Significance
A large number of nucleotides is continuously damaged in every cell. Damaged nucleotides stall replicative DNA polymerases on template strands. Resulting replication blockage is released by two alternative pathways, error-free template switching (TS) and error-prone translesion DNA synthesis (TLS). TLS plays a major role in converting DNA damage to mutations, and thus, the relative usage of TLS over TS determines the frequency of mutagenesis. The relative usage is controlled by post-translational modification of proliferating cell nuclear antigen (PCNA), which physically interacts with DNA synthesis enzymes, in Saccharomyces cerevisiae. Environmental DNA damage induces ubiquitylation of PCNA and promotes TLS. We here show that SUMOylation of PCNA ensures the release of replication blockage by error-free TS pathways and prevents mutagenesis during the physiological cell cycle.
Keywords: PIAS1, PIAS4, PCNA, SUMOylation, template switching (TS)
Abstract
DNA damage tolerance (DDT) releases replication blockage caused by damaged nucleotides on template strands employing two alternative pathways, error-prone translesion DNA synthesis (TLS) and error-free template switch (TS). Lys164 of proliferating cell nuclear antigen (PCNA) is SUMOylated during the physiological cell cycle. To explore the role for SUMOylation of PCNA in DDT, we characterized chicken DT40 and human TK6 B cells deficient in the PIAS1 and PIAS4 small ubiquitin-like modifier (SUMO) E3 ligases. DT40 cells have a unique advantage in the phenotypic analysis of DDT as they continuously diversify their immunoglobulin (Ig) variable genes by TLS and TS [Ig gene conversion (GC)], both relieving replication blocks at abasic sites without accompanying by DNA breakage. Remarkably, PIAS1−/−/PIAS4−/− cells displayed a multifold decrease in SUMOylation of PCNA at Lys164 and over a 90% decrease in the rate of TS. Likewise, PIAS1−/−/PIAS4−/− TK6 cells showed a shift of DDT from TS to TLS at a chemosynthetic UV lesion inserted into the genomic DNA. The PCNAK164R/K164R mutation caused a ∼90% decrease in the rate of Ig GC and no additional impact on PIAS1−/−/PIAS4−/− cells. This epistatic relationship between the PCNAK164R/K164R and the PIAS1−/−/PIAS4−/− mutations suggests that PIAS1 and PIAS4 promote TS mainly through SUMOylation of PCNA at Lys164. This idea is further supported by the data that overexpression of a PCNA-SUMO1 chimeric protein restores defects in TS in PIAS1−/−/PIAS4−/− cells. In conclusion, SUMOylation of PCNA at Lys164 promoted by PIAS1 and PIAS4 ensures the error-free release of replication blockage during physiological DNA replication in metazoan cells.
Homologous recombination (HR) plays an essential role in DNA replication by repairing double-strand breaks (DSBs) and mediating the bypass of lesions following replication fork stalling (1, 2). HR repairs DSBs induced during DNA replication by anticancer agents, camptothecin (topoisomerase I inhibitor) and olaparib, the poly[ADP ribose]polymerase inhibitor (3, 4). In addition, HR contributes to a DNA-damage-tolerance (DDT) pathway, reactivation of stalled replication forks, and filling in of discontinuities remaining in the replicating DNA without actually repairing DNA lesions that cause replication blockage (5, 6). In this context, HR facilitates transient switching of replication primers from the damaged template strand to the newly synthesized sister chromatid (7–12) via a mechanism that remains very poorly understood even in Saccharomyces cerevisiae (13). Template switch (TS) by HR as well as HR-mediated DSB repair occasionally involves a crossover between the two sisters, leading to sister chromatid exchange (SCE) (14–16). In addition to TS by HR, replication blockage is released by translesion DNA synthesis (TLS), a process by which DNA damage is bypassed by employing error-prone TLS DNA polymerases. TLS and TS seem to be compensatory for each other in DDT because of synthetic lethality of cells deficient in both the Polζ TLS polymerase and the Rad54 HR factor (17, 18). The loss of the Polη TLS polymerase causes increases in the number of UV-induced SCEs, suggesting that TS partially compensates for TLS over UV lesions (19, 20).
The two major DDT pathways, error-free TS and error-prone TLS, are controlled by ubiquitylation and SUMOylation of PCNA. Molecular mechanisms for this control have been established in S. cerevisiae, whereas it remains elusive whether the same control mechanisms operate in metazoan cells. Activation of TLS depends on monoubiquitylation by the Rad18 ubiquitylation enzyme at a highly conserved lysine (K) residue of PCNA, a DNA clamp that acts as a processivity factor for replicative DNA polymerase δ in S. cerevisiae (21, 22). Error-free TS is facilitated by polyubiquitylation of PCNA by Ubc13 ubiquitin-conjugating enzyme and Rad5 ubiquitin ligase in S. cerevisiae (11, 21, 23, 24). It remains unclear whether the polyubiquitylation activates TS also in metazoan cells as the polyubiquitylation of PCNA by Ubc13 activates TLS rather than TS in Schizosaccharomyces pombe (25). A recent report suggests that polyubiquitylation is not required for efficient TS in mammalian cells (26). SUMOylation of PCNA suppresses abnormal recombination in S. cerevisiae (27, 28). Human cell lines and the chicken DT40 cell line show SUMOylation of PCNA (29–31). Thus, an important unsolved question is the role of PCNA SUMOylation in metazoan cells.
Although the phenotypic assay for accurately evaluating TS in S. cerevisiae has been well established (11), there is no standard phenotypic assay for evaluating TS in mammalian cells, the absence of which has hampered analysis of this DDT pathway. A recently developed bioassay, the piggyBlock assay, now allows for accurately measuring the relative ratio between TS and TLS events at chemically generated lesions, such as thymine-thymine (TT)-cyclobutane-pyrimidine-dimer (TT-CPD) UV damage, in the context of chromosomal DNA (32, 33). In addition to the piggyBlock assay, the chicken DT40 B lymphocyte cell line provides a unique opportunity to accurately measure the usage of the two major DDT events: release of replication blockage by TS and TLS (figure S1 in ref. 34). The DT40 cell line offers this unique opportunity as it diversifies its Ig variable (Ig V) genes during in vitro culture (35) by releasing replication blockage at sites of activation-induced deaminase- (AID-) induced abasic sites. This release occurs via TS and TLS leading to templated Ig gene conversion (GC) and nontemplated single base substitutions (Ig V hypermutation), respectively (34, 36, 37). Ig GC dominates over the nontemplated mutagenesis during diversification of Ig V in DT40 cells, however, defects in HR factors BRCA1, BRCA2, and XRCC2/3 cause a marked shift from Ig GC to TLS-dependent Ig V hypermutation (38–40). Thus, Ig GC and TLS share the same substrate—replication forks stalled at abasic sites without accompanying DNA breakage. To summarize, one can accurately examine the actual usage of TS and TLS at stalled replication forks by counting the number of individual Ig V diversification events during in vitro culture of DT40 cells.
