Significance
Sex determination is one of the most fundamental but also extraordinary plastic processes in nature. Many different master sex-determining genes have been characterized in vertebrates, and most of them are known to fulfill essential functions during sexual development and thus are already tightly linked to the process that they now govern. Only one exception is currently known: the salmonid master sex-determining gene (sdY), which arose from the duplication of an immune-related gene. Here we show that SdY prevents female differentiation by interacting and blocking the action of a key ovarian differentiation factor. These results suggest that the evolution of unusual vertebrate master sex determination genes is strongly constrained by their ability to interact with the canonical gonadal differentiation pathway.
Keywords: sex determination, Forkhead box proteins, sex differentiation, fish, evolution
Abstract
Evolutionary novelties require rewiring of transcriptional networks and/or the evolution of new gene functions. Sex determination (SD), one of the most plastic evolutionary processes, requires such novelties. Studies on the evolution of vertebrate SD revealed that new master SD genes are generally recruited from genes involved in the downstream SD regulatory genetic network. Only a single exception to this rule is currently known in vertebrates: the intriguing case of the salmonid master SD gene (sdY), which arose from duplication of an immune-related gene. This exception immediately posed the question of how a gene outside from the classical sex differentiation cascade could acquire its function as a male SD gene. Here we show that SdY became integrated in the classical vertebrate sex differentiation cascade by interacting with the Forkhead box domain of the female-determining transcription factor, Foxl2. In the presence of Foxl2, SdY is translocated to the nucleus where the SdY:Foxl2 complex prevents activation of the aromatase (cyp19a1a) promoter in cooperation with Nr5a1 (Sf1). Hence, by blocking a positive loop of regulation needed for the synthesis of estrogens in the early differentiating gonad, SdY disrupts a preset female differentiation pathway, consequently allowing testicular differentiation to proceed. These results also suggest that the evolution of unusual vertebrate master sex determination genes recruited from outside the classical pathway like sdY is strongly constrained by their ability to interact with the canonical gonadal differentiation pathway.
Sexual development is a fundamental process that shapes animal morphology, physiology, and behavior. The development of the undifferentiated embryonic gonad toward a testis or an ovary is regulated by a complex network of genes, where the initial triggers for male or female sex differentiation can come from the environment or the genome (1). A great number of studies revealed that the chromosomal, molecular, and cellular mechanisms of genetic sex determination (SD) are highly variable (1–4). It is now particularly clear that SD mechanisms evolved frequently and independently, leading to a high turnover of the genes governing sexual development (1, 5), even between closely related organisms. For instance, the therian sex-determining gene SRY is not found in other vertebrates (6), and recent studies have identified many different master SD genes in birds, amphibians, and fish (7–14). In these species, known members of the downstream regulatory sex differentiation network usurped the position at the top of the sex determination cascade to become the master SD gene. However, not all downstream sex differentiation genes are equally able to take the lead as sexual master switches, and currently, only the genes encoding transcription factors Sox3 and Dmrt1 and several components of TGF-β signaling have been identified as master SD genes in vertebrates. This frequent reuse of the same SD genes led to the hypothesis that there are limited options in becoming a master sex-determining gene, which can be met by only a very limited number of genes from the sex differentiation network. However, this “limited option” hypothesis (15) was challenged by the discovery of the unusual salmonid sex-determining gene (16, 17). This gene, called sdY for “sexually dimorphic on the Y,” turned out to be a duplicated and truncated version of a gene encoding IFN regulatory factor 9 (irf9), which functions in the immune response of vertebrates. Upon IFN binding to its receptor and activation of STAT signaling, IRF9 complexes with both STAT1 and STAT2 in the cytoplasm and then translocates to the nucleus to activate effector genes of the antiviral response through its DNA-binding domain (18). IRF9 nonimmunity roles have been described in neurons and liver and heart pathophysiology (19), but so far no evidence implicates it in sex determination or sex differentiation processes. The birth of such a master SD gene recruited from outside of the classical sex differentiation cascade raises the intriguing question of its functional evolution and how this unusual SD gene determines sex. Did SdY evolve a new function to be able to interact directly with the classical gonadal sex differentiation cascade, or does it use part of its ancestral pathway, that is, the IFN immune-related response, for its action?
Results and Discussion
During evolution of SdY, the DNA-binding domain and the nuclear localization signals of Irf9 were lost, while the protein–protein interaction domain (IRF association domain, or IAD) was preserved and underwent some sequence diversification (16). To test the hypothesis that the IAD domain of SdY still functions in protein binding, we first performed molecular modeling and found that the 3D structure of SdY strongly overlaps the IAD domain of IRF proteins (Fig. 1A). As the IAD domain is the only domain of known function predicted from the primary sequence of the sdY gene (16, 17), we hypothesized that SdY could still exert its function based on protein–protein interactions. We thus searched for SdY interacting proteins using a yeast two-hybrid (Y2H) screen with SdY used as bait and with a rainbow trout prey cDNA library prepared from late differentiating testes sampled when sdY expression is still high (16). Among the 46 different putative interacting proteins there were none of the known Irf9 partners like Stat1 or Stat2. Instead, we found a very strong enrichment of many members of the Forkhead box (FOX) family (11 FOX proteins, SI Appendix, Tables S1–S3). The Forkhead box, a highly conserved DNA-binding domain (20) common to all FOX proteins, was identified as the minimum domain needed for an effective interaction with SdY (Fig. 1B and SI Appendix, Table S3). Interestingly, among all of the FOX proteins interacting with SdY in yeast, we found the well-known female sex differentiation protein Foxl2 (21, 22). Taking into account the importance of Foxl2 in vertebrate sex differentiation, we reasoned that this would be an interesting and biologically relevant SdY partner. We then explored the interaction of SdY with trout Foxl2 in a direct yeast interaction assay and confirmed that SdY and Foxl2 can interact together (SI Appendix, Fig. S1).
Fig. 1.
SdY conserves the structure of the IRF protein–protein interaction domain and interacts with the Forkhead box domain of Fox proteins. (A) SdY shares structural homologies with IAD, a protein–protein interaction domain. The structure of SdY (in gray) was modeled using the crystal structure of IRF5 as a template (in green). This SdY structure reveals eight β-sheets forming a β-sandwich and three α-helices that are highly conserved with IRF5. (B) SdY interacts in yeast with Fox proteins through their highly conserved DNA-binding domain. The alignments of the SdY-Fox interacting clone sequences (gray lines) delineate the minimum domain or selected interacting domain needed for an effective interaction with SdY in yeast, which is the Forkhead box domain (110 aa, black lines). The 11 Fox proteins characterized in the Y2H screen are represented by open cylinders with numbers of interacting clones indicated on the right side.
