Significance
There has been recent interest in fast-growing microbes to accelerate discoveries in medicine, biology, and biotechnology. Additionally, microbes with rapid-growth properties are of significant industrial interest as a chassis for bioproduction. While such microbes have been identified, the determinants of their rapid growth remain poorly understood. In this study, we have elucidated the molecular basis for the rapid growth of Synechococcus 2973, a fast-growing oxygenic photosynthetic organism, and applied these insights to significantly enhance the growth rate of a related model cyanobacterium, Synechococcus 7942. Such engineered photosynthetic microbes are expected to open the doors for significantly higher productivity under sustainable conditions.
Keywords: cyanobacteria, Synechococcus, photosynthesis, growth
Abstract
Cyanobacteria are emerging as attractive organisms for sustainable bioproduction. We previously described Synechococcus elongatus UTEX 2973 as the fastest growing cyanobacterium known. Synechococcus 2973 exhibits high light tolerance and an increased photosynthetic rate and produces biomass at three times the rate of its close relative, the model strain Synechococcus elongatus 7942. The two strains differ at 55 genetic loci, and some of these loci must contain the genetic determinants of rapid photoautotrophic growth and improved photosynthetic rate. Using CRISPR/Cpf1, we performed a comprehensive mutational analysis of Synechococcus 2973 and identified three specific genes, atpA, ppnK, and rpaA, with SNPs that confer rapid growth. The fast-growth–associated allele of each gene was then used to replace the wild-type alleles in Synechococcus 7942. Upon incorporation, each allele successively increased the growth rate of Synechococcus 7942; remarkably, inclusion of all three alleles drastically reduced the doubling time from 6.8 to 2.3 hours. Further analysis revealed that our engineering effort doubled the photosynthetic productivity of Synechococcus 7942. We also determined that the fast-growth–associated allele of atpA yielded an ATP synthase with higher specific activity, while that of ppnK encoded a NAD+ kinase with significantly improved kinetics. The rpaA SNPs cause broad changes in the transcriptional profile, as this gene is the master output regulator of the circadian clock. This pioneering study has revealed the molecular basis for rapid growth, demonstrating that limited genetic changes can dramatically improve the growth rate of a microbe by as much as threefold.
Cyanobacteria are oxygenic photosynthetic prokaryotes that generate biomass using only CO2 and sunlight as carbon and energy sources, respectively. For this reason, these organisms show great potential as carbon-neutral platforms to produce biofuels and other chemical feedstocks as alternatives to petrochemicals. In the last two decades, many cyanobacterial strains have been engineered to produce alcohols, aldehydes and ketones, fatty acids, isoprene and terpenes, ethylene, sugars, organic acids, and long-chain alka(e)nes (1). Cyanobacteria have several advantages over other photosynthetic organisms such as eukaryotic algae and plants including faster growth, more efficient solar energy conversion, higher biomass accumulation, and facile genetic manipulation (2). Unfortunately, cyanobacterial production systems have not been deployed yet because of the low production titers for commonly employed model organisms such as Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942. For cyanobacteria to compete with heterotrophic bacteria as bioproduction platforms, their photosynthetic CO2 fixation rates need to be significantly increased.
We have recently described Synechococcus elongatus UTEX 2973 as a promising candidate as a viable cyanobacterial production platform (3, 4). Unlike many model strains, this organism is capable of biomass production at rates that are comparable to heterotrophs such as the yeast Saccharomyces cerevisiae. Interestingly, this organism is almost genetically identical to the model cyanobacterium Synechococcus 7942. The two strains have an identical gene set differing by only 53 SNPs, a 7.5-kb deletion, and a 188-kb inversion (3). Despite the high degree of genetic similarity, the two strains are phenotypically quite different. Synechococcus 2973 grows at elevated temperature and achieves a doubling time of 1.5 h, compared with 4.9 h for Synechococcus 7942 (4). Additionally, Synechococcus 2973 is tolerant of high light, growing well under light intensities exceeding that of natural sunlight (2,000 μmol⋅m−2⋅s−1). In contrast, Synechococcus 7942 becomes severely photoinhibited at intensities less than half that. Relative to Synechococcus 7942, Synechococcus 2973 also exhibits several adjustments to its photosynthetic apparatus including decreased phycobilisome and increased Photosystem I (PSI) cytochrome f and plastocyanin contents. Such differences lead to the alleviation of a photosynthetic bottleneck, resulting in an increased capacity for photosynthetic electron flux. This in turn supports the utilization of higher light intensities by Synechococcus 2973 while producing more ATP and NADPH for carbon fixation and, ultimately, the faster growth rate of this cyanobacterium (4).
The extremely close relatedness of these two cyanobacterial strains provides an excellent system to investigate how a slower-growing, less productive model organism can be transitioned into a fast-growing and highly productive substrain. Among the small number of genetic differences between the two strains must lie the molecular determinants of the photosynthetic improvements in Synechococcus 2973 relative to the widely studied model cyanobacterium Synechococcus 7942. Among the 53 SNPs, 35 result in single amino acid substitutions in various proteins, while the remaining SNPs are in noncoding regions. Thus, the SNP-containing genes are alternate alleles in the two organisms. Identification and characterization of such universal fast-growth–associated alleles could unlock improvements in other model and industrially relevant cyanobacteria.
Results
Identification of SNPs Resulting in Rapid Growth.
Based on the available annotations, it was not immediately apparent which SNPs give rise to the phenotypic differences that we previously reported in the two strains (4). Therefore, we performed a comprehensive mutational analysis of Synechococcus 2973 to determine which of the 55 genetic differences between that strain and Synechococcus 7942 are critical for rapid photoautotrophic growth and high light tolerance. We reasoned that a combination of SNPs would be required for rapid growth, so that engineering individual SNPs into Synechococcus 7942 would not generate the desired phenotypes. However, if an SNP is part of the combination that leads to rapid growth, then reversion of that SNP to the version in the slow-growing strain would break up the combination, resulting in the loss of the rapid-growth phenotype. Following this rationale, we used CRISPR/Cpf1 gene editing (5) to convert each SNP in Synechococcus 2973 to the corresponding version found in Synechococcus 7942. The 7-kb deletion in Synechococcus 2973 also may have contained a factor that regulates growth. Hence, we replaced the genes lost in that deletion to determine the effect of the deletion on growth as well.
The plasmid pUH24 in Synechococcus 2973 contains 16 of the 55 differences between the two strains; however, this plasmid has previously been shown to be dispensable in related strains without a phenotype (6). This allowed us to take a shortcut in our mutational analysis. The plasmid was cured from Synechococcus 2973 using CRISPR/Cpf1, and the cured strain grew at the same rate as the parental strain (SI Appendix, Fig. S1A). Hence, none of the SNPs on the plasmid contributes to rapid growth. This allowed us to eliminate 16 of the 55 differences as factors that lead to rapid autotrophic growth. Next, we addressed the genes in the deletion region by restoring all of the genes and their full operons into neutral site I (7) in Synechococcus 2973. We also used CRISPR/Cpf1 gene editing to recreate the deletion in Synechococcus 7942. Growth analysis revealed that the deletion did not result in the loss of a factor that regulates growth, as Synechococcus 2973 with the deletion restored and Synechococcus 7942 with the deletion recreated grew at the same rates as their parental strains (SI Appendix, Fig. S1B).
Next, we made a large series of mutants with the CRISPR/Cpf1 system, each with a different markerless single-nucleotide change, to individually convert each of the remaining 39 chromosomally encoded SNPs in Synechococcus 2973 to its corresponding allele in Synechococcus 7942. In one case a gene (rpaA) contained both coding and promoter SNPs. Here, the coding and noncoding SNPs were converted individually as well as in combination. Also, in the Synechococcus 2973 23S rRNA there are two SNPS relative to the sequence in Synechococcus 7942. Since there are two identical rRNA operons in the genome, we were not able to target one specifically. Thus, the two rRNA SNPs were not converted in this study. We also did not make reversions of SNPs that resulted in silent mutations.