We here investigated the role of the PIAS1 and PIAS4 small ubiquitin-like modifier (SUMO) E3 ligase enzymes in TS by generating PIAS1−/−/PIAS4−/− cells from the chicken DT40 and human TK6 B lymphocyte lines. The piggyBlock assay indicates that PIAS1−/−/PIAS4−/− mutation decreased the usage of TS relative to TLS in TK6 cells. The PIAS1−/−/PIAS4−/− mutation increased UV sensitivity in TK6 cells deficient in Polη (POLH−/− cells), the major TLS polymerase of bypassing UV damage but not in wild-type cells. These observations suggest that PIAS1 and PIAS4 promote TS and thereby compensate for a defect in TLS. Moreover, both the PIAS1−/−/PIAS4−/− and the PCNAK164R/K164R mutations result in approximately 10-fold decreases in the rate of Ig GC with the PIAS1−/−/PIAS4−/− mutation showing an epistatic relationship with PCNAK164R/K164R mutations, suggesting that the SUMOylation of PCNA at the K164 residue by PIAS1 and PIAS4 promotes TS. This idea is supported by the data that PIAS1−/−/PIAS4−/− cells are deficient in SUMOylation of PCNA at the K164 residue. Furthermore, ectopic expression of a PCNA-SUMO1 chimeric protein significantly reversed defects in TS in PIAS1−/−/PIAS4−/− DT40 cells, including UV-induced SCE and Ig GC. In conclusion, SUMOylation of PCNA at Lys164 ensures the release of replication blockage by an error-free DDT pathway and prevents mutagenesis caused by error-prone TLS polymerases during the cell cycle.
Results
PIAS1−/−/PIAS4−/− Chicken DT40 and Human TK6 Cells Are Capable of Performing HR-Dependent DSB Repair.
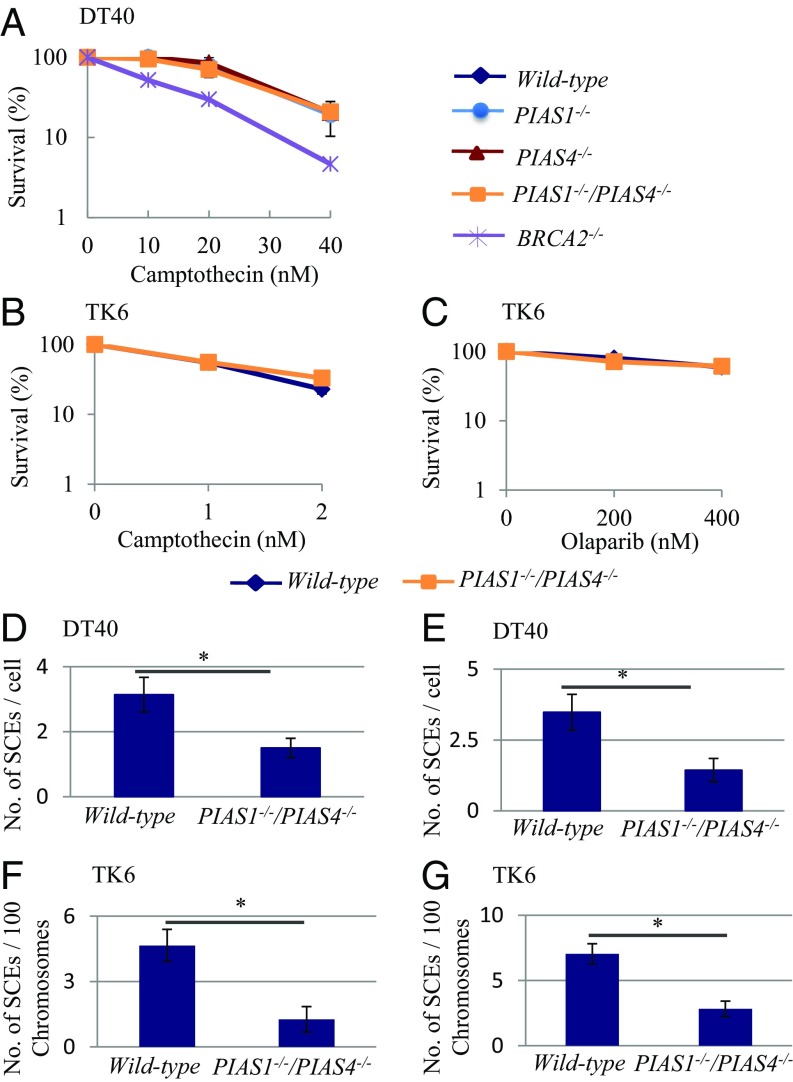
HR plays two major roles in the maintenance of DNA replication, repair of DSBs formed at single-strand breaks (SSBs) in the template strand and TS following replication stalling at damaged bases. To evaluate the former role, we generated PIAS1−/−, PIAS4−/− and PIAS1−/−/PIAS4−/− DT40 cells (SI Appendix, Fig. S1) and measured the sensitivity of these cell lines to killing by camptothecin, which generates one-end breaks during DNA replication. The three mutant clones showed no prominent sensitivity to camptothecin (Fig. 1A). We also generated XPA−/−, POLH−/−, and PIAS1−/−/PIAS4−/− cell lines from the human TK6 B cell line (SI Appendix, Fig. S2). The PIAS1−/−/PIAS4−/− TK6 cells showed the same tolerance to camptothecin as did the wild-type cells (Fig. 1B). Likewise, the PIAS1−/−/PIAS4−/− TK6 cells were tolerant to olaparib, the poly[ADP ribose]polymerase inhibitor that strongly inhibits SSB repair (Fig. 1C). Considering the dominant role played by HR in repairing DSBs induced by camptothecin and olaparib (3, 4), neither PIAS1 nor PIAS4 may be required for efficiently performing HR-dependent DSB repair. We conclude that the PIAS1−/−/PIAS4−/− mutation does not compromise the capability of general HR.