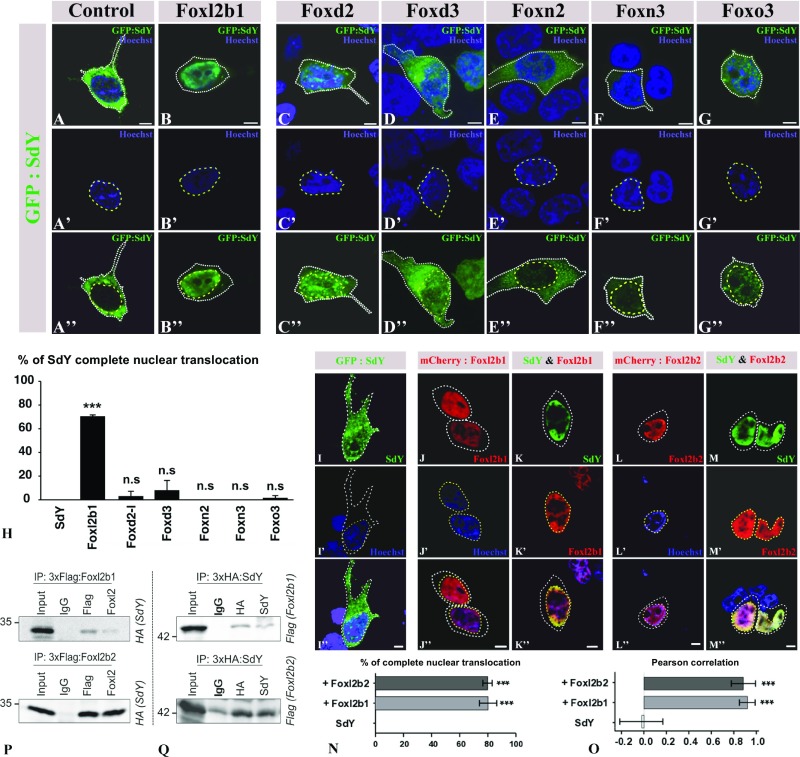
To better characterize this interaction, an in vitro approach was developed using cell transfection assays. In HEK 293T cells transfected only with sdY plasmid, SdY protein was localized predominantly in the cytoplasm (Fig. 2 A–A″ and SI Appendix, Fig. S2). However, when cotransfected along with Foxl2, SdY was completely translocated into the nucleus (Fig. 2 B–B″ and SI Appendix, Fig. S2). Such a complete SdY nuclear translocation was observed only with fish Foxl2 proteins (Fig. 2H), including the two rainbow trout paralogous gene products (Foxl2b1 and Foxl2b2) resulting from the salmonid whole-genome duplication (23) (Fig. 2 I–O) and the medaka, Oryzias latipes, Foxl2 (SI Appendix, Figs. S3 and S4). No complete nuclear translocation was observed with some other rainbow trout Fox proteins (Fig. 2 C–H and SI Appendix, Fig. S2), with some mammalian Foxl2, that is, mouse and goat (SI Appendix, Fig. S3), and with the rainbow trout Foxl2b2 containing a modified mouselike Forkhead box domain (SI Appendix, Fig. S4). This complete nuclear relocalization of SdY with trout and medaka Foxl2s indicated some specific protein–protein interaction and that this interaction required the conformation of a fish Forkhead domain. This interaction was also confirmed in vitro by co-immunoprecipitation experiments (Fig. 2 P and Q and SI Appendix, Fig. S5A) and in vivo by showing that SdY was also translocated into the nucleus following coinjection with Foxl2 in medaka embryos (SI Appendix, Fig. S5 B and C).
Fig. 2.
SdY interacts with Foxl2, resulting in its nuclear translocation. (A–H) GFP:SdY alone (A–A″) and GFP:SdY in combination with different trout Fox proteins, Foxl2 (Foxl2b1) (B–B″), Foxd2 (C–C″), Foxd3 (D–D″), Foxn2 (E–E″), Foxn3 (F–F″), Foxo3 (G–G″), were cotransfected in HEK 293T cells (delimited by white dotted lines). GFP:SdY is translocated in the nucleus (delimited by yellow dotted lines and stained in blue with Hoechst staining) only in the presence of Foxl2 (B–B″). (Scale bar, 5 μm.) (H) Percentage of transfected cells (mean ± standard deviation on 200 cells) in which SdY is completely translocated in the nucleus after three independent cotransfection experiments with different trout Fox proteins. Significant differences compared with SdY alone were calculated using an unpaired two-tailed Student’s t test, ***P < 0.001; ns, nonsignificant. (I–O) Foxl2b1 and Foxl2b2 are both able to drive SdY complete nuclear translocation (delimited by yellow dotted lines and stained in blue with Hoechst staining). Confocal images of HEK 293T cells (delimited by white dotted lines) transiently transfected with sdY (I–I″), mCherry:Foxl2b1 alone (J–J″), SdY and mCherry:Foxl2b1 (K–K″), mCherry:Foxl2b2 alone (L–L″), SdY and mCherry:Foxl2b2 (M–M″). (Scale bar, 10 µm.) (N) Quantitative analysis in the presence or absence of Foxl2b1 and Foxl2b2. Percentage of complete SdY nuclear translocation (mean ± standard deviation on 100 cells) after three independent cotransfection experiments. Statistical significances compared with SdY alone were calculated using an unpaired two-tailed Student’s t test. (O) SdY colocalizes with Foxl2 in the nucleus. SdY, SdY-Foxl2b1, and SdY-Foxl2b2 colocalizations were measured in the nucleus for SdY (n = 5), SdY and Foxl2b1 (n = 5), and SdY and Foxl2b2 (n = 5) with Pearson’s correlation. Statistical significance was calculated using an unpaired two-tailed Student’s t test, ***P < 0.001. (P and Q) SdY binds with Foxl2 in co-immunoprecipitation (IP) experiments. HEK 293T cells were transiently transfected with expression plasmids for SdY fused to a hemagglutinin tag (3xHA:SdY) and for Foxl2 fused to a 3xFlag tag (3xFlag:Foxl2b1 or 3xFlag:Foxl2b2). Whole-cell lysates were used for immunoprecipitation with anti-Flag or anti-Foxl2 (P) and with anti-HA or anti-SdY (Q) followed by immunoblotting with the appropriate antibodies. Input represents 10% whole-cell lysate. IgG mouse antibody was used as the control. In P, 3xFlag:Foxl2b1 or 3xFlag:Foxl2b2 was immunoprecipitated with either Flag (Top) or FoxL2 (Bottom) antibodies followed by immunoblotting with an antibody against the HA tag to reveal the interaction with 3xHA:SdY (SdY). In Q, 3xHA:SdY was immunoprecipitated with an HA or SdY antibody, followed by immunoblotting with an antibody against the Flag tag to reveal 3xFlag:Foxl2b1 (Foxl2b1) (Top) or 3xFlag:Foxl2b2 (Foxl2b2) (Bottom).
