Significance
We have developed a fluorescence resonance energy transfer (FRET) biosensor to visualize in single live cells the histone H3 Lys-9 trimethylation (H3K9me3) dynamics. Together with another FRET biosensor for simultaneous monitoring of the neighboring histone H3 Ser-10 phosphorylation (H3S10p) in the same cell, we found an anticorrelation between the dynamics of H3K9me3 and H3S10p during cell cycles, with H3S10p facilitating the decrease of H3K9me3 at onset of mitosis. This decrease of H3K9me3 is accompanied by dissolution of heterochromatin structures before chromatin condensation and nuclear envelope dissolution. The coordinated regulation of H3S10p and H3K9me3 may enhance access of remodeling complexes to the chromatin. Our results hence provide insights on how histone modifications and chromatin structures are coordinated to regulate mitosis.
Keywords: histone modifications, chromatin reorganization, FRET biosensors
Abstract
The dramatic reorganization of chromatin during mitosis is perhaps one of the most fundamental of all cell processes. It remains unclear how epigenetic histone modifications, despite their crucial roles in regulating chromatin architectures, are dynamically coordinated with chromatin reorganization in controlling this process. We have developed and characterized biosensors with high sensitivity and specificity based on fluorescence resonance energy transfer (FRET). These biosensors were incorporated into nucleosomes to visualize histone H3 Lys-9 trimethylation (H3K9me3) and histone H3 Ser-10 phosphorylation (H3S10p) simultaneously in the same live cell. We observed an anticorrelated coupling in time between H3K9me3 and H3S10p in a single live cell during mitosis. A transient increase of H3S10p during mitosis is accompanied by a decrease of H3K9me3 that recovers before the restoration of H3S10p upon mitotic exit. We further showed that H3S10p is causatively critical for the decrease of H3K9me3 and the consequent reduction of heterochromatin structure, leading to the subsequent global chromatin reorganization and nuclear envelope dissolution as a cell enters mitosis. These results suggest a tight coupling of H3S10p and H3K9me3 dynamics in the regulation of heterochromatin dissolution before a global chromatin reorganization during mitosis.
Posttranslational modifications of histone proteins have profound roles in the regulation of gene expressions (1). Dynamic histone modifications have also been implicated in the regulation of chromosomal architectures (2). In a cell cycle, the packing and unpacking of chromosomes correlate with dynamic histone modifications (3–5). For example, H3S10 phosphorylation can trigger the recruitment of histone deacetylase HST2 to deacetylate H4K16 and allow its interaction with H2A for chromatin condensation in Saccharomyces cerevisiae (6). However, it remains an enigma on how histone modulations are coordinated and how the dynamic histone modification governs the chromatin organization and cell cycle progression in live mammalian cells.
The trimethylation of histone H3 on lysine 9 (H3K9me3), catalyzed by evolutionarily conserved site-specific methyltransferases such as SUV39H1/2 and SETDB1/2 (7), is the hallmark of the heterochromatin (8, 9). Previous reports suggested that the level of H3K9me3 might increase in G2 and mitotic phases and decline at centromeres upon the completion of cell division (10, 11). Recent genome-wide analysis of H3K9me3 during cell cycle (12) and the observation of histone K9 demethylation KDM4C on mitotic chromatin point to an overall decrease of H3K9me3 in the nucleus during mitosis (13). This paradox in observations may be attributed to the epigenetic heterogeneity across different individual cells, as well as the lack of precise and sensitive detecting tools in monitoring the dynamic H3K9me3 signals at subcellular regions (14–16).
The complexity of H3K9me3 regulation during cell cycles is further increased by the potential of crosstalk between H3K9 and its neighboring residue H3S10, which is phosphorylated by Aurora kinase B in late G2/early prophase (17). Indeed, H3S10p has been shown to cause chromosome condensation and segregation during cell mitosis (18, 19). The biochemical results utilizing proteins purified from an average of cell populations suggest that H3S10p may cause the chromatic release of heterochromatin protein 1 (HP1) and its associated histone methyltransferases such as SUV39H1/2 (20–22). However, the interrelation between H3S10p and H3K9me3 in live cells is subjected to additional layers of regulation involving H3K14 acetylation and potentially other factors (23, 24).
