric rotation toward the right side of the embryo, which imparts the directionality to cardiac looping (Fig. 1A). We first determined whether cells from the chick heart tube before and during looping are intrinsically chiral. To effectively determine the chirality of less-adhesive cells, we successfully developed an in vitro 3D system, in which cells were cultured at the interface of two layers of Matrigel of different concentrations (Fig. 1B). The cells exhibited persistent rotational motion with a coherent directionality, which is consistent with the previous findings on 2D micropatterns (27). In this study, we isolated cells from the chick cardiac tube before the onset of looping (HH9) until the completion of looping (HH11) and confirmed the cell phenotype with anti–MF-20 (a myocardial marker) staining (Fig. 1 B and C). Utilizing the 3D chirality assay, we categorized the motion of cells as undergoing rotation, no rotation, and complex rotation (for cells undergoing rotation in a different plane or switching the direction of rotation). The rotating cells were further classified as either clockwise or counterclockwise, depending on the directional bias. We found that chick cardiac cells from HH9, HH10, and HH11 cardiac tubes exhibited a dominant clockwise bias of rotation, and therefore the cells were intrinsically chiral before and during looping (Fig. 1 D and E). It is important to note that the percentage of cells undergoing clockwise rotation was highest (73%) at HH10 with a very strong statistical significance, the stage at which the chicken embryonic cardiac tube begins the asymmetric looping. We also analyzed the rotational bias of cells isolated from a single left-looped heart arising due to spontaneous inversion in chicken embryos, and interestingly, those cells exhibited significant counterclockwise rotational motion in vitro with 69% of the rotating cells undergoing counterclockwise rotation, demonstrating the correlation between this intrinsic rotational bias of cardiac cells and the directionality of cardiac looping (SI Appendix, Table S1). These data, taken together, indicate that cardiac cells exhibit intrinsic clockwise chirality during the embryonic looping stages.
Fig. 1.
Chick cardiac cells before and during C looping exhibit dominant CW rotation in vitro. (A) Confocal images of the chicken embryonic heart between HH9 and HH11. Blue arrow indicates the rightward direction of cardiac rotation at HH10. Red, F-actin (Phalloidin); green, anti-ZO1; blue, nucleus (Hoechst). (Scale bar: 200 µm.) A, anterior; E, endocardium; L, left; M, myocardium; OV, omphalomesenteric veins; P, posterior; R, right; and SPL, splanchnopleure. (B) Overview of culture of cardiac cells at the interface of a bilayered Matrigel system. Cells were isolated from the chicken embryonic heart tube during the looping stages, followed by a brief culture in the bilayered Matrigel system before analyzing the directionality of rotation using time-lapse microscopy. (C) MF20 antibody staining showing the abundance of myocardial cells in Matrigel during 3D culture. (Scale bar: 150 µm.) (D) Percentage of cardiac cells isolated from chicken embryos between HH9 and HH11 undergoing CW, CCW, no rotation, complex rotation, and rotation. (E) Table denoting total number of cells from different experiments undergoing CW, CCW, no rotation, complex rotation, and rotation. Boldface type indicates a dominant bias. *P < 0.05, ***P < 0.001; ns, nonsignificant.
Activation of PKC Signaling Reverses Intrinsic Chiral Rotational Bias of Cardiac Cells and the Directionality of Cardiac Looping.
Next, we wanted to identify molecular signaling pathways that regulate the inherent chiral rotation of cardiac cells. We screened for compounds from a library of common drugs that cause congenital laterality defects (SI Appendix, Fig. S2) or birth defects in LR asymmetry. We focused on methoxamine (α1-adrenergic receptor agonist), thalidomide (immunomodulatory and anticancer drug), paroxetine (antidepressant, selective serotonin reuptake inhibitor), lidocaine (anesthetic, numbing agent), and nitrofurazone (antibiotic), covering the major classes of drugs that cause congenital laterality defects (30). We performed a dose-dependent study of the effect of these drugs on the intrinsic chirality of cells isolated from chicken embryonic heart tubes during looping stages (HH9–HH10) (SI Appendix, Fig. S2). We found that thalidomide, paroxetine, and methoxamine increased the percentage of counterclockwise (CCW) cells, suggesting a potential role of PKC in cardiac cell chirality (31–34). Moreover, our work from the 2D micropatterning system with clockwise cells such as human umbilical vein endothelial cell (HUVEC) lines (27) demonstrated that the activation of PKC signaling reverses chirality of endothelial cells from clockwise (CW) to CCW (35). Based on these results we identified PKC signaling as a promising candidate for regulating the chirality of cardiac cells.
Therefore, we next determined whether activation of PKC signaling is sufficient to reverse the intrinsic chirality of cardiac cells. We treated cells isolated from chick cardiac tube at HH10 with 12-O-tetradecanoylphorbol-13-acetate (TPA) (a potent small-molecule activator of PKC signaling) in the 3D bilayer Matrigel culture system and found that TPA treatment at a very low dose significantly switches the rotational bias from CW dominance (73% CW vs. 27% CCW) to CCW dominance (31% CW vs. 69% CCW) (Fig. 2 A and B). This result confirms that activation of PKC signaling is sufficient to reverse the intrinsic chirality of cardiac cells isolated during C-looping stages. We additionally tested Bryostatin 1 and Indolactam V, two other activators of PKC signaling (SI Appendix, Fig. S3) (36, 37). Consistent with our previous results, application of 10 μM Bryostatin 1 reverses the chirality of chick cardiac cells from CW to CCW (SI Appendix, Fig. S3 A and B). Indolactam V treatment of chick cardiac cells similarly results in a significant increase of CCW rotation in the treated cardiac cells compared with the controls (SI Appendix, Fig. S3 C and D). However, the effect of Indolactam V treatment was less pronounced compared with TPA treatment, possibly due to relatively low potency of Indolactam V compared with TPA, as observed in our previous studies and literature (38).
Fig. 2.
Activation of PKC signaling reverses intrinsic chiral rotational bias of cardiac cells and reverses the directionality of cardiac looping. (A) Percentage of CW, CCW, no rotation, complex rotation, and rotation in cardiac cells isolated from HH10 chicken embryos cultured in the 3D bilayered Matrigel system with and without TPA treatment. (B) Table summarizing the number of cells with and without TPA treatment. (C) Images of HH11 chicken embryo cultured with and without TPA categorized into each group. Blue outline shows rightward heart looping and red outline shows left looping, respectively. (D) Percentage of right-, left-, and no-looped hearts in control and TPA-treated chicken embryos. *P < 0.05, **P < 0.01, ***P < 0.001; ns, nonsignificant.
To relate PKC activation directly with cardiac looping, we assessed the activation of PKC signaling in early straight heart tubes by staining HH9 chicken embryos with phospho-PKC-α antibody. We observed phospho-PKC-α–positive cells in the ventral myocardium before cardiac looping (SI Appendix, Fig. S4). More interestingly, our results show that the left side of the heart tube contains a higher amount of phospho-PKC-α compared with the right side of the heart tube (SI Appendix, Fig. S4).