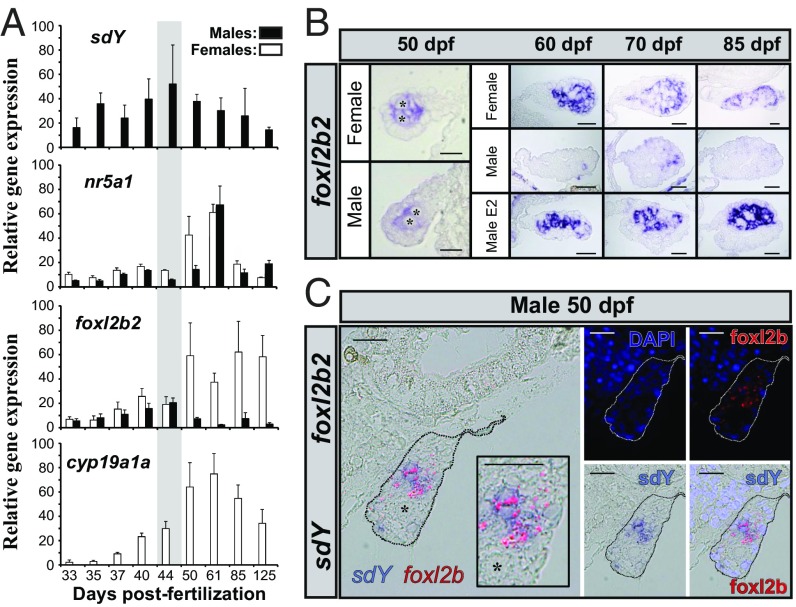
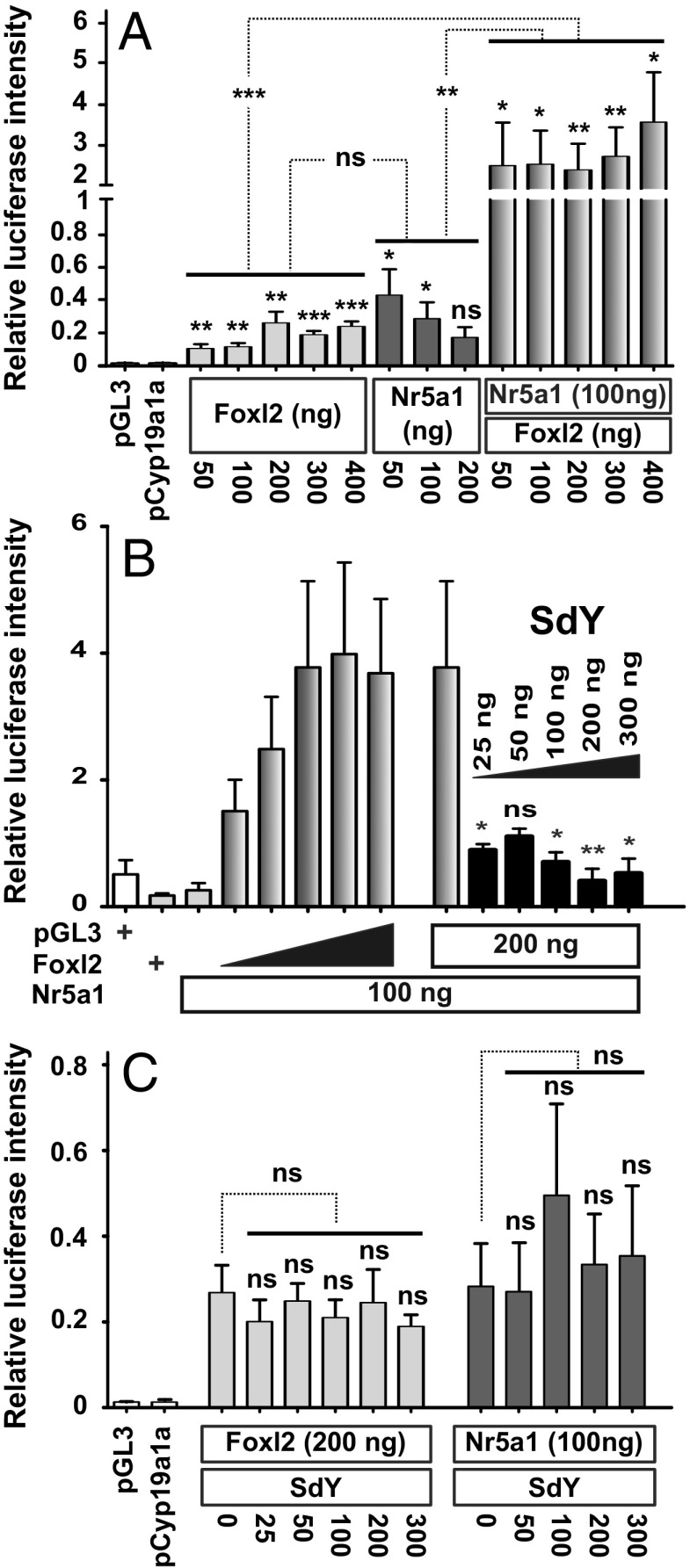
To obtain further insights into the physiological relevance of the SdY and Foxl2 interaction in vivo, a gene expression time course of foxl2, nr5a1, and sdY genes in differentiating trout gonads was performed. In agreement with its male-determining role, sdY expression was detected only in male gonads, with a peak of expression around 45 d postfertilization (dpf) (Fig. 3A). In contrast, foxl2b1, foxl2b2, and nr5a1 were not expressed in a sexually dimorphic fashion before the time point at which sdY peaks in males; after this time point foxl2b1 and foxl2b2 are markedly up-regulated in females and down-regulated in males (Fig. 3 A and B and SI Appendix, Fig. S6). We also explored expression of gonadal aromatase (cyp19a1a), as this gene is a well-known direct target of Foxl2 (24). We found that cyp19a1a is expressed only in female gonads and its expression parallels the expression of the trout foxl2 genes (Fig. 3A). These cyp19a1a, nr5a1, and foxl2 expression patterns are consistent with the critical role of Foxl2 in the up-regulation of Cyp19a1/cyp19a1a (24, 25), in cooperation with Nr5a1 (steroidogenic factor 1, Sf1) for driving ovarian differentiation (26). In addition, sdY and foxl2 are colocalized in some somatic cells of the early differentiating gonad in the male rainbow trout (Fig. 3C), suggesting that they could interact to modulate cyp19a1a expression. Interestingly, trout foxl2 genes are also strongly and positively regulated by estrogens (Fig. 3B), the steroid end products of the aromatase enzyme (21). This points to a positive regulatory loop with Foxl2 inducing cyp19a1a expression and thus increasing estrogen synthesis that will, in return, stimulate the expression of foxl2 (21). Taking into account the pivotal role of Cyp19a1a and estrogens in fish ovarian differentiation (27) and our results on a specific interaction of SdY with Foxl2, we proposed that SdY exerts its sex-determining function by suppressing this positive regulatory loop through its interaction with Foxl2. To evaluate this, we first confirmed, using a luciferase reporter assay, that activation of the medaka cyp19a1a promoter requires the presence of both Foxl2 and Nr5a1, which work in synergy to activate cyp19a1a (25) (Fig. 4A). We then tested the effect of SdY on the transcriptional activity of the cyp19a1a promoter and demonstrated that SdY strongly represses the synergistic Foxl2- and Nr5a1-induced cyp19a1a expression (Fig. 4B) but not the Foxl2-alone- or Nr5a1-alone-induced cyp19a1a expression (Fig. 4C).