Growth analysis of the series of mutants revealed four coding SNPs and one noncoding SNP across three genes that when converted resulted in decreased growth rate (Table 1). The SNPs that we thus identified as the genetic factors for rapid photoautotrophic growth result in the following amino acid changes. First, a Y252C replacement in AtpA, the alpha subunit of ATP synthase (8), results in the change of a surface Cys in Synechococcus 7942 to a Tyr in Synechococcus 2973 (SI Appendix, Fig. S2A). This conversion may cause a loss of redox regulation of ATP synthase in Synechococcus 2973 (9). Second, a D260E replacement in the NAD+ kinase PpnK (10) results in the change of a Glu in Synechococcus 7942 to an Asp in Synechococcus 2973. Although both these amino acids are negatively charged, the side chain of Glu is one carbon longer. Structural analysis of PpnK revealed that this residue is located in the active site with the negatively charged side chain being electrostatically repelled by the phosphates on NAD+ or ATP (SI Appendix, Fig. S2B). The shorter, negatively charged side chain of D260 in PpnK of Synechococcus 2973 is positioned closer to the negatively charged phosphate on NAD, which affects enzyme kinetics (Table 2). Finally, in the master output regulator of the circadian clock RpaA (11), we identified SNPs resulting in R121Q and K134E replacements. Additionally, the promoter of rpaA in Synechococcus 2973 contains a 7-bp deletion that falls 102 bp upstream of the translational start codon. Both coding SNPs in rpaA are in an unordered loop linking the response receiver domain to the helix-turn-helix DNA-binding domain. As shown in SI Appendix, Fig. S3, both the coding and promoter SNPs in Synechococcus 2973 are necessary for its rapid growth.
Table 1.
Growth rates of Synechococcus 2973 with various SNPs reverted to the allele in Synechococcus 7942
| Nucleotide position | Locus tag | Gene function | Doubling time, h | % change in growth rate |
| 2973-WT | — | Control | 2.1 ± 0.2 | N/A |
| 7942-WT | — | Control | 8.5 ± 1.1 | N/A |
| pUH24 | — | Cure 2973 of plasmid | 2.1 ± 0.0 | 0 |
| 126938 | M744_705 | Hypothetical protein | 2.0 ± 0.0 | 4 |
| 236706* | M744_1335 | ATP synthase F0F1 alpha | 3.1 ± 0.5 | −49 |
| 474883† | — | Noncoding 101 bp upstream of rpaA | 2.1 ± 0.2 | 0 |
| 475352† | M744_2605 | rpaA, SNP I, circadian response regulator | 2.0 ± 0.0 | 4 |
| 475390† | M744_2605 | rpaA, SNP II, circadian response regulator | 2.0 ± 0.0 | 4 |
| 610804 | M744_3335 | Manganese ABC transporter ATP-binding | 2.1 ± 0.2 | 0 |
| 705129 | M744_3855 | Guanylate cyclase | 2.1 ± 0.2 | 0 |
| 891346* | M744_4780 | PpnK – NAD+ kinase | 4.5 ± 0.2 | −117 |
| 1042862 | — | Noncoding, 362 bp upstream of ycfA | 2.1 ± 0.3 | 0 |
| 1080351 | M744_5865 | Hypothetical protein | 2.1 ± 0.0 | 0 |
| 1113358 | M744_6025 | Molecular chaperone DnaK | 2.1 ± 0.5 | 0 |
| 1222741 | M744_6570 | Hydrolase | 2.1 ± 0.2 | 0 |
| 1222972 | M744_6570 | Hydrolase | 2.1 ± 0.2 | 0 |
| 1237531 | M744_6650 | CTP synthetase | 2.1 ± 0.1 | 0 |
| 1238113 | M744_6650 | CTP synthetase | 2.2 ± 0.4 | −4 |
| 1273424 | M744_6850 | Chorismate mutase | 2.5 ± 0.6 | −16 |
| 1533430 | — | Noncoding, 120 bp upstream of COP23 | 2.1 ± 0.3 | 0 |
| 1619501 | M744_8615 | DNA-directed RNA polymerase β subunit | 2.3 ± 0.1 | −9 |
| 1718274 | M744_11685 | Anthranilate synthase | 1.9 ± 0.2 | 7 |
| 1741647 | — | Noncoding, downstream of two genes | 2 ± 0.2 | 4 |
| 1749120 | — | Noncoding, 21 bp upstream of porin | 2.2 ± 0.2 | −4 |
| 2139224 | — | Noncoding, 130 bp upstream of histone like protein | 2.2 ± 0.0 | −4 |
| 2249045 | M744_12130 | Long-chain fatty acid CoA ligase | 2.1 ± 0.1 | 0 |
| 2339502 | M744_12130 | Long-chain fatty acid CoA ligase | 2.0 ± 0.2 | 0 |
| 2347222 | — | Noncoding, 37 bp upstream of hypothetical | 2.2 ± 0.2 | −4 |
| 2348957 | — | Noncoding, 26 bp upstream of hypothetical | 2.1 ± 0.3 | 0 |
| 2347664–2347220 | — | Noncoding, insertion upstream of hypothetical | 2.1 ± 0.3 | 0 |
| 2364720 | M744_12285 | Glutamate synthase | 2.1 ± 0.1 | 0 |
| 2608863 | M744_13540 | Photosystem I assembly protein | 2.0 ± 0.0 | 4 |
| Multiple* | M744_2605/noncoding | rpaA, SNP I SNPII, noncoding promoter SNP combined | 3.4 ± 0.3 | −61 |
Significant difference from Synechococcus 2973 controls: 95% CI (1.87, 2.33). N/A, not applicable.
SNPs that significantly slowed growth.
SNPs that did not individually slow growth but had an effect when combined.
Table 2.
Enzyme kinetics of the Synechococcus 2973 allele of NAD+ kinase compared with the Synechococcus 7942 allele
| Allele | NAD+ | ATP | ||||
| KM, mM | Vmax, nmol⋅min−1⋅mg−1 | Kcat, min−1 | KM, mM | Vmax, nmol⋅min−1⋅mg−1 | Kcat, min−1 | |
| 2973 NADK | 0.25 ± 0.06 | 625 ± 79 | 20.8 ± 2.6 | 0.25+/0.05 | 625 ± 84 | 20.8 ± 2.2 |
| 7942 NADK | 0.52 ± 0.08 | 434 ± 21 | 14.4 ± 0.8 | 0.37+/0.04 | 370 ± 42 | 12.3 ± 0.6 |
Kinetic parameters for NAD+ and ATP are shown as indicated.
Engineering Synechococcus 7942 for Rapid Photoautotrophic Growth.
Next, we used the CRISPR/Cpf1 system to engineer the SNPs that we identified as the genetic factors for rapid autotrophic growth into the slower-growing Synechococcus 7942 to verify that we could engineer rapid growth of this cyanobacterium. First, each of the three genes was individually converted to the Synechococcus 2973 version to determine if the mutations can independently increase growth rate. Growth analysis revealed that the mutations in both atpA and ppnK can independently increase the growth rate of Synechococcus 7942 (Fig. 1 A and C). Introduction of the rpaA promoter SNP or the pair of coding SNPs did not yield an increase in growth rate (SI Appendix, Fig. S3). The individual coding SNPs were not studied separately because they lie only 39 bp apart and it was difficult to change one without changing the other; thus, in this case we refer to the alternative alleles of rpaA to indicate that both coding SNPs are always changed together. However, when the pair of coding and promoter SNPs were combined, the growth rate of Synechococcus 7942 increased significantly (Fig. 1 A and C). This indicates that each gene/allele that we identified in our mutational analysis works independently to increase the growth rate of Synechococcus 7942. Furthermore, the fact that both the coding and promoter SNPs in rpaA are required for accelerated growth indicates that changes in the expression as well as in the activity of this regulator are required to achieve fast growth.
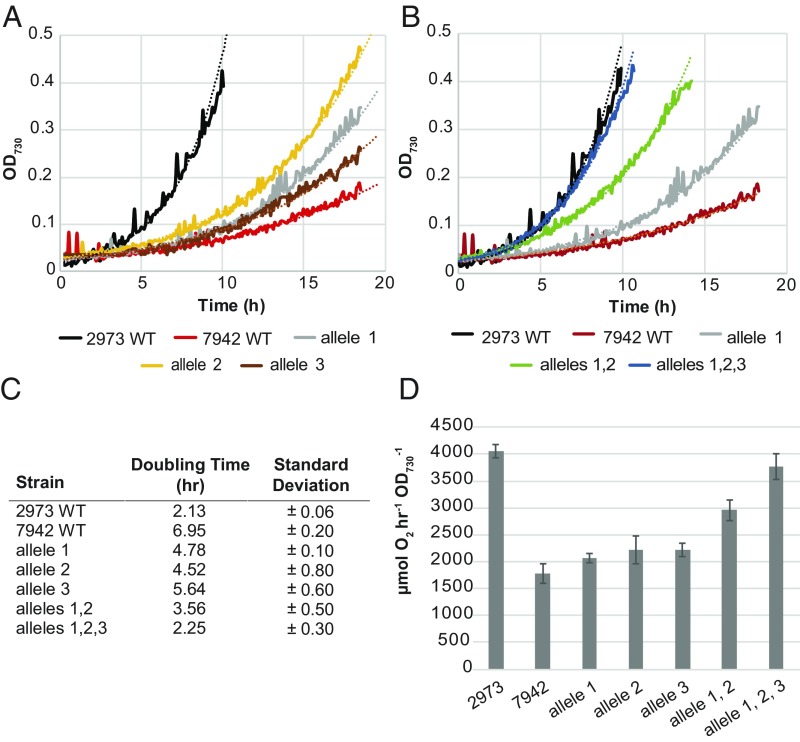
Fig. 1.