Fig. 1.
SUMO E3 ligase mutants displayed a reduced number of SCE events. (A) Sensitivity of DT40 mutants to camptothecin. The viability of cells was measured at 48 h. (B and C) Sensitivity of TK6 mutants to camptothecin (B) and olaparib (C). The viability of cells was measured by colony formation in the methylcellulose. (D–G) SCE induced by 0.25 J/m2 UV (D) and 10 μM cisplatin (E) in the chicken DT40 cells and 0.25 J/m2 UV (F) and 2 μM cisplatin (G) in the human TK6 cells. At least 50 mitotic cells were analyzed per condition in each experiment. The histogram shows subtracted numbers of mean values for SCEs before the exposure from SCEs after the exposure. The error bars indicate SEM. Statistical analyses were performed by Student’s t test (*P < 0.01).
Significant Decreases in the Rate of SCE in PIAS1−/−/PIAS4−/− Cells.
We addressed whether or not SUMOylation by PIAS1 and PIAS4 contributed to HR in the context of DDT. To this end, we measured SCE induced by UV and cisdiamminedichloroplatinum (II) (cisplatin). Cisplatin generates a considerably larger number of intrastrand cross-links than interstrand cross-links and effectively stalls the progression of large numbers of replication forks as does UV-induced lesions. Spontaneously arising SCE events tended to occur less frequently in PIAS1−/−/PIAS4−/− DT40 cells than in wild-type cells (Fig. 1 D and E and SI Appendix, Fig. S3). SCE events induced by UV and cisplatin were a few times less in PIAS1−/−/PIAS4−/− DT40 cells than in the wild-type control. Likewise, PIAS1−/−/PIAS4−/− TK6 cells showed a three times decrease in the induced SCEs compared with wild-type cells (Fig. 1 F and G and SI Appendix, Fig. S3). These data support the hypothesis that PIAS1 and PIAS4 facilitate TS by HR to release replication blockage.
PIAS1−/−/PIAS4−/− and POLH−/− Mutations Cause a Synergistic Increase in Sensitivity to Cisplatin and UV in TK6 Cells.
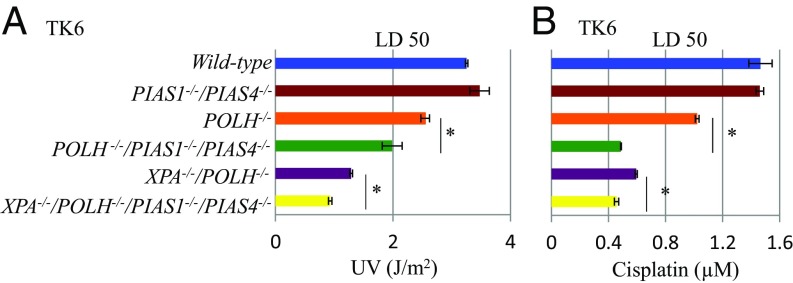
If PIAS1 and PIAS4 control the usage of TS without affecting the capability of HR, PIAS1−/−/PIAS4−/− cells might show sensitivity to UV and cisplatin when TLS is not fully functional. PIAS1−/−/PIAS4−/− cells showed no detectable sensitivity to UV or cisplatin, whereas POLH−/− cells showed modest sensitivity to both UV and cisplatin (Fig. 2). Remarkably, PIAS1−/−/PIAS4−/−/POLH−/− cells showed considerably higher sensitivity to UV and cisplatin than POLH−/− cells (Fig. 2). Likewise, disruption of PIAS1−/−/PIAS4−/− in POLH−/−/XPA−/− cells further increased their sensitivity to cisplatin and UV (Fig. 2). These observations indicate that PIAS1 and PIAS4 can contribute to cellular tolerance to cisplatin and UV when the capability of TLS is reduced as seen in the increase in the number of UV-induced SCE in Polη-deficient cells (19, 20). Collectively, PIAS1 and PIAS4 may substitute for the loss of Polη by enhancing the usage of TS.
Fig. 2.
PIAS1−/−/PIAS4−/− and POLH−/− mutations cause a synergistic increase in the sensitivity of TK6 cells to cisplatin and UV. (A and B) Clonogenic cell survival assay following exposure of indicated cell lines to UV (A) and cisplatin (B). Lethal dose 50% (LD50) is the dose of DNA-damaging agents that reduces cellular survival to 50% relative to cells nontreated with DNA-damaging agents. The error bars show the SD of the mean of, at least, three independent experiments. Statistical analyses were performed by Student’s t test (*P < 0.01).
Significant Decrease in TS Relative to TLS in PIAS1−/−/PIAS4−/− TK6 Cells.
We next investigated whether PIAS1 and PIAS4 altered the ratio of TS versus TLS at UV damage CPD using the piggyBlock assay. To this end, we inserted a CPD into the piggyBlock transposon-based vector (32), transfected the CPD-carrying vector into the TK6 cells, immediately performed limiting dilution, and selected clones having randomly integrated vector DNA with puromycin (SI Appendix, Fig. S4A). To avoid elimination of the integrated CPD by nucleotide excision repair, we performed experiments in the XPA−/−/POLH−/− background (41).
We analyzed individual clones having randomly integrated the CPD-carrying vector. These clones were mosaics as the cells within the clone inherit either the Watson or the Crick strand of the parental integrant (SI Appendix, Fig. S4 A and B). Thus, in this assay, release of the replication block at the CPD site by error-free TS and by TLS can be distinguished as the CPD-containing TpT is placed opposite a GpC. TS would result in GpC at the CPD site, whereas TLS would insert ApA (accurate TLS) or other bases (inaccurate TLS) at the site [note, insertion of GpC opposite the T-T CPD would be unusual (42)]. Using this approach, we also observed a decrease in the ratio of TS relative to TLS by more than 50% in PIAS1−/−/PIAS4−/−/ POLH−/−/XPA−/− cells in comparison with POLH−/−/XPA−/− cells (SI Appendix, Fig. S4C). To summarize, the significant decreases in both the frequency of the SCE (Fig. 1) and the ratio of TS relative to TLS (SI Appendix, Fig. S4) indicate that PIAS1 and PIAS4 regulate the two DDT pathways by enhancing the relative usage of error-free TS.