Fig. 3.
The gonadal expression patterns of sdY, foxl2, nr5a1, and cyp19a1a (A and B) are in agreement with a repressive effect of SdY on the Foxl2 positive regulation of the aromatase promoter (C). (A) Gene expression profiles of sdY, nr5a1, foxl2b2, and cyp19a1a in male and female gonads from 33 to 125 dpf. All values represent the mean ± standard deviation of three biological replicates (in percentage of the highest measured value). The gray area highlights the period before the sexually dimorphic expression of trout foxl2 genes during gonadal differentiation. (B) Gonadal localization of foxl2b2 transcripts (NBT/BCIP signal in blue) in male and female gonads. foxl2b2 is expressed in somatic cells of both female and male gonads at 50 dpf and only in female gonads at later stages (60, 70, and 85 dpf). In males fed with estrogens (male E2) foxl2b2 is strongly up-regulated compared with control males quickly after (60 dpf) the application of the treatment (55 dpf). (Scale bar, 20 µm.) (C) Colocalization by double in situ hybridization of sdY (NBT/BCIP signal in blue) and foxl2b2 (HNPP/Fast Red signal in red fluorescence) in somatic cells of a rainbow trout male differentiating gonad at 50 dpf. Cell nuclei are shown in the dark-field panels stained with DAPI either with or without the HNPP fluorescent detection of foxl2b2. Germ cells are shown by an asterisk. (Scale bar, 20 µm.)
Fig. 4.
SdY prevents the Foxl2/Nr5a1 positive regulation of the cyp19a1a promoter. Medaka cyp19a1a promoter activity (pGL3-cyp19a1a promoter coupled to a firefly luciferase) was measured using a luciferase reporter assay after HEK 293T cell cotransfection with either medaka nr5a1 and/or foxl2 expression plasmids and variable concentrations of the rainbow trout sdY expression plasmid. All results were calculated as the mean ± SEM of three biological replicates in two independent experiments. (A) Foxl2 and Nr5a1 act in synergy to induce cyp19a1a expression. Medaka cyp19a1a luciferase assay with variable concentrations of foxl2 (50–400 ng) and nr5a1 (50–200 ng) or 100 ng of nr5a1 with variable concentrations of foxl2 (50–400 ng). Statistical significances of luciferase activity within treatments were tested using an unpaired two-tailed Student’s t test. Effects of foxl2 alone and nr5a1 alone compared with their synergistic effect (shown by asterisks on the dotted lines joining the different groups) were tested by a one-way ANOVA with a post hoc Dunnett test. (B) SdY prevents Foxl2/Nr5a1 positive regulation of the cyp19a1a promoter. Medaka cyp19a1a luciferase assay with variable concentrations of foxl2 (50–400 ng) combined with a fixed concentration of nr5a1 (100 ng) and a fixed concentration of foxl2 (200 ng) and nr5a1 (100 ng) combined with variable concentrations of sdY (25–300 ng). Empty plasmid control (pGL3) and Foxl2 alone (200 ng) are depicted by a + sign. Statistical significances of luciferase activity were tested using a Mann–Whitney U test. (C) SdY does not repress cyp19a1a promoter expression induced by medaka Foxl2 alone or Nr5a1 alone. Medaka cyp19a1a luciferase assay with fixed foxl2 (200 ng) or nr5a1 (100 ng) concentrations combined with variable concentrations of sdY (25–300 ng). Statistical significances of luciferase activity within treatments were tested using an unpaired two-tailed Student’s t test. The effect of foxl2 or nr5a1 alone compared with foxl2 or nr5a1 and variable concentrations of sdY was tested by a one-way ANOVA with post hoc Tukey tests. ***P < 0.001; **P < 0.01; *P < 0.1; ns, P > 0.05 (nonsignificant).
In our attempt to understand how irf9, a transcriptional regulator of the immune system, has evolved into a master SD gene whose expression is necessary and sufficient to drive testicular differentiation (16), we obtained multiple sets of evidence supporting the hypothesis that SdY exerts its sex-determining function by interacting with the female-determining factor Foxl2. An interaction of SdY with Foxl2 was not anticipated, but some Forkhead box proteins such as the FOXO proteins have been shown to have direct protein–protein interactions with a variety of unrelated transcription factors (28). In zebrafish the Foxo3b protein is even able to repress the transcriptional activity of irf genes by a direct IRF/FOX protein–protein interaction (29). This may suggest that SdY acquired its ability to specifically bind to fish Foxl2 proteins based on a preexisting possibility of interaction of Irf9 with the FOX protein.
