Significance
Hepatocellular carcinoma (HCC) is a heterogeneous and deadly form of liver cancer. Here, we stratified and characterized HCC tumors by applying graph and control theory concepts to the topology of genome-scale metabolic networks. We identified three HCC subtypes with distinct differences in metabolic and signaling pathways and clinical survival and validated our results by performing additional experiments. We further identified HCC subtype-specific genes pivotal in controlling the entire metabolism and discovered genes that can be targeted for development of efficient treatment strategies for specific HCC subtypes. Our systems-level analyses provided a systematic way for characterization of HCC subtypes and identification of drug targets for effective treatment of HCC patients.
Keywords: hepatocellular carcinoma, biological networks, personalized medicine, genome-scale metabolic models, systems biology
Abstract
Hepatocellular carcinoma (HCC) is one of the most frequent forms of liver cancer, and effective treatment methods are limited due to tumor heterogeneity. There is a great need for comprehensive approaches to stratify HCC patients, gain biological insights into subtypes, and ultimately identify effective therapeutic targets. We stratified HCC patients and characterized each subtype using transcriptomics data, genome-scale metabolic networks and network topology/controllability analysis. This comprehensive systems-level analysis identified three distinct subtypes with substantial differences in metabolic and signaling pathways reflecting at genomic, transcriptomic, and proteomic levels. These subtypes showed large differences in clinical survival associated with altered kynurenine metabolism, WNT/β-catenin–associated lipid metabolism, and PI3K/AKT/mTOR signaling. Integrative analyses indicated that the three subtypes rely on alternative enzymes (e.g., ACSS1/ACSS2/ACSS3, PKM/PKLR, ALDOB/ALDOA, MTHFD1L/MTHFD2/MTHFD1) to catalyze the same reactions. Based on systems-level analysis, we identified 8 to 28 subtype-specific genes with pivotal roles in controlling the metabolic network and predicted that these genes may be targeted for development of treatment strategies for HCC subtypes by performing in silico analysis. To validate our predictions, we performed experiments using HepG2 cells under normoxic and hypoxic conditions and observed opposite expression patterns between genes expressed in high/moderate/low-survival tumor groups in response to hypoxia, reflecting activated hypoxic behavior in patients with poor survival. In conclusion, our analyses showed that the heterogeneous HCC tumors can be stratified using a metabolic network-driven approach, which may also be applied to other cancer types, and this stratification may have clinical implications to drive the development of precision medicine.
Hepatocellular carcinoma (HCC) is a prevalent form of liver cancer and the third-leading cause of cancer-related worldwide mortality with an increasing prevalence globally (1). Due to its large heterogeneity, a complete understanding of the molecular mechanisms underlying HCC onset and progression remains elusive. Comprehensive approaches capable of incorporating intertumor variability, while providing biological insights, are thus of great need for revealing the underlying molecular mechanisms of HCC progression, characterization of HCC subtypes, and identifying therapeutic targets for development of effective treatment strategies for specific patient groups.
Systems biology approaches have been employed to characterize the tumors and study the altered biological processes (2–5). Characterizations of HCC using omics data including genomics, transcriptomics, proteomics, and metabolomics are currently available (6–12). This integrative analysis enabled the identification of markers associated with recurrence and poor prognosis (13–15). Moreover, genome-scale metabolic models (GEMs), collections of biochemical reactions, and associated enzymes and transporters have been successfully used to characterize the metabolism of HCC, as well as identify drug targets for HCC patients (11, 16–18). For instance, HCC tumors have been stratified based on the utilization of acetate (11). Analysis of HCC metabolism has also led to identification of potential anticancer metabolite analogs that would not be toxic for noncancerous liver tissues (16). These observations indicated the vital need for integrating large-scale omics data and systems-level analyses. However, while these methods implicitly consider metabolic network structure, they do not permit stratifying tumors based on network heterogeneity itself, and instead rely on identification of key genes/metabolites and tumor stratification based on their expression levels. In turn, topology-driven network analyses, including protein–protein interaction, signaling, and transcriptional regulatory and metabolic networks (19–21), provide an alternative view for characterizing tumors. For instance, network analysis enabled the identification of essential genes from a lethality perspective, as well as those capable and indispensable for controlling networks (4, 22–25). However, topology-driven methods do not take into account biological functionality, one important strength in GEM analyses.
While molecular classifications and identification of gene signatures have undoubtedly provided important contributions for revealing the underlying molecular mechanism involved in the occurrence of HCC (12, 26, 27), a function-driven yet comprehensive characterization of HCC was lacking. Here, we used an unsupervised and systematic method to take advantage of the directed relationships in tumor metabolism. We integrated multiomics data with metabolic network-based analysis to introduce a whole network-driven stratification of HCC tumors. We observed consistent tumor stratification across different datasets consisting of hundreds of HCC tumors. Through the utilization of the entire metabolic network for tumor stratification, our method relied on the network topology rather than the known signatures or molecular features. Importantly, though we have only considered the metabolic network information, different HCC subtypes displayed substantial differences at the metabolic, signaling pathways as well as clinical survival. Additionally, we identified HCC subtype-specific therapeutic targets that have important roles in controlling the cancer network but not in noncancerous liver samples. Finally, we experimentally observed that expression of genes associated with good and poor prognosis tumors shows opposite responses under hypoxic conditions.
Results
Functional Gene–Gene Networks for Characterizing Metabolic Heterogeneity in HCC.
We retrieved The Cancer Genome Atlas (TCGA) transcriptomics and clinical data for 369 HCC individuals, along with 50 matched noncancer liver samples from the Genomic Data Commons portal (12). We split this transcriptomics dataset into two parts: a test set, consisting of 186 patients with detailed clinical information for clinical and signature data analysis, and a validation set, consisting of 183 patients with detailed clinical information. We integrated the transcriptomics data in the test set with an HCC-specific genome-scale metabolic model (16) to generate patient-specific HCC and nontumor liver GEMs (HMR2; SI Appendix, Materials and Methods). After excluding 10 nonfunctional GEMs (<6%), we constructed personalized directed “functional” gene–gene networks (fGGNs), a novel approach introduced here for revealing the importance of a metabolic gene in HCC (SI Appendix, Figs. S1 and S2) inspired by a previous approach (28) (details are in SI Appendix, Materials and Methods). Throughout, fGGNs present gene–gene–directed networks (thus detailing a clear signal flow between genes), where genes (enzymes) are nodes connected by edges if a metabolite product of a gene’s reaction serves as a substrate for the reaction driven by the other gene, or if the two genes are required for the reaction to occur (SI Appendix, Fig. S2).