To date, it remains unclear on how H3K9me3 and H3S10p are dynamically coordinated to regulate chromatin reorganization and cell cycle dynamics in a single live cell. Here we have engineered and characterized highly sensitive and specific biosensors based on fluorescence resonance energy transfer (FRET) to simultaneously visualize the dynamics of H3S10p and H3K9me3 during mitosis in single cells. We observed a dynamic, anticorrelated coupling between H3S10p and H3K9me3 during a cell cycle. We further demonstrated that this coupling between these two histone modifications constitutes a regulatory mechanism underlying the reduction of heterochromatin-like structures, possibly enhancing the accessibility and hence recruitment of chromatin remodelers, before the global reorganization and condensation of chromatin at the mitosis entrance.
Results
Engineering a FRET-Based Reporter (H3K9me3 Biosensor) with High Sensitivity and Specificity.
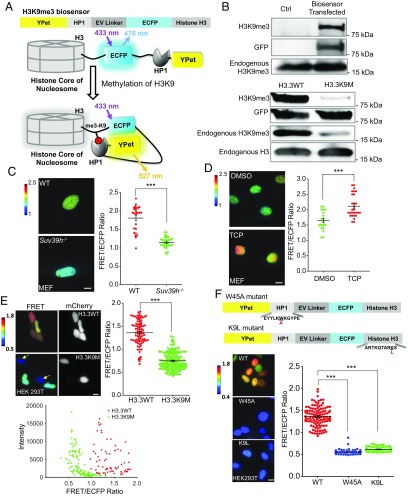
We first engineered a highly sensitive and specific FRET reporter (H3K9me3 biosensor) containing a full-length histone H3, a sensitive FRET pair that includes an enhanced cyan FP (ECFP) as a donor and a yellow FP variant (YPet) as an acceptor for energy transfer (25), a flexible EV linker, and a chromodomain of heterochromatin protein 1 (HP1) (residues 21–76 of HP1) (26, 27) (Fig. 1A). In the resting state, the H3K9me3 biosensor is expected to have an open conformation with low FRET signals. When H3K9 is trimethylated, HP1 can bind to H3K9me3, resulting in a strong FRET signal. We showed that trimethylation can occur on the exogenous H3K9me3 biosensor in the same manner as on endogenous H3K9 (Fig. 1B). The mutation Lys9Met (K9M) in histone H3.3 (H3.3K9M) can promote global loss of H3K9me3 through sequestration and inhibition of SAM-bound SET domain methyltransferase (28). Accordingly, we observed that H3.3K9M expression caused a significant reduction of trimethylation on both H3K9me3 biosensor and endogenous H3K9 (Fig. 1B). This result verifies that the trimethylation of H3K9me3 biosensor is regulated similarly as the endogenous H3K9.
Fig. 1.