Growth of Synechococcus 2973, Synechococcus 7942, and mutant strains. (A) Representative growth curves of Synechococcus 7942 with different SNPs from Synechococcus 2973 incorporated individually. (B) Representative growth curves of Synechococcus 7942 with SNPs from Synechococcus 2973 incorporated in combinations. (C) Doubling times of both Synechococcus strains and mutants thereof (n = 3). (D) Whole-chain oxygen evolution of both wild-type strains and Synechococcus 7942 with various allele combinations from Synechococcus 2973 (n = 3). Allele 1, ATP synthase; allele 2, NAD+ kinase; allele 3, both coding SNPs and promoter deletion in rpaA. Growth was performed in a multicultivator bioreactor with 5% CO2, 900 µmol⋅m−2⋅s−1 light, 38 °C.
Finally, we combined all five SNPs into the same strain of Synechococcus 7942 to produce a markerless 5× mutant. As each successive gene was mutated to the Synechococcus 2973 version, we observed an increase in growth rate (Fig. 1 B and C). Remarkably, when all five SNPs were engineered into the same strain, we achieved a growth rate (2.2-h doubling time) which is comparable to that of Synechococcus 2973 (Fig. 1 B and C). This indicates that we had identified the complete complement of SNPs that converts the slow-growing, light-sensitive Synechococcus 7942 strain into a fast-growing, high-light–tolerant strain similar to Synechococcus 2973. It is interesting that two of the three genes, atpA and ppnK, are directly involved in energy metabolism, which suggests that energy availability rather than some other factor is the key determinant of the photoautotrophic growth rate of Synechococcus 7942. It is noteworthy that this small combination of SNPs converted Synechococcus 7942 into a high-light–tolerant strain that exhibits a 2.2-h doubling time at 900 μmol⋅m−2⋅s−1 light. Also, at relatively lower light intensities (250 μmol⋅m−2⋅s−1), the two wild-type Synechococcus strains and Synechococcus 7942 with all three converted alleles grew at about the same rate (SI Appendix, Fig. S4). However, under higher light intensities (900 μmol⋅m−2⋅s−1), the growth rates of these strains were markedly different. Increasing the light intensity to above 250 μmol⋅m−2⋅s−1 causes photoinhibition in Synechococcus 7942, which slowed growth; both Synechococcus 2973 and the fast-growing mutant of Synechococcus 7942 were able to overcome photoinhibition and harness the increased light energy to accelerate growth (SI Appendix, Fig. S4) as well as photosynthetic rate (see below). This phenotypic difference was also reported in our previous work in more detail (4).
To enable rapid growth, Synechococcus 2973 exhibits photosynthetic rates that are twofold higher than that of Synechococcus 7942 (on a cellular basis) (4). We investigated whether the changes that we engineered into Synechococcus 7942 increased its photosynthetic rate as well. We compared rates of whole-chain oxygen evolution on a per OD basis rather than a per chlorophyll basis because the chlorophyll content of Synechococcus 2973 is twofold higher than that of Synechococcus 7942 (4), which obscures the observable differences in photosynthetic rates. We found that the photosynthetic rate of Synechococcus 7942 more than doubled, from 1,750 μmol O2⋅h−1⋅OD730−1 to 3,785 μmol O2⋅h−1⋅OD730−1, when all three alleles (five SNPs) were changed to their corresponding Synechococcus 2973 versions (Fig. 1D). Interestingly, incorporation of individual SNPs showed little effect; a significant effect was observed only when both the Synechococcus 2973 ATP synthase and NAD+ kinase alleles were introduced together. The full benefit to oxygen evolution became apparent only when all three alleles were combined. Together, the increased growth rate and increased rates of O2 evolution indicate that the three Synechococcus 2973 alleles worked in combination to dramatically improve the photosynthetic productivity of the model cyanobacterium Synechococcus 7942. We previously reported that Synechococcus 2973 exhibits altered pigment content relative to Synechococcus 7942 (4). An absorbance scan of the 5× mutant revealed that it has now taken on the characteristic spectrum of Synechococcus 2973 with an increased chlorophyll peak (682 nm) and decreased phycobilisome peak (625 nm) (SI Appendix, Fig. S4C). In addition to altered pigment content, we noted differences in energy transfer through the phycobilisomes in Synechococcus 2973 relative to Synechococcus 7942. Synechococcus 2973 exhibits reduced fluorescence from phycocyanin (PC) at 77K, which suggests increased efficiency of energy transfer through PC (4). The 77K fluorescence spectrum of the 5× mutant revealed that it exhibits a photophysiological phenotype similar to Synechococcus 2973 (SI Appendix, Fig. S4D).
Comparisons of NADK Alleles.
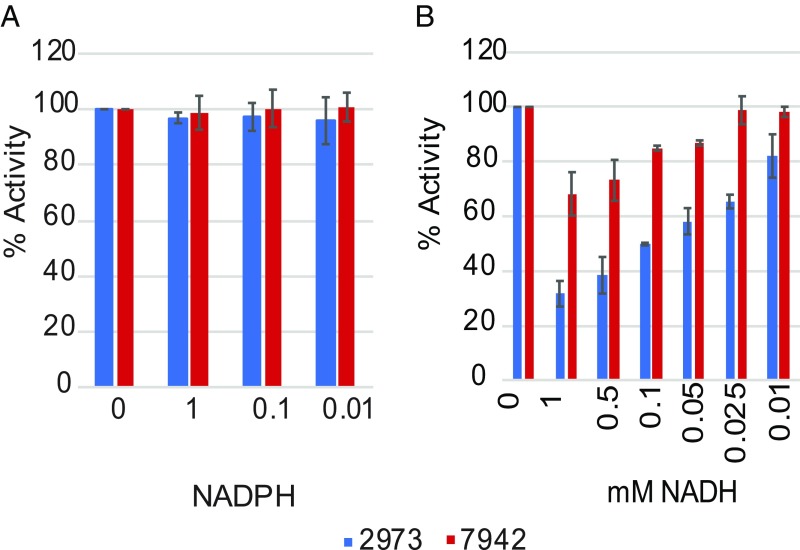
As described above, the SNP resulting in the D260E change in NAD+ kinase significantly increased the growth rate of Synechococcus 7942, but this substitution near the active site leads to a subtle change since both residues are negatively charged. We therefore investigated in more detail how this replacement affects the enzyme’s kinetics. Both variants of the NADK were overexpressed and purified from Escherichia coli. We then examined the kinetics of substrate binding for both substrates, ATP and NAD. Both substrates bind in the same pocket with which D260 interacts. Thus, this SNP may affect the binding affinity of one or both of the substrates (12, 13). Additionally, NADH and NADPH act as competitive inhibitors of NADK by also binding at that site (14). Therefore, we compared the inhibitory effects of NADH and NADPH in the two versions of the NADK as well.
We determined that the Synechococcus 2973 version of the enzyme has a significantly higher affinity for both substrates, as indicated by the lower KM toward both NAD and ATP (Table 2 and SI Appendix, Fig. S5). In addition, the Synechococcus 2973 version of the enzyme displays a significantly higher Vmax and Kcat toward both NAD and ATP (Table 2). Together, these findings indicate that the D260E substitution in PpnK generates a more active enzyme, thus increasing NADP+ production in Synechococcus 2973. Heterotrophic NADK genes are inhibited by both NADH and NADPH (14). However, we observed no inhibition of the cyanobacterial NADK by NADPH for either version of the protein (Fig. 2A). The high tolerance of the cyanobacterial version of the enzyme to NADPH is likely required due to the large amounts of NADPH that are produced during the light reactions of photosynthesis. Interestingly, we observed a large difference in inhibition by NADH for the two versions of the enzyme (Fig. 2B). The Synechococcus 2973 version of the enzyme is strongly inhibited by NADH, but the Synechococcus 7942 version of the enzyme is relatively tolerant to NADH. It appears that in Synechococcus 2973 NADP+ production is increased only if the NADH pool is kept very small. We recently reported that the fast-growth phenotype of Synechococcus 2973 is a result of increased titers of PSI, cytochrome f, and plastocyanin, enabling a higher level of photosynthetic electron flux (4). A more active NADK is complementary to those findings, as increased NADK activity would yield increased titers of NADP+, the final acceptor in the photosynthetic electron transport chain. This in turn enables efficient electron flow, generating more reducing power in the form of NADPH.