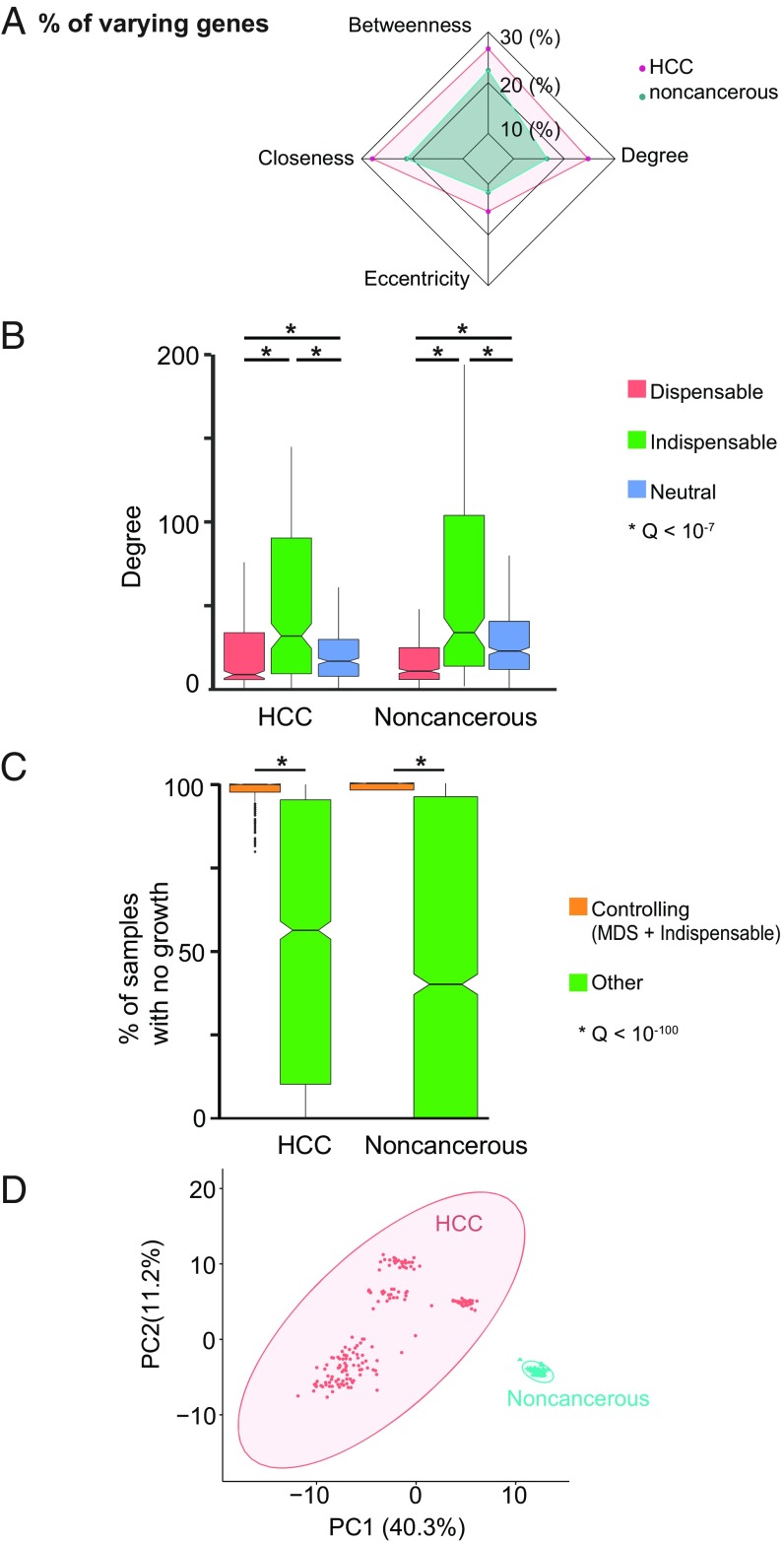
After validation of fGGNs against randomly generated networks (SI Appendix, Fig. S3A), we compared heterogeneity across patients by testing fGGN similarity within and between HCC versus noncancer liver. We investigated genes based on their network centrality, given that central nodes tend to act as hubs with higher biological importance (21, 24, 29, 30). For instance, we evaluated (Dataset S1) their degree centrality (number of genes associated with every gene), betweenness centrality (number of gene-connecting shortest paths that pass through a gene), eccentricity centrality (maximum distance between a gene and other genes), and closeness centrality (length of the shortest path between a gene and all other genes). Comparison of these scores within HCC and noncancer groups indicated that the former group is substantially more heterogeneous than the latter, where the median node absolute deviation for each of the parameters tends to be larger in HCC compared with nontumor samples (Fig. 1 A and B). In turn, between-group comparison showed substantial differences between HCC and nontumor samples at the network level (SI Appendix, Fig. S3B). Overall, all tested parameters showed that noncancer fGGNs are more similar to each other in comparison with HCC fGGNs at the network level.
Fig. 1.
Network-based approaches to identify driver genes involved in progression of HCC. (A) Radar plot of median node absolute deviation for betweenness, closeness, degree, and eccentricity indicates a larger variability in the HCC vs. noncancer networks. (B) Relation of degree centrality and controllability in cancer and noncancerous samples. Groups of genes were classified based on their dispensability (indispensable, dispensable, neutral) when identified in each category in >80% of the fGGNs for HCC and noncancerous networks. Indispensable genes tend to be more central than neutral or dispensable, in both HCC and noncancerous tissues (for all six comparisons, Q < 10−7, Mann–Whitney U test). For each group (indispensable vs. dispensable vs. neutral), we observed no statistical differences in degrees of HCC vs. noncancerous for the three tested comparisons (Q > 0.2). (C) Silencing of controlling genes leads to lethality in >95% of HCCs (vs. <50% for silencing of other genes). In noncancerous samples, silencing either kills all or none of the samples, where all controlling genes lead to lethality (48% for other genes). Both comparisons show statistically significant differences (Q < 10−100, Mann–Whitney U test). (D) Principal component analysis of cancer and noncancer for controllability of fGNNs. Ellipses indicate 95% confidence regions (one outlier is identified at this confidence level).
We then identified genes that are pivotal in controlling the full networks using a network controllability approach. For instance, based on control theory, one may define the minimum driver node sets (MDSs) as those nodes that influence the dynamics of a directed network as previously defined (22, 31). Based on this notion, nodes (genes) may be classified as indispensable, neutral, and dispensable, namely those whose removal from the network respectively increase, do not change, or decrease the minimum number of MDSs. One prime example of their importance was shown for indispensable proteins, which are commonly targeted by disease-causing mutations (22). Importantly, in a network, a gene may be classified either as indispensable, neutral, or dispensable based on its role in affecting the MDS number. Here, we took advantage of the curated, directed, and comprehensive features of GEMs to characterize the gene–gene relationship which captures metabolic associations and their functionality. We identified MDSs and node dispensability at the gene–gene level by using fGGNs, and further defined “controlling genes” as those with a pivotal importance in controlling network dynamics, that is, MDSs or indispensable genes. The approach used here to systematically and functionally characterize metabolism may also be extended to other cancers and diseases.
We found 224 and 313 genes in HCC and noncancerous fGGNs, respectively, which identified as MDS in >80% of the networks. These genes are associated with transport reactions, fatty acid metabolism, oxidative phosphorylation, nucleotide metabolism, and carnitine shuttle (Dataset S2). Among the MDS genes, 85 and 68 are exclusive to HCC and noncancerous networks, respectively.
Next, we identified 188 indispensable genes in ≥80% of the HCC fGGNs, whereas 248 indispensable genes in ≥80% of the noncancerous networks. Our observations indicated that indispensable genes tend to be connected to a higher number of genes (i.e., higher degree), indicative of high centrality in both HCC and noncancerous networks (Q < 10−7, Mann–Whitney U test; Fig. 1C). In HCC, indispensable genes are connected to 32 other genes (median degree), as opposed to neutral and dispensable genes, which respectively show median degrees of 17 and 9, respectively. We also found qualitatively similar observations in noncancerous networks for genes that are indispensable (median degree 34), neutral (median degree 22), and dispensable (median degree 11). Our analysis suggested that indispensable genes tend to be involved in a higher number of reactions (Dataset S2), thus displaying higher importance for controlling network dynamics. However, dispensable and indispensable genes may show similar degrees, indicating that not all highly connected genes (i.e., hubs) have network-controlling properties. Nevertheless, most indispensable genes are more central in both HCC and noncancerous networks. Among the 412 and 429 controlling genes (MDSs and indispensable) in HCC and noncancerous fGGNs, we identified 116 HCC-specific and 133 noncancerous-specific controlling genes. Reaction-level dispensability and controllability are detailed in Dataset S2.
We performed in silico gene essentiality analysis using personalized GEMs. Essentiality analysis of all 2,892 metabolic genes in GEMs showed that >95% of HCC samples have not grown when MDSs or indispensable genes are silenced, much higher than the observed fractions of silencing of other genes (<50%) (Fig. 1C). Based on the controllability and MDS classifications, we also observed clear separation of HCC and noncancer fGGNs as indicated by principal component analysis (Fig. 1D), otherwise not achieved when solely considering gene expression (SI Appendix, Fig. S3C). These observations showed that despite the high heterogeneity expressed at the gene level in HCC, network analyses identified distinct and important genes that may be used to efficiently stratify HCC and noncancer samples based on network controllability.
Network-Based Stratification Reveals Biological and Clinical Survival Differences in HCC.
After identifying the general features of fGGNs in terms of gene centrality, dispensability, and controllability, we used these concepts for in-depth characterizations of HCC and HCC subtypes. We stratified and characterized the HCC subtypes using the complete metabolic network as described above. To do so, we introduced the utilization of fGGNs to stratify tumors based on gene expression data and techniques previously employed to stratify tumors based on somatic mutations (32). We combined the personalized fGGNs into a single generic fGGN, which is representative of the features of all 186 patients and consists of 1,972 metabolic genes (SI Appendix, Materials and Methods), and used this generic fGGN for stratification of HCC patients. Through integration of patient transcriptomics data with the generic fGGN and employment of network smoothing to spread the influence of each expression profile on the neighborhood of the network, we generated expression profiles that reflect the fGGN structure. These expression profiles were subsequently stratified using nonnegative matrix factorization.