Significant Decreases in the Rate of Ig Gene Conversion in the PIAS1 and PIAS4 Mutants.
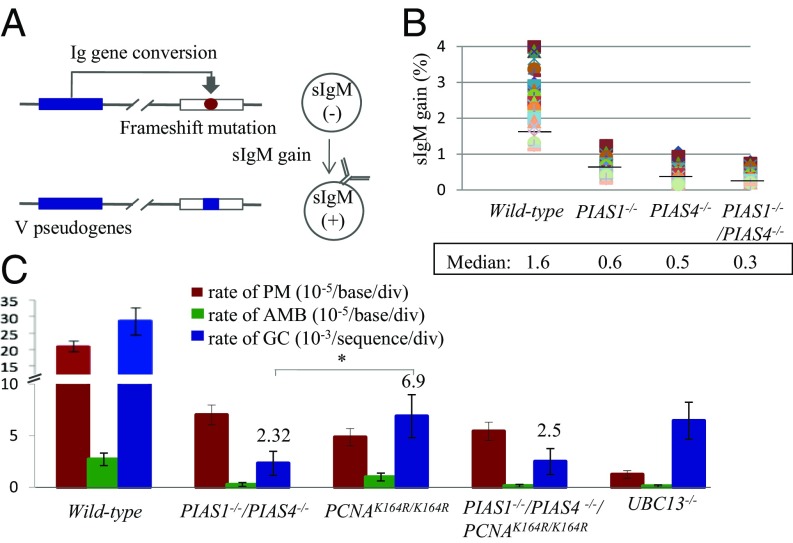
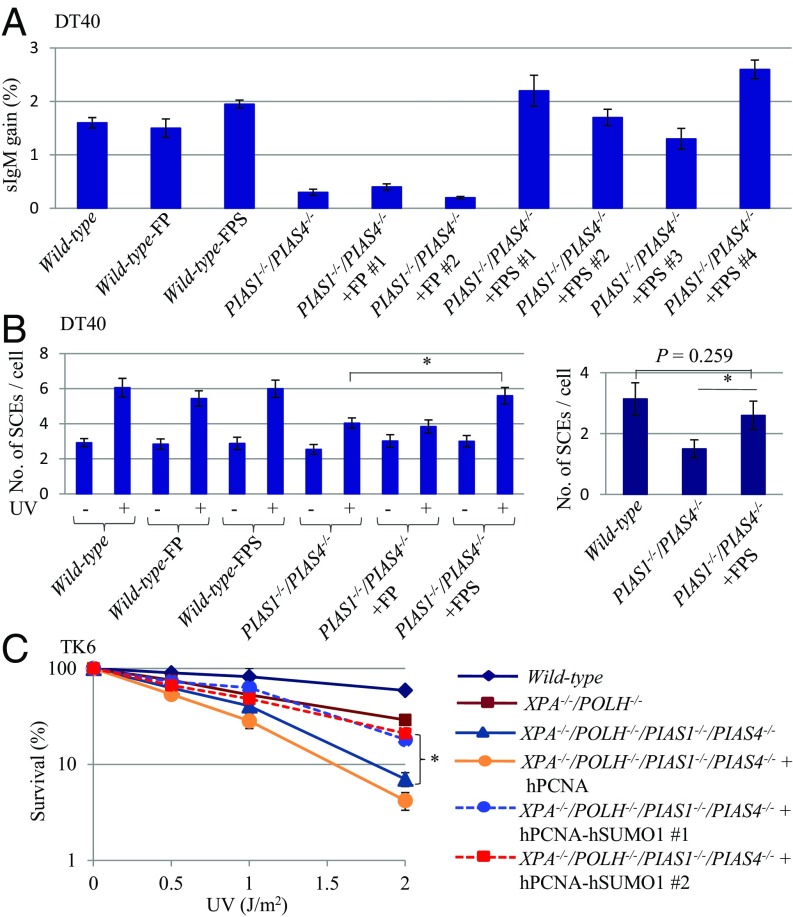
TS and TLS are responsible for Ig V diversification by Ig GC and nontemplated single base substitutions in chicken B lymphocytes (figure S1 in ref. 34). The rate of Ig GC was measured using two methods: by measuring the expression of surface IgM (sIgM) in DT40 subclones (Fig. 3 A and B) and by examining Ig V sequences (Fig. 3C and SI Appendix, Fig. S6). In the first method, we disrupted the PIAS1 and PIAS4 genes in the DT40 cells that carry a defined frameshift mutation in the light-chain Vλ gene (35). GC occurring at the frameshift mutation eliminates the mutation, leading to reexpression of sIgM (Fig. 3A). The fraction of sIgM-positive cells in 36 subclones from each genotype was measured after 3-wk clonal expansion. The median value of the fraction of sIgM-positive cells was 1.6% for wild-type, 0.61% for PIAS1−/−, 0.47% for PIAS4−/−, and 0.34% for PIAS1−/−/PIAS4−/− cells (Fig. 3B). We conclude that Ig GC was greatly compromised in the PIAS1 and PIAS4 mutant cells.
Fig. 3.
Analysis of the Ig GC rate in DT40 B cell clones. (A) Diagram of the Ig GC assay. The frameshift mutation caused by the insertion of G in the rearranged Vλ segment in the parent cells is indicated by the red circle. GC events using an upstream ΨV donor segment can eliminate the frameshift mutation, resulting in a gain in sIgM expression. The ΨV segment is indicated by the blue box. (B) The proportion of sIgM-positive cells was determined in 36 parallel cultures derived from single sIgM-negative parental cells after clonal expansion (3 wk). Median percentages are noted below each data set. (C) Rates of Ig GC and nontemplated mutation in AID-expressing clones carrying the indicated genotypes. The y axis represents the rate of nontemplated mutations (10−5/base/div), ambiguous mutation (10−5/base/div), and gene conversion (10−3/sequence/div). The error bars indicate SEM. Statistical analyses were performed by Student’s t test (*P < 0.01).