Development and characterization of H3K9me3 biosensor. (A) A schematic drawing of the H3K9me3 biosensor and its activation mechanism. (B) Immunoblots of methylation on the H3K9me3 biosensor and endogenous H3, in control HEK 293T cells (Upper) and in these cells expressing WT or mutant (K9M) H3.3 (Lower). (C) FRET ratiometric images of H3K9me3 biosensor expressed in WT mouse embryonic fibroblasts (WT MEFs) and those lacking the methyltransferase Suv39h1 and Suv39h2 (Suv39h−/− MEFs) (Left). The scatterplot represents the FRET/ECFP ratios of the H3K9me3 biosensor in individual cells of WT and Suv39h−/− MEFs (Right), ***P < 0.001 (Student’s t test, n = 25, 30). (D) The FRET ratiometric images of the H3K9me3 biosensor in MEFs with DMSO or 5 μM TCP treatment for 24 h (Left). The scatterplot represents the FRET/ECFP ratio of the H3K9me3 biosensor in individual cells of DMSO and TCP groups (Right), ***P < 0.001 (Student’s t test, n = 20, 28). (E) (Upper Left) The FRET ratiometric images of HEK 293T cells expressing the H3K9me3 biosensor together with either mCherry-H3.3WT or mCherry-H3.3K9M (the yellow arrows point to cells expressing a high level of mCherry-H3.3K9M and hence displaying a low level of H3K9me3). (Upper Right) The scatterplot represents the FRET/ECFP ratio of H3K9me3 biosensor in HEK 293T cells coexpressing mCherry-H3.3WT (H3.3WT) or mCherry-H3.3K9M (H3.3K9M), ***P < 0.001 (Student’s t test, n = 86, 158). (Lower) The dot plot indicates that the FRET/ECFP ratio of H3K9me3 biosensor is affected by the expression of H3.3WT or H3.3K9M but not by the levels of their expressions. (F) (Upper) Schematic drawing of H3K9me3 biosensor mutants, either with the binding domain HP1 mutated (W45A) or with the H3 lysine 9 site mutated (K9L). (Lower Left) The FRET ratiometric images in HEK293T cells expressing the biosensor or its mutants (W45A or K9L), respectively. (Lower Right) The scatterplot represents the FRET/ECFP ratios in HEK293T cells expressing various biosensors as indicated, ***P < 0.001 (Student’s t test, n = 114, 86, 93). The color scale bars represent the range of emission ratio, with cold and hot colors indicating low and high levels of H3K9me3, respectively. (Scale bar, 10 μm.)
The specificity of H3K9me3 biosensor was further verified in WT mouse embryonic fibroblasts (MEFs) and in knockout MEFs deficient for both methyltransferases Suv39h1/2 (Suv39h−/−). The FRET signal of the biosensor and the highlighted heterochromatin-like structures in WT MEFs were significantly suppressed in Suv39h−/− cells (Fig. 1C). In contrast, a demethylase inhibitor Tranylcypromine (TCP), which is capable of promoting the histone methylation by suppressing demethylase, increased the biosensor FRET signal (Fig. 1D and SI Appendix, Fig. S1). We also introduced the H3K9me3 biosensor with either mCherry-H3.3WT or mCherry-H3.3K9M into human HEK 293T cells. The biosensor FRET signals were drastically reduced in cells expressing mCherry-H3.3K9M but not mCherry-H3.3WT (Fig. 1E). Furthermore, human induced pluripotent stem cells and human embryonic stem cells that are known to have lower levels of H3K9me3 than fibroblasts (29) showed significantly lower FRET signals (SI Appendix, Fig. S2). Altogether, these results suggest that our biosensor can faithfully report on the dynamic H3K9me3 status in different cell types. The negative mutations with W45A disrupting HP1 binding capability or with K9L eliminating the H3K9 methylation site in the biosensor also completely abolished the detection capability of the biosensor in both mouse and human cells (Fig. 1F and SI Appendix, Fig. S3), verifying the specificity and activation mechanism of H3K9me3 biosensor as we designed.
The Dynamic Nature of H3K9me3 During Mitosis.