Fig. 2.
NAD+ kinase activity in the presence of various concentrations of competitive inhibitors. (A) Activity with added NADPH. (B) Activity with added NADH (n = 3).
Comparison of ATP Synthase Alleles.
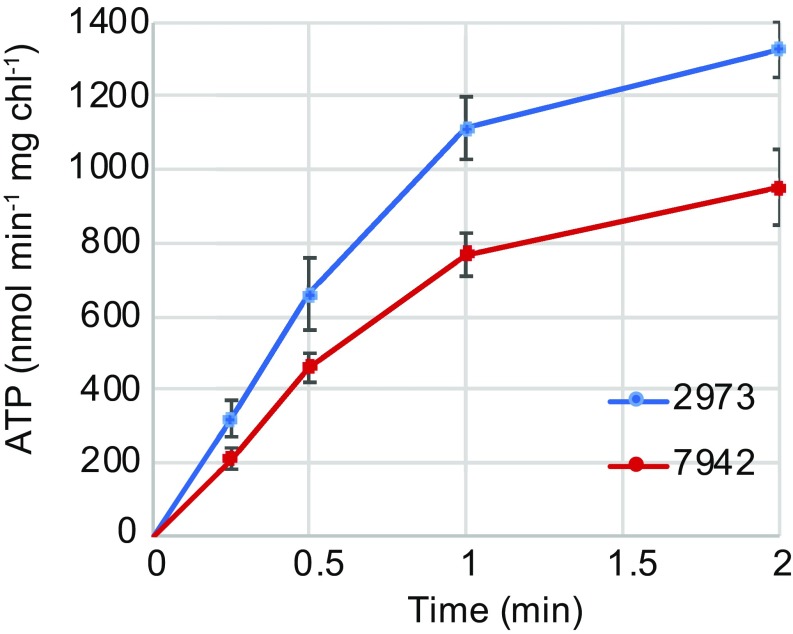
Next, we sought to compare the ATP synthase activities of the two different alleles of this important enzyme. To accomplish this, we prepared well-coupled membrane vesicles from wild-type Synechococcus 2973 and a mutant thereof in which the ATP synthase had been converted to the Synechococcus 7942 version of the allele (15, 16). As described above, Synechococcus 2973 has higher titers of photosynthetic electron transfer components (4), so that light-driven ATP production may not reflect the innate differences between the two versions of ATP synthase. Hence, we used a classic acid–base transition to energize the membranes, generating an identical proton gradient in both samples (17, 18) and bypassing other differences in photophysiology. Membrane preparations were equilibrated in a low-pH succinate buffer and then diluted into a higher-pH reaction buffer to generate a proton gradient across the membrane. After the ΔpH was generated, samples were collected at various time intervals (Materials and Methods).
We determined that immediately after dilution into the base stage, a proton motive force (PMF) was generated resulting in ATP production. The membranes containing the Synechococcus 2973 allele of ATP synthase produced ATP at a rate of 1,113 nmol⋅min−1⋅mg Chl−1 while membranes containing the Synechococcus 7942 version of the allele produced ATP at a rate of 773 nmol⋅min−1⋅mg Chl−1 (Fig. 3). No ATP synthesis was observed in the presence of the uncoupler carbonyl cyanide m-chlorophenyl hydrazine (CCCP). We concluded that the Synechococcus 2973 allele of the ATP synthase has a higher rate of ATP synthesis when placed under the same PMF as the Synechococcus 7942 allele. Clearly, energy supply limits growth in Synechococcus 7942, as engineering this strain with an improved ATP synthase or with NAD+ kinase individually increased the growth rate, and engineering both improved alleles into the strain resulted in an additive effect (Fig. 1).
Fig. 3.
ATP synthase activity of the Synechococcus 2973 allele vs. the Synechococcus 7942 allele (n = 3).
Transcriptome Profiles of Wild-Type and SNP Interconversion Mutants.
We performed global RNA-sequencing (RNA-seq) analysis of all SNP mutants exhibiting a growth phenotype as well as both wild-type strains (Dataset S1). Our goal was to survey the transcriptional differences between the two wild-type strains and confirm that these differences are recapitulated by the mutations that shift one strain’s phenotype to that of the other. First, we compared the differences in gene expression based on normalized transcripts per million (TPM) in the two wild-type strains. We chose a 1.5-fold difference in expression as the cutoff for differentially expressed genes. Thus, genes showing less than a 1.5-fold difference were considered comparable in expression level, while genes showing more than a 1.5-fold difference were considered differentially expressed. A pathway enrichment analysis was also performed to determine metabolic pathways that were overrepresented in the up-regulated and down-regulated gene sets (SI Appendix, Fig. S6 and Dataset S2). We found that in Synechococcus 2973 only 15% of the genes show at least 1.5-fold higher expression, while 51% of the genes show at least 1.5-fold lower expression (Dataset S3). This finding suggests that Synechococcus 2973 focuses more of its resources on a few specific processes. In particular, Synechococcus 2973 exhibited increased nutrient capture, as iron, phosphate, nitrate, sulfate, and bicarbonate transporters were more highly expressed in this strain. The enrichment analysis showed a significant overrepresentation of the ABC transporter category (P = 0.009) among the up-regulated gene sets. We found that the psbA2 gene was expressed at higher levels in Synechococcus 2973, which may aid in Photosystem II (PSII) turnover under high-light conditions. We also found an up-regulation in photosynthetic carbon fixation (P = 0.0086) and gluconeogenesis (P = 0.0005) in Synechococcus 2973, as genes such as RuBP carboxylase (EC: 4.1.1.39), phosphoglycerate kinase (EC: 2.7.2.3), fructose-1,6-bisphosphatase (EC: 3.1.3.11), and sedoheptulose-1,7-bisphosphatase (EC: 3.1.3.37) were up-regulated. These could help fuel the higher need for carbon in Synechococcus 2973, which requires 0.502 g carbon per gram of biomass as opposed to 0.497 g carbon per gram of biomass for Synechococcus 7942 (3). In addition, we observed increased expression of the translation machinery (P = 0.0018) in Synechococcus 2973, as numerous ribosomal proteins, translation elongation factors, and ribosome recycling factors were more highly expressed. In Synechococcus 2973, NAD(P) transhydrogenase was down-regulated, which may serve to maintain a higher NADPH/NADH ratio. Interestingly, per our enrichment analysis, the nicotinate and nicotinamide metabolism category was the only significantly down-regulated pathway (P = 0.0055) in Synechococcus 2973.
Genes for several sigma factors, including sigG, sigB, rpoE, sigA, and sigF, were also down-regulated in Synechococcus 2973, which may be why most of the genes showed lower expression in this strain. Recombination and repair genes, including ruvC, recJ, recG, recO, mutT, recF, recQ, recN, and mutL, were also down-regulated in this strain. Synechococcus 2973 exhibited decreased expression of cytochrome c oxidase suggesting less active respiration in this strain. Glycogen synthase was up-regulated and glycogen-degrading genes were down-regulated in this strain, leading to the previously reported high glycogen levels (4). The clock genes kaiA, kaiB, and kaiC were down-regulated in Synechococcus 2973. Interestingly, it grows well during a diurnal cycle (5), suggesting clock function is dampened but not lost.
Among other significantly up-regulated pathways are Cys and Met metabolism (P = 0.0123) and the biosynthesis of amino acids (P = 0.0392). This is further corroborated by the biomass composition of the two organisms: Synechococcus 2973 cells have a higher amino acid content than Synechococcus 7942 cells (53% vs. 40.9%) (3).