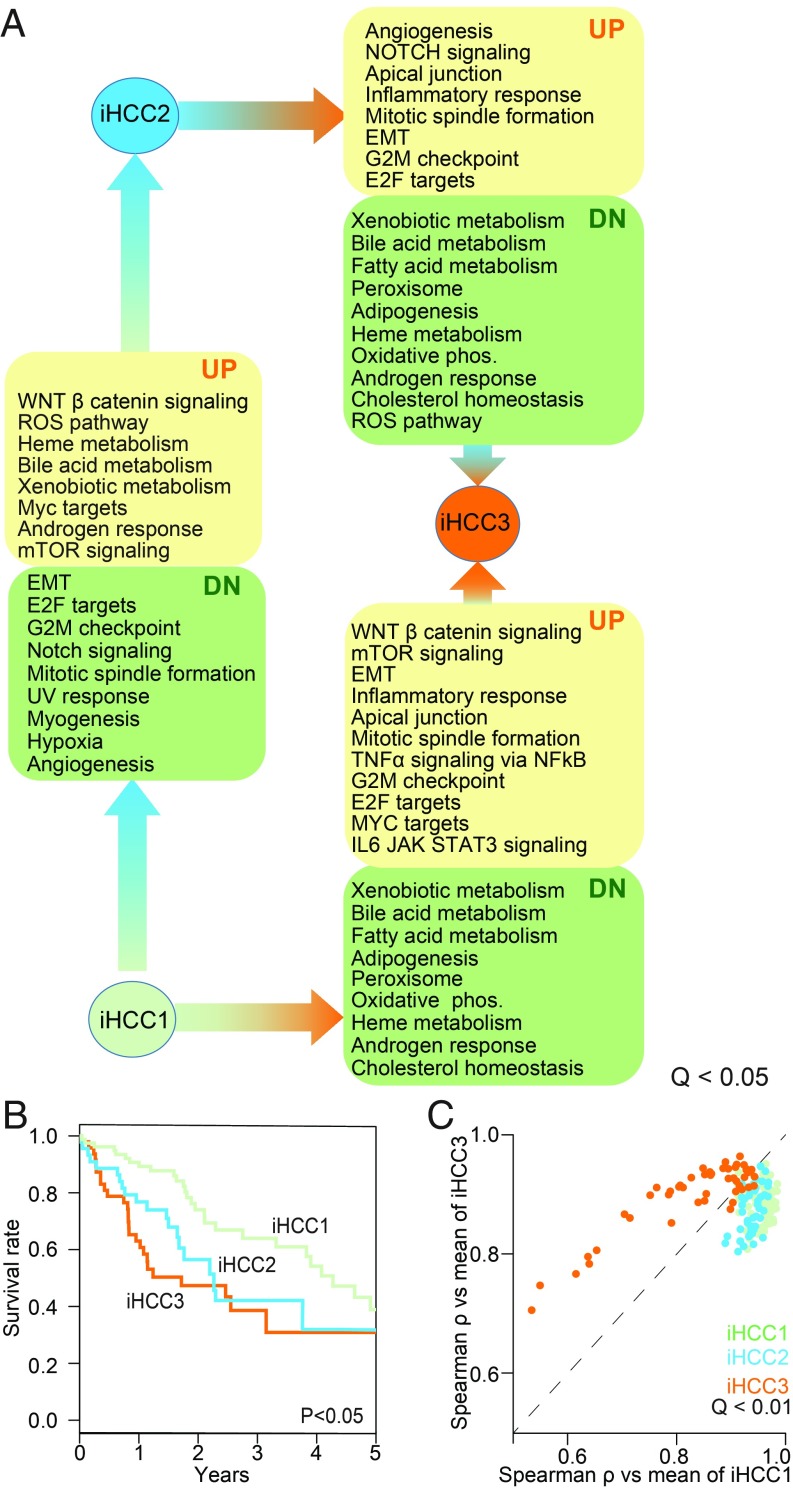
We identified an optimum number of three HCC subtypes (SI Appendix, Fig. S3D), each consisting of 85, 49, and 52 patients with substantial gene expression, biological process, and clinical survival differences (Fig. 2 and Dataset S3). These subtypes are henceforth termed iHCC1, iHCC2, and iHCC3. We also performed tumor stratification using Recon3D (33) as a reference model rather than HMR2-derived (34, 35) stratification. We obtained relatively good agreement with the Recon 3D stratification: 81, 83, and 75% of samples are respectively categorized as iHCC1, iHCC2, and iHCC3 by both HMR2 and Recon3D (SI Appendix, Fig. S4), even though Recon3D does not include ∼37% of the metabolic genes in HMR2.
Fig. 2.
Network-based approaches identified three different HCC subtypes. (A) Gene set-enriched biological processes (Q < 0.05) in different HCC subtypes including iHCC1, iHCC2, and iHCC3. Arrows indicate direction of change (e.g., iHCC2 shows up-regulated heme metabolism compared with iHCC1). (B) Kaplan–Meier survival analysis shows significant differences in patient survival between the three HCC subtypes (iHCC1 > iHCC2 > iHCC3). (C) Correlation plot between tumors and mean gene expression in iHCC1 and iHCC3 (Q < 0.01) showed that iHCC2 tumors tend to be more similar to iHCC1 than iHCC3. This is reinforced by the higher Euclidean distance between iHCC3 fGGNs and other fGGNs in this or the two other subtypes, compared with distances within or between iHCC1 and iHCC2 (SI Appendix, Fig. S6). EMT, epithelial-to-mesenchymal transition; ROS, reactive oxygen species.
We performed differential expression analysis based on the RNA-sequencing data of HCC subtypes and identified 2,409 differentially expressed genes between iHCC2 versus iHCC3, 2,318 genes between iHCC1 versus iHCC3, and 1,115 genes between iHCC1 versus iHCC2 (Q < 0.05; SI Appendix, Dataset S3). Cancer hallmark gene set enrichment analysis (36) highlighted significant differences in hallmarks of cancer (Q < 0.01; Fig. 2A). For instance, iHCC3 displayed up-regulation of biological processes associated with E2F targets, mTOR, MYC, inflammatory response, mitosis, G2M checkpoint, and DNA repair compared with iHCC1/iHCC2. iHCC2 also showed activation of the WNT/β-catenin pathway. We also found that biological processes associated with mitosis and the cell cycle are down-regulated in iHCC2 compared with iHCC1/iHCC3 and inflammation is up-regulated in iHCC1/iHCC3.
Among the differentially expressed genes between the low- and high-survival iHCC3 and iHCC1 groups, we identified several prognostic markers (SI Appendix, Fig. S5 and Dataset S3). For instance, compared with iHCC3, tumors from iHCC1 displayed up-regulated expression of 64 favorable prognostic markers and down-regulated expression of 45 unfavorable prognostic markers (Q < 0.05; SI Appendix, Fig. S5), among the 469 metabolic genes previously identified as prognostic markers in liver cancer (15). In turn, iHCC2 showed mixed up- and down-regulation of these prognostic markers. iHCC3 tumors additionally presented down-regulated expression of 123 (out of 157) liver-specific genes (Q < 0.05; Dataset S3), up-regulation of genes associated with previously identified immune signatures (37), and metastasization such as HIF1α, IL1, TNFα, NFκB, and TGFβ (Dataset S3).
We also observed that survival differences of the three groups are consistent with expression of prognostic markers, where iHCC1 presents the highest survival rate, followed by iHCC2, and iHCC3 (log-rank P < 0.001; Fig. 2B). Though differences are observed between the three groups, iHCC3 tumors are markedly distinct from those of iHCC2 and iHCC1. We found that a larger number of genes are differentially expressed between iHCC3 and iHCC1/iHCC2 compared with iHCC1 versus iHCC2, and several cancer hallmarks are simultaneously enriched in iHCC3 in comparison with either iHCC1 or iHCC2 (Fig. 2A). For instance, iHCC3 tumors presented down-regulation of oxidative phosphorylation, fatty acid metabolism, and adipogenesis, and up-regulation of DNA repair, G2M checkpoint, epithelial-to-mesenchymal transition, and inflammation compared with iHCC1 or iHCC2 (Dataset S3).
We also performed gene set enrichment analysis using Piano (36) and biological processes terms retrieved from the Molecular Signatures Database (MSigDB) to reveal iHCC subtype-specific responses (Dataset S4). For instance, iHCC1 displayed up-regulated tryptophan and indole metabolism and down-regulated noncoding RNA metabolism and ribosome biogenesis (Q < 0.05) compared with tumors of iHCC2 and iHCC3. Tumors of iHCC2 displayed (Q < 0.05) up-regulated heme metabolism, glutamine metabolism, drug metabolism and transport, and oxidative demethylation, but down-regulated cell development and G protein-coupled receptor signaling, compared with iHCC3 and iHCC1. Tumors of iHCC3 displayed the largest changes in biological processes compared with iHCC1 or iHCC2, with up-regulation of multiple processes associated with cell proliferation, cell-cycle progression and mitosis, development, chromosome segregation, cytoskeleton organization, immune response, DNA replication, and recombination (Q < 0.05). In turn, iHCC3 displayed down-regulated fatty acid β-oxidation, lipid oxidation, small-molecule biosynthesis and catabolism, metabolism of several amino acids including glycine, glutamate, glutamine, serine, and aspartate, drug catabolism, and response to xenobiotic stimulus (Dataset S4). Consistent with the substantial differences between iHCC3 and the two other tumor groups, iHCC2 tumors displayed similar metabolic behavior to those of iHCC1 (Dataset S5), and their gene expression is more similar to those of iHCC1 than to those of iHCC3 (Fig. 2B; mean Spearman’s ρ ∼ 0.9 iHCC1 vs. iHCC2, <0.8 iHCC3 vs. iHCC1 or iHCC2). Comparison of personalized fGGNs in each subtype further supported the results of our analysis (SI Appendix, Fig. S6).