To evaluate the role of PIAS1 and PIAS4 in Ig GC on a nucleotide sequence level, we ectopically expressed AID in DT40 clones, expanded subclones for 2 wk, and examined the nucleotide sequences of the Ig V region in individual subclones (SI Appendix, Fig. S6). The ectopic expression of AID generated an excess amount of AID-mediated lesions, and induced Ig V nontemplated single base substitutions [point mutations (PMs) in Fig. 3C and SI Appendix, Fig. S6] in addition to Ig GC events (GC in Fig. 3C and SI Appendix, Fig. S6). PIAS1−/−/PIAS4−/− cells displayed a decrease of more than 10 times in the rate of Ig GC compared with wild-type cells (Fig. 3C and SI Appendix, Fig. S6); this great decrease is in agreement with the data of sIgM reexpression (Fig. 3B). An approximately three times decrease in the rate of TLS-dependent Ig V diversification in PIAS1−/−/PIAS4−/− cells indicates the potential role of the PCNA SUMOylation in TLS by DNA polymerases other than Polη (43) (Fig. 3C and SI Appendix, Fig. S6). The ratio of TS-dependent Ig V diversification relative to the TLS-dependent one was decreased by four times in PIAS1−/−/PIAS4−/− cells in comparison with wild-type cells. The degree of this decrease was in the same order of magnitude as the two times decrease in the ratio of TS relative to TLS evaluated by the piggyBlock assay (SI Appendix, Fig. S4). Taken together, PIAS1 and PIAS4 are required for efficient TS-mediated Ig V diversification.
An Epistatic Relationship Between PIAS1−/−/PIAS4−/− and PCNAK164R/K164R Mutations in Ig GC.
Unlike Ubc13 of S. cerevisiae, Ubc13 of metazoan cells plays an important role in HR-mediated DSB repair (44, 45). Remarkably, UBC13−/− DT40 cells displayed defective gap filling following UV irradiation (44) as seen in Polη-deficient cells. Moreover, they displayed a more than 10 times decrease in the rate of TLS-mediated Ig V hypermutation (PMs in Fig. 3C and SI Appendix, Fig. S6) in comparison with the wild-type cells. These observations suggest that TS might be controlled by a mechanism other than polyubiquitylation of PCNA. Unexpectedly, DT40 cells carrying the Lysine164 to Arginine mutation of PCNA (PCNAK164R/K164R mutation) (46) showed a 90% decrease in Ig GC in addition to a decrease in TLS-dependent hypermutation at the Ig Vλ gene (Fig. 3C and SI Appendix, Fig. S6) as reported previously (46). This observation led us to hypothesize that SUMOylation at K164 of PCNA by PIAS1 and PIAS4 might be required for efficient Ig GC.
To test the hypothesis, we explored a genetic interaction between the PCNAK164R/K164R and the PIAS1−/−/PIAS4−/− mutations. To this end, the K164R mutation was inserted in the PCNA gene of PIAS1−/−/PIAS4−/− cells (SI Appendix, Fig. S5 A and B). The resulting PIAS1−/−/PIAS4−/−/PCNAK164R/K164R cells were then examined for the rate of Ig GC. The frequency of Ig gene conversion was very similar between the PIAS1−/−/PIAS4−/− and the PIAS1−/−/PIAS4−/−/PCNAK164R/K164R clones with these clones showing a two times lower rate of Ig GC in comparison with the PCNAK164R/K164R clone (Fig. 3C and SI Appendix, Fig. S6). Thus, the PIAS1−/−/PIAS4−/− mutation has an epistatic relationship with the PCNAK164R/K164R mutation in terms of Ig GC. The epistatic relationship supports the idea that the SUMOylation at K164 of PCNA is promoted by PIAS1 and PIAS4 in the up-regulation of TS by HR.
SUMOylation of PCNA by PIAS1 and PIAS4.
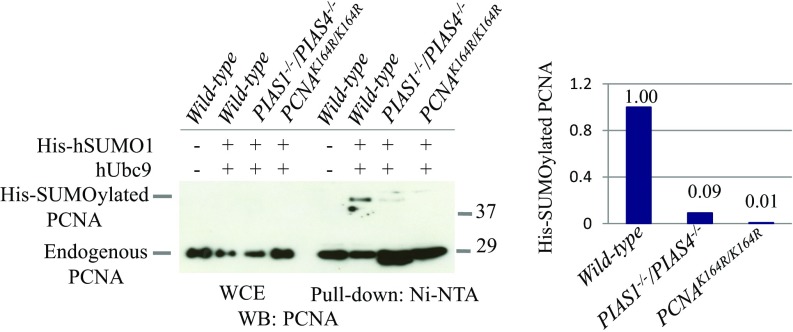
We investigated whether PIAS1 and PIAS4 performed SUMOylation at the K164 residue of PCNA. To detect the SUMOylation of PCNA, we transfected an expression construct encoding 6His-tagged human SUMO1 into cells by retroviral infection (SI Appendix, Fig. S7) (47), isolated cellular extracts in the denaturing condition, and pulled down his-tagged SUMOylated proteins by the nickel column. To enhance SUMOylation, we also ectopically expressed the human Ubc9 E2 conjugating enzyme (48). We confirmed the identity of bands corresponding to SUMOylated PCNA at the K164 residue by comparing the Western blot analysis of wild-type and PCNAK164R/K164R cells (Fig. 4). We analyzed cells having no exogenous genotoxic stress since previous studies reported that treatment of wild-type cells with an alkylating agent or UV has no detectable impact on the SUMOylation of PCNA (30, 31). We also confirmed that the SUMOylation of PCNA is damage independent (SI Appendix, Fig. S5C) and the signal for SUMOylated PCNA was seen in wild-type but not PCNAK164R/K164R cells (Fig. 4). This signal was significantly reduced in PIAS1−/−/PIAS4−/− cells in comparison with wild-type cells, indicating the role played by PIAS1 and PIAS4 in the SUMOylation of PCNA (Fig. 4). We then investigated whether SUMOylation of PCNA by PIAS1 and PIAS4 has any effect on the ubiquitylation of PCNA. The extent of PCNA ubiquitylation is very similar between the wild-type and the PIAS1−/−/PIAS4−/− cells (SI Appendix, Fig. S5D).
Fig. 4.
SUMOylation of PCNA by PIAS1 and PIAS4 E3 ligases. DT40 cells stably transfected with His-tagged hSUMO1 were analyzed by pull-down assay using Ni-NTA magnetic beads. Whole cell lysates and lysates after pulled down using Ni-NTA magnetic beads are shown on the Left. Quantification of SUMOylated PCNA and unmodified PCNA by pull-down experiments were shown on the Right.