For each SNP displaying a growth phenotype, we made pairwise comparisons in a reciprocal fashion between both wild-type strains possessing both versions of each allele. In particular, we compared the change in expression between wild-type Synechococcus 2973 and the same strain with each SNP converted to the Synechococcus 7942 allele and between wild-type Synechococcus 7942 and the same strain with each SNP converted to the Synechococcus 2973 allele. Importantly, we identified genes that exhibited a 1.5-fold change in one direction and also a 1.5-fold change in the opposite direction in the reciprocal mutant as genes whose expression is specifically affected by a given SNP. We found that the ATP synthase and NAD+ kinase SNPs had little effect on global gene expression, as only 1.6% and 3.9% of the genes, respectively, showed a 1.5-fold change in one direction and a 1.5-fold change in the opposite direction in the reciprocal mutant (Datasets S4 and S5). As expected, the global clock output regulator rpaA caused a much more significant change in the transcriptome with 17% of the genes (447 genes) showing at least a 1.5-fold change in expression in both mutants compared with their wild types (Dataset S6). Although about one-sixth of the genome showed altered expression between the two rpaA alleles, it is likely that only a few of these genes play a role in the rapid-growth phenotype. Notably, multiple sigma factors, glycogen debranching enzyme, hydrogenase, recombination and repair factors, clock genes, cytochrome c oxidase, transcription factors, and NAD(P) transhydrogenase were all down-regulated in strains with the Synechococcus 2973 allele of rpaA compared with strains with the Synechococcus 7942 allele. The gene that shows the single largest decrease in expression in a Synechococcus 2973 allele of rpaA background is rpaA itself. Introduction of the Synechococcus 2973 rpaA allele into Synechococcus 7942 caused a 134-fold drop in the expression of rpaA itself, suggesting that the 7-bp promoter deletion significantly reduces the expression of rpaA.
As we incorporated SNPs to transition the slow-growing Synechococcus 7942 into the fast-growing Synechococcus 2973, we used RNA-seq to monitor the changes in the transcriptome of Synechococcus 7942 as well. For the two wild-type strains, only 889 genes (33%) of the transcripts are within 1.5-fold expression of each other in either direction, which is a testament to the large phenotypic differences observed between the two strains (Table 3 and Dataset S3). After engineering the Synechococcus 2973 SNPs into Synechococcus 7942, individually and in combination, we compared the transcriptomes of the resulting mutants with wild-type Synechococcus 2973. We found that the transcriptome of any single mutant does not resemble that of Synechococcus 2973, because only 837 (31%), 1,119 (41%), and 347 (13%) transcripts had expression levels similar to those of Synechococcus 2973 in the ATP synthase, NAD kinase, or rpaA conversion strains, respectively (Table 3 and Dataset S7). Upon the addition of the NAD kinase SNP into the strain containing the ATP synthase SNP (2× mutant), transcriptome similarities with Synechococcus 2973 increased only slightly, to 1,160 genes (Table 3 and Dataset S7). Finally, we compared the Synechococcus 7942 5× mutant in which all three genes (atpA, ppnK, and rpaA) had been engineered to contain the Synechococcus 2973 SNPs. Here we found that the expression of 2,377 transcripts (88%) was within 1.5-fold of that observed in wild-type Synechococcus 2973 (Table 3 and Dataset S7). Thus, when all three alleles are incorporated into Synechococcus 7942, it takes on the transcriptional profile of Synechococcus 2973, as most of the genes (88%) show comparable expression in the 5× mutant of Synechococcus 7942 and wild-type Synechococcus 2973 (Table 3, column 5), whereas only 33% of the genes show comparable expression in the two wild-type strains (Table 3, column 1). Therefore, we were able to monitor the transition of the slower-growing Synechococcus 7942 into the fast-growing Synechococcus 2973 both phenotypically and by systems-level transcriptome-wide expression profiles.
Table 3.
Comparison of the transcriptional profile of Synechococcus 7942 containing various substituted SNPs from Synechococcus 2973 with both wild-type strains
| SNP engineered into Synechococcus 7942 | ||||||
| Genes with <1.5-fold expression levels of | Wild-type 7942 (%) | ATP synthase (%) | NAD kinase (%) | rpaA (%) | 2× (%) | 5× (%) |
| 2973 | 889 (33) | 837 (31) | 1,119 (41) | 347 (13) | 1,124 (41) | 2,377 (88) |
| 7942 | 2,691 (100) | 2,440 (90) | 1,187 (44) | 410 (15) | 2,010 (74) | 972 (36) |
The table shows the number of genes that exhibit a <1.5-fold difference, indicating that they have comparable expression in the two strains. 2×, Synechococcus 2973 ATP synthase and NAD+ kinase alleles engineered into Synechococcus 7942. 5×, Synechococcus 2973 ATP synthase, NAD+ kinase, and rpaA alleles engineered into Synechococcus 7942.
Comparison of SNP Alleles Across Diverse Cyanobacteria.
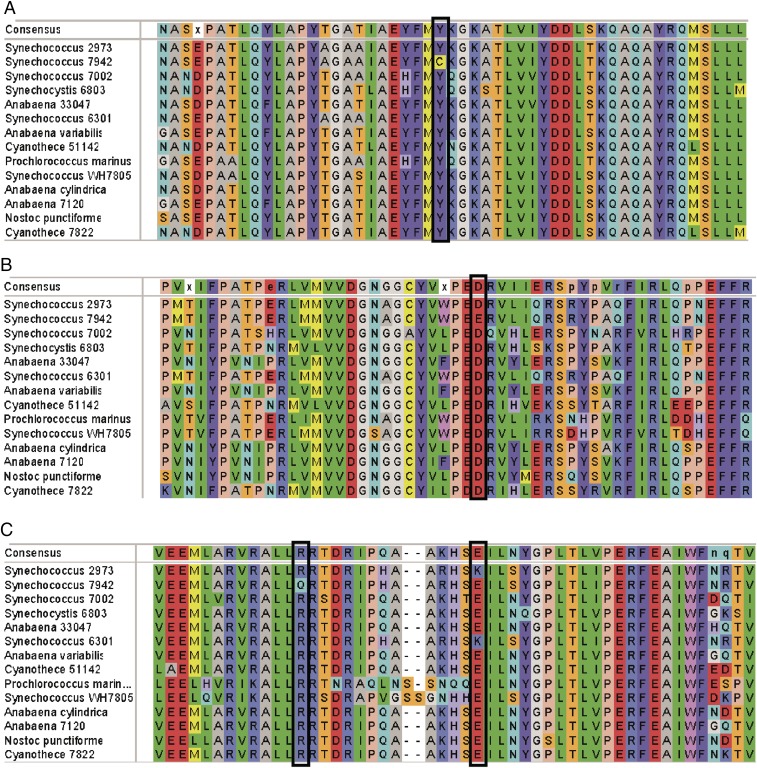
Finally, we were interested in the distribution of the alleles of these highly conserved genes across diverse species of cyanobacteria with various growth rates. We aligned the sequences of the three encoded proteins in the two Synechococcus strains and other commonly studied cyanobacteria (Fig. 4). What we found was striking: In each case, the SNP is in a conserved residue. Interestingly, in three of the four cases, the residue is conserved in Synechococcus 2973 and all other cyanobacteria examined, irrespective of their growth rate. In fact, Synechococcus 7942 appears to be the outlier among other cyanobacteria at these loci. We can only conclude that these SNPs did not evolve specifically for rapid growth but instead they produce the canonical amino acid for their respective proteins in Synechococcus 2973. Therefore, it appears that rather than Synechococcus 2973 being a fast-growing mutant of Synechococcus 7942, Synechococcus 2973 has the more typical allele for these three proteins. Furthermore, Synechococcus 7942 seems to have acquired mutations at these three loci that slow down growth, impair the photosynthetic rate, and impart sensitivity to high light.
Fig. 4.
Local alignments of the SNPs that are involved in rapid growth in various cyanobacteria. (A) ATP synthase. (B) NAD+ kinase. (C) RpaA. The SNPs in Synechococcus 7942 and Synechococcus 2973 are indicated by boxes.