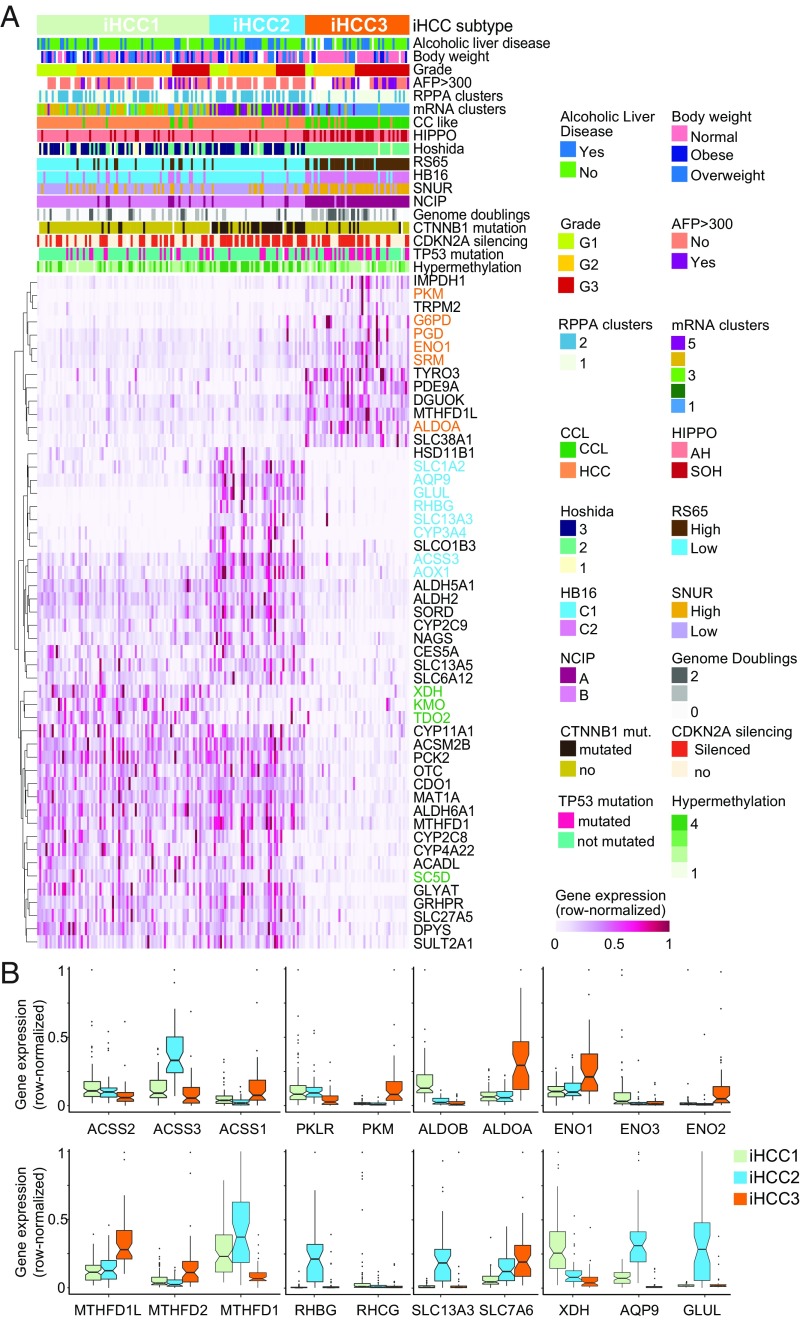
Importantly, our stratification method highlighted several “stratifying” genes whose expression is substantially different between the three iHCC groups. This is the case of XDH, KMO, TDO2, and SC5D in iHCC1; GLUL, AQP9, RHBG, SLC1A2, SLC13A3, ACSS3, AOX1, and CYP3A4 in iHCC2; and PKM, G6PD, PGD, ENO1, SRM, and ALDOA in iHCC3 (Fig. 3 A and B and SI Appendix, Fig. S7). Other genes such as MTHFD1, ALDH6A1, and ACSM2B are similar in both iHCC1 and iHCC2 but differ significantly in comparison with iHCC3.
Fig. 3.
HCC tumors stratified based on metabolic network analysis. (A) We determined three novel HCC subtypes and show the stratifying genes in iHCC1 (green), iHCC2 (cyan), and iHCC3 (orange). (B) Expression of stratifying genes and enzymes catalyzing the same reactions in the three iHCC groups is shown.
The Association Between Metabolic, Wnt/β-Catenin, and PI3K/AKT/mTOR Signaling Pathways.
The above results indicated that the iHCC subgroups present specific features at the survival recurrence signature, gene expression, prognostic marker, and metabolic level identified solely based on network analysis. These tumors are also differentially associated with known HCC properties such as HIPPO signature, hypermethylation, DNA copy number, cholangiocarcinoma-like traits (6), RS65 gene-based risk scores (38), and HB16 signature (7) (Fig. 3A and Dataset S3). For instance, 84% of iHCC2 subjects are men (vs. ∼50% in other iHCCs), and about half of the patients in iHCC2 and iHCC3 displayed alcoholic liver disease, much higher than the <25% observed in iHCC1 (Q < 0.01). Additionally, iHCC2 tumors also showed lower genome doubling, higher hypermethylation, and CDKN2 silencing (Fig. 3A; Q < 10−4), and all iHCC2 tumors showed α-fetoprotein (AFP) <300 ng/mL. iHCC1 and iHCC2 tumors are associated (Q < 10−4, χ2 test) with markers of hepatocyte differentiation (>54% tumors display Hoshida 3) (8) and maturity (>79% HB16 C1). In turn, no iHCC3 tumors showed differentiation markers (0% Hoshida 3) and instead are associated with known markers of low survival (Q < 0.05, χ2 test; Fig. 3A and Dataset S3) including National Cancer Institute proliferation (NCIP) subtype score A (>96%), high recurrence risk Seoul National University recurrence (SNUR) subtype (>76%) (14), and high expression of recurrence risk marker CD24 (log fold change ∼2.55 for comparison vs. iHCC1, Q < 0.00085). The lower survival and predominance of aggressive tumors in iHCC3 are associated with the significantly (Q < 0.02, χ2 test) larger proportion of advanced tumors in this group [>51% grade (G) 3, <49% G1 and G2] compared with iHCC2 (30% G3, <70% G1 and G2) or iHCC1 (<22% G3, >77% G1 and G2). iHCC2 also showed altered cytochrome P450 and xenobiotic metabolism in comparison with the two other clusters (SI Appendix, Figs. S8 and S9).
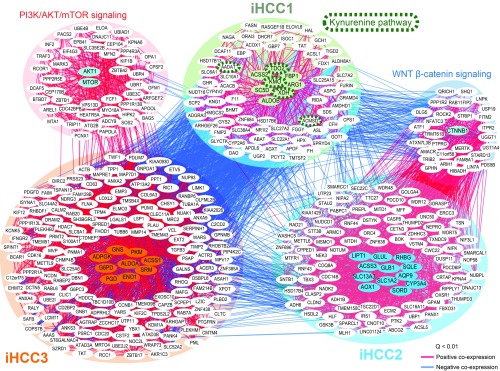
Interestingly, we had several observations associate altered Wnt/β-catenin and PI3K/AKT/mTOR signaling with the novel iHCC phenotypes described here. Most iHCC3 tumors were associated with MYC and AKT activation, as indicated by the high incidence of Hoshida 2 (in 96% of tumors; Fig. 3A). Additionally, we identified the top 25 genes coexpressed with stratifying/controlling genes in each iHCC for 360 TCGA tumors, and observed positive coexpression of AKT1 and MTOR and stratifying/controlling genes in iHCC3 and their coexpressed genes (Pearson’s r > 0.32, Q < 0.01; Fig. 4). AKT1 and MTOR were negatively coexpressed with stratifying/controlling genes in iHCC1 and iHCC2. In turn, Hoshida signatures were not substantially different between iHCC1 and iHCC2 (22 and 11% Hoshida 1, respectively, Q > 0.3, χ2 test).