Next, we examined whether PKC activation can reverse the directionality of cardiac C looping. We treated chicken embryos in an ex ovo liquid-sandwich culture system with a varying dosage of TPA added to the culture medium at the HH9 minus stage, just before the formation of the straight heart tube (Fig. 2 C and D). In the control embryos, ∼80% of the heart tubes were right looped, whereas in the experimental embryos the dominant direction of looping was leftward (55%) compared with rightward (10%) (Fig. 2D). Therefore, with TPA treatment, there was a 70% decrease in the percentage of embryos undergoing rightward looping and 55% increase in leftward looping compared with control, both of which were statistically significant (Fig. 2D). Our results demonstrate that the activation of PKC signaling is sufficient to reverse the directionality of chick cardiac looping. Taken together, we demonstrated that a molecular signaling pathway that reverses intrinsic cardiac cell chirality reverses the directionality of cardiac looping.
We next tested the effect of formin inhibition by using SMIFH2 (a small molecule inhibitor) on the directionality of cardiac looping (39). Formin is known to mediate chirality at the actin cytoskeleton level (26) and regulate the establishment of LR asymmetry during development in pond snails and frogs (40). A dose-dependent treatment of SMIFH2 on chicken embryos at the HH9 minus stage caused an increase in no-looped and leftward-looped hearts (SI Appendix, Fig. S5). Based on all of the data taken together, we conclude that modulation of intrinsic cell chirality results in alterations in the directionality of cardiac looping.
The Golgi Complex Exhibits an Overall Rightward Polarization in the Ventral Myocardium with a Stronger Bias on the Right Side Before Cardiac Looping.
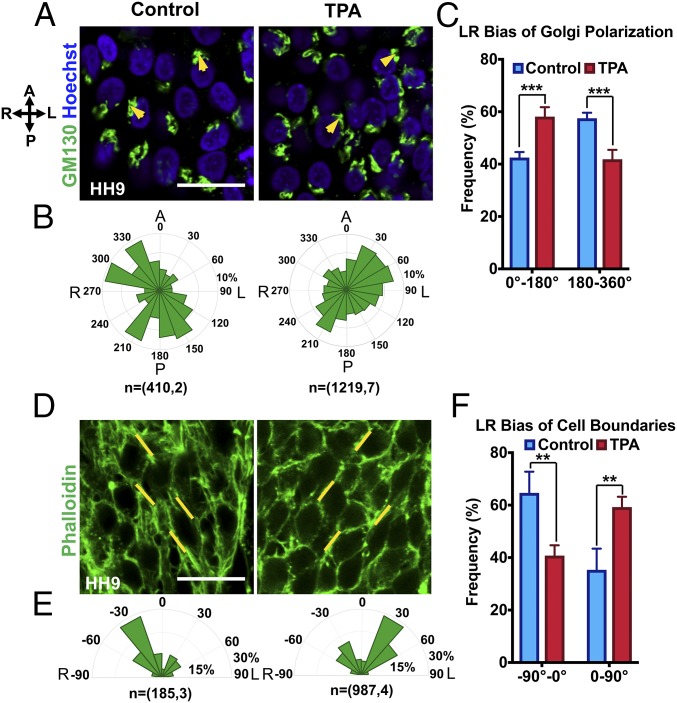
To analyze LR polarity in the myocardium before looping, we examined the polarization of the Golgi apparatus within individual myocardial cells (Fig. 3 A–E and SI Appendix, Fig. S6) in the ventral myocardium. Fluorescently labeled Golgi apparatus with anti-Golgi (GM130) antibody was used to determine Golgi positioning relative to the nucleus (labeled with Hoechst) with respect to the embryonic anterior–posterior (AP) axis (Fig. 3 A and B and SI Appendix, Fig. S6 A–D). LR polarity of the Golgi was determined by the angle between a vector drawn from the centroid of the nucleus to the centroid of the Golgi and the embryonic AP axis (Fig. 3B). The Golgi polarization was defined as anterior left, posterior left, posterior right, or anterior right (Fig. 3C). We observed a preferential polarization of the Golgi toward the embryonic right in the overall ventral myocardium at HH9 with the majority of the angles falling between 180° and 360° (Fig. 3 D and E). Overall, these results indicate that the myocardium before cardiac looping has an overall right bias of Golgi polarization, which could be indicative of directional cell alignment or directional collective cell migration, causing the heart to rotate toward the right.
Fig. 3.
Golgi complex exhibits an overall rightward polarization in the ventral myocardium with a stronger bias on the right side before cardiac looping. (A) Confocal images of Golgi (anti-GM130, green) and nucleus (Hoechst, blue) in the right and the left ventral myocardium, respectively, at HH9. (Scale bar: 20 µm.) (B) Confocal images of Golgi (anti-GM130, green) and nucleus (Hoechst, blue) showing the vector depicting LR bias of the Golgi. (Scale bar: 5 µm.) (C) Color schematic designating the directional regions of Golgi polarization in the embryonic myocardium with respect to the nucleus and embryonic axes. (D and E) Quantification of Golgi polarization with respect to the nucleus along the anterior–posterior and right–left axes. Overall, the Golgi is preferentially polarized toward the right side of the embryo at HH9 before cardiac rotation starts. Right side, left side, and whole refer to the right, left, and entire ventral myocardium, respectively. (F) Cells from the right half of the heart tube exhibit dominant CW rotational bias whereas cells from the left half exhibit randomized rotational bias. Shown is the percentage of cardiac cells isolated from right and left sides of the cardiac tube at HH9 undergoing CW, CCW, no rotation, complex rotation, and rotation. (G) Table denoting the number of cardiac cells from different experiments undergoing CW, CCW, no rotation, complex rotation, and rotation. (H) Activation of Nodal signaling reverses intrinsic chiral rotational bias of cardiac cells whereas inhibition of Nodal signaling has no effect. Shown is the percentage of CW, CCW, no rotation, complex rotation, and rotation in cardiac cells isolated from HH9–HH11 chicken embryos cultured in the 3D bilayer Matrigel system with and without Nodal (1 μg/mL) and SB431542 (75 μM) treatment. (I) Table showing the number of cells with and without Nodal and SB431542 treatment. Boldface type indicates dominant bias. A, anterior; P, posterior. *P < 0.05, ***P < 0.001; ns, nonsignificant.
Intriguingly, we also observed a position-specific bias of the Golgi LR polarity in the myocardium. Cells in the right ventral myocardium (while cardiac fusion is ongoing) at HH9 exhibited a very dominant anterior-rightward bias of Golgi polarization from early HH9 (Fig. 3 D and E and SI Appendix, Fig. S6). Whereas the cells in the left ventral myocardium had no obvious AP polarization and had a relatively randomized LR polarization with a slight left bias. To determine whether there is any position-specific difference in intrinsic chirality of the cells in the straight heart tube at HH9, we separated the right and left halves of the cardiac tube, isolated cells, and analyzed in vitro rotational bias (Fig. 3 F and G). Consistent with the biases of Golgi polarization, cells from the right half exhibited significantly dominant CW rotation (63%) in vitro whereas the cells of the left side had a comparable percentage of cells undergoing CW and CCW rotation with a slightly higher (14%) percentage of cells undergoing CCW rotation (Fig. 3 F and G). These results suggest that whereas cells from the right half of the heart tube showed dominant CW rotational behavior, cells from the left half showed relatively randomized rotational behavior. Overall, these results indicate that cells in the embryonic heart during the looping stages are intrinsically chiral and the dominant chirality of the cardiac tube could originate from cells located in the right half of the cardiac tube.