Discussion
Synechococcus 2973 differs from Synechococcus 7942 by only 53 SNPs, a 7-kb deletion, and a 177-kb inversion. However, Synechococcus 2973 produces biomass more than twice as fast as Synechococcus 7942 and exhibits significantly higher photosynthetic rates in conjunction with high light tolerance. We performed a comprehensive mutational analysis on the SNPs in Synechococcus 2973 and identified three genetic loci that lead to its remarkable phenotypes. Just five SNPs in three genes (atpA, ppnK, and rpaA) are responsible for the differences reported between the two strains. We introduced these SNPs into Synechococcus 7942 and found that it gained high light tolerance with a commensurate increase in growth rate, making it comparable to Synechococcus 2973. Therefore, we altered the genome of Synechococcus 7942 at five loci and successfully decreased its doubling time from 6.8 h to 2.2 h, a 309% increase in growth rate under optimal conditions. Commensurate with the increase in growth rate, the photosynthetic rate was also doubled in the fast-growing mutant. Two of the three genes implicated in the rapid growth of Synechococcus 2973 (ATP synthase and NAD+ kinase) are involved in generating the cell’s energy currency. In this study, we demonstrated that the Synechococcus 2973 versions of these enzymes exhibit improved kinetics, resulting in increased ATP production as well as an increased supply of NADP+ to accept electrons from the photosynthetic electron transfer chain (ETC). In fact, Abernathy et al. (19) recently reported that Synechococcus 2973 has higher titers of NADPH and a higher energy charge (ATP/ADP ratio) than Synechococcus 7942. Furthermore, in a recent computational study of Synechococcus 2973 Mueller et al. (20) flagged both ATP synthase and NAD kinase SNPs as possible contributors to rapid growth and predicted that increased activities of these enzymes would increase growth rate. Many have widely speculated that Rubisco activity is the limiting factor for the carbon-fixation rate (21, 22). Our results here suggest that the energy supply limits the carbon-fixation rate, at least in Synechococcus 7942. This is supported by our previous comparison of the photophysiology in the two strains in which we uncovered a photosynthetic bottleneck in the ETC of Synechococcus 7942 that depresses the rate of electron flow, leading to light sensitivity, a decreased carbon-fixation rate, and slower growth (4). The current study provides the molecular underpinnings of the improved photosynthetic ETC in Synechococcus 2973. In this strain, the NAD+ kinase displays a higher rate of catalysis for the phosphorylation of NAD+ to NADP+, which should generate a larger pool of NADP+ to be reduced by the photosynthetic ETC. Such a change supports the increased rate of electron transfer observed in Synechococcus 2973, thus generating an increased level of NADPH, the central reducing power for biosynthesis in these photosynthetic cells. Previously, it was determined that Synechococcus 2973 has increased titers of cytochrome f, plastocyanin, and PSI to support a higher level of photosynthetic electron flux (4); here we show that increasing the supply of the terminal electron acceptor NADP+ is also important for increased growth.
Such an enhancement in the reducing power in Synechococcus 2973 is complemented by an ATP synthase with higher specific activity. Increased ETC activity would serve to generate increased PMF. The increased activity of ATP synthase would serve to utilize the PMF faster and generate substantially more ATP. It is not coincidental that increased ATP and NADPH production in Synechococcus 2973 drastically increases its photosynthetic carbon fixation, as these are the two main energy suppliers for that process. Our findings in this study and previous work (4) indicate that increased energy production in the form of ATP and NADPH by Synechococcus 2973 is directly responsible for this strain’s capacity for rapid autotrophic growth.
Inclusion of only the atpA and ppnK SNPs in Synechococcus 7942 was not sufficient to result in rapid photoautotrophic growth under high light. We also had to include two coding SNPs and one promoter SNP in the master output regulator of the circadian clock, rpaA, to recapitulate the rapid-growth phenotype, which suggests that both a change in the expression and a change in the activity of this important protein are important components. RpaA is a global transcriptional regulator that binds to the promoters of more than 100 genes, linking the core clock Kai oscillator to global transcriptional rhythms (23). Deletion of rpaA abrogates the clock and locks the cells in a dawn-like state, while overexpression of phosphomimetic RpaA closes the cell-division gate and locks the cells in a dusk-like state (24). When the Synechococcus 2973 rpaA allele was introduced into Synechococcus 7942, the gene showing the largest decrease in expression was rpaA itself, likely due to the 7-bp promoter deletion. Interestingly, however, rpaA-null mutants do not grow during diurnal light–dark cycles (24, 25). We have previously shown that Synechococcus 2973 grows well during such a diurnal cycle (3), indicating that the promoter deletion significantly reduces expression of rpaA but does not generate a null phenotype.
RpaA sits on top of a large regulon that produces complex changes in gene expression with variations in timing and response to different levels of RpaA (23). Markson et al. noted that some genes are transiently activated by low levels of RpaA but become repressed by high levels of this regulator. In general, RpaA is responsible for enhancing the expression of subjective dusk genes while repressing the expression of subjective dawn genes. Thus, we hypothesize that the large down-regulation of rpaA by the Synechococcus 2973 allele serves to dampen the circadian amplitude in this strain. Furthermore, this allele presumably locks the cells in a partial dawn-like state during which they are poised to divide. Ashby and Mullineaux (26) observed that rpaA mutants display an increased efficiency of energy transfer from phycobilisomes to PSII, which links rpaA expression to photosynthetic efficiency as well. The same group reported an increased PSI/PSII ratio and decreased phycobilisome content in an rpaA mutant of Synechocystis 6803; both are changes that we identified as leading to improved photosynthetic rates in Synechococcus 2973 (26). Furthermore, Puszynska and O’Shea (27) reported that an rpaA-null mutant accumulates very little glycogen during the day. In contrast, we found significantly elevated glycogen content in Synechococcus 2973 (4). Also, it was reported that the rpaA-null mutant shows decreased expression of glycolytic enzymes. However, our RNA-seq analysis found increased expression of genes for glycolytic enzymes in Synechococcus 2973. Therefore, due to the complex nature of the rpaA regulon, we find that some changes in expression caused by the Synechococcus 2973 allele of rpaA resemble the deletion mutant, while other changes resemble an rpaA-overexpression strain. Unfortunately, due to the wide-ranging pleiotropic effects observed from the two rpaA alleles, it is challenging to determine the exact role (or, more likely, the many roles) of the Synechococcus 2973 rpaA allele in the fast-growth phenotype. Introduction of this allele into Synechococcus 7942 generated a transcriptional profile that looked like neither parental strain, and not until all three alleles were combined was the Synechococcus 2973 transcriptional profile recreated. This indicates a higher order of interaction among these alleles and suggests that the Synechococcus 2973 rpaA allele has a different effect on the organism depending on which version of the atpA and ppnK alleles it possesses. Further work will be needed to determine how the altered energy balance generated by the Synechococcus 2973 ATP synthase and NAD+ kinase alleles is sensed by the circadian clock system and subsequently alters the complex transcriptional profiles mediated by RpaA. Admittedly, due to the lack of replicates, our high-level survey of the transcriptional differences between the two strains and the mutants thereof is limited in the statistical power that is needed to dive deeper into the pleiotropic effects caused by the rpaA alleles and their interactions with the other alleles. The complex interactions between energy capture and utilization with the circadian clock are certainly an area of interest that demands more in-depth analysis in the future.
It seems farfetched that five SNPs can convert a slow-growing light-sensitive strain into a high-light–tolerant fast-growing strain if the entire proteome of Synechococcus 7942 was wired for slower growth and metabolism. We hypothesize that Synechococcus 7942 is already metabolically wired for rapid photoautotrophic growth and that at some time in the recent past it lost the ability to grow rapidly while adapting to a low-light lifestyle. By returning the five mutant loci to the canonical version existing among cyanobacteria, we could unlock the full potential of Synechococcus 7942.
Materials and Methods
Strains and Culture Conditions.
Synechococcus 2973 and Synechococcus 7942 were maintained on BG11 agar plates at 38 °C with 125 μmol⋅m−2⋅s−1 light. Both strains were routinely grown with shaking in liquid BG11 medium at 38 °C with 500 μmol⋅m−2⋅s−1 light and 1% CO2 in an AlgaeTron AG230 growth chamber (Photon Systems Instruments). E. coli strains were grown in LB medium with 50 μg/mL kanamycin. To determine growth parameters, strains were grown in BG11 liquid at 38 °C with 900 μmol⋅m−2⋅s−1 light and 5% CO2 in a MC-1000 multicultivator bioreactor (Photon Systems Instruments). In this case, strains were diluted 1:100 from a growing culture into the multicultivator chamber, were grown for 24 h to adapt them to the bioreactor, and were diluted to an OD730 of 0.025 to begin the growth experiment. Growth rates (K′) were calculated using Microsoft Excel by fitting an exponential curve to the logarithmic section of the growth data (typically OD730 <0.3) and using the slope, m, as K′ (y = kemx). Doubling times were then calculated as ln (2)/K′.
Construction of Cyanobacterial Strains.
