Commuting patterns influence housing and job choices
Beijing subway map. Image courtesy of iStock/LordRunar.
Few longitudinal studies have examined links between job and housing dynamics and commuting patterns of urban residents. Jie Huang et al. (pp. 12710–12715) conducted a 7-year study that tracked transit smartcards of 4,248 commuters in Beijing, China. Each cardholder commuted by subway at least four days each week. The authors found that individuals whose commutes to the workplace were longer than 45 minutes changed residences to reduce travel times. Conversely, individuals with commutes shorter than 45 minutes increased travel times for better jobs and housing. The authors also examined the commuting patterns, residential profiles, and socioeconomic statuses of four groups of individuals. Stayers, individuals who remained at the same residence and workplace over the study period, were more likely than other groups to have shorter commutes and to be homeowners with middle- to high-range incomes. Home movers, individuals who changed residences but remained at the same workplace, had middle-range incomes and upgraded from tenancy to ownership, which lengthened commutes. Switchers, individuals who changed both residences and workplaces, tended to move into more expensive homes, which increased commute times. Job hoppers, individuals who frequently changed workplaces but stayed at the same residence, had long commutes and affordable housing and tended to be migrants with temporary employment and low wages. According to the authors, travel times can influence job and housing choices. — M.S.
Genes underlying cyanobacterial growth
Cyanobacteria can be used to produce biofuels and other chemical feedstocks, but cyanobacterial production systems have not been deployed yet because of the low growth and photosynthetic rates of commonly used model organisms. To address this challenge, Justin Ungerer et al. (pp. E11761–E11770) examined genetic differences between the model cyanobacterium Synechococcus elongatus PCC 7942 and the relative Synechococcus 2973, which is almost genetically identical but has twice the photosynthetic rate and produces biomass three times more rapidly. The authors performed a comprehensive mutational analysis of Synechococcus 2973 using the genome-editing tool CRISPR/Cpf1. The analysis revealed that five single nucleotide polymorphisms (SNPs)—variations in single building blocks of DNA—in three genes, atpA, ppnK, and rpaA, are responsible for the rapid growth and photosynthetic rates of this strain. Whereas atpA and ppnK are known to be involved in energy metabolism, rpaA is regarded as the master output regulator of the circadian clock. When the authors used CRISPR/Cpf1 to introduce the five SNPs into Synechococcus 7942, the photosynthetic rate of this strain doubled and the growth rate more than tripled, becoming comparable to that of Synechococcus 2973. According to the authors, uncovering the molecular basis of rapid growth in cyanobacteria could yield biotechnological gains. — J.W.
Primate colonization of the Caribbean
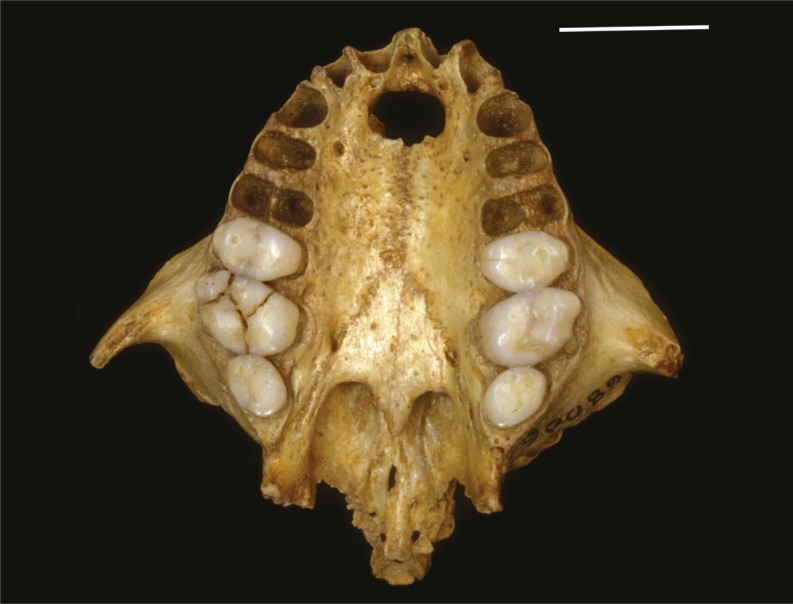
Xenothrix full palate.