Fig. 4.
Coexpression analysis highlights the association between stratifying and controlling genes in iHCC subtypes. Stratifying and controlling genes for iHCC1, iHCC2, and iHCC3 and their top 25 coexpressed genes are included. Coexpression between iHCCs was determined based on TCGA transcriptomics data of 369 HCC tumor samples. We additionally included AKT1 and MTOR, transcription factors involved in PI3K/AKT/mTOR signaling, and CTNNB1, which encodes for the transcription factor β-catenin in the Wnt signaling pathway. Edges indicate positive (red) or negative (blue) Pearson correlations (Q < 0.01).
The following five observations suggested a strong association between disturbed Wnt signaling and the iHCC2 phenotype. First, 75% of iHCC2 tumors showed mutations in CTNNB1, a gene that codes for β-catenin in the Wnt pathway (Fig. 3A), substantially higher than the <13% observed in iHCC1 and iHCC3 (Q < 10−5, χ2 test). Second, iHCC2 tumors also showed up-regulated expression of β-catenin target genes, for instance glutamine synthetase GLUL, glutamate transporter SLC1A2, and ornithine aminotransferase (SI Appendix, Fig. S7). Third, coexpression analysis indicated that stratifying/controlling genes in iHCC2 and their coexpressed genes are positively coexpressed with CTNNB1 (Pearson’s r > 0.32, Q < 0.01; Fig. 4). This is not observed in the case of iHCC3/iHCC1 genes, which are negatively coexpressed with AKT1 or MTOR (Pearson’s r < −0.2, Q < 0.01). Fourth, the association between Wnt signaling in iHCC2 and AKT activation in iHCC3 was also identified using an independent dataset of 91 HCC microarray samples and associated immunohistochemistry (SI Appendix, Fig. S10). Associations between different HCC tumors and IFN, proliferation (PI3K/AKT activation), CTNNB1 phosphorylation/mutation (i.e., Wnt signaling), or chromosome 7 polysomy were previously identified (10). Using the authors’ previously defined classes (Gene Expression Omnibus accession no. GSE9843), we observe that all tumors with CTNNB1-phosphorylating activation and mutation showed high expression of iHCC2-stratifying genes. Additionally, tumors showing RPSA, AKT, or IGFR activation showed high expression of iHCC3-stratifying genes, thus reinforcing the relationship between PI3K/AKT/mTOR signaling activation and iHCC3. Tumor stratification based on iHCC-stratifying genes showed differential distribution in the HCC subgroups previously identified (10) (Dataset S6). Lastly, a transcriptomics dataset with four HCC samples displaying CTNNB1 mutation (27) showed high expression of many iHCC2-stratifying genes including GLUL, RHBG, SLC13A3, and ACSS3 (SI Appendix, Fig. S11). These observations thus indicated distinct genomic features for the iHCC2 and iHCC3 phenotypes, respectively associated with aberrant Wnt signaling and PI3K/AKT/mTOR signaling activation. Interestingly, three stratifying genes (TDO2, KMO, XDH) and two coexpressed genes (AADAT, ACMSO) are involved with the kynurenine pathway (KP) (Fig. 4), a metabolic pathway leading to NAD+ production and associated with tryptophan metabolism (39). iHCC1 also shows up-regulated tryptophan metabolism in comparison with the two other iHCC groups (Dataset S4).
Together with the above observations, the validations in three independent datasets at the genomic, transcriptomic, and proteomic level (10, 13, 26, 27, 40) additionally reinforced our confidence in the stratifying genes and survival differences in iHCC1, iHCC2, and iHCC3. Specifically, the metabolic network-derived antagonistic expression of stratifying genes identified in 186 HCC tumor transcriptomics data is consistently observed in (i) a validation transcriptomics dataset of 183 HCC tumors attained from TCGA (SI Appendix, Fig. S12A); (ii) a microarray dataset consisting of 221 HCC samples (SI Appendix, Fig. S12B); (iii) coexpression analysis of 369 HCC tumors from TCGA (Fig. 4); (iv) a microarray dataset comprising 91 HCC tumors (SI Appendix, Fig. S10); and (v) a comparison of CTNNB1 mutant versus a noncancerous transcriptomics set (SI Appendix, Fig. S11). Additionally, survival analysis performed on the validation TCGA dataset or an additional dataset (13, 26) (SI Appendix, Fig. S12) was consistent with the observed survival differences in iHCC1 > iHCC2 > iHCC3 (Fig. 2B).
Alternative Metabolic Differences Between HCC Subtypes.
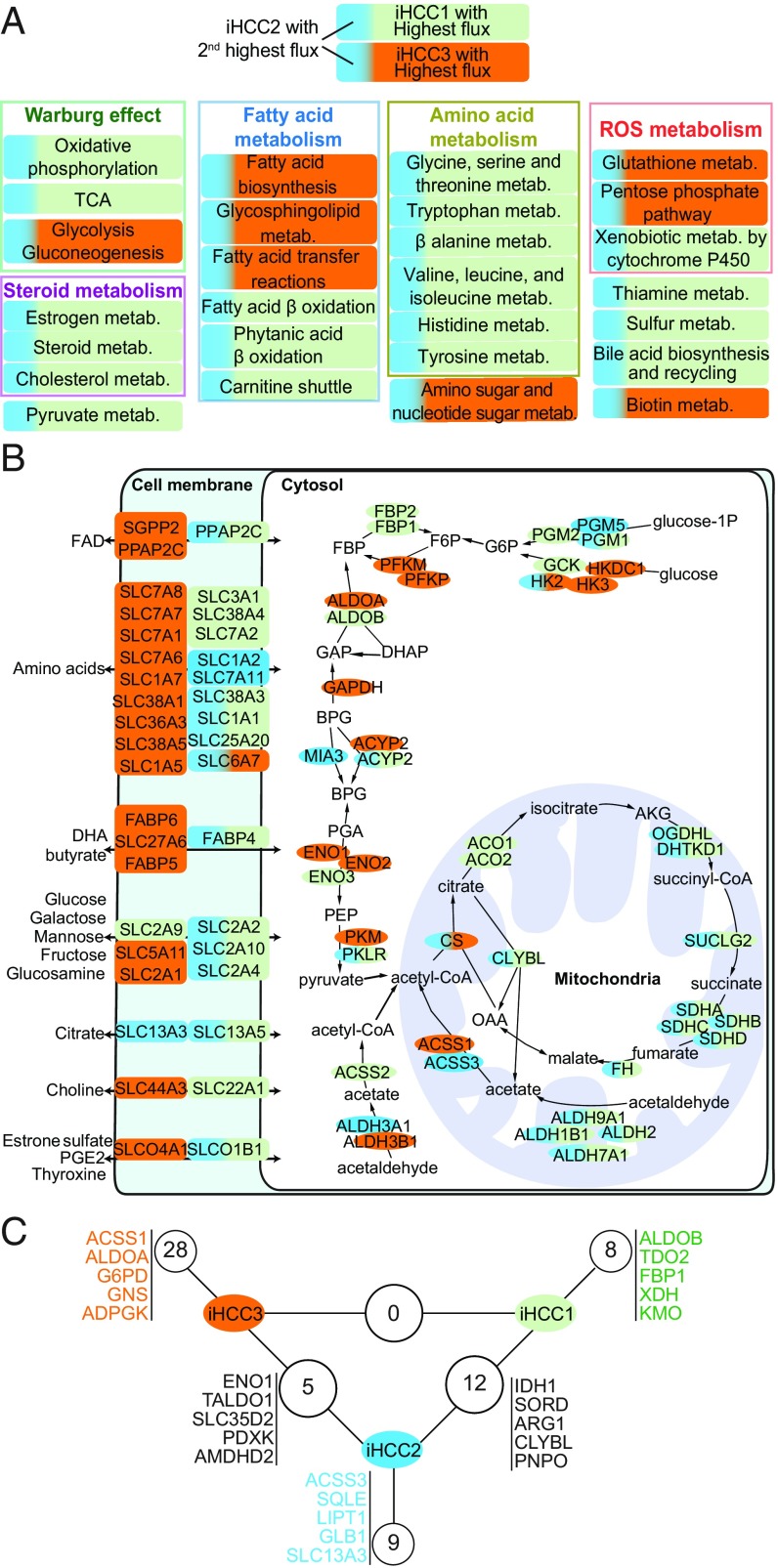
We further identified metabolic differences between iHCC1, iHCC2, and iHCC3 at a pathway- and reaction-centered level using GEMs. GEMs were generated for each cluster through MADE (41) and TIGER (42), using as input the differentially expressed genes and considering maximization of biomass as an objective function. We found that fluxes in each of the models (Fig. 5A) were consistent with the hallmarks of cancer identified above (Fig. 2A) and with the expression data mapped onto Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways (Dataset S3), as well as with the substantial metabolic differences between iHCC3 and iHCC1 or iHCC2. Specifically, iHCC3 GEMs showed lower fluxes in metabolism of amino acids, cofactors and coenzymes, pyruvate, fatty acid oxidation, carnitine shuttle, steroids, and oxidation phosphorylation compared with iHCC1/iHCC2, and lower in iHCC2 than iHCC1. Compared with iHCC1/iHCC2, iHCC3 showed higher glycolytic but lower citric acid cycle (TCA) fluxes consistent with a strong Warburg effect, as well as higher fluxes of fatty acid biosynthesis.