Since we observed a distinction in the overall chiral bias of myocardial cells between the right and left halves of the cardiac tube, we next asked whether global embryonic LR signals regulate cardiac cell chirality. We examined the effect of application of the Nodal protein and the small molecule Nodal inhibitor (SB431542) on the intrinsic chirality of cells isolated from chicken embryonic heart tubes during looping stages (HH9–HH11) (Fig. 3 H and I). Our results demonstrate that Nodal signaling reverses the chirality of cardiac cells from CW to CCW whereas inhibition of Nodal signaling has no effect on the CW chirality of cardiac cells. This is in agreement with our previous findings that the cells in the right myocardium originating from the Nodal negative mesoderm exhibit dominant CW chirality, whereas the cells in the left myocardium, having contributions from the Nodal-positive mesoderm, exhibit more randomized cellular bias. Additionally, we also examined the effect of BMP signaling on the chiral bias of cardiac cells. Interestingly, inhibition of BMP signaling by the application of small molecule inhibitor LDN193189 does not affect the chiral bias of cardiac cells isolated from heart tubes before the looping stages at HH9, whereas BMP4 application randomizes the chiral bias of cardiac cells at very high concentrations (SI Appendix, Fig. S7). BMP4 at low, physiologically relevant doses, on the other hand, does not affect the chirality of cardiac cells (SI Appendix, Fig. S7). Our results taken together suggest that perhaps right-sided BMP signaling is not instructive for the establishment of cardiac cell chirality but rather provides a permissive environment for the maintenance of the CW chirality of the cardiac cells.
Cell Boundaries in the Ventral Myocardium Exhibit a Dominant Rightward Bias Before Cardiac Looping.
To understand the origin of chiral bias before the onset of cardiac looping, we examined whether there is a resultant LR asymmetric cell alignment in the developing myocardium in addition to the Golgi LR polarization. We hypothesized that the cells in the myocardium exhibit LR polarity in cell shape before the onset of cardiac looping, which is displayed by LR asymmetric cell alignment with respect to the embryonic AP axis. To determine LR asymmetric cell alignment, the morphology of the myocardial cells was examined at stages between HH9 minus and HH11 (Fig. 4 and SI Appendix, Fig. S8). Anti-MF20 and anti-Nkx2.5 antibodies were used to confirm cardiac tissues.
Fig. 4.
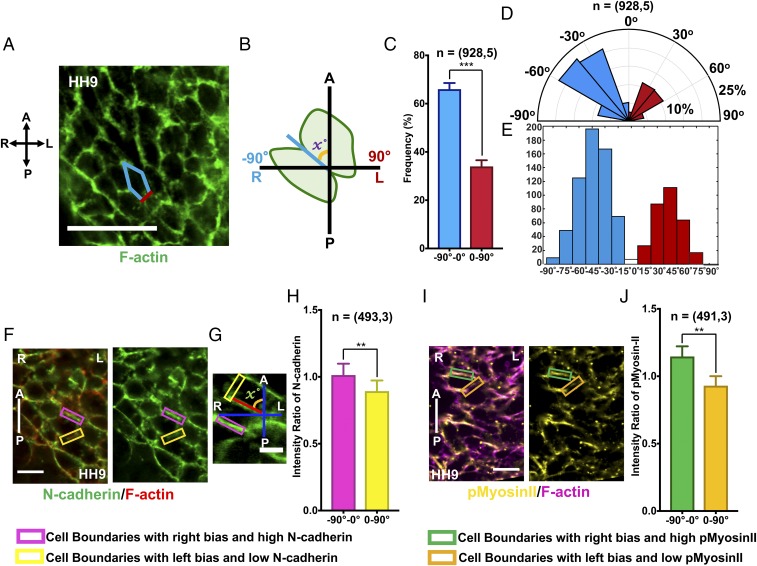
Cell boundaries in the ventral myocardium exhibit a dominant rightward bias of alignment, and N-cadherin and myosin II are enriched LR asymmetrically on cell boundaries before cardiac looping. (A) Confocal image of the chick ventral myocardium at HH9, with Phalloidin (F-actin) staining to visualize cell boundaries. (Scale bar: 50 µm.) (B) Schematic of quantification of the alignment of a cell boundary with respect to the AP axis. Right bias or left bias is determined by the value of x. The angle −90° to 0° is considered as a rightward bias of cell alignment and 0°–90° as a leftward bias. (C) Percentage of cell boundaries in the ventral myocardium at HH9 with angles between −90° to 0° and 0°–90°. (D) Circular histogram of angle distribution of cell boundaries in the ventral myocardium at HH9. (E) Linear histogram of angle distribution of cell boundaries in the ventral myocardium at HH9. Blue denotes cell boundaries with a right bias and red those with a left bias. (F) Confocal images of the chick ventral myocardium at HH9, with N-cadherin (green) and F-actin (Phalloidin, red) staining. (Scale bar: 10 µm.) (G) Magnified images of cells within the ventral myocardium to depict cell boundaries with a right bias and a left bias with respective N-cadherin expression. (Scale bar: 5 µm.) (H) Normalized N-cadherin intensity on cell boundaries in the ventral myocardium at HH9 with angles between −90° to 0° and 0°–90°. (I) Confocal image of the chick ventral myocardium at HH9, with p-Myosin-II (yellow) and F-actin (Phalloidin, magenta) staining. Magnified images of cells within the ventral myocardium depict cell boundaries with right bias and left bias with respective p-Myosin-II expression. (Scale bar: 10 µm.) (J) Normalized p-Myosin-II intensity on cell boundaries in the ventral myocardium at HH9 with angles between −0° to 90° and 0°–90°, respectively. n = (number of cell boundaries, number of embryos). A, anterior; L, left; P, posterior; R, right. **P < 0.01, ***P < 0.001.
Using quantitative analysis of confocal images in ImageJ, we mapped the cell alignment of different regions of myocardium before and during rotation with respect to the embryonic AP and LR axes (Fig. 4 A–E and SI Appendix, Fig. S8). At HH9, ventral myocardial (VM) cells adopt a prominent LR asymmetric shape with respect to the AP axis (Fig. 4A). The majority of the myocardial cells appeared to be aligned toward the right side of the embryo with respect to the vertical AP axis. We measured the angle between cell boundaries (visualized by cortical actin in Fig. 4 A–E) and the AP axis of the heart tube to determine right or left bias (Fig. 4 A–E). Cell boundaries with alignment angles between −90° and 0° were defined to have a right bias, whereas angles between 0° and 90° were categorized as a left bias (Fig. 4B). Just before cardiac looping at HH9, ventral myocardial cells exhibited a dominant rightward bias with the majority of the cell boundaries having an alignment angle between 0° and −90° (Fig. 4 C–E). This rightward bias in the VM disappears by HH10 when the cardiac rotation starts and the cell boundaries are mostly perpendicular to the AP axis (SI Appendix, Fig. S9). These results together demonstrate that the cells of the myocardium have a LR-biased planar cell shape that arises transiently before cardiac rotation. We propose that the appearance of right-biased Golgi polarization and chiral cell shapes in the VM at HH9 marks the onset of symmetry breaking at the cellular level before cardiac C looping.
N-Cadherin and Myosin II Are Enriched on Cell Boundaries with a Right Bias in the Ventral Myocardium Before Cardiac Looping.