All point mutations were introduced with our previously reported CRISPR/Cpf1 system based on the pSL2680 vector (5). Briefly, an editing vector was introduced into the wild-type strain through triparental mating (28). Methods for building editing vectors are described in SI Appendix, SI Methods. The resulting colonies were repatched three times onto BG11 supplemented with 10 μg/mL kanamycin to allow the editing to take place. Mutants were then verified by amplifying a region of chromosomal DNA with primers located outside the homology region that exists on the editing plasmid followed by Sanger sequencing. Once a mutant was verified, the colony was grown up to stationary phase in BG11 without antibiotics (∼50 µmol⋅m−2⋅s−1, 38 °C) and then was diluted 1:1,000 into BG11 without antibiotics and was grown to stationary phase again to allow the editing plasmid to be cured. The resulting culture was plated as a dilution series on BG11 plates to obtain single colonies. One hundred colonies were patched onto BG11 with 10 μg/mL kanamycin and then onto BG11 without antibiotics to screen for kanamycin-sensitive patches that had lost the editing plasmid, resulting in a clean, markerless mutation.
Oxygen Evolution.
Cultures were grown in a multicultivator bioreactor with 5% CO2, 900 μmol⋅m−2⋅s−1 light, 38 °C. Cells were harvested during log phase and adjusted based on equal cell numbers. Light-induced oxygen evolution was measured for 1 min at 38 °C under saturating light (1,500 μmol⋅m−2⋅s−1) using a custom-built Clark- type electrode. Data were processed in Microsoft Excel.
Overexpression and Purification of PpnK.
This NAD kinase was purified following a protocol modified from Grose et al. (14). Additional information on overexpression and purification of Ppnk is located in SI Appendix, SI Methods.
NAD Kinase Assay.
A typical NADK assay reaction consisted of 200 μL of 5× reaction buffer [500 mM Tris⋅HCl (pH 7.1), 5 mM MgCl2], either 20 µL of 50 mM NAD and a variable amount of 50 mM ATP or a variable amount of 50 mM NAD with 20 µL of 50 mM ATP, 30 μg NADK, and water to 1 mL (29, 30). The concentration of either NAD or ATP was varied over a series of reactions while the other was held at 1 mM. Reactions were allowed to proceed for 30 min at 37 °C and were stopped by the addition of 10 µL of 0.5 M EDTA. The reactions were then deproteinated by spinning them through a 10-K molecular weight cutoff concentrator column, and the flowthrough was subjected to analysis by HPLC. KM, Vmax, and Kcat were calculated from the Lineweaver–Burke plots as follows: Vmax was calculated as 1/y-intercept. Km was calculated as (slope × Vmax). Kcat was calculated as Vmax/enzyme concentration.
HPLC Analysis of Nucleotides.
HPLC analysis was performed according to ref. 31 with slight modifications. Our HPLC analysis was able to detect ATP, ADP, NAD+, NADH, NADP+, and NADPH simultaneously. Nucleotide analyses was carried out throughout this work on an Agilent 1200 series HPLC system equipped with a quaternary pump, degasser, diode array detector, and a Peltier-cooled auto-sampler set at 4 °C with separation on a ZORBAX XDB-C18 rapid-resolution (4.6 × 250 mm, 3.5 μm) column. A 100-μL injection was used, and nucleotides were detected via absorbance at 254 nm. The HPLC was run under isocratic conditions at a flow rate of 2 mL/min with the mobile phase consisting of 0.15 M phosphate-citrate buffer, pH 6.8. Chromatographic analysis was performed for 5 min at 15 °C followed by an increase to 45 °C for 25 min. Standard nucleotides were prepared from 10 mM stock solutions in 1× reaction buffer. Calibration was carried out by injection of a standard mixture of nucleotides used at different concentrations. The data was analyzed with Agilent Chemstation for Windows.
Membrane Preparations.
Membranes were isolated via osmotic shock followed by differential centrifugation (15, 16). We harvested 200 mL of cells from stationary-phase cultures of either wild-type Synechococcus 2973 or SL2770 (wild-type 2973 with the 7942 ATP synthase allele) strains. Cells were resuspended in 25 mL hypertonic buffer (0.5 M sorbitol, 1 M sucrose, 10 mM Tricine, 10 mM MgCl2, 5 mM NaH2PO4, 2.5 mM K2HPO4, 2 mg/mL lysozyme, 1 mM PMSF, pH 7.8) and were incubated for 1 h at 37 °C. The cells were then pelleted by centrifugation for 10 min at 8,500 × g, 4 °C. Osmotic lysis was then carried out by resuspending the pellet in 30 mL osmotic lysis buffer (10 mM Tricine, 10 mM MgCl2, 1 mM PMSF, pH 7.8) and incubating for 5 min on ice. Large debris and unbroken cells were removed by centrifugation for 5 min at 1,000 × g. Membranes were then pelleted by centrifugation for 30 min at 40,000 × g, 4 °C. Membranes were resuspended to an equal chlorophyll amount in acid buffer (10 mM succinic acid, 2 mM MgCI2, 10 mM KCI, 2 mM NaH2PO4, 10 mM Tricine, pH 6.0).
ATP Synthase Activity.
A membrane sample was diluted to 50 μg/mL chlorophyll in acid buffer (10 mM succinic acid, 2 mM MgCI2, 10 mM KCI, 2 mM NaH2PO4, 10 mM Tricine, pH 6.0) and incubated for 5 min in the dark at room temperature to equilibrate and adapt to the dark. The membranes were then diluted 1:1 into basic buffer (10 mM KCl, 2 mM MgCl2, 2 mM NaH2PO4, 200 mM Na-Tricine, 2 mM ADP, pH 8.6). Samples of 100 μL were taken immediately after the dilution as well as at different intervals and were quenched into an equal volume of 2.5% TCA to stop the reaction. The reaction was then neutralized with 5 µL of 1 M Tris/6 M KOH. The cell debris was removed by centrifugation for 5 min at 13,000 × g. The supernatant was then diluted 1:5, and ATP was quantitated using a luciferase-based ATP detection kit (Abcam). Luminescence was measured on a Synergy Mx plate reader (BioTek). The data was collected using BioTek Gen5 software. A blank consisting of a reaction without membranes was performed as control for ATP contamination of the ADP reagent. The ATP measured from such a blank was subtracted from all subsequent measurements.
RNA-Seq Analysis.
RNA was isolated using RNAwiz reagent (Ambion) according to the manufacturer’s instructions and was quantified on a NanoDrop ND-1000 instrument (NanoDrop Technologies). Library preparation, RNA-seq, and data normalization were performed by the Genome Technology Access Center at Washington University in St. Louis. Library preparation was performed with 1 µg of total RNA, and integrity was determined using an Agilent bioanalyzer. Ribosomal RNA was removed by a hybridization method using Ribo-Zero rNA removal (Bacteria) kits (Illumina). mRNA was then fragmented in buffer containing 40 mM Tris acetate (pH 8.2), 100 mM potassium acetate, and 30 mM magnesium acetate and heating to 94° for 150 s. mRNA was reverse transcribed to yield cDNA using SuperScript III RT enzyme (Life Technologies) and random hexamers. A second-strand reaction was performed to yield ds-cDNA. cDNA was blunt ended, had an A base added to the 3′ ends, and then had Illumina sequencing adapters ligated to the ends. Ligated fragments were then amplified for 15 cycles using primers incorporating unique index tags. Fragments were sequenced on an Illumina HiSeq 2500 instrument using single reads extending 50 bases. RNA-seq reads were aligned to the Ensembl top-level assembly with STAR version 2.0.4b. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:featureCount version 1.4.5. Transcript counts were produced by Sailfish version 0.6.3. Sequencing performance was assessed for total number of aligned reads, total number of uniquely aligned reads, genes and transcripts detected, ribosomal fraction known junction saturation, and read distribution over known gene models with RSeQC version 2.3. All gene-level and transcript counts were then imported into the R/Bioconductor package EdgeR, and trimmed mean of M-values (TMM) normalization size factors were calculated to adjust for samples’ differences in library size. Ribosomal features as well as any feature not expressed in at least the smallest condition size minus one sample were excluded from further analysis, and TMM size factors were recalculated to created effective TMM size factors. The TMM size factors and the matrix of counts were then imported into R/Bioconductor package Limma, and weighted likelihoods based on the observed mean-variance relationship of every gene/transcript and sample were then calculated for all samples using the voomWithQualityWeights function. Performance of the samples was assessed with a spearman correlation matrix and multidimensional scaling plots. Gene/transcript performance was assessed with plots of residual SD of every gene to its average log-count with a robustly fitted trend line of the residuals. Generalized linear models were then created to test for gene/transcript-level differential expression. Output RNA-seq files were analyzed using custom Python scripts. Genomic identifiers, gene annotations, and functional descriptions were paired with corresponding expression values by ordering data outputs by chromosomal location and systematically pairing them with Synechococcus 2973 information from the University of Iowa’s alignable tight genomic clusters (ATGC) website based on National Center for Biotechnology Information protein accession and version identifiers. An additional script was written to identify genes from reciprocal mutant pairs that show at least a 1.5-fold TPM change in opposite directions. TPM values were extracted from the annotated data sets, and ratios between wild-type and mutant expression values were calculated. Loci that exhibited a 1.5-fold TPM expression change when the wild-type Synechococcus 7942 allele was replaced with the wild-type Synechococcus 2973 allele, and vice versa, were output to secondary files. Some gene annotations and functions in the final dataset were manually updated based on information from the IMG/MER website (https://img.jgi.doe.gov/).