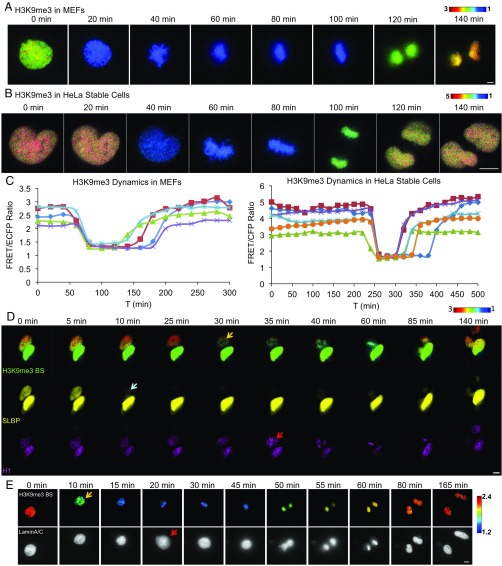
We noticed that in an asynchronous cell population, individual cells displayed different levels of FRET signals (Fig. 1F), implying that cell cycle status may affect the H3K9 trimethylation and hence FRET signals of the H3K9me3 biosensor. We hence tracked the H3K9me3 dynamics in single live cells. As shown in Fig. 2 A–C and SI Appendix, Fig. S4, this biosensor revealed a dynamic H3K9me3 during cell cycle progression in both mouse MEFs and human HeLa cells: the FRET signals decreased at the entrance of mitosis and recovered quickly upon the mitosis exit (Movie S1, MEF, and Movie S2, HeLa). The FRET dynamics could not be observed with the negative mutants of H3K9me3 biosensor (K9L and W45A) (SI Appendix, Fig. S5), indicating that the dynamic H3K9me3 signals during mitosis detected by our H3K9me3 biosensor are attributed to the H3K9 trimethylation and its ensuing recruitment of HP1 as we designed but not to the nonspecific global structural changes of the chromatin during cell cycles. We have also quantitatively compared our H3K9me3 biosensor with a previous published H3K9 trimethylation reporter (PP reporter) (27) side by side in tracking cell cycle progresses. A sixfold higher sensitivity was observed for our biosensor over the PP reporter (SI Appendix, Fig. S6). Although highly sensitive, our biosensor did not cause any detectable perturbation of cell cycle progression in host cells (SI Appendix, Fig. S7). This is probably because there are 30–60 million copies of endogenous histone H3 in each single cell (30), and the exogenously expressed histone H3 fused in the biosensor represents only a minor fraction, hence causing minimal perturbation of the host cells.
Fig. 2.
The dynamic nature of H3K9me3 during mitosis. (A) FRET/ECFP ratio images of the H3K9me3 biosensor in a representative MEF going through mitosis. The MEFs were transiently transfected with the H3K9me3 biosensor plasmid. (B) FRET/ECFP ratio images of the H3K9me3 biosensor in a representative HeLa cell going through mitosis. The CRISPR-generated stable HeLa cells were used for dynamic tracking. (C) Time courses of the raw FRET/ECFP ratio of the H3K9me3 biosensor in MEFs (Left; n = 5) and in HeLa cells (Right; n = 6) going through mitosis. (D) Tracking of cell cycle stages in HeLa cells via time-lapse imaging of H3K9me3 biosensor (H3K9me3 BS, ratiometric images; Upper), mOrange2-SLBP (SLBP, yellow; Middle), and H1-mMaroon1 (H1, purple; Lower). The arrow in each channel indicates the time of changes of the corresponding signals. (E) The FRET/ECFP ratio images (Upper row) and mCherry fluorescence images (Lower row) in HeLa cells expressing H3K9me3 biosensor and mCherry-LaminA/C. Arrows indicate the time points of the histone demethylation and the laminA/C dissolution. The color bars represent the range of emission ratio of H3K9me3, with cold and hot colors indicating low and high levels, respectively. (Scale bar, 10 μm.)
H3K9me3 Reduction Occurs in G2 Phase.
To more precisely identify the timing of demethylation event during the cell cycle, the H3K9me3 biosensor was applied together in the same cell with degradable mOrange2-SLBP (18-126) and H1-mMaroon1, which are markers for S/G2 transition and M phase, respectively (31). Time-lapse imaging revealed that the reduction of H3K9me3 level visualized by our FRET biosensor occurs at the end of G2 phase (Fig. 2D), with the mOrange2-SLBP disappearing first followed by the demethylation event before the cell entry into mitosis as reflected by the condensation of mMaroon1-fused H1. Expressing H3K9me3 biosensor with mCherry-LaminA/C in the same HeLa cells further revealed that the H3K9me3 reduction accompanied by the dissociation of heterochromatin structures from the LaminA/C nuclear envelope occurs about 10 min before the dissolution of nuclear envelope and hence the diffusion of LaminA/C into the cytoplasm (Fig. 2E). Thus, our biosensor revealed that H3K9me3 reduction occurs in G2 phase before global chromatin reorganization and nuclear envelope dissolution.
Dynamic Coupling Between H3K9me3 and H3S10p Revealed by Simultaneous Dual-FRET Imaging.