The PCNA-SUMO1 Hybrid Protein Restores TS of PIAS1−/−/PIAS4−/− Cells.
We tested whether or not the SUMOylation of PCNA facilitates TS. To this end, we employed a PCNA-SUMO1 chimeric protein, C-terminal fusion of SUMO1 (SI Appendix, Figs. S8 and S9). The ectopic expression of PCNA-SUMO1 inhibits DSB formation in the cells during a treatment with replication-blocking alkylating agents (31). We tested the effect of PCNA-SUMO1 on TS by using the following three phenotypic assays: Ig GC, UV-induced SCE in DT40 cells, and UV sensitivity of TK6 cells. Ectopic expression of PCNA-SUMO1 or PCNA had no detectable impact on the rate of Ig GC in wild-type cells evaluated by measuring the reexpression of sIgM (Fig. 5A and SI Appendix, Fig. S8C). Remarkably, the expression of PCNA-SUMO1 but not PCNA restored the rate of Ig GC in PIAS1−/−/PIAS4−/− DT40 cells to a nearly normal level. Likewise, the expression of PCNA-SUMO1 but not PCNA restored the frequency of UV-induced SCE in PIAS1−/−/PIAS4−/− DT40 cells (Fig. 5B and SI Appendix, Fig. S8D). We assessed the effect of PCNA-SUMO1 expression on UV sensitivity of TK6 cells. The presence of PCNA-SUMO1 significantly increased cellular tolerance to UV in PIAS1−/−/PIAS4−/−/POLΗ−/−/XPA−/− cells but not in POLH−/−/XPA−/− cells (Fig. 5C). To explore the involvement of interactions between SUMO1 and SUMO-interacting motifs (SIMs) of potential downstream molecules in DDT, we introduced mutations in the SUMO1 part of the fusion protein which mutations compromise interactions of SUMO1 with SIMs (49). The mutated SUMO-PCNA chimeric proteins failed to increase cellular tolerance to UV in PIAS1−/−/PIAS4−/−/POLΗ−/−/XPA−/− cells (SI Appendix, Fig. S10). Collectively, PCNA-SUMO1 is capable of suppressing a defect in TS in both chicken DT40 and human TK6 cells. To summarize, our current paper revealed the critical role for the SUMOylation of PCNA by PIAS1 and PIAS4 in the promotion of TS by HR.
Fig. 5.
Ectopic expression of hPCNA-hSUMO1 reverses the mutant phenotype of PIAS1−/−/PIAS4−/− cells. (A) The proportion of sIgM-positive DT40 cells was determined as in Fig. 3B. The error bars indicate SEM. (B) The number of SCEs induced by 0.25 J/m2 UV in the DT40 cells was determined as in Fig. 1A. The error bars indicate SEM. (C) Clonogenic cell survival assay following exposure of hPCNA-hSUMO1 expressing XPA−/−/POLH−/− and PIAS1−/−PIAS4−/−/XPA−/−/POLH−/− TK6 cells to UV. The x axis represents the dose of the indicated DNA-damaging agent on a linear scale; the y axis represents the survival fraction on a logarithmic scale. The error bars indicate SD. FP: Flag-hPCNA. FPS: Flag-hPCNA-hSUMO1. Statistical analyses were performed by Student’s t test (*P < 0.01).
Discussion
We here show that PIAS1 and PIAS4 facilitate TS by HR in human TK6 and chicken DT40 B cell lines. We evaluated TS by employing four phenotypic assays, (i) SCE-induced by cisplatin and UV (Fig. 1), (ii) UV sensitivity of PIAS1−/−/PIAS4−/−, POLH−/−, and PIAS1−/−/PIAS4−/−/POLH−/− cells (Fig. 2A), (iii) a transposon-based TS substrate carrying GpC opposite the T-T CPD (SI Appendix, Fig. S4), and (iv) Ig GC (Fig. 3). These four phenotypic assays consistently showed that the loss of PIAS1 and PIAS4 caused decreases in the frequency of TS. Observed lack of SUMOylation at K164 of PCNA in vivo in the absence of PIAS1 and PIAS4 (Fig. 4) indicates that these two enzymes promote the SUMOylation event. We also show that PIAS1 and PIAS4 facilitate TS in DT40 cells, most likely through SUMOylation of PCNA at the K164 residue. An epistatic relationship between the PCNAK164R and the PIAS1−/−/PIAS4−/− mutations in Ig GC suggests a functional relationship between the PCNA SUMOylation and the TS-dependent Ig diversification. This idea is further supported by the data that the ectopic expression of PCNA-SUMO1 reversed the defective TS of PIAS1−/−/PIAS4−/− cells (Fig. 5 and SI Appendix, Fig. S8). We conclude that the SUMOylation at K164 of PCNA by PIAS1 and PIAS4 ensures the preferential usage of TS over TLS in DDT, preventing mutagenesis by TLS during physiological DNA replication.
Yeast genetic studies have indicated that HR facilitates TS (11, 50). We propose that PIAS1 and PIAS4 promote TS but not HR-dependent DSB repair during DNA replication. First, PIAS1−/−/PIAS4−/− DT40 and TK6 cells underwent significant decreases in SCEs induced by UV and cisplatin (Fig. 1). Moreover, PIAS1−/−/PIAS4−/− cells displayed over 90% decreases in the rate of TS-mediated Ig V diversification (Fig. 3). Second, although HR-deficient cells are sensitive to UV and cisplatin (51, 52), the PIAS1−/−/PIAS4−/− mutation increases the sensitivity to cisplatin and UV only in the POLH−/− TK6 cells but not in the wild-type cells (Fig. 2). Considering a substantial functional overlap between TLS and TS, these sensitivity data also support the role of PIAS1 and PIAS4 in the promotion of TS. Collectively, we propose that PIAS1 and PIAS4 shift the usage of the two DDT pathways from TLS to TS without affecting the efficiency of general HR.