Pathway Enrichment Analysis.
To identify the differentially expressed genes, a log twofold change cutoff of 1.5 was chosen. This identified 76 up-regulated genes and 596 down-regulated genes in Synechococcus 2973. Pathway categories were extracted from Kyoto Encyclopedia of Genes and Genomes (KEGG) (32), and enrichment analysis was carried out using the R-package goseq (33). The Wallenius’ noncentral hypergeometric distribution was used to calculate the P value for the presence of a pathway in up- and down-regulated gene sets. The overrepresentation of a pathway in the up- and down-regulated set of genes was considered statistically significant if the corresponding P value for that pathway was less than 0.05.
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation Grant MCB-1546840 (to H.B.P. and C.D.M.). K.E.W. is supported by a William H. Danforth Fellowship at Washington University in St. Louis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 12547.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814912115/-/DCSupplemental.
References
- 1.Lai MC, Lan EI. Advances in metabolic engineering of cyanobacteria for photosynthetic biochemical production. Metabolites. 2015;5:636–658. doi: 10.3390/metabo5040636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knoot CJ, Ungerer J, Wangikar PP, Pakrasi HB. Cyanobacteria: Promising biocatalysts for sustainable chemical production. J Biol Chem. 2018;293:5044–5052. doi: 10.1074/jbc.R117.815886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ungerer J, Lin P-C, Chen H-Y, Pakrasi HB. Adjustments to photosystem stoichiometry and electron transfer proteins are key to the remarkably fast growth of the cyanobacterium Synechococcus elongatus UTEX 2973. MBio. 2018;9:e02327-17. doi: 10.1128/mBio.02327-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ungerer J, Pakrasi HB. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci Rep. 2016;6:39681. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau RH, Doolittle WF. Covalently closed circular DNAs in closely related unicellular cyanobacteria. J Bacteriol. 1979;137:648–652. doi: 10.1128/jb.137.1.648-652.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McEwen JT, Machado IMP, Connor MR, Atsumi S. Engineering Synechococcus elongatus PCC 7942 for continuous growth under diurnal conditions. Appl Environ Microbiol. 2013;79:1668–1675. doi: 10.1128/AEM.03326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarn DF, Whitaker RA, Alam J, Vrba JM, Curtis SE. Genes encoding the alpha, gamma, delta, and four F0 subunits of ATP synthase constitute an operon in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1988;170:3448–3458. doi: 10.1128/jb.170.8.3448-3458.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SB, Murray CI, Chung HS, Van Eyk JE. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc Med. 2013;23:14–18. doi: 10.1016/j.tcm.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casey WT, et al. The effect of polyphosphate kinase gene deletion on polyhydroxyalkanoate accumulation and carbon metabolism in Pseudomonas putida KT2440. Environ Microbiol Rep. 2013;5:740–746. doi: 10.1111/1758-2229.12076. [DOI] [PubMed] [Google Scholar]
- 11.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poncet-Montange G, Assairi L, Arold S, Pochet S, Labesse G. NAD kinases use substrate-assisted catalysis for specific recognition of NAD. J Biol Chem. 2007;282:33925–33934. doi: 10.1074/jbc.M701394200. [DOI] [PubMed] [Google Scholar]
- 13.Kawai S, Murata K. Structure and function of NAD kinase and NADP phosphatase: Key enzymes that regulate the intracellular balance of NAD(H) and NADP(H) Biosci Biotechnol Biochem. 2008;72:919–930. doi: 10.1271/bbb.70738. [DOI] [PubMed] [Google Scholar]
- 14.Grose JH, Joss L, Velick SF, Roth JR. Evidence that feedback inhibition of NAD kinase controls responses to oxidative stress. Proc Natl Acad Sci USA. 2006;103:7601–7606. doi: 10.1073/pnas.0602494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholts MJC, Aardewijn P, Van Walraven HS. Membrane vesicles from Synechocystis 6803 showing proton and electron transport and high ATP synthase activities. Photosynth Res. 1996;47:301–305. doi: 10.1007/BF02184291. [DOI] [PubMed] [Google Scholar]
- 16.Krab K, Bakels RHA, Scholts MJC, van Walraven HS. Activation of the H+-ATP synthase in thylakoid vesicles from the cyanobacterium Synechococcus 6716 by ΔµH+. Including a comparison with chloroplasts, and introducing a new method to calibrate light-induced ΔµH+ Biochim Biophys Acta. 1993;1141:197–205. [Google Scholar]
- 17.Jagendorf AT, Uribe E. ATP formation caused by acid-base transition of spinach chloroplasts. Proc Natl Acad Sci USA. 1966;55:170–177. doi: 10.1073/pnas.55.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Walraven HS, Hollander EE, Scholts MJC, Kraayenhof R. The H+/ATP ratio of the ATP synthase from the cyanobacterium Synechococcus 6716 varies with growth temperature and light intensity. Biochim Biophys Acta. 1997;1318:217–224. [Google Scholar]
- 19.Abernathy MH, et al. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol Biofuels. 2017;10:273. doi: 10.1186/s13068-017-0958-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller TJ, Ungerer JL, Pakrasi HB, Maranas CD. Identifying the metabolic differences of a fast-growth phenotype in Synechococcus UTEX 2973. Sci Rep. 2017;7:41569. doi: 10.1038/srep41569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zorz JK, et al. The RUBISCO to photosystem II ratio limits the maximum photosynthetic rate in picocyanobacteria. Life (Basel) 2015;5:403–417. doi: 10.3390/life5010403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ort DR, et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA. 2015;112:8529–8536. doi: 10.1073/pnas.1424031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markson JS, Piechura JR, Puszynska AM, O’Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci USA. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diamond S, et al. Redox crisis underlies conditional light-dark lethality in cyanobacterial mutants that lack the circadian regulator, RpaA. Proc Natl Acad Sci USA. 2017;114:E580–E589. doi: 10.1073/pnas.1613078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashby MK, Mullineaux CW. Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol Lett. 1999;181:253–260. doi: 10.1111/j.1574-6968.1999.tb08852.x. [DOI] [PubMed] [Google Scholar]
- 27.Puszynska AM, O’Shea EK. Switching of metabolic programs in response to light availability is an essential function of the cyanobacterial circadian output pathway. eLife. 2017;6:e23210. doi: 10.7554/eLife.23210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clerico EM, Ditty JL, Golden SS. Specialized techniques for site-directed mutagenesis in cyanobacteria. In: Rosato E, editor. Circadian Rhythms: Methods and Protocols. Humana Press; Totowa, NJ: 2007. pp. 155–171. [DOI] [PubMed] [Google Scholar]
- 29.Kawai S, Mori S, Mukai T, Hashimoto W, Murata K. Molecular characterization of Escherichia coli NAD kinase. Eur J Biochem. 2001;268:4359–4365. doi: 10.1046/j.1432-1327.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 30.Bieganowski P, Seidle HF, Wojcik M, Brenner C. Synthetic lethal and biochemical analyses of NAD and NADH kinases in Saccharomyces cerevisiae establish separation of cellular functions. J Biol Chem. 2006;281:22439–22445. doi: 10.1074/jbc.M513919200. [DOI] [PubMed] [Google Scholar]
- 31.Aso Y, Gotoh S, Yamasaki N. A HPLC method for simultaneous analysis of NAD(P)+ and NAD(P)H-its application to the study of spinach ferredoxin: NADP+ reductase-catalyzed transhydrogenation. Agric Biol Chem. 1989;53:1635–1639. [Google Scholar]
- 32.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.