The specific interaction of SdY with Foxl2 carries SdY into the nucleus, where it can prevent, by blocking the synergistic action of Nr5a1 (Sf1) and Foxl2, the implementation of a positive regulatory loop controlling the expression of cyp19a1a. By doing so very early in the differentiation process of the gonads, i.e., long before implementation of the cyp19a1a loop of regulation in females (Fig. 3A), SdY would completely prevent estrogen production in the differentiating male gonads. The absence of estrogen production subsequently triggers masculinization as it has been demonstrated in many fish species, including rainbow trout, using treatments with an aromatase inhibitor (30) or, more recently, by direct inactivation of the cyp19a1a gene (26, 31, 32). Such masculinization following the blockade of estrogen production is even effective in adult females (27), showing that estrogens in fish are needed not only for ovarian differentiation but also for ovarian maintenance. However, the fact that foxl2b1 and foxl2b2 gene expression is not down-regulated in the male gonad before 45–50 dpf suggests that the inhibition of the positive regulatory loop between cyp19a1a expression, estrogen production, and foxl2 gene expression is not active at these early testicular differentiation stages. This absence of inhibition of foxl2 expression in the early male differentiating gonad could suggest that there is additional regulation of this positive loop or that expression of foxl2 is not sensitive to estrogen at these early developmental stages. Such a mechanism of action through the blockage of cyp19a1a and estrogen production assigns to SdY an activity as an antiovarian determining factor directly preventing the ovarian differentiation pathway instead of activating the male pathway. However, it cannot be totally excluded that SdY, besides suppressing the female pathway, may also affect directly the activation of the male pathway. Nevertheless, known important male developmental actors such as Dmrt1, Amh, and Sox9 (1) were not identified in the Y2H screen, and this along with their late expression during rainbow trout male gonad development (33) compared with sdY expression suggests that they are not direct interacting partners of SdY.
In summary, we provide strong evidence that SdY determines sex in rainbow trout not by using part of its ancestral Irf9 pathway but by directly interacting with Foxl2, an important member of the classical gonadal sex differentiation cascade. This suggests that innovation at the top of the vertebrate sex determination cascade may be constrained because novel master SD genes have to cope with the regulation of the conserved vertebrate sex differentiation cascade. The “limited option” hypothesis is mainly based on the idea that only a small subset of genes and chromosomes, because they are better at doing the job, would be independently and repeatedly selected as new vertebrate master SD genes (15). We now propose that the limited option is actually more constrained by the conservation of the sex differentiation pathway and that evolution of SD genes may include some innovations like SdY if these unusual SD genes evolved a means to build an interface with the sex differentiation cascade.
Materials and Methods
Protein Structure Prediction.
Three-dimensional homology modeling of SdY was predicted with the software SWISS-MODEL (https://swissmodel.expasy.org/) using the structure of the dimeric IRF5 (PDB ID 3DSH) transactivation domain (34) as a template. The resulting model was obtained by the superposition of the template and SdY. The 3D views of SdY were made with PyMOL (molecular graphics system, version 1.7.4; Schrödinger).
Yeast Two-Hybrid Screen.
Yeast two-hybrid screening was performed by Hybrigenics Services (https://www.hybrigenics-services.com). The coding sequence for SdY (amino acids 1–215) (GenBank accession number GI:392583258) was PCR amplified and cloned into pB27_A as a C-terminal fusion to LexA (N-LexA-SdY-C) and into pB66_A as a C-terminal fusion to the Gal4 DNA-binding domain (N-Gal4-SdY-C). These constructs were checked by sequencing and used as baits to screen a random-primed Oncorhynchus mykiss immature male gonad (gonads sampled around 75 dpf) cDNA library; 112 million interactions were tested with pB27_SdY, and 71.3 million interactions were tested with p66_SdY, leading to the detection of 24 and 178 processed clones, respectively. The prey fragments of these 202 positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. Each fragment corresponding to an interacting protein was identified using GenBank release 192 (National Center for Biotechnology Information). The common sequence shared by all prey fragments of the same protein defines the selected interacting domain containing all of the structural determinants required for a given interaction to occur. A confidence score [predicted biological score (PBS)] that outlines the reliability of the interaction is given to each interaction as previously described (35). PBS scores were divided into four categories, from A (highest confidence) to D (lowest confidence).
Genomic DNA Extraction.
To clone rainbow trout foxl2b1 and foxl2b2, genomic DNA was extracted from liver tissue with lysis buffer (10 mM Tris·HCl, 100 mm EDTA, 0.5% SDS, and 20 µg/mL RNase A). Proteinase K was added to 150 mg/mL, and the sample was incubated at 56 °C overnight. A double-extraction phenol–chloroform (1:1) followed by chloroform–isoamyl alcohol (24:1) extraction was done. DNA precipitation was performed with an equal volume of isopropanol (1:1). The precipitate was pelleted by centrifugation at 16,000 × g and washed twice in 70% ethanol, dried at room temperature, and dissolved in 2 mL of distilled water.
Cloning.
Plasmids and primers used are listed in the SI Appendix, Tables S4 and S5. The coding sequence of SdY was amplified from the psdy:sdy-pcry:cfp plasmid (16) and inserted into pCS2+. From this plasmid, the PCR-amplified fragment was inserted into pCS2+:HA:mCherry (gift from Manfred Gessler, University of Wuerzburg, Wuerzburg, Germany), pCS2+-emGFP, and pCS2+-3xHA expression plasmids. Rainbow trout foxl2b1 and foxl2b2 and medaka foxl2 (Ol-foxl2) were cloned by PCR amplification on genomic DNA and inserted in pCS2+ plasmid, pCS2+:HA:mCherry, and pCS2+:3xFlag between the EcoRI-XhoI restriction sites. The constructs 3xHA-pCS2+ and 3xFlag-pCS2+ were obtained by concatemerization of three single HA sequences (3xHA) or three single FLAG sequences (3xFLAG) flanked by HindIII restrictions sites. pCS2+-emGFP:SdY was obtained by inserting a PCR-amplified fragment corresponding to emGFP in frame into the EcoRI site. To explore the hypothesis that SdY could be able to interact with all Fox proteins through an interaction with their highly conserved Forkhead domain, we selected different rainbow trout Fox proteins from an EST resource collection in which ESTs were cloned into a CMV expression (pCMV-Sport6) plasmid (36). Five trout cDNA clones encoding for Foxd2, Foxd3, Foxl3, Foxn3, and Foxn2 were found to have a complete open coding frame, including Foxn3 and Foxn2, which were identified as SdY partners in the Y2H screen. The rainbow trout Foxl2b2 sequence with threonine 64 and threonine 79 replaced by adenines (T64A; T79A) was synthesized (Genscript) by replacing in the foxl2b2 cDNA sequence the threonine codon sequence ACC or ACT with an adenine codon sequence GCC at amino acid positions 64 and 79, respectively. The corresponding sequence was inserted in a pcDNA3.1 plasmid between the EcoR1 and XhoI restriction enzyme sites and checked by sequencing.
Cell Culture.