The molecular mechanism for activation of TS by PCNA polyubiquitylation is poorly understood in S. cerevisiae (6). Likewise, it is unclear how PCNA SUMOylation affects TS in DT40 cells. SUMOylated PCNA might provide a platform for HR factors including the Rad51 recombinase since purified mammalian RAD51 interacts noncovalently with SUMO (53). SUMOylated PCNA may also recruit poly[ADP ribose]polymerase 1 (PARP1) and ELG1/ATAD5, and thereby activate TS. PARP1 is required for efficient Ig gene conversion in DT40 cells (54). Polymers formed by SUMOylation and PARylation cooperate to achieve robust responses (55). ELG1/ATAD5 has a SIM and physically interacts with both ubiquitin-specific protease 1 and PCNAs that are localized at replication forks stalled at damaged template strands (56, 57). ELG1 stably associating with SUMOylated PCNA may increase the relative usage of TS by decreasing the level of PCNA ubiquitination. PIAS1 and PIAS4 can activate TS by SUMOylating molecules other than the K164 residue of PCNA because PIAS1−/−/PIAS4−/− cells showed a more severe phenotype in Ig gene conversion than did PCNAK164R/K164R DT40 cells (Fig. 3C and SI Appendix, Fig. S6). PIAS1 and PIAS4 might SUMOylate PARP1 since SUMOylated PARP-1 is constitutively associated with chromatin (58). Another question is the role for PIAS1 and PIAS4 in PCNA SUMOylation, as they are dispensable for in vitro SUMOylation of PCNA as previously reported (31). Unidentified molecules might facilitate an interaction between PCNA and these E3 ligases in vivo, which is supported by a recent finding indicating a noncatalytic role of Rad18 in SUMOylation of Polη (59).
The role of PCNA polyubiquitylation at the K164 residue seems to be distinctly different among S. cerevisiae, S. pombe, and metazoan cells since Ubc13 significantly promotes TLS in S. pombe (25) and DT40 cells (Fig. 3C and SI Appendix, Fig. S6) but not in S. cerevisiae (6). This is supported by the data that RNF8, a ubiquitin ligase, play a role with Ubc13 in PCNA polyubiquitylation (60) and are required for efficient TLS-mediated Ig V diversification in DT40 cells (34). The role of PCNA SUMOylation at the K164 residue might also differ between S. cerevisiae and metazoan cells. The PCNA SUMOylation suppresses crossover-type HR by recruiting the Srs2 DNA helicase, an antirecombinase, at the site of stalled replication forks in S. cerevisiae (27, 61). Likewise, mammalian C12orf48/PARI (PARPBP), a potential Srs2 functional ortholog, suppresses HR-mediated DSB repair during DNA replication in both human and chicken cell lines (62). The data suggest an important role played by interactions between SUMOylated PCNA and PARI in suppressing TS as well as HR-mediated DSB repair. Nonetheless, a recent report indicated that interactions of Srs2 with Rad51 and PCNA-SUMO promote noncrossover-type HR in S. cerevisiae (63). We show that the primary role of PCNA SUMOylation at the K164 residue is to facilitate TS by HR. Considering the potential role of the PCNA SUMOylation in TLS as well as TS in DT40 cells (Fig. 3C and SI Appendix, Fig. S6), the regulation of DDT by PCNA SUMOylation is more complex in metazoan cells in comparison with S. cerevisiae.
Our paper sheds light on the distinct roles played by TS and TLS in DDT. PIAS1−/−/PIAS4−/− mutation increases the sensitivity to cisplatin and UV only in a TLS-deficient background but not in a wild-type one (Fig. 2). Moreover, RAD18−/− but not PIAS1−/−/PIAS4−/− cells show sensitivity to cisplatin or UV. These observations suggest that TLS is preferentially used over TS in DDT to repair excess amounts of DNA lesions caused by environmental factors. This idea agrees with the data that treatment of cells with DNA-damaging agents trigger the monoubiquitylation of PCNA (64, 65) but do not enhance the SUMOylation of PCNA (SI Appendix, Fig. S5C) (30, 31). Taken together, we propose that the SUMOylation contributes to a dominant usage of TS in DDT during the normal cell cycle. By contrast, the monoubiquitylation of PCNA by Rad18 (64, 65) is to counteract against excess numbers of DNA lesions induced by environmental mutagens, including UV and chemotherapeutic agents by up-regulating TLS.
Materials and Methods
Measurement of SCE Levels.
Colony-Survival Assay.
Cell sensitivity to DNA-damaging agents was evaluated by counting colony formation in methylcellulose plates as described previously (67).
Nucleotide Sequence Analysis of AID-Induced Ig V Diversification.
Ectopic expression of AID by retroviral infection and analysis of Ig V diversification was carried out as described previously (29, 41, 34).
Supplementary Material
Acknowledgments
We thank Dr. Lajos Haracska (Hungarian Academy of Sciences, Hungary) for the Flag-PCNA-SUMO1 plasmid. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sport and Culture (KAKENHI 25650006, 23221005, and 16H06306) (to S.T.). This work was also supported by grants from the Takeda Research Foundation (to H.S.) and Japan Society for the Promotion of Science Core-to-Core Program, A. Advanced Research Networks (to S.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716349115/-/DCSupplemental.