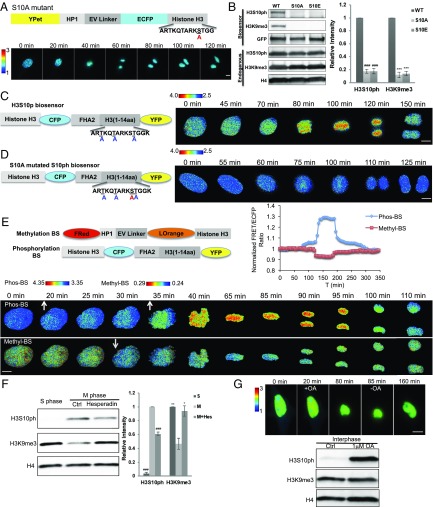
Because H3S10 positioned immediately next to H3K9 can be phosphorylated during mitosis and may affect H3K9me3 (21), we further investigated the dynamic coupling between H3S10p and H3K9me3 during mitosis. When the serine in H3S10 was mutated to nonphosphorylatable Alanine (S10A) or Glutamic acid (S10E), the FRET signals in the H3K9me3 biosensor remained low and unchanged during the cell cycle (Fig. 3A and SI Appendix, Fig. S8). Neither H3S10p nor H3K9me3 signals can be observed from the S10A or S10E biosensor groups, although the H3S10p and H3K9me3 levels of the endogenous H3 remained unchanged comparing to the control cells (Fig. 3B). In contrast to the effects of S10A and S10E, K9L mutation did not eliminate H3S10p level on the biosensor (SI Appendix, Fig. S9). Therefore, H3S10p and H3K9me3 may be coupled during cell cycle progression, with H3S10p dominating the regulation of H3K9me3.
Fig. 3.
An overall anticorrelation between methylation and phosphorylation during mitosis visualized by dual FRET imaging. (A) A schematic drawing of S10A mutant of H3K9me3 biosensor and its ratiometric images in MEFs. (B) (Left) Western blot analysis of H3S10p and H3K9me3 of the H3K9me3 biosensor (WT) and its S10A or S10E mutant, as well as the endogenous H3 in HEK 293T cells. (Right) Bar graphs represent the mean ± SEM of the H3S10p and H3K9me3 signals. ###P < 0.001, ***P < 0.001, compared with WT group (Student’s t test, n = 3). (C and D) (Left) A schematic drawing of the H3S10p biosensor (C) or its S10A mutant (D). (Right) The corresponding FRET/ECFP ratio images of the biosensors obtained from a HeLa cell going through mitosis. (E) Schematic drawings of the domain structures of H3K9me3 and H3S10p biosensors for dual FRET imaging (Upper Left). The time courses of the emission ratio of H3S10p (blue) and H3K9me3 (red) biosensors in a single HeLa cell (Upper Right). (Lower) The FRET/ECFP ratio images of H3S10p (Phos-BS; Upper) and H3K9me3 biosensor (Methyl-BS; Lower) in a single representative HeLa cell going through mitosis. (F) (Left) Western blot analysis of endogenous H3S10p and H3K9me3 from double thymidine-synchronized (S-phase) or nocodazole-arrested (M-phase) HeLa cells with or without 200 nM hesperadin treatment. (Right) Bar graphs represent mean ± SEM of the H3S10p and H3K9me3 signals. ###P < 0.001, *P < 0.05, **P < 0.01, compared with the M-phase control group (Student’s t test, n = 3). (G) (Upper) The representative FRET/ECFP ratio images in an interphase HeLa cell expressing H3K9me3 biosensor before and after 1 μM Okadaic Acid (OA) treatment followed by its washout. (Lower) The corresponding endogenous H3S10p and H3K9me3 levels were analyzed by Western blot for interphase HeLa cells treated with DMSO (Ctrl) or 1 μM OA. The color bars represent the range of emission ratio, with cold and hot colors indicating low and high levels of H3K9me3, respectively. (Scale bar, 10 μm.)