HEK 293T cells were cultured and maintained in DMEM (PAN Biotech), supplemented with 10% FCS (PAN Biotech) and 1% penicillin–streptomycin (PAN Biotech) at 37 °C with 5% CO2. Transfections for HEK 293T cells were performed by incubating cells with polyethylenimine (PEI) (100 mg/mL PEI diluted 1:100 in 150 mM NaCl) and respective plasmids (10 µg for 10-cm dishes, 2 µg for six-well plates) for 6–8 h in fresh medium. Then, the medium was discarded, and fresh medium was added.
Rainbow trout gonadal (RTG2) cells were cultured and maintained in L15 medium, 20 mM glutamine (PAN Biotech), supplemented with 10% FCS (PAN Biotech) and 1% penicillin–streptomycin (PAN Biotech), at 20 °C in an atmosphere of air. For transfection, RTG2 cells were detached by Trypsin-EDTA (P0781; Sigma-Aldrich) and pelleted by centrifugation at 1,000 × g for 5 min, washed once with medium and once with PBS. The pellet was drained and resuspended in solution V (Amaxa Kit) at a density of 106 cells/mL; 2 µg of plasmid were added to the suspension. After mixing, the suspension was transferred to a cuvette (Kit V; Amaxa). Program D-23 was used to electroporate the cells. After transfection, cells were immediately transferred to six-well plates filled with medium. All experiments were performed 72 h after transfection.
Immunofluorescence.
HEK 293T cells were seeded in a 6-well plate containing coverslips. After pCS2+-meGFP-SdY and pCS2+-mCherry-Foxl2b1 (or pCS2+-3xFLAG:Foxl2b2 or pCS2+-3xFLAG-Ol-Foxl2) cotransfection for 48 h, cells were fixed in 4% fresh paraformaldehyde for 15 min, extensively washed, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. Then cells were blocked with 1% BSA for 20 min. The primary antibody (SI Appendix, Table S6) was incubated overnight at 4 °C. After extensive washing with PBS, cells were incubated with Alexa 488 or Alexa 594 conjugated secondary antibodies in 1% BSA for 1 h, followed by Hoechst 33342 (Invitrogen) staining for 5 min (1 μg/mL final concentration). Cells were mounted using Mowiol 4-88 (Roth). Confocal images were acquired using a Nikon Eclipse C1 laser-scanning microscope (Nikon), fitted with a 60× Nikon objective (PL APO, 1.4 N.A.), and Nikon image software. Images were collected at 1,024 × 1,024 pixel resolution. The stained cells were optically sectioned in the z axis. The step size in the z axis varied from 0.2 to 0.25 mm to obtain 50 slices per imaged file. All experiments were independently repeated at least three times.
Colocalization Analyses.
The Nikon NIS-Elements imaging analysis software was used for the colocalization analyses. Confocal images of double-stained sections were first subjected to background correction. SdY nuclear translocation was counted as complete translocation when the majority of the GFP:SdY signal was found in the nucleus. Pearson’s correlation was calculated and used to obtain the colocalization values as percentages of SdY overlapping with Foxl2b1 or Foxl2b2 for a minimum of five cells (n = 5). The Pearson’s coefficient values were defined as very strong colocalization between 0.85 and 1, strong colocalization between 0.5 and 0.85, and weak or no colocalization between −1 and 0.5.
Co-Immunoprecipitation.
HEK-293T cells were transfected with pCS2+-3xHA:SdY and pCS2+-3xFlag-Foxl2b1(or pCS2+-3xFlag:Foxl2b2 or pCS2+-3xFlag-Ol-Foxl2) constructs to be assessed for their ability to co-immunoprecipitate. After 48 h cells were scraped and resuspended in 50 µL lysis buffer [20 mM Hepes (pH 7.8), 500 mM NaCl, 5 mM MgCl2, 5 mM KCl, 0.1% deoxycholate, 0.5% Nonidet-P40, 10 mg/mL aprotinin, 10 mg/mL leupeptin, 200 mM Na3VO4, 1 mM phenylmethanesulfonyl fluoride, and 100 mM NaF]. Cells were incubated in lysis buffer for 30 min at 4 °C and then cleared by high-speed centrifugation for 20 min. After Bradford protein concentration measurement, HNTG buffer (20 mM Hepes pH 7.5; 150 mM NaCl; 10% glycerol; 0.1% Triton X-100) was added (1:1) to 250 µg of the whole-cell lysate. After preclearing with IgG antibodies for 1 h at 4 °C, whole-cell lysates were used for immunoprecipitation with the corresponding antibodies. One microgram of anti-Flag, anti-HA, or IgG antibody was added to 500 µL of cell lysate or 5 µg of anti-SdY or anti-FoxL2 (SI Appendix, Table S6) and then incubated at 4 °C overnight. After the addition of washed protein G agarose beads (Pierce, 20398), incubation in HNTG buffer was continued for another 2 h. Immunoprecipitates were washed (five times with centrifugation at 1,000 × g, supernatant discarded, HNTG lysis buffer added) and eluted with SDS/PAGE loading buffer by boiling for 10 min. Co-immunoprecipitation was detected by standard Western blot analysis procedure.
Western Blotting.
Cells were lysed in a Hepes-based lysis buffer [20 mM Hepes (pH 7.8), 500 mM NaCl, 5 mM MgCl2, 5 mM KCl, 0.1% deoxycholate, 0.5% Nonidet-P40, 10 mg/mL aprotinin, 10 mg/mL leupeptin, 200 mM Na3VO4, 1 mM phenylmethanesulfonyl fluoride, and 100 mM NaF] for 3 h. Cells debris was pelleted by centrifugation for 15 min at 16,000 × g. Cell lysate protein concentration was measured with a Bradford assay (Cary 50 Spectrophotometer; Varian). The protein lysates (30–50 μg) were resolved by SDS/PAGE on 12% Tris·glycine gels followed by transfer to nitrocellulose membranes. Unspecific binding was blocked with 5% BSA in Tris buffered saline with Tween-20 (TBST) [10 mM Tris (pH 7.9); 150 mM NaCl; 0.1% Tween] for 1 h at room temperature. Incubation with primary antibodies was performed overnight at 4 °C. After three washes with TBST, HRP conjugated antibodies were incubated with blocking solution for 1 h. Following the washes, membranes were incubated with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) for 1 min. The signal from the membrane was detected using a Photo Image Station 4000MM (Kodak). At least two independent experiments were performed, and representative protein blot images are shown.
Quantitative PCR.