References
- 1.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 2.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y, O’Connor MJ, de Bono J, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 4.Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boiteux S, Jinks-Robertson S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics. 2013;193:1025–1064. doi: 10.1534/genetics.112.145219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branzei D, Szakal B. DNA damage tolerance by recombination: Molecular pathways and DNA structures. DNA Repair (Amst) 2016;44:68–75. doi: 10.1016/j.dnarep.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres-Ramos CA, Prakash S, Prakash L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:2419–2426. doi: 10.1128/MCB.22.7.2419-2426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blastyák A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30:684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izhar L, et al. Analysis of strand transfer and template switching mechanisms of DNA gap repair by homologous recombination in Escherichia coli: Predominance of strand transfer. J Mol Biol. 2008;381:803–809. doi: 10.1016/j.jmb.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 12.Giannattasio M, et al. Visualization of recombination-mediated damage bypass by template switching. Nat Struct Mol Biol. 2014;21:884–892. doi: 10.1038/nsmb.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coïc E, Feldman T, Landman AS, Haber JE. Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Genetics. 2008;179:199–211. doi: 10.1534/genetics.108.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoda E, et al. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochegger H, Sonoda E, Takeda S. Post-replication repair in DT40 cells: Translesion polymerases versus recombinases. Bioessays. 2004;26:151–158. doi: 10.1002/bies.10403. [DOI] [PubMed] [Google Scholar]
- 16.Wechsler T, Newman S, West SC. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita YM, et al. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 2002;21:5558–5566. doi: 10.1093/emboj/cdf534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonoda E, et al. Multiple roles of Rev3, the catalytic subunit of polζ in maintaining genome stability in vertebrates. EMBO J. 2003;22:3188–3197. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleaver JE, et al. Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res. 1999;59:1102–1108. [PubMed] [Google Scholar]
- 20.De Weerd-Kastelein EA, Keijzer W, Rainaldi G, Bootsma D. Induction of sister chromatid exchanges in xeroderma pigmentosum cells after exposure to ultraviolet light. Mutat Res. 1977;45:253–261. doi: 10.1016/0027-5107(77)90025-2. [DOI] [PubMed] [Google Scholar]
- 21.Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 22.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Lawrence CW. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc Natl Acad Sci USA. 2005;102:15954–15959. doi: 10.1073/pnas.0504586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulon S, et al. Rad8Rad5/Mms2-Ubc13 ubiquitin ligase complex controls translesion synthesis in fission yeast. EMBO J. 2010;29:2048–2058. doi: 10.1038/emboj.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gervai JZ, Gálicza J, Szeltner Z, Zámborszky J, Szüts D. A genetic study based on PCNA-ubiquitin fusions reveals no requirement for PCNA polyubiquitylation in DNA damage tolerance. DNA Repair (Amst) 2017;54:46–54. doi: 10.1016/j.dnarep.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 28.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Sale JE, Calandrini DM, Takata M, Takeda S, Neuberger MS. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 30.Arakawa H, et al. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gali H, et al. Role of SUMO modification of human PCNA at stalled replication fork. Nucleic Acids Res. 2012;40:6049–6059. doi: 10.1093/nar/gks256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen IS, et al. DNA lesion identity drives choice of damage tolerance pathway in murine cell chromosomes. Nucleic Acids Res. 2015;43:1637–1645. doi: 10.1093/nar/gku1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livneh Z, et al. High-resolution genomic assays provide insight into the division of labor between TLS and HDR in mammalian replication of damaged DNA. DNA Repair (Amst) 2016;44:59–67. doi: 10.1016/j.dnarep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 34.Mohiuddin, et al. The role of HERC2 and RNF8 ubiquitin E3 ligases in the promotion of translesion DNA synthesis in the chicken DT40 cell line. DNA Repair (Amst) 2016;40:67–76. doi: 10.1016/j.dnarep.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buerstedde JM, et al. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sale JE. Immunoglobulin diversification in DT40: A model for vertebrate DNA damage tolerance. DNA Repair (Amst) 2004;3:693–702. doi: 10.1016/j.dnarep.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 37.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sale JE. Competition, collaboration and coordination–Determining how cells bypass DNA damage. J Cell Sci. 2012;125:1633–1643. doi: 10.1242/jcs.094748. [DOI] [PubMed] [Google Scholar]
- 39.Hatanaka A, et al. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol Cell Biol. 2005;25:1124–1134. doi: 10.1128/MCB.25.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longerich S, Orelli BJ, Martin RW, Bishop DK, Storb U. Brca1 in immunoglobulin gene conversion and somatic hypermutation. DNA Repair (Amst) 2008;7:253–266. doi: 10.1016/j.dnarep.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirota K, et al. In vivo evidence for translesion synthesis by the replicative DNA polymerase δ. Nucleic Acids Res. 2016;44:7242–7250. doi: 10.1093/nar/gkw439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbs PEM, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohzaki M, et al. DNA polymerases ν and θ are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao GY, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 45.Jackson SP, Durocher D. Review regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Saberi A, et al. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol Cell Biol. 2008;28:6113–6122. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohiuddin, et al. A novel genotoxicity assay of carbon nanotubes using functional macrophage receptor with collagenous structure (MARCO)-expressing chicken B lymphocytes. Arch Toxicol. 2014;88:145–160. doi: 10.1007/s00204-013-1084-7. [DOI] [PubMed] [Google Scholar]
- 48.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 50.Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell. 2010;38:649–661. doi: 10.1016/j.molcel.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nojima K, et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11711. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- 52.Eppink B, et al. The response of mammalian cells to UV-light reveals Rad54-dependent and independent pathways of homologous recombination. DNA Repair (Amst) 2011;10:1095–1105. doi: 10.1016/j.dnarep.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 53.Ouyang KJ, et al. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paddock MN, Buelow BD, Takeda S, Scharenberg AM. The BRCT domain of PARP-1 is required for immunoglobulin gene conversion. PLoS Biol. 2010;8:e1000428. doi: 10.1371/journal.pbio.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pellegrino S, Altmeyer M. Interplay between ubiquitin, SUMO, and poly(ADP-Ribose) in the cellular response to genotoxic stress. Front Genet. 2016;7:63. doi: 10.3389/fgene.2016.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee KY, et al. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J Biol Chem. 2010;285:10362–10369. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang K, et al. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev. 2011;25:1847–1858. doi: 10.1101/gad.17020911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zilio N, et al. DNA-dependent SUMO modification of PARP-1. DNA Repair (Amst) 2013;12:761–773. doi: 10.1016/j.dnarep.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Despras E, et al. Rad18-dependent SUMOylation of human specialized DNA polymerase eta is required to prevent under-replicated DNA. Nat Commun. 2016;7:13326. doi: 10.1038/ncomms13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, et al. PCNA is ubiquitinated by RNF8. Cell Cycle. 2008;7:3399–3404. doi: 10.4161/cc.7.21.6949. [DOI] [PubMed] [Google Scholar]
- 61.Burkovics P, et al. Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 2013;32:742–755. doi: 10.1038/emboj.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moldovan GL, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miura T, Shibata T, Kusano K. Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity. Proc Natl Acad Sci USA. 2013;110:16067–16072. doi: 10.1073/pnas.1303111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe K, et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–3896. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kikuchi K, et al. Structure-specific endonucleases xpf and mus81 play overlapping but essential roles in DNA repair by homologous recombination. Cancer Res. 2013;73:4362–4371. doi: 10.1158/0008-5472.CAN-12-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keka IS, et al. Smarcal1 promotes double-strand-break repair by nonhomologous end-joining. Nucleic Acids Res. 2015;43:6359–6372. doi: 10.1093/nar/gkv621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.