To understand whether the LR asymmetric cell shapes are concomitant with the presence of the LR asymmetric distribution of cell adhesion proteins, we analyzed N-cadherin expression along the cell boundaries in the ventral myocardium at HH9 (Fig. 4 F–H). N-cadherin is the primary cell–cell adhesion molecule expressed in the developing heart tube during early morphogenesis and is visible throughout the myocardium during tubular stages. We observed an enrichment of N-cadherin in the cell boundaries that makes an angle between −90° and 0° (right bias) with the AP axis in the myocardium (Fig. 4 F and G, pink rectangles). The normalized N-cadherin intensity ratio was consistently higher on cell boundaries that form an angle between −90° and 0° (right bias, pink rectangles, Fig. 4H, 1.1) compared with cell boundaries that form an angle between 0° and −90° (left bias, yellow rectangles, Fig. 4H, 0.93) with a statistical significance. The higher intensity of N-cadherin on the right-biased cell boundaries was consistent between right and left VM (SI Appendix, Fig. S10). In accordance with the previous findings, the dominance of right-biased cell boundaries is stronger in the right VM than in the left VM (SI Appendix, Fig. S10).
Asymmetric enrichment of N-cadherin is often accompanied by similarly polarized myosin II expression. To determine whether there is a LR-polarized myosin II expression on the VM before cardiac looping, we used immunostaining with phospho-myosin II on chicken embryos. As expected, we observed an enrichment of myosin II on the right-biased cell boundaries of the myocardium (Fig. 4I, green rectangles). The normalized myosin-II intensity ratio was higher on cell boundaries that form an angle between −90° and 0° (right bias, green rectangles, Fig. 4J, 1.15) compared with cell boundaries that form an angle between 0° and −90° (left bias, orange rectangles, Fig. 4J, 0.93). The right-biased enrichment of myosin II was also consistent between the right and the left myocardium. However, similar to the N-cadherin results, the right-biased enrichment was stronger in the right myocardium compared with the left myocardium (SI Appendix, Fig. S11).
These results taken together show that there is a LR-polarized distribution of N-cadherin and myosin II concomitant with the appearance of cell chirality in the myocardium at HH9 before the onset of asymmetric cardiac looping.
Activation of PKC Signaling Reverses Biases of Golgi Polarization, Cell Shape, and the Intracellular Chirality Marker in the Myocardium.
Finally, we wanted to investigate whether the intrinsic LR bias of cardiac cells mediates chiral cell shapes and LR Golgi polarization before the onset of cardiac looping. For this purpose, we examined cell chirality in the myocardium in the embryos after TPA treatment at the HH9 minus stage (Fig. 5). In DMSO control embryos, we observed a dominant rightward bias of Golgi polarization as evident by higher Golgi polarization angles falling between 180ο and 360ο (right biased, 57.5%) compared with between 0ο and 180ο (left biased, 42.5%) (Fig. 5 A–C). In contrast, the bias of Golgi polarization was reversed in the VM after TPA treatment (Fig. 5 A–C). The majority of the polarization angles were between 0ο and 180ο (left biased, 58%) compared with between 180ο and 360ο (right biased, 42%). This indicates that there was a reversal of LR polarization of Golgi in the VM of chicken embryos after PKC activation. Next, we examined the effect of PKC activation on the occurrence of right-biased cell shapes in the VM at HH9 (Fig. 5 D–F). Concomitant with the Golgi polarization results, we observed a reversal of LR alignment of cell boundaries from right bias to left bias (Fig. 5 D–F). In controls, the majority of the cell boundaries were between −90° and 0° (right biased, 65%) compared with between 0° and 90° (left biased, 35%). Comparatively, in embryos that had undergone PKC activation, dominant bias angles of cell boundaries were between 0° and 90° (left biased, 59%) compared with between −90° and 0° (right biased, 41%) (Fig. 5 D–F). We further assessed the effect of PKC activation on the right and the left myocardium (SI Appendix, Fig. S12). A similar extent of reversal of Golgi LR polarization and LR bias of cell boundaries was observed between the left and the right myocardium (SI Appendix, Fig. S12). These results suggest that chirality of cell shapes was reversed in the PKC-activated embryos from right biased to left biased.
Fig. 5.
Activation of PKC signaling reverses cell chirality in the myocardium before cardiac looping. (A) Confocal images of Golgi (anti-GM130, green) and nucleus (Hoechst, blue) in the ventral myocardium of control and TPA (a PKC activator)-treated embryos at HH9. (B) Circular histogram of angle distribution of Golgi polarization in the ventral myocardium at HH9 in control and TPA-treated groups. n = (number of cells, number of embryos). (C) Quantification of Golgi polarization with respect to the nucleus along the anterior–posterior and right–left axes for control and TPA-treated embryos. (D) Confocal images of the chick ventral myocardium at HH9 for control and TPA-treated chicken embryos, with Phalloidin (F-actin) staining to visualize cell boundaries. (E) Circular histogram of angle distribution of cell boundaries in the ventral myocardium at HH9 for control and TPA-treated embryos. n = (number of cell boundaries, number of embryos). (F) Percentage of cell boundaries in the ventral myocardium at HH9 with angles between −90° to 0° and 0°–90° for control and TPA-treated embryos. **P < 0.01, ***P < 0.001. A, anterior; L, left; P, posterior; R, right. (Scale bars: 20 µm.)
Taken together, these data suggest that PKC activation reverses cell chirality in the myocardium, leading to reversal of directionality of cardiac looping. We have already demonstrated that PKC activation also reverses the bias of intrinsic chiral rotation of chick cardiac cells during the looping stages. Therefore, these results indicate that intrinsic cellular chirality regulates LR symmetry in the myocardium before cardiac looping through mediating LR polarization of Golgi and chiral cell shapes. To confirm that PKC activation reverses chirality within the cells of the VM in vivo during cardiac looping, we used LR bias of the cell centroid with respect to the nuclear–Golgi axis as an intracellular chirality marker (SI Appendix, Fig. S13 A–E). We defined left- or right-biased cells, depending on the positioning of the cell centroid on the left or right of a straight-line vector drawn from the centroid of the nucleus to the centroid of the Golgi, respectively (SI Appendix, Fig. S13E). This bias of the cells was reversed in the PKC-activated embryos, compared with the untreated control (SI Appendix, Fig. S13F). This could be indicative of a reversal of internal LR asymmetry within the myocardial cells. Therefore, we propose that reversing intrinsic cell chirality via activation of PKC signaling switches directionality of cardiac looping through reversal of cell chirality in the myocardium. Overall, these results strongly suggest that intrinsic cellular chirality of the myocardial cells instructs the directionality of rightward rotation of the embryonic cardiac tube.
Discussion
Our study demonstrates that intrinsic cell chirality regulates the directionality of cardiac looping, which is essential for proper cardiac morphogenesis (4, 6). We show the LR symmetry breaking at the cell level in the cardiac tube before the onset of asymmetric looping, as observed by LR biases in the positioning of the Golgi complex and the distributions N-cadherin and myosin II in the myocardial cells. The reversal of cell chirality switches the cellular LR asymmetry as well as the looping directionality of developing cardiac tubes, suggesting the critical role of intrinsic cell chirality in cardiac looping.