Expression levels of sdY, foxl2b1, foxl2b2, nr5a1, and cyp19a1a were measured by qPCR as previously described (37). Gonads (15–20 pairs of gonads per time points) were sampled in triplicate at 33, 35, 37, 40, 44, 50, 61, 85, and 125 dpf in both genetic all-male (XY) and all-female (XX) populations of rainbow trout. Total RNA was extracted using the RNAqueous-Micro Kit (Ambion, Life Technologies) for the 33- to 50-dpf samples and the RNAqueous-Kit (Ambion, Life Technologies) for the 61- to 125-dpf samples. All samples were then treated with the TURBO DNA-free Kit (Ambion, Life Technologies) to remove any leftover genomic DNA. Reverse transcriptions were carried out using 150 ng of total RNA as the starting material with the Ovation RNA Amplification System (NuGEN Technologies), following the manufacturer’s recommendations. Quantitative PCR was performed using the StepOnePlus system (Applied Biosystems) using 4 µL of reverse-transcribed cDNA (single tube quantification per sample, with three biological replicates for each sex and time point) diluted to 1:90, the Fast SYBR Green Master Mix (Applied Biosystem), and 600 nmol of each primer listed in SI Appendix, Table S7. The enzyme was activated for 20 s at 95 °C, followed by 40 cycles of denaturation at 95 °C for 5 s and annealing and elongation at 60 °C for 30 s.
Whole-Mount in Situ Hybridization.
In situ hybridization was performed as previously described (38). RNA probes were produced from PCR products obtained by amplification of foxl2b2. Ten nanograms of the PCR product were used as a template for digoxigenin-labeled RNA probe synthesis using digoxigenin 11-UTP (Roche Diagnostics Corp.) and T3 or T7 RNA polymerase (Promega) following standard protocols. Whole-mount in situ hybridization was carried out using an in situ Pro, Intavis AG robotic station. Male and female embryos were fixed overnight in 4% paraformaldehyde at 4 °C, dehydrated in 100% methanol, and stored at −20 °C. Before in situ hybridization they were rehydrated, permeabilized by proteinase K treatment (25 µg/mL, 30 min, at room temperature), and postfixed (4% paraformaldehyde and glutaraldehyde 0.2%, for 20 min). Prehybridization and hybridization media contained 50% formamide, 5XSSC, 0.1% Tween 20, 0.005% heparin, 0.1 mg/mL tRNA. Hybridization was carried out at 65 °C for 16 h. After posthybridization washes, embryos were incubated in blocking buffer (PBS/Triton 0.1%/Tween 20 0.2%, containing 2% serum) for 2 h before the addition of the alkaline phosphatase coupled anti-digoxigenin antibody (1:2,000; Roche Diagnostics Corp.) for 6 h. After washing, the color reaction was performed in the presence of nitro-blue tetrazolium/5-Bromo-4-chloro-3′-indoly phosphate (NBT/BCIP) (Roche). Briefly, dehydration and paraffin infiltration were performed in a Citadel 1000 tissue processor (Shandon). Dehydrated tissues were embedded in plastic molds in paraffin using a HistoEmbedder (TBS88; Medite). Each embedded sample was sectioned 5 µm thick on a MICRO HM355 (Thermo Fisher Scientific).
Colocalization of sdY and foxl2 by in Situ Hybridization.
Fifty-dpf male rainbow trout were fixed in Bouin’s fixative at 4 °C. One hour after fixation and dehydration, the tissues were embedded in paraffin and cut into 4-μm sections. Antisense sdY and foxl2b2 RNA probes were synthesized using in vitro transcription with a fluorescein RNA labeling mix (Roche) and a DIG RNA labeling mix (Roche), respectively. Sections were deparaffinized, rehydrated, and treated with 1% H2O2 in TBST buffer at room temperature for 30 min and 1 μg/mL proteinase K (Roche) at 37 °C for 13 min. After the enzymatic treatment, sections were dehydrated with ethanol and chloroform and then hybridized with sdY and foxl2 probes simultaneously at 60 °C for 18 h. Fluorescein was visualized by using an anti-fluorescein–alkaline phosphatase (anti-fluorescein–AP) Fab fragment (Roche) (1:1,000) and the HNPP Fluorescent Detection Set (Roche). Digoxigenin (DIG) was visualized by using an anti-digoxigenin–AP Fab fragment (Roche) (1:500) and NBT/BCIP. Before the DIG visualization, alkaline phosphatase was inactivated in 0.1 M glycine/0.1% Tween 20 at room temperature for 30 min. DAPI staining was performed to visualize nuclei.
Luciferase Assay.
HEK 293T cells were transfected using PEI with the following plasmids: 0.3 µg of pGL3-OlaCyp19a1a sequence (kindly provided by D. Wang Deshou, Key Laboratory of Aquatic Science of Chongqing, School of Life Sciences, Southwest University, Chongqing, China), 0.05–0.4 µg of pCS2+-SdY expression plasmid, 0.05–0.4 µg of pCS2+-OlaFoxl2, 0.1 µg of pcDNA3.1-OlaNr5a1, and 0.1 µg of plasmid thymidine kinase-Renilla used for calibration. Each experiment was performed with a 1.0-µg final amount. Adjustments were made with empty plasmid (pCS2+) accordingly. Firefly luciferase and Renilla luciferase readings were obtained using the Dual-Luciferase Reporter Assay System (Promega) and LUMAT LB 9501 luminometer (Berthold Technologies).
One-by-one Yeast Two-Hybrid Assay.
Diploid cells containing the same bait construct of the yeast two-hybrid assay (p66_SdY) and a prey plasmid construct coding for Foxl2b2 cloned in frame with the activation domain of GAL4 (p14-N-GAL4-Foxl2b1-C) were mated and spotted on selective media. The medium lacking tryptophan and leucine was used as a control for the yeast growth test and to check for the presence of the bait or the prey. The assay is based on the histidine reporter gene. A triple-negative medium (tryptophan, leucine, and histidine) selects yeast growth if interaction occurs. Interaction pairs were tested at decreasing concentrations (10−1, 10−2, 10−3, 10−4) from two independent clones. An inhibitor of the histidine gene product 3-AT was used to increase stringency at four different concentrations (1, 5, 10, 50 mM).
RNA Injections.
For injections capped RNA GFP-sdY, mCherry-foxl2b2, Olafoxl2 from pCS2+-meGFP:SdY, pCS2+-HA:mCherry:Foxl2b2, and pCS2+-OlaFoxl2, respectively, was transcribed from linearized pCS2+ plasmid using the SP6/T3/T7 m MESSAGE mMACHINE Kit (Ambion). One nanoliter was injected into the cytoplasm of one-cell stage Medaka embryos.
Statistical Analysis.