Fig. 5.
iHCC subgroups rely on alternative enzymes catalyzing the same reactions and display specific synthetic lethal genes. (A) Flux balance analysis performed on iHCC-specific models shows that iHCC1 or iHCC3 displays the highest reaction fluxes, followed by iHCC2. The predominant color in each box shows the iHCC subtype that displays the highest flux. (B) Metabolic genes involved in transport, glycolysis, and the TCA colored according to expression in each subgroup. (C) Numbers of synthetic lethal genes found in iHCC subgroups are shown, highlighting five synthetic lethal genes per subgroup. No synthetic lethal genes are simultaneously identified in iHCC1 and iHCC3, but several are found between iHCC2 and the other subgroups.
Additionally, differences in the gene expression indicated that the three iHCC groups rely on alternative enzymes for catalyzing the same reactions (Fig. 3B and 5B). For instance, acetate is converted to acetyl-CoA by acetyl-CoA synthases; the reaction is catalyzed by ACSS1, ACSS2, and ACSS3. iHCC1 expresses ACSS2, localized in the cytoplasm, whereas iHCC2 and iHCC3 express ACSS3 and ACSS1, respectively, which are localized in the mitochondria (SI Appendix, Fig. S13). Cleavage of fructose-1,6-bisphosphate is catalyzed by aldolase and the reaction is catalyzed by ALDOA, ALDOB, and ALDOC. While iHCC1 highly expresses the liver-specific ALDOB, iHCC3 highly expresses the nonspecific ALDOA, and iHCC2 shows similar expression for both genes. Alternative utilization of pyruvate is also observed. iHCC1/iHCC2 show high expression of the liver-specific PKLR, whereas iHCC3 shows high expression of PKM. PKLR has recently been identified as a liver-specific target for effective treatment of fatty liver disease and HCC (43). Our analysis enabled the identification of the right patient population where PKLR or PKM inhibitors can be used for effective treatment of patients.
Glucokinase in iHCC1 is switched to hexokinase 2 in iHCC3 whereas ENO3 is substituted for ENO1. Additionally, PFKFB4, HSD17B6, and GLYATL2 in iHCC3 are switched to PFKFB1, HSD17B1, and GLYAT in iHCC1 and iHCC2, respectively. We also had similar observations of expression of genes that encode for aldehyde dehydrogenases (e.g., ALDH1B1, ALDH9A1, ALDH2, ALDH3A2, ALDH3B1), among others (SI Appendix, Fig. S13). A number of amino acids, sugars, cofactors, and hormone transporters are also differentially expressed between the three clusters, and in particular between iHCC3 and iHCC1/iHCC2 (Fig. 5B). These observations translated into distinct central metabolism, particularly between the high- and low-survival groups including iHCC1 and iHCC3, while iHCC2 shared many of these properties with iHCC1. Several membrane transporters including amino acid, glucose and monosaccharide, choline, butyrate, and citrate transporters also showed substantial switching between iHCC1 and iHCC3.
Controlling genes are also differentially expressed between the three HCC subtypes (Dataset S6), indicating different controllability metabolic behavior between them. For instance, in fatty acid elongation, ELOVL6 is a controlling gene in iHCC2 but ELOVL5 is a controlling gene in iHCC1 and iHCC3. In glycerolipid metabolism, PLA2G12B is a controlling gene in iHCC1, PLA2G16 is a controlling gene in iHCC3, and both are controlling genes in iHCC2. In purine and pyrimidine metabolism, NME6 and NT5E are controlling genes in iHCC3 but not in iHCC1/iHCC2, which showed another NME as a controlling gene. In glycolysis, PKLR and BPGM are controlling genes in iHCC1/iHCC2, but PKM and PGAM1 are controlling genes in iHCC3. In histidine metabolism, NAA15 and SLC40A1 are controlling genes in iHCC1 and SLC11A2 is a controlling gene in iHCC2, whereas NAA30 and SLC40A1 are controlling genes in iHCC3.
We showed the importance of controlling genes. Next, we performed synthetic lethality analysis for those stratifying and controlling genes that were found just in HCC networks. This enabled the identification of several potential therapeutic targets. Among controlling and stratifying genes, we find 8, 9, and 28 subtype-specific genes in iHCC1, iHCC2, and iHCC3, respectively, whose in silico knockout leads to lethality in their respective subtype but not in the others (Fig. 5C). Among these, we identified ALDOB, TDO2, and KMO in iHCC1, ACSS3, SQLE, and LIPT1 in iHCC2, and ACSS1, ALDOA, and G6PD in iHCC3. Knockout of 12 controlling/stratifying genes simultaneously leads to lethality in iHCC1 and iHCC2 and includes IDH1, SORD, and ARG1. Several of these synthetic lethal genes are known targets of DrugBank drugs, including some Food and Drug Administration (FDA)-approved drugs (Dataset S7). These observations pointed toward several HCC-specific potential therapeutic targets that may be used to target low- (iHCC3), intermediate- (iHCC2), and high- (iHCC1) survival groups. We identified FDA-approved drugs and kinase inhibitors, among other drugs, whose known targets include these genes (Dataset S7).
Poor Survival-Associated Genes Showed Hypoxic Behavior.
Several stratifying genes in the three iHCC groups are associated with redox metabolism (Fig. 3 and SI Appendix, Fig. S10) such as G6PD, PKM, and ALDOA in iHCC3, or ALDH2 and MTHFD1 in iHCC1 and iHCC2. Gene expression differences translated into altered redox metabolism (Fig. 2A) and antioxidant defenses (e.g., catalase or glutathione-based H2O2 scavenging; Dataset S5). Additionally, we have previously observed stratification of HCC samples based on acetate metabolism and hypoxia (11). Indeed, the expression of HIF1A is substantially higher in iHCC3 than in iHCC1 or iHCC2 (log2 fold change ∼ 1, Q < 0.05).