Intrinsic cell chirality is a property of the cell dependent on phenotype and enables polarized cells to prefer right or left bias during collective migration or rotation. Previously, phenotype-based intrinsic cell chirality has been implicated in multicellular morphogenesis on 2D micropatterns (15, 16, 20, 27–29, 41). Recently, we developed a platform for determining the chirality of less-adhesive cells in 3D culture, by analyzing the directionality of coherent angular motion, when cultured at the interface of two layers of Matrigel of different concentrations. Here we show that cells in the cardiac tube before the onset of looping exhibit a dominant CW rotational bias. Previously, we demonstrated that epithelial spheroids derived from the single rotating cells retain the coherent angular motion and the directionality of the single cell. It is tempting to speculate whether a similar scenario is applicable in the context of cardiac rotation, in which the top–bottom/z-axis polarity of the cardiac cells could be related to the AP polarity of the cardiac tube and the CW rotational direction correlates that to the rotational bias of the individual cardiac cells.
The CW chiral bias of the cardiac cells and the rightward cell alignment and Golgi polarization in the VM are evident from the early HH9 stage before cardiac fusion and before the onset of cardiac contraction. The rightward cell alignment is transient and disappears when the rotation begins (SI Appendix, Fig. S9). This finding is in accordance with the Drosophila embryonic hindgut and genitalia chiral rotation, where cell shapes exhibit transient LR polarity as the cells align with a left or right bias with respect to the AP axis which mediates the directionality of rotation (18, 19, 21).
One of the key findings of this study is that PKC activation reverses the handedness of cardiac looping and correspondingly the chirality of the cardiac cells. This is supported by the fact that PKC activators such as TPA and Indolactam V switch the chirality of endothelial cells from CW to CCW (35). One of the promising PKC isoform candidates is PKC-α which mediates the switch of cell chirality in endothelial cells. Interestingly, our results show the presence of activated PKC-α in the cardiac tube before the onset of cardiac looping. Further investigation is needed about which isoforms mediate the switch in chirality in cardiac cells and the role of endogenous PKC signaling during cardiac looping.
It is intriguing to note that our results suggest that the chirality of the overall cardiac tube possibly originates from the right side. Both in our in vitro rotation data and in the in vivo myocardial chirality results, the right side is more chiral with a dominant CW bias whereas the left side is comparatively randomized. This is supported by a previous study on chicken embryos indicating that the cardiac looping directionality could originate from actin alignment in the right caudal myocardium (42, 43). Very recently, it has been suggested that LR asymmetric cell movement and forces originating primarily from the right side and controlled by PRRX1 signaling in chick, Prrx1a in zebrafish, and SNAIL1 in mouse direct cardiac laterality (11). On a similar note, the right and the left side of the embryonic heart tube have differential heparanase activity with the left side having higher heparanase activity. Heparanase cleaves syndecan 4, activating PKC-α (44, 45). Therefore, the left side of the heart could have higher levels of PKC-α activation than the right side. Indeed, in agreement with this, we found relatively higher expression of phospho-PKC-α in the left half of the myocardium than in the right half, showing that there are higher levels of PKC activation in the left myocardium compared with the right. This is also supported by our results that cells in the right side of the heart have CW chirality whereas cells in the left side have a slight bias toward CCW chirality. Injecting heparanase on the right side reverses the looping direction, similar to our results of activation of PKC signaling reversing the looping directionality of the cardiac tube. In summary, relating cardiac laterality to myocardial cell chirality predominantly originating from the right side could explain the cellular basis of a right-sided biochemical signaling pathway driving cardiac laterality.
Recent studies have suggested that an actomyosin-dependent mechanism regulates cardiac looping. Our results show that myosin II along with N-cadherin shows preferential enrichment along the cell boundaries that have a right bias. One of the predominant mechanisms proposed to explain cardiac laterality is LR asymmetric actomyosin contractions (14, 17, 19, 21, 46). Similarly, during LR asymmetric hindgut rotation in Drosophila embryos, DE-Cad and MyoID enrichment on the cell boundaries with a bias between −90° and 0° seems to restrict the expansion of cell boundaries, introduce LR asymmetrical cortical tension, and facilitate directional tissue rotation (19). During Drosophila genitalia rotation, in which epithelial tissue rotates clockwise around the genitalia, MyoID regulates LR asymmetric polarized enrichment of myosin II, leading to LR-biased junction remodeling and LR asymmetric cell intercalation resulting in directional collective cellular movement (21). We propose that before cardiac looping, similar LR polarization of N-cadherin and myosin II on cell boundaries could lead to LR asymmetric cellular contraction and junctional remodeling, driving directional tissue rotation. However, further studies have to be undertaken to elucidate the precise role of LR-biased enrichment of N-cadherin and myosin II in myocardium before the onset of cardiac looping.
In this study, we have also addressed how cell chirality relates to the global molecular signaling pathways establishing embryonic laterality such as Nodal (8, 47) and BMP (11, 48). Our results show that application of Nodal protein switches the chirality of cardiac cells from CW to CCW. Interestingly, the chick cardiac cells retain their CW bias even after the inhibition of Nodal signaling. This is in accordance with our results where the right side of the heart tube, originating from Nodal negative right lateral plate mesoderm, exhibits dominant CW chirality. Whereas the left side of the heart, originating from the Nodal positive side, shows more randomized chiral behavior with a slightly higher percentage of cells undergoing CCW rotation. Our results also suggest that BMP signaling is not required for the maintenance of CCW chiral bias of cardiac cells and possibly plays a permissive rather than an instructive role in the establishment of cell chirality. However, it is possible that downstream effectors of BMP signaling such as PRRX1 are instructive during the establishment and maintenance of cardiac chirality. The dose-dependent role of BMP4 signaling in regulating cell chirality is similar to previous findings of a dose- and time-dependent effect of BMP4 on cardiac laterality during development (49–51). For example, injection of a high concentration of BMP4 to the right side of the zebrafish embryos reverses cardiac asymmetries (51). In summary, the myocardial chirality could be established during the initial differentiation of cardiac mesoderm but regulated in a position-specific manner by the embryonic LR signals. Recently it has been shown that myo1d is upstream of the Nodal cascade in Xenopus and zebrafish and is the unifying feature of LR asymmetries across phyla (52–54). Combining these data with our results shows that there is possibly an interplay between cell chirality and the global embryonic LR signals, where early on during development, cell chirality regulates the establishment of the embryonic LR axis, which in turn regulates the position-specific variation in cell chirality of cardiac mesoderm, regulating the future directionality of cardiac looping.
In summary, our study introduces tissue intrinsic cell chirality as a biophysical and cellular basis of cardiac laterality and possibly as a universal mechanism regulating directional tissue rotation during vertebrate embryogenesis.
Materials and Methods
Chicken Embryos.
Fertilized chicken eggs (Charles River Laboratories) were incubated to the desired stages at 38.5 °C in a humidified incubator in accordance with protocols approved by the Institutional Review Board at Rensselaer Polytechnic Institute. Embryos were staged according to the HH table of normal stages (55) and harvested in normal saline solution.
Cell Isolation from Chick Heart Tubes.
Heart tubes were dissected out from chicken embryos in ice-cold Leibovitz’s L15 medium (Gibco) supplemented with 1% penicillin–streptomycin (Sigma-Aldrich), between stages HH9 and HH12, using a flame-sharpened tungsten needle. The tissues were immediately digested with 0.25% Trypsin/EDTA (Gibco) solution (prewarmed at 37 °C) or Accutase (Sigma-Aldrich) at 37 °C. Every 4 min, the suspension was pipetted up and down several times to facilitate digestion and homogeneity; this was repeated three times for a total of 12 min. Freshly prepared Neurobasal Medium (Gibco) supplemented with B27 (Gibco), 5% FBS (VWR), 1% penicillin–streptomycin (Sigma-Aldrich), and 1% GlutaMax (Gibco) was added to neutralize the Trypsin solution and stop digestion. A cell pellet was obtained by centrifuging at 300 × g for 5 min. Finally, the pellet was resuspended in the supplemented prewarmed Neurobasal Medium before seeding within the bilayered Matrigel for 3D chirality analysis.