Most of the data were analyzed using an unpaired Student’s t test. A Mann–Whitney U test was then used to compare the median value when the Kolmogorov–Smirnov test was negative. Statistical analysis of multiple groups was performed using an ANOVA one-way test with post hoc Dunnett or Tukey tests for multiple comparison (a control group compared with an experimental group or a control sample compared to an experimental group, respectively). All statistical analyses were performed with GraphPad Prism (version 5; GraphPad Software). Significant differences are symbolized in figures by asterisks if P < 0.001 (***), P < 0.05 (**), or P < 0.01 (*) or indicated by ns if not significant.
Supplementary Material
Acknowledgments
We acknowledge Wang Deshou for the medaka cyp19a1a promoter and Reiner Veitia for helpful discussions. This work was supported by Agence Nationale de la Recherche (ANR) Grant ANR-11-BSV7-0016 (SDS project) and grants from the Deutsche Forschungsgemeinschaft (Scha408/12-1, 10-1; to M.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.L.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803826115/-/DCSupplemental.
References
- 1.Herpin A, Schartl M. Plasticity of gene-regulatory networks controlling sex determination: Of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 2015;16:1260–1274. doi: 10.15252/embr.201540667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graves JAM. How to evolve new vertebrate sex determining genes. Dev Dyn. 2013;242:354–359. doi: 10.1002/dvdy.23887. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y-T, Capel B. Cell fate commitment during mammalian sex determination. Curr Opin Genet Dev. 2015;32:144–152. doi: 10.1016/j.gde.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Windley SP, Wilhelm D. Signaling pathways involved in mammalian sex determination and gonad development. Sex Dev. 2015;9:297–315. doi: 10.1159/000444065. [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi K, Hamaguchi S. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn. 2013;242:339–353. doi: 10.1002/dvdy.23927. [DOI] [PubMed] [Google Scholar]
- 6.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- 7.Matsuda M, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 8.Nanda I, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci USA. 2002;99:11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 10.Kamiya T, et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu) PLoS Genet. 2012;8:e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattori RS, et al. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci USA. 2012;109:2955–2959. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S, et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet. 2014;46:253–260. doi: 10.1038/ng.2890. [DOI] [PubMed] [Google Scholar]
- 14.Myosho T, et al. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics. 2012;191:163–170. doi: 10.1534/genetics.111.137497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall Graves JA, Peichel CL. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010;11:205. doi: 10.1186/gb-2010-11-4-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano A, et al. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr Biol. 2012;22:1423–1428. doi: 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 17.Yano A, et al. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol Appl. 2013;6:486–496. doi: 10.1111/eva.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci USA. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu W-L, Sun Y. Interferon regulatory factor 9 plays a dual function in health and disease. J Hepatol. 2015;62:1446. doi: 10.1016/j.jhep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 20.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 21.Baron D, et al. An evolutionary and functional analysis of FoxL2 in rainbow trout gonad differentiation. J Mol Endocrinol. 2004;33:705–715. doi: 10.1677/jme.1.01566. [DOI] [PubMed] [Google Scholar]
- 22.Boulanger L, et al. FOXL2 is a female sex-determining gene in the goat. Curr Biol. 2014;24:404–408. doi: 10.1016/j.cub.2013.12.039. [DOI] [PubMed] [Google Scholar]
- 23.Bertho S, et al. Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex Dev. 2016;10:111–129. doi: 10.1159/000447611. [DOI] [PubMed] [Google Scholar]
- 24.Pannetier M, et al. FOXL2 activates P450 aromatase gene transcription: Towards a better characterization of the early steps of mammalian ovarian development. J Mol Endocrinol. 2006;36:399–413. doi: 10.1677/jme.1.01947. [DOI] [PubMed] [Google Scholar]
- 25.Wang D-S, et al. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol Endocrinol. 2007;21:712–725. doi: 10.1210/me.2006-0248. [DOI] [PubMed] [Google Scholar]
- 26.Li M-H, et al. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology. 2013;154:4814–4825. doi: 10.1210/en.2013-1451. [DOI] [PubMed] [Google Scholar]
- 27.Guiguen Y, Fostier A, Piferrer F, Chang C-F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen Comp Endocrinol. 2010;165:352–366. doi: 10.1016/j.ygcen.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 28.van der Vos KE, Coffer PJ. FOXO-binding partners: It takes two to tango. Oncogene. 2008;27:2289–2299. doi: 10.1038/onc.2008.22. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Cai X, Zhang D, Xu C, Xiao W. Zebrafish foxo3b negatively regulates antiviral response through suppressing the transactivity of irf3 and irf7. J Immunol. 2016;197:4736–4749. doi: 10.4049/jimmunol.1601187. [DOI] [PubMed] [Google Scholar]
- 30.Guiguen Y, et al. Involvement of estrogens in the process of sex differentiation in two fish species: The rainbow trout (Oncorhynchus mykiss) and a tilapia (Oreochromis niloticus) Mol Reprod Dev. 1999;54:154–162. doi: 10.1002/(SICI)1098-2795(199910)54:2<154::AID-MRD7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 31.Dranow DB, et al. Bmp15 is an Oocyte-produced signal required for maintenance of the adult female sexual phenotype in zebrafish. PLoS Genet. 2016;12:e1006323. doi: 10.1371/journal.pgen.1006323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau ES-W, Zhang Z, Qin M, Ge W. Knockout of zebrafish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci Rep. 2016;6:37357. doi: 10.1038/srep37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vizziano D, Randuineau G, Baron D, Cauty C, Guiguen Y. Characterization of early molecular sex differentiation in rainbow trout, Oncorhynchus mykiss. Dev Dyn. 2007;236:2198–2206. doi: 10.1002/dvdy.21212. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15:1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Formstecher E, et al. Protein interaction mapping: A Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rexroad CE, 3rd, et al. Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenet Genome Res. 2003;102:347–354. doi: 10.1159/000075773. [DOI] [PubMed] [Google Scholar]
- 37.Marivin E, et al. Sex hormone-binding globulins characterization and gonadal gene expression during sex differentiation in the rainbow trout, Oncorhynchus mykiss. Mol Reprod Dev. 2014;81:757–765. doi: 10.1002/mrd.22344. [DOI] [PubMed] [Google Scholar]
- 38.Yano A, Nicol B, Guerin A, Guiguen Y. The duplicated rainbow trout (Oncorhynchus mykiss) T-box transcription factors 1, tbx1a and tbx1b, are up-regulated during testicular development. Mol Reprod Dev. 2011;78:172–180. doi: 10.1002/mrd.21279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.