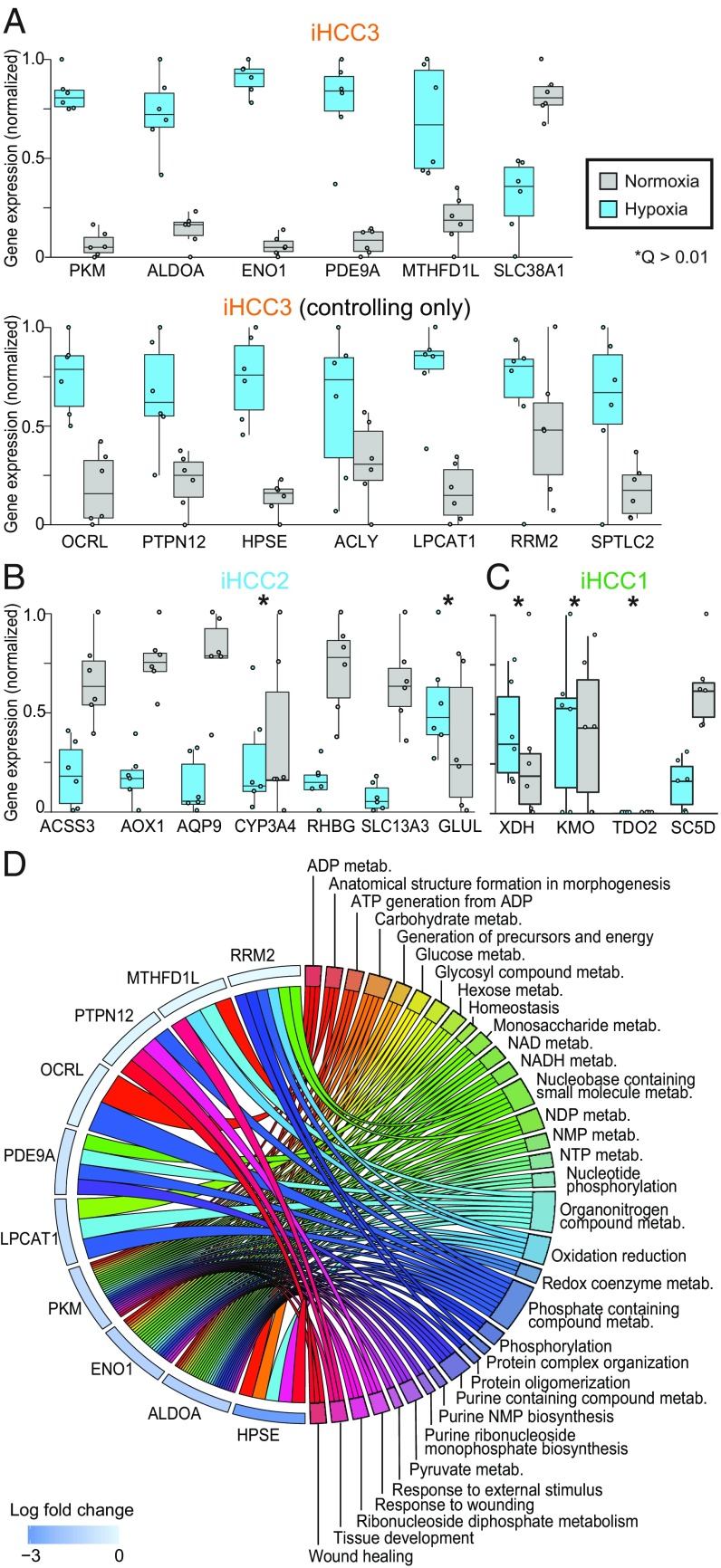
We performed a transcriptomics analysis of HepG2 cells grown under hypoxia and normoxia (SI Appendix, Fig. S14). We found (SI Appendix, Fig. S14C; Q < 0.01) that differentially expressed genes are associated with responses to stress and oxygen, NADH, ADP, and RNA metabolism. We found that immune system and tissue development biological processes are up-regulated under hypoxia. In turn, DNA metabolism, replication and repair, and cell cycle-related processes are up-regulated under normoxia. Further, among the differentially expressed genes, the expression of the stratifying genes PKM, ALDOA, MTHFD1L, ENO1, and PDE9A and controlling genes in iHCC3 is significantly higher under hypoxic than normoxic conditions (Q < 0.01; Fig. 6A). Interestingly, stratifying and controlling genes in iHCC2 or iHCC1 are either unchanged or show down-regulated expression under hypoxia (Fig. 6 B and C, respectively). Additionally, among the 28 controlling genes exclusive to iHCC3 (Fig. 5C), we find that the expression of OCRL, PTPN12, HPSE, ACLY, LPCAT1, RRM2, and SPTLC2 is up-regulated under hypoxia (Fig. 6A). Among those stratifying/controlling genes up-regulated under hypoxia in iHCC3, we found multiple up-regulated biological processes that also involve these genes, including those involved in energetic, carbohydrate, and nucleotide metabolism and tissue development (Fig. 6D). These observations indicate that stratifying and controlling genes in iHCC3 and iHCC1/iHCC2 tumors respond strongly and antagonistically to hypoxic behavior.
Fig. 6.
Stratifying and controlling genes in iHCC3 show specific responses to hypoxia. HepG2 cell lines were grown under normoxic or hypoxic conditions (n = 6 per condition) and transcriptomics data were generated. Expression of stratifying and controlling genes in iHCC3 (A), iHCC2 (B), and iHCC1 (C), and gene association with enriched biological processes (D; Q < 0.05). All genes with the exception of CYP3A4, GLUL, XDH, KMO, and TDO2 (Q > 0.01) are differentially expressed between hypoxia and normoxia. NMP, NDP, and NTP indicate nucleoside mono, di-, and triphosphate, respectively.
Discussion
Given the high heterogeneity in HCC, previous studies stratified HCC patients through unsupervised clustering of tumors based on genomics and transcriptomics data (6, 7, 13). This led to the valuable discovery of many patient group-specific differences such as cholangiocarcinoma-like feature traits (6), hepatic stem-like phenotypes (7), signaling differences (8–10), recurrence risk (14, 38), and metabolism (11). In our study, we introduced an fGGN-based characterization and stratification of HCC patients. We combined objective-dependent and -independent approaches to perform a functional metabolic network-based stratification of hundreds of HCC patients. Our analyses combined genomics, transcriptomics, proteomics, and clinical data across four datasets with hundreds of HCC tumors, in silico analysis including genome-scale metabolic modeling, gene silencing, coexpression and network analysis, and additional cell-line experiments. We identified distinct differences in metabolic and signaling pathways and in clinical survival between three major HCC subtypes, iHCC1 to iHCC3. The three iHCCs presented 18 metabolic genes highly expressed by one group but not the others, namely stratifying genes. Our analyses showed that these genes can be consistently used for stratifying HCC tumors in independent cohorts. We have additionally identified 32 controlling genes, those that display pivotal roles in controlling networks and whose targeting would lead to lethality in one of the HCC subtypes.
Tumors in the low-survival and progressive (higher-grade) iHCC3 group showed several signatures of low survival and high recurrence (13, 14) and high expression of markers of poor survival (15). In turn, the high- and moderate-survival groups iHCC1 and iHCC2 were associated with markers of low recurrence, high survival, hepatocyte differentiation, and maturity (8, 14, 38). iHCC1, iHCC2, and iHCC3, respectively, displayed high expression of genes involved in acetate metabolism, ACSS2, ACSS3, and ACSS1. These observations are consistent with our previous analyses which showed that HCC tumors can be stratified based on acetate metabolism (11). ACSS2 is highly expressed by healthy liver tissue (11), consistent with the high expression displayed by the high-survival iHCC1 group. On the other hand, ACSS1 is highly expressed in iHCC3, consistent with previous observations in a low-survival group (11). Finally, we identified that iHCC2 displays high expression of ACSS3, unlike other iHCCs.
In turn, iHCC3 tumors showed high expression of ACSS1 and are associated with hypoxic environments. Experiments with HepG2 cells additionally showed strong and opposing responses to hypoxia by different iHCCs. Stratifying and controlling genes in iHCC3 are up-regulated under hypoxia compared with normoxia. This is opposed by those in iHCC1 and iHCC2, which are either unchanged or show decreased expression under hypoxic conditions. These observations indicated opposing hypoxic responses under low and high survival.
iHCC1 is the tumor group with the highest survival, and showed a high inflammation response compared with iHCC2. Interestingly, several stratifying genes or their coexpressed genes are involved in the KP. This pathway is found upstream of NAD biosynthesis, and is the main tryptophan sink in the cell (39). Several of its genes are up-regulated in adipose tissue in obesity (44, 45), consistent with the observation that 56% of patients in iHCC1 are obese or overweight. Several metabolites of the KP have been associated with inflammatory and immune responses, for instance in the induction of cytokines and macrophage-induced chemokines (46). Interestingly, KP overactivation has been observed in type 2 diabetes (T2D) and is one of the T2D-driving mechanisms observed in prediabetic patients (47). The observation that T2D is one of the risk factors for HCC (47) raises the possibility that the iHCC1 phenotype is associated with T2D, unlike other iHCCs. This is reinforced by the observation that the genes TDO, KMO, AADAT and ACMSD, and IL6R, stratifying genes in iHCC1 or their coexpressed genes, are up-regulated in obesity or T2D (47). Finally, both T2D and obesity showed activated fatty acid oxidation, similar to iHCC1, which displayed the highest fatty acid oxidation of the three iHCCs, suggesting a potential association between those diseases and iHCC1 tumors. Overall, iHCC1 showed the highest fluxes in metabolism of amino acids, cofactors and coenzymes, pyruvate, fatty acid oxidation, carnitine shuttle, steroids, TCA, and oxidative phosphorylation.
iHCC2 showed higher similarity to iHCC1 compared with iHCC3, but also exhibited specific features including lower fatty acid biosynthesis and high glutamine metabolism, and β-catenin–associated up-regulated fatty acid oxidation. One of the main features of iHCC2 tumors is the association with β-catenin pathway alterations. CTNNB1 encodes for β-catenin, which is mutated in ∼20% of HCCs (48) (SI Appendix, Fig. S15). Mutations in this gene are associated with increased concentration of nuclear β-catenin and its target genes (e.g., glutamine synthetase GLUL and glutamate transporter SLC1A2) and lower patient survival (49, 50). Glutamine synthetase is involved in ammonia detoxification, and β-catenin–controlled induction of GLUL leads to autophagy in HCC (51). β-Catenin controls mitochondrial homeostasis by regulating the TCA and fatty acid oxidation, and protects against alcohol-induced liver injury or ethanol-induced metabolic stress (SI Appendix, Fig. S16) (52). This is consistent with the overactivation of the pathways involved in detoxification, that is, drug and xenobiotic metabolism in comparison with other subtypes. β-Catenin also regulates the expression of acetaldehyde dehydrogenases (e.g., ALDH2, ALDH3A1, and ALDH3A2) (52), thus controlling TCA fluxes, as well as the stratifying gene acyl-CoA oxidase (AOX1), which is involved in fatty acid β-oxidation (53). Our modeling analyses additionally indicated that iHCC2 showed low fatty acid biosynthesis fluxes, consistent with the negative regulation of this process by β-catenin. Sorafenib, a drug that targets expression of liver-related Wnt-target GLUL and leads to higher sensitization in HCC tumors with high β-catenin activation (54), thus presents as a potential drug in iHCC2 but not in the other tumor groups.