Rotational Analysis of Cardiac Cells in a 3D Matrigel Gradient System.
The preparation of the base layer of growth factor-reduced Matrigel (Corning) was coordinated with the digestion of the chick cardiac cells, so that the digested cells could be immediately seeded on the Matrigel base layer. The bilayered Matrigel system was formed using an eight-chambered glass slide (Ibidi) consisting of a base layer of 100% Matrigel and a top layer of 2% Matrigel. First, a bottom layer of 100% Matrigel was prepared by pipetting 50 μL of ice-cold Matrigel into the individual chamber. A pipette tip was gently used to spread the Matrigel on the bottom of the well to achieve an even bottom layer. The prepared bottom layer of 100% Matrigel was incubated at 37 οC for 20 min for solidification. Freshly isolated chick cardiac cells were then seeded onto the Matrigel base layer at a density of 6,000 cells/cm2 and were allowed to attach for 2 h in Neurobasal Medium (with B27 and 5% FBS and 1% Glutamax and 1% penicillin–streptomycin). The medium was replaced with ice-cold 2% Matrigel in fresh cold Neurobasal Medium (with or without drugs) to form the top layer. The pipette tips were prechilled to ensure that the 2% Matrigel did not solidify while pipetting onto the wells. The cells were cultured at 37 °C and 5% CO2 for at least 16 h.
After 1 d of culture time-lapse imaging was performed using an inverted Zeiss Axio Observer.Z1 or a Keyence BZ-x700 microscope for analyzing single-cell/tissue rotation. Phase-contrast time-lapse images were gathered with a 10× objective for at least 2 h with an interval of 1 min. The cells were categorized as undergoing rotation (planar) or no rotation or complex rotation (out of plane or switching directionality). The direction of planar cellular rotation was classified into either CW or CCW.
Immunostaining and Imaging of Chicken Embryos.
Chicken embryos were fixed in 4% paraformaldehyde (PFA) for 1 h at room temperature. Embryos were washed once in 1× PBS and three times in 1× PBST (1× PBS + 0.1% TritonX-100 + 2% BSA) for 5 min each. Next, the embryos were blocked in 1× PBST with 10% goat serum for 1 h. Primary antibodies were applied for 24–48 h at 4οC. The following primary antibodies were used: anti-ZO-1 (1:50, ab59720; Abcam), anti-MF20 (1:10, MF 20; Developmental Studies Hybridoma Bank), anti-Nkx2.5 (1:100, ab35842; Abcam), anti-GM130 (1:100, BDB610822; Fisher Scientific), anti–N-cadherin (1:20, 6B3; Developmental Studies Hybridoma Bank), anti–phospho-myosin II (1:50, 3671S; Cell Signaling Technology) and anti–phospho-PKC-α (Thr-638, 1:100, 44-962G; ThermoFisher Scientific). The slides were washed several times in 1× PBS, followed by three 1× PBST washes, each for 5 min. Alexa Fluor 488/568 secondary antibodies (1:200 in 1× PBST; ThermoFisher Scientific) were applied for 2 h at room temperature in a humidified chamber followed by several 1× PBS and 1× PBT washes of 5 min each. Alexa Fluor 488/568 Phalloidin (1:40 in 1× PBST; ThermoFisher Scientific) was applied with the secondary antibodies for visualization of actin. The embryos were then incubated with Hoechst 33342 (H3570, 200 μg/mL in 1× PBS; ThermoFisher Scientific) for 30 min. A final wash with 1× PBS for 5 min was followed by serial washes in 50% glycerol/PBS and 90% glycerol/PBS for clearing and imaging. The embryos were mounted using SlowFade Diamond AntiFade reagent (S36963; ThermoFisher Scientific). All imaging was performed using an inverted Zeiss LSM 510 confocal microscope. Image processing and analysis were performed using Fiji (ImageJ).
Immunostaining of Chick Cardiac Cells Embedded in Matrigel.
Cardiac cells in Matrigel were fixed with a prewarmed mixture of 4% PFA and 1% glutaraldehyde for 15 min. This was followed by quenching in sodium borohydride (1 mg/mL; Sigma Aldrich) before proceeding with normal immunostaining protocol as described above. All of the steps were performed at room temperature.
Chicken Embryo Culture and Drug Treatments.
Chicken embryos were cultured using either a sandwich liquid culture method (56) or an EC culture method as described previously (57). For the liquid culture of chicken embryos, drugs were added directly into the prewarmed media (Neurobasal supplemented with B27, 5% FBS, 1% penicillin–streptomycin, and 1% GlutaMax) at HH9 minus stage. TPA (10–50 nM, P8139; Sigma-Aldrich), Bryostatin 1 (10 nM–10 μM, IB7431; Sigma-Aldrich), and Indolactam V (10 nM–500 μM, I0661; Sigma-Aldrich), three common cell-permeable, small-molecule activators of PKC, were used in our experiments (36, 37, 58, 59). For chicken embryo liquid culture experiments, a range of concentrations of TPA from 0.5 μM to 1 μM was used. SMIFH2 [Inhibitor of formin homology 2 (FH2) domain, 4401; Tocris Bioscience], a small-molecule inhibitor of formin, was used in a concentration range between 5 μM and 100 μM (39). For drug screening in the 3D chirality assay with other chemicals that affect embryonic laterality, a range of concentrations was used. The chemicals used were thalidomide (5–100 μg/mL, 06-521-00; Fisher Scientific), lidocaine (1–100 μM, 30-575-0; Fisher Scientific), methoxamine hydrochloride (100–500 μM, M6524; Sigma-Aldrich), paroxetine hydrochloride (1–100 μM, P9623; Sigma-Aldrich), nitrofurazone (0.1–10 μg/mL, 31706; Sigma-Aldrich), human recombinant Nodal (1 μg/mL, 3218-ND-025; R&D Systems), SB431542 (75 μM, S4317; Sigma-Aldrich), LDN193189 (10 mM–1 μM, SML0559; Sigma-Aldrich), and BMP4 (1–500 ng/mL, 314-BP-010; R&D Systems). The control media contained equivalent amounts of the respective solvents (DMSO, HCl, or water).
Quantitative Image Analysis of LR Bias of Golgi and Cell Boundaries in Chick Myocardium.
All of the confocal micrographs for analysis were displayed in the same manner so the orientation of the embryonic axes (AP, dorsal–ventral, right–left) was consistent between images. Similarly, the images were rotated so that the AP axis aligned vertically in each image. LR polarization of the Golgi apparatus (labeled with anti-GM130) within the individual myocardial cells was measured by determining the angle between a vector drawn from the centroid of the nucleus to the centroid of the Golgi and the embryonic AP axis. A custom-written program in MATLAB was used to draw the angular histogram depicting Golgi polarization as anterior left, posterior left, posterior right, or anterior right. For quantification of LR asymmetric cell alignment, image processing was performed as described above. The angle between cell boundaries (visualized by Phalloidin or anti-ZO1) and the vertical embryonic AP axis was measured to determine right or left bias. The frequency and distribution of alignment of cell boundaries were depicted in histograms.