These findings prompted us to engineer another FRET biosensor capable of monitoring H3S10p (H3S10p biosensor), which consists of an ECFP/YPet-based FRET pair, with FHA2 as a phosphoserine-binding domain and histone H3 peptide as a substrate (consisting of 1–14 residues with three nonspecific threonine sites mutated which leaves S10 as the only phosphorylatable site). A full-length histone H3 was fused at N terminus of the biosensor to target the biosensor at nucleosomes (32) (Fig. 3C). Among several different designs (SI Appendix, Fig. S10), this H3S10p biosensor allowed the dynamic tracking of H3S10p during cell cycle progression in HeLa cells, with a gradual and moderate increase occurring at late G2 phase and a sharp rise at the early stage of the mitosis followed by a recovery decreasing process upon mitosis exit (Fig. 3C). The negative S10A mutant at the substrate sequence, but not at the H3 tag, of the H3S10p biosensor abolished the FRET dynamics during the cell cycle (Fig. 3D and SI Appendix, Fig. S11). Consistent with these findings, the FRET signals of the H3S10p biosensor can be increased by the phosphatase inhibitor okadaic acid (OA) and suppressed by the Aurora B kinase inhibitor hesperadin (SI Appendix, Fig. S12). These results verified that the FRET response of H3S10p is due to the specific phosphorylation at the substrate of the biosensor as we designed. Fusing H3 at the C terminus of the H3S10p biosensor also reported a dynamic FRET change during the cell cycle progression (SI Appendix, Fig. S13A), which is unaffected by the S10A mutation at the H3 tag (SI Appendix, Fig. S13B). These results suggest that the reporting function of H3S10p biosensor is dependent on the substrate phosphorylation but not on the H3 fusion tag. The phosphorylation changes in the substrate of the H3S10p biosensor are similar to the endogenous histone H3 during cell cycles of HEK 293T cells (SI Appendix, Fig. S14), verifying the specificity of our H3S10p biosensor in monitoring the endogenous H3S10 phosphorylation. Taken together, these results suggest that the FRET dynamics monitored by the H3S10p biosensor can reflect the designed substrate phosphorylation that mimics the endogenous H3S10p in the host cells.
To simultaneously visualize H3S10p and H3K9me3 utilizing two FRET pairs with distinct colors in a single live cell, we replaced the FRET pair in the H3K9me3 biosensor with LSSmOrange and FusionRed, which are spectrally compatible for dual FRET imaging with the ECFP and YPet used in the H3S10p biosensor, allowing us to use the same excitation light for both sensors in the same cell (29, 33). The results clearly indicate a temporal correlation in the dynamics between H3K9me3 and H3S10p during mitosis in the same cell (SI Appendix, Fig. S3E): H3S10p gradually increases during the late G2 phase before a sharp rise at the entrance into mitosis and then remains at a high level during mitosis until a rapid downfall at the mitosis exit, followed by a gradual return to the basal level after mitosis. The sharp increase in H3S10p dynamics at mitosis followed by a fall is accompanied by a concomitant decrease and recovery of H3K9me3 in the same cell during mitosis. This detected H3S10 phosphorylation is accompanied by a decrease of H3K9me3 and then a chromatin condensation during mitosis in the same HeLa cell. These results further support the role of H3S10p in leading the reduction of H3K9me3, before the global reorganization of chromatin at the entrance of mitosis.
This overall anticorrelation in time between H3K9me3 and H3S10p in single HeLa cells was also confirmed by immunoblotting of endogenous H3 tail averaged over cell populations (Fig. 3F), with low H3S10p and high H3K9me3 at interphase but high H3S10p and low H3K9me3 at mitosis. Consistent with these findings, the suppression of Aurora kinase B and hence H3S10p by hesperadin in mitotic cells caused a decrease of H3S10ph and an increase of H3K9me3 (Fig. 3F). Because both H3S10p and H3K9me3 changed during mitosis, we further examined whether the FRET changes of the H3K9me3 biosensor could be falsely caused by the H3S10p-induced repulsion of the HP1 chromodomain in the H3K9me3 biosensor without H3K9me3 changes. Suppressing the phosphatases in HeLa cells at interphase with OA to promote H3S10p levels indeed triggered a marked increase of H3S10p and a significant change in cell morphology but did not cause detectable FRET changes in the H3K9me3 biosensor (Fig. 3G and SI Appendix, Fig. S15). This result indicates that the FRET signals of H3K9me3 biosensor indeed reflect a decrease of H3K9me3 during mitosis, consistent with previous findings that H3S10p alone cannot repulse HP1 binding in cells (23), although H3S10p can negatively regulate the HP1 binding in vitro with purified proteins in solutions (21, 22).