Finally, iHCC3 tumors were associated with multiple features of malignant tumors, including hypoxic behavior, epithelial-to-mesenchymal transition, higher fluxes in fatty acid biosynthesis, and a strong Warburg effect. For instance, TGF-β, HIF-α, and NF-κB genes associated with hypoxic response, metastasis, and malignancy are up-regulated in iHCC3, and iHCC3 shares several signature activities of metastatic tissues (55). One of the main features of this tumor group is the association with PI3K/AKT/mTOR signaling activation. It also showed downstream activation of asparagine synthetase (ASNS), glycolysis, and the pentose phosphate pathway by PI3K/AKT/mTOR signaling (56), consistent with our observation in iHCC3 (SI Appendix, Fig. S16). Up-regulation of ASNS in iHCC3 is significantly correlated with metastatic potential, and overexpression of ASNS promotes metastatic progression (57). Drugs targeting PI3K/AKT/mTOR signaling or these processes, such as l-asparaginase, rapamycin, or their analogs, thus arise as potential therapeutics for the treatment of iHCC3 but not the other iHCCs.
Overall, these observations highlighted distinct differences in metabolic and signaling pathways in HCC tumors that stem from the high intertumor heterogeneity and were associated with patient survival. In silico predictions enabled identification of several HCC subtype-specific potential therapeutic gene targets that offer full control over the metabolic network. Revealing the mechanistic differences between HCC subtypes, together with the identification of HCC subtype-specific drug targets, may foster the development of efficient treatment strategies and precision medicine for HCC patients.
Materials and Methods
All of the materials and methods in this study are detailed in SI Appendix, Materials and Methods, including gene expression retrieval, processing, and validation datasets; generation of personalized and subtype GEMs; construction of personalized and generic directed functional gene–gene networks; centrality and controllability of personalized fGGNs; identification of controlling genes through in silico gene silencing analysis; network-based stratification of fGGNs; generation of fGGNs and tumor stratification based on Recon3D; differential expression and gene set enrichment analysis; KEGG pathway analysis; coexpression analysis; identification of potential drugs targeting synthetic lethal genes; validation; hypoxia experiments in HepG2 cells; and statistics.
Supplementary Material
Acknowledgments
This work was financially supported by the Knut and Alice Wallenberg Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807305115/-/DCSupplemental.
References
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Mardinoglu A, Boren J, Smith U, Uhlen M, Nielsen J. Systems biology in hepatology: Approaches and applications. Nat Rev Gastroenterol Hepatol. 2018;15:365–377. doi: 10.1038/s41575-018-0007-8. [DOI] [PubMed] [Google Scholar]
- 3.O’Day E, et al. Are we there yet? How and when specific biotechnologies will improve human health. Biotechnol J. 2018:e1800195. doi: 10.1002/biot.201800195. [DOI] [PubMed] [Google Scholar]
- 4.Najafi A, Bidkhori G, Bozorgmehr JH, Koch I, Masoudi-Nejad A. Genome scale modeling in systems biology: Algorithms and resources. Curr Genomics. 2014;15:130–159. doi: 10.2174/1389202915666140319002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benfeitas R, Uhlen M, Nielsen J, Mardinoglu A. New challenges to study heterogeneity in cancer redox metabolism. Front Cell Dev Biol. 2017;5:65. doi: 10.3389/fcell.2017.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo HG, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–3041. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairo S, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sohn BH, et al. Inactivation of Hippo pathway is significantly associated with poor prognosis in hepatocellular carcinoma. Clin Cancer Res. 2016;22:1256–1264. doi: 10.1158/1078-0432.CCR-15-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang DY, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Björnson E, et al. Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 2015;13:2014–2026. doi: 10.1016/j.celrep.2015.10.045. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JS, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 14.Woo HG, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 15.Uhlen M, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 16.Agren R, et al. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10:721. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bidkhori G, et al. Metabolic network-based identification and prioritization of anticancer targets based on expression data in hepatocellular carcinoma. Front Physiol. 2018;9:916. doi: 10.3389/fphys.2018.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv W, et al. The drug target genes show higher evolutionary conservation than non-target genes. Oncotarget. 2016;7:4961–4971. doi: 10.18632/oncotarget.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guney E, Menche J, Vidal M, Barábasi AL. Network-based in silico drug efficacy screening. Nat Commun. 2016;7:10331. doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barabási AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinayagam A, et al. Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc Natl Acad Sci USA. 2016;113:4976–4981. doi: 10.1073/pnas.1603992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Z, Zhao C, Di Z, Wang WX, Lai YC. Exact controllability of complex networks. Nat Commun. 2013;4:2447. doi: 10.1038/ncomms3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JS, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 27.Ding X, et al. Transcriptomic characterization of hepatocellular carcinoma with CTNNB1 mutation. PLoS One. 2014;9:e95307. doi: 10.1371/journal.pone.0095307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barabási AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 30.Junker BH, Schreiber F. Analysis of Biological Networks. John Wiley & Sons; Hoboken, NJ: 2011. [Google Scholar]
- 31.Liu YY, Slotine JJ, Barabási AL. Controllability of complex networks. Nature. 2011;473:167–173. doi: 10.1038/nature10011. [DOI] [PubMed] [Google Scholar]
- 32.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunk E, et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36:272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mardinoglu A, et al. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol Syst Biol. 2013;9:649. doi: 10.1038/msb.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mardinoglu A, et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:3083. doi: 10.1038/ncomms4083. [DOI] [PubMed] [Google Scholar]
- 36.Väremo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SM, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badawy AA. Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int J Tryptophan Res. 2017;10:1178646917691938. doi: 10.1177/1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinstein JN, et al. Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen PA, Papin JA. Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics. 2011;27:541–547. doi: 10.1093/bioinformatics/btq702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen PA, Lutz KA, Papin JA. TIGER: Toolbox for integrating genome-scale metabolic models, expression data, and transcriptional regulatory networks. BMC Syst Biol. 2011;5:147. doi: 10.1186/1752-0509-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, et al. Network analyses identify liver-specific targets for treating liver diseases. Mol Syst Biol. 2017;13:938. doi: 10.15252/msb.20177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Favennec M, et al. The kynurenine pathway is activated in human obesity and shifted toward kynurenine monooxygenase activation. Obesity (Silver Spring) 2015;23:2066–2074. doi: 10.1002/oby.21199. [DOI] [PubMed] [Google Scholar]
- 45.Moriya C, Satoh H. Teneligliptin decreases uric acid levels by reducing xanthine dehydrogenase expression in white adipose tissue of male Wistar rats. J Diabetes Res. 2016;2016:3201534. doi: 10.1155/2016/3201534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 47.Oxenkrug GF. Increased plasma levels of xanthurenic and kynurenic acids in type 2 diabetes. Mol Neurobiol. 2015;52:805–810. doi: 10.1007/s12035-015-9232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forbes SA, et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YD, et al. Genetic alterations of Wnt signaling pathway-associated genes in hepatocellular carcinoma. J Gastroenterol Hepatol. 2008;23:110–118. doi: 10.1111/j.1440-1746.2007.05250.x. [DOI] [PubMed] [Google Scholar]
- 50.Zucman-Rossi J, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 51.Sohn BH, Park IY, Shin JH, Yim SY, Lee JS. Glutamine synthetase mediates sorafenib sensitivity in β-catenin-active hepatocellular carcinoma cells. Exp Mol Med. 2018;50:e421. doi: 10.1038/emm.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu S, et al. β-Catenin is essential for ethanol metabolism and protection against alcohol-mediated liver steatosis in mice. Hepatology. 2012;55:931–940. doi: 10.1002/hep.24766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehwald N, et al. β-Catenin regulates hepatic mitochondrial function and energy balance in mice. Gastroenterology. 2012;143:754–764. doi: 10.1053/j.gastro.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 54.Lachenmayer A, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res. 2012;18:4997–5007. doi: 10.1158/1078-0432.CCR-11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knott SRV, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;554:378–381. doi: 10.1038/nature25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.