N-Cadherin and p-Myosin-II Intensity Measurements.
Image processing and quantification of alignment of cell boundaries were performed as described in the previous section. Mean signal intensity of N-cadherin and p-Myosin-II was calculated using Fiji. The intensity of each cell boundary was normalized to the average intensity of all of the cell boundaries before comparison for directional biases.
3D Reconstruction of Chicken Embryonic Hearts.
Multiphoton (Zeiss LSM 510) image stacks of anti–MF20-stained chicken embryos were stitched together using the Grid/Collection Stitching plugin in Fiji. Three-dimensional surface rendering of the embryonic heart was performed using Imaris (Bitplane) software.
Image Analysis for Intracellular Chirality in Chick Myocardium.
LR bias of the cell centroid with respect to the nuclear–Golgi axis in the myocardium was used as an intracellular chirality marker. Confocal images of control and TPA-treated embryos stained with Phalloidin (F-actin), anti-GM130 (Golgi), and Hoechst (nucleus) were used for analysis. Phalloidin staining was used at first for manual segmentation of cell boundaries, followed by marking the position of the cell centroid in those images using Fiji. A straight-line vector was drawn from the center of the nucleus to the center of the Golgi. Left (L)- or right (R)-biased cells were identified and marked based on the positioning of the cell centroid on the left or the right of the nucleus–Golgi vector, respectively. The cells with the cell centroid overlapping with the nucleus–Golgi axis were considered as nonbiased or neutral (N).
Statistics.
JMP 13 (SAS), Prism (GraphPad), and MATLAB (MathWorks) were used for statistical analysis. Data were presented as mean ± SEM unless indicated otherwise. Statistical analysis was performed by two-way ANOVA or two-tailed t test (for two-sample comparisons). For statistical analysis of dominance of CW and CCW rings/rotation, the binomial cumulative function in MATLAB was used. Significant differences were accepted at P < 0.05 for all experiments.
Data Availability.
All data that support the findings of this study are available within the main text and SI Appendix of this paper.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (Office of Director/National Institute of Child Health and Development Grant DP2HD083961), the National Science Foundation [Faculty Early Career Development Program (CAREER) Grant CMMI-1254656], the American Heart Association (Grant 13SDG17230047), and the March of Dimes (Grant MOD 5-FY14-111) for funding. L.Q.W. is a Pew Scholar in Biomedical Sciences (Grant PEW 00026185), supported by the Pew Charitable Trusts.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808052115/-/DCSupplemental.
References
- 1.Bayraktar M, Männer J. Cardiac looping may be driven by compressive loads resulting from unequal growth of the heart and pericardial cavity. Observations on a physical simulation model. Front Physiol. 2014;5:112. doi: 10.3389/fphys.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeHaan RL, Gottlieb SH. The electrical activity of embryonic chick heart cells isolated in tissue culture singly or in interconnected cell sheets. J Gen Physiol. 1968;52:643–665. doi: 10.1085/jgp.52.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manasek FJ. Embryonic development of the heart. I. A light and electron microscopic study of myocardial development in the early chick embryo. J Morphol. 1968;125:329–365. doi: 10.1002/jmor.1051250306. [DOI] [PubMed] [Google Scholar]
- 4.Männer J. On rotation, torsion, lateralization, and handedness of the embryonic heart loop: New insights from a simulation model for the heart loop of chick embryos. Anat Rec A Discov Mol Cell Evol Biol. 2004;278:481–492. doi: 10.1002/ar.a.20036. [DOI] [PubMed] [Google Scholar]
- 5.Taber LA. Biophysical mechanisms of cardiac looping. Int J Dev Biol. 2006;50:323–332. doi: 10.1387/ijdb.052045lt. [DOI] [PubMed] [Google Scholar]
- 6.Francis RJ, Christopher A, Devine WA, Ostrowski L, Lo C. Congenital heart disease and the specification of left-right asymmetry. Am J Physiol Heart Circ Physiol. 2012;302:H2102–H2111. doi: 10.1152/ajpheart.01118.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyle C, Brown NA, Wolpert L. Development of left/right handedness in the chick heart. Development. 1992;115:1071–1078. doi: 10.1242/dev.115.4.1071. [DOI] [PubMed] [Google Scholar]
- 8.Lenhart KF, Holtzman NG, Williams JR, Burdine RD. Integration of nodal and BMP signals in the heart requires FoxH1 to create left-right differences in cell migration rates that direct cardiac asymmetry. PLoS Genet. 2013;9:e1003109. doi: 10.1371/journal.pgen.1003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linask KK, Yu X, Chen Y, Han MD. Directionality of heart looping: Effects of Pitx2c misexpression on flectin asymmetry and midline structures. Dev Biol. 2002;246:407–417. doi: 10.1006/dbio.2002.0661. [DOI] [PubMed] [Google Scholar]
- 10.Logan M, Pagán-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 11.Ocaña OH, et al. A right-handed signalling pathway drives heart looping in vertebrates. Nature. 2017;549:86–90. doi: 10.1038/nature23454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taber LA, Voronov DA, Ramasubramanian A. The role of mechanical forces in the torsional component of cardiac looping. Ann N Y Acad Sci. 2010;1188:103–110. doi: 10.1111/j.1749-6632.2009.05089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voronov DA, Alford PW, Xu G, Taber LA. The role of mechanical forces in dextral rotation during cardiac looping in the chick embryo. Dev Biol. 2004;272:339–350. doi: 10.1016/j.ydbio.2004.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Noël ES, et al. A Nodal-independent and tissue-intrinsic mechanism controls heart-looping chirality. Nat Commun. 2013;4:2754. doi: 10.1038/ncomms3754. [DOI] [PubMed] [Google Scholar]
- 15.Inaki M, Liu J, Matsuno K. Cell chirality: Its origin and roles in left-right asymmetric development. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150403. doi: 10.1098/rstb.2015.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDowell G, Rajadurai S, Levin M. From cytoskeletal dynamics to organ asymmetry: A nonlinear, regulative pathway underlies left-right patterning. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150409. doi: 10.1098/rstb.2015.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naganathan SR, Middelkoop TC, Fürthauer S, Grill SW. Actomyosin-driven left-right asymmetry: From molecular torques to chiral self organization. Curr Opin Cell Biol. 2016;38:24–30. doi: 10.1016/j.ceb.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Sato K, Hiraiwa T, Shibata T. Cell chirality induces collective cell migration in epithelial sheets. Phys Rev Lett. 2015;115:188102. doi: 10.1103/PhysRevLett.115.188102. [DOI] [PubMed] [Google Scholar]
- 19.Taniguchi K, et al. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science. 2011;333:339–341. doi: 10.1126/science.1200940. [DOI] [PubMed] [Google Scholar]