Researchers have unearthed fossil evidence of four extinct primate species in the Caribbean, but the history of the primates’ arrival and diversification throughout the region is unclear. Roseina Woods et al. (pp. 12769–12774) extracted ancient DNA under adverse preservation conditions from Xenothrix mcgregori, a monkey species found in Jamaica. Previously thought to possibly lie outside extant groups of New World monkeys, this species is instead a member of the Callicebinae, or titi monkeys. Despite its morphological dissimilarity to other members of this subfamily, which themselves exhibit close similarity to each other, Xenothrix is a sister genus to Cheracebus monkeys and thus lies deep within the modern titi monkey radiation. The authors suggest that the divergence in the Xenothrix body plan may have occurred after colonization of the Caribbean islands, and that the species may have arrived to find multiple vacant niches ready for adaptive colonization. Because of the likely timing of divergence between Xenothrix and Cheracebus, around 11 million years ago, the authors suggest that Xenothrix may have traveled over water to reach Jamaica and represents one of at least two primate colonizations of the Caribbean islands. According to the authors, the results suggest that adaptation can significantly shape species morphology in novel environments, even when species come from morphologically conservative families. — P.G.
Measuring the basic reproduction number in real-world scenarios
The basic reproduction number, a fundamental concept in epidemiology, is defined as the average number of secondary cases generated by a typical infectious individual in a fully susceptible population. Quan-Hui Liu et al. (pp. 12680–12685) set out to assess the measurability of the basic reproduction number in real-world epidemic scenarios. Using detailed sociodemographic data, the authors generated two multiplex networks describing the contact patterns of a subset of Italian and Dutch populations. When the authors simulated the progression of a simple influenza transmission model on these networks, they found that the clustering of contacts typical of real-world human populations led to considerable variability of the basic reproduction number. The authors tested the robustness of the results by modeling the 2009 influenza pandemic in Italy and provided a plausible explanation for the patterns observed based on the intrinsic structure of human contact networks. The findings highlight the challenges of measuring the basic reproduction number in realistic populations. According to the authors, the basic reproduction number may not be an ideal descriptor of epidemic dynamics in populations featuring realistic contact networks. — S.R.
Bioluminescent tools from fungi

Neonothopanus gardneri growing on the base of the babassu palm tree in Brazil.
Bioluminescence, a natural process in which the enzyme luciferase oxidizes the compound luciferin to emit light, is widespread among microbes, fungi, and animals. Researchers have unraveled the biosynthetic pathway of bacterial luciferin and used engineered luciferases as molecular reporters. However, a complete pathway to synthesize luciferin and bestow bioluminescence on eukaryotes has not been engineered. Alexey Kotlobay, Karen Sarkisyan, Yuliana Mokrushina, Marina Marcet-Houben, Ekaterina Serebrovskaya, et al. (pp. 12728–12732) uncovered the set of enzymes that allows the bioluminescent fungus Neonothopanus nambi to emit light. The bioluminescent system includes a luciferase and three enzymes that enable biosynthesis of fungal luciferin from the metabolite caffeic acid. The authors report that insertion of the fungal luciferase gene along with two other genes into the yeast Pichia pastoris makes the yeast cells spontaneously emit green light. Further, the authors demonstrated that fungal luciferase can serve as a bioluminescent reporter in amphibian embryos and human cancer cells, among other cell types. According to the authors, the fungal bioluminescent system could help create spontaneously glowing plants and animals for use in biomedical imaging and landscape design, among other potential applications. — P.N.