To find further evidence of H3K9 demethylation during mitosis, we developed a sensor to study the interaction between the SUV39H1 methyltransferase and H3 by replacing the HP1 domain in the H3K9me3 biosensor with full-length SUV39H1. The dynamics of SUV39H1 biosensor were visualized in Suv39h−/− MEFs. SUV39H1 dissociates from histone H3 when cells enter mitosis and reassociates with histone H3 when cells exit mitosis; these findings support the decrease of H3K9me3 during mitosis (SI Appendix, Fig. S16).
H3S10p Governs the Demethylation of H3K9me3 at the Mitotic Entrance.
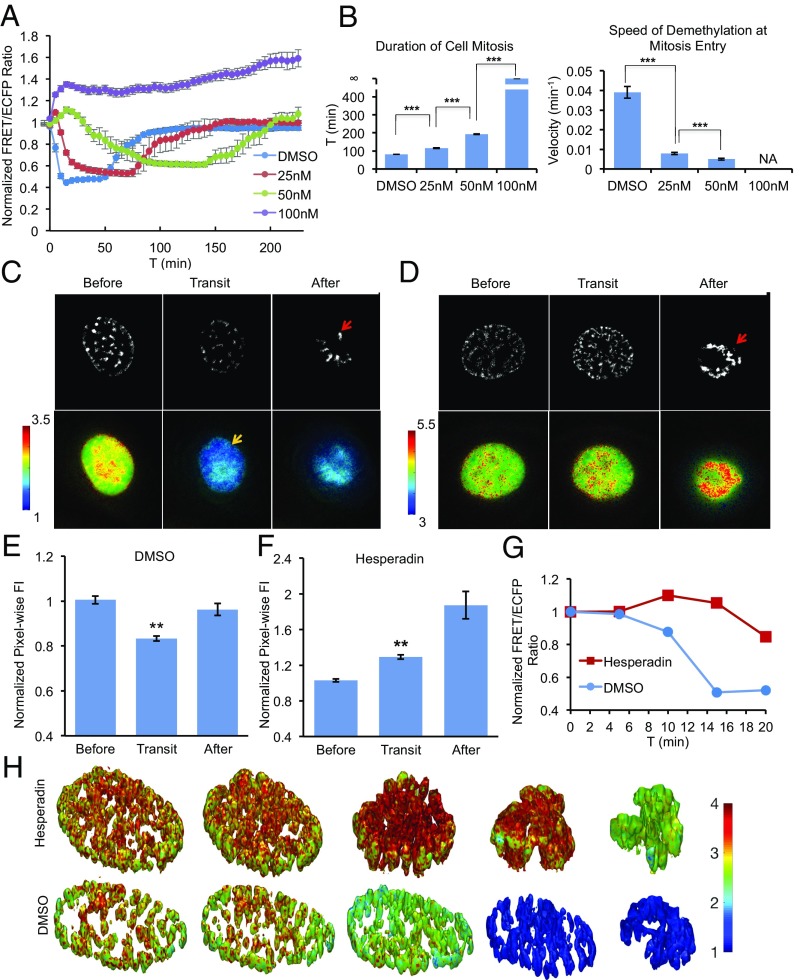
To further examine the causal relationship between H3S10p and H3K9me3, different dosages of Aurora B kinase inhibitor hesperadin (DMSO for 0, 25, 50, and 100 nM) were applied to investigate the effect of different levels of H3S10p inhibition on H3K9me3 dynamics during cell mitosis. The results revealed that as the hesperadin concentration increased, the speed of H3K9me3 demethylation at the mitosis entrance slowed down proportionally, and this was accompanied by a corresponding lengthening of the mitosis duration (Fig. 4 A and B). It is interesting to note a transient increase of FRET signals at moderate levels of hesperadin treatment, reflecting the rise of H3K9me3 before the cells slowly entered mitosis, whereas a high dose of hesperadin caused a sustained rise of H3K3me3 and mitotic arrest (Fig. 4A). These results suggest that the level of H3S10p can govern and precisely tune the H3K9me3 demethylation dynamics and hence mitotic duration. In contrast, the basal level and dynamics of H3S10p were not changed significantly in either Suv39h−/− MEFs or TCP-treated HeLa cells, although the H3K9me3 levels in these cells are significantly different from the control groups (SI Appendix, Fig. S17). These results suggest that H3S10p governs the demethylation of H3K9me3 at mitotic entrance but not vice versa.
Fig. 4.
H3S10p governs the H3K9me3 demethylation and chromatin reorganization/condensation. (A) Time courses of normalized FRET/ECFP ratio in double thymidine-arrested HeLa cells expressing the H3K9me3 biosensor and treated with different concentrations of hesperadin (DMSO = 0, 25, 50, and 100 nM). Hesperadin was added to the culture 1 h before the start of imaging. (B) Bar graphs represent mean ± SEM of the mitosis duration and demethylation speed at mitotic entrance under different hesperadin treatments. . ***P < 0.001 (Student’s t test, n ≥ 3). NA, not applicable. (C and D) The representative FRET/ECFP ratio images and the projected 2D clusters of heterochromatin-like structures in HeLa cells expressing H3K9me3 biosensor and treated with (C) DMSO or (D) 50 nM hesperadin before entering mitosis (before), at G2-M transition (transit), and after entering mitosis (after), as indicated (red arrows indicate the global reorganization of chromatin structures, and the yellow arrow indicates the decrease of FRET ratio). (E and F) Bar graphs represent mean ± SEM of pixelwise fluorescence intensities of heterochromatin clusters in DMSO- and 50 nM hesperadin-treated groups. **P ≤ 0.005, indicates statistically significant difference between the indicated group and other groups. (Student’s t test, n = 4. There are 142 clusters of before phase, 115 clusters of transit phase, and 46 clusters of after phase analyzed in the bar graph for the DMSO group, and there are 60 clusters of before phase, 69 clusters of transit phase, and 21 clusters of after phase analyzed in the bar graph for the hesperadin group.) (G) The time courses of the H3K9me3 biosensor (red, hesperadin; blue, DMSO) and (H) representative images of 3D heterochromatin-like clusters in HeLa cells treated with DMSO or 50 nM hesperadin at mitosis entrance. The color bars represent the range of emission ratio, with cold and hot colors indicating low and high levels of H3K9me3, respectively.
H3K9me3 Demethylation Leads to Dissolution of Heterochromatin-Like Structures at the Mitotic Entrance.
H3K9me3 recruits HP1 (7, 34, 35) to cause the ensuing phase separations (36, 37), and is regarded as a hallmark of heterochromatin structures. Because H3K9me3 demethylation occurs earlier than the global reorganization of chromatin structures and nuclear envelope dissolution during mitosis (Fig. 2 D and E), we examined the role of H3K9me3 dynamics in regulating the reorganization process of chromatin during mitosis. In control cells, there was a decrease of H3K9me3 and heterochromatin-like structures, highlighted by the clusters identified by our quantitative analysis on the intensity distribution of the FRET biosensor (Materials and Methods), before the global condensation of the mitotic chromatin (Fig. 4 C and E). However, in the hesperadin-treated cells with inhibited H3S10p, no significant decrease in heterochromatin-like structures or H3K9me3 can be observed before chromatin reorganization and condensation (Fig. 4 D and F). As such, H3S10p may govern H3K9me3 demethylation in causing the dissolution of heterochromatin-like structures to allow the access of chromatin remodeling complexes and facilitate a global reorganization of chromatin structures. The inhibition of H3S10p can suppress the H3K9me3 reduction and heterochromatin dissolution, thus slowing down the mitotic process. This conclusion is further supported by our reconstruction of the dynamic heterochromatin-like structures from 3D imaging deconvolution at the mitosis entrance with or without hesperadin treatment (Fig. 4 G and H).
Mathematical Model.