- 20.Wan LQ, Chin AS, Worley KE, Ray P. Cell chirality: Emergence of asymmetry from cell culture. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150413. doi: 10.1098/rstb.2015.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato K, et al. Left-right asymmetric cell intercalation drives directional collective cell movement in epithelial morphogenesis. Nat Commun. 2015;6:10074. doi: 10.1038/ncomms10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McSheene JC, Burdine RD. Examining the establishment of cellular axes using intrinsic chirality. Proc Natl Acad Sci USA. 2011;108:12191–12192. doi: 10.1073/pnas.1109367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raymond MJ, Jr, et al. Multiaxial polarity determines individual cellular and nuclear chirality. Cell Mol Bioeng. 2017;10:63–74. doi: 10.1007/s12195-016-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymond MJ, Jr, Ray P, Kaur G, Singh AV, Wan LQ. Cellular and nuclear alignment analysis for determining epithelial cell chirality. Ann Biomed Eng. 2016;44:1475–1486. doi: 10.1007/s10439-015-1431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamada A, Igarashi M. Revealing chiral cell motility by 3D Riesz transform-differential interference contrast microscopy and computational kinematic analysis. Nat Commun. 2017;8:2194. doi: 10.1038/s41467-017-02193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tee YH, et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol. 2015;17:445–457. doi: 10.1038/ncb3137. [DOI] [PubMed] [Google Scholar]
- 27.Wan LQ, et al. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc Natl Acad Sci USA. 2011;108:12295–12300. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, et al. Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci USA. 2007;104:9296–9300. doi: 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamanaka H, Kondo S. Rotating pigment cells exhibit an intrinsic chirality. Genes Cells. 2015;20:29–35. doi: 10.1111/gtc.12194. [DOI] [PubMed] [Google Scholar]
- 30.Levin M. Left-right asymmetry in embryonic development: A comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Deng XF, Mulay S, Varma DR. Role of Ca(2+)-independent PKC in alpha 1-adrenoceptor-mediated inotropic responses of neonatal rat hearts. Am J Physiol. 1997;273:H1113–H1118. doi: 10.1152/ajpheart.1997.273.3.H1113. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Wang LU, Bian H, Zhou X, Ruan C. Effects of paroxetine on spatial memory function and protein kinase C expression in a rat model of depression. Exp Ther Med. 2015;10:1489–1492. doi: 10.3892/etm.2015.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liedtke CM, Cole T, Ikebe M. Differential activation of PKC-delta and -zeta by alpha 1-adrenergic stimulation in human airway epithelial cells. Am J Physiol. 1997;273:C937–C943. doi: 10.1152/ajpcell.1997.273.3.C937. [DOI] [PubMed] [Google Scholar]
- 34.Meierhofer C, Dunzendorfer S, Wiedermann CJ. Protein kinase C-dependent effects on leukocyte migration of thalidomide. J Infect Dis. 1999;180:216–219. doi: 10.1086/314842. [DOI] [PubMed] [Google Scholar]
- 35.Fan J, Ray P, Lu Y, Kaur G, Schwarz JJ, Wan LQ. Cell chirality regulates intercellular junctions and endothelial permeability. Sci Adv. 2018;4:eaat2111. doi: 10.1126/sciadv.aat2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heikkilä J, Akerman KE. (-)-Indolactam V activates protein kinase C and induces changes in muscarinic receptor functions in SH-SY5Y human neuroblastoma cells. Biochem Biophys Res Commun. 1989;162:1207–1213. doi: 10.1016/0006-291x(89)90802-4. [DOI] [PubMed] [Google Scholar]
- 37.Stone RM, Sariban E, Pettit GR, Kufe DW. Bryostatin 1 activates protein kinase C and induces monocytic differentiation of HL-60 cells. Blood. 1988;72:208–213. [PubMed] [Google Scholar]
- 38.Geiges D, et al. Activation of protein kinase C subtypes alpha, gamma, delta, epsilon, zeta, and eta by tumor-promoting and nontumor-promoting agents. Biochem Pharmacol. 1997;53:865–875. doi: 10.1016/s0006-2952(96)00885-4. [DOI] [PubMed] [Google Scholar]
- 39.Rizvi SA, et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davison A, et al. Formin is associated with left-right asymmetry in the pond snail and the frog. Curr Biol. 2016;26:654–660. doi: 10.1016/j.cub.2015.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, et al. Nanowire magnetoscope reveals a cellular torque with left-right bias. ACS Nano. 2016;10:7409–7417. doi: 10.1021/acsnano.6b01142. [DOI] [PubMed] [Google Scholar]
- 42.Itasaki N, Nakamura H, Sumida H, Yasuda M. Actin bundles on the right side in the caudal part of the heart tube play a role in dextro-looping in the embryonic chick heart. Anat Embryol (Berl) 1991;183:29–39. doi: 10.1007/BF00185832. [DOI] [PubMed] [Google Scholar]
- 43.Itasaki N, Nakamura H, Yasuda M. Changes in the arrangement of actin bundles during heart looping in the chick embryo. Anat Embryol (Berl) 1989;180:413–420. doi: 10.1007/BF00305115. [DOI] [PubMed] [Google Scholar]
- 44.Keum E, et al. Syndecan-4 regulates localization, activity and stability of protein kinase C-alpha. Biochem J. 2004;378:1007–1014. doi: 10.1042/BJ20031734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue X, Schultheiss TM, McKenzie EA, Rosenberg RD. Role of heparan sulfate in dextral heart looping in chick. Glycobiology. 2004;14:745–755. doi: 10.1093/glycob/cwh083. [DOI] [PubMed] [Google Scholar]
- 46.Davis A, et al. Stomach curvature is generated by left-right asymmetric gut morphogenesis. Development. 2017;144:1477–1483. doi: 10.1242/dev.143701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grimes DT, Burdine RD. Left-right patterning: Breaking symmetry to asymmetric morphogenesis. Trends Genet. 2017;33:616–628. doi: 10.1016/j.tig.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith KA, et al. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell. 2008;14:287–297. doi: 10.1016/j.devcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Chen JN, et al. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- 50.Chocron S, Verhoeven MC, Rentzsch F, Hammerschmidt M, Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Schilling TF, Concordet JP, Ingham PW. Regulation of left-right asymmetries in the zebrafish by Shh and BMP4. Dev Biol. 1999;210:277–287. doi: 10.1006/dbio.1999.9214. [DOI] [PubMed] [Google Scholar]
- 52.Juan T, et al. Myosin1D is an evolutionarily conserved regulator of animal left-right asymmetry. Nat Commun. 2018;9:1942. doi: 10.1038/s41467-018-04284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saydmohammed M, et al. Vertebrate myosin 1d regulates left-right organizer morphogenesis and laterality. Nat Commun. 2018;9:3381. doi: 10.1038/s41467-018-05866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tingler M, et al. A conserved role of the unconventional myosin 1d in laterality determination. Curr Biol. 2018;28:810–816.e3. doi: 10.1016/j.cub.2018.01.075. [DOI] [PubMed] [Google Scholar]
- 55.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 56.Filas BA, Varner VD, Voronov DA, Taber LA. Tracking morphogenetic tissue deformations in the early chick embryo. J Vis Exp. 2011:e3129. doi: 10.3791/3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapman SC, Collignon J, Schoenwolf GC, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 58.Geraldes P, King GL. Activation of protein kinase C isoforms and its impact on diabetic complications. Circ Res. 2010;106:1319–1331. doi: 10.1161/CIRCRESAHA.110.217117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.González MI, Bannerman PG, Robinson MB. Phorbol myristate acetate-dependent interaction of protein kinase Calpha and the neuronal glutamate transporter EAAC1. J Neurosci. 2003;23:5589–5593. doi: 10.1523/JNEUROSCI.23-13-05589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available within the main text and SI Appendix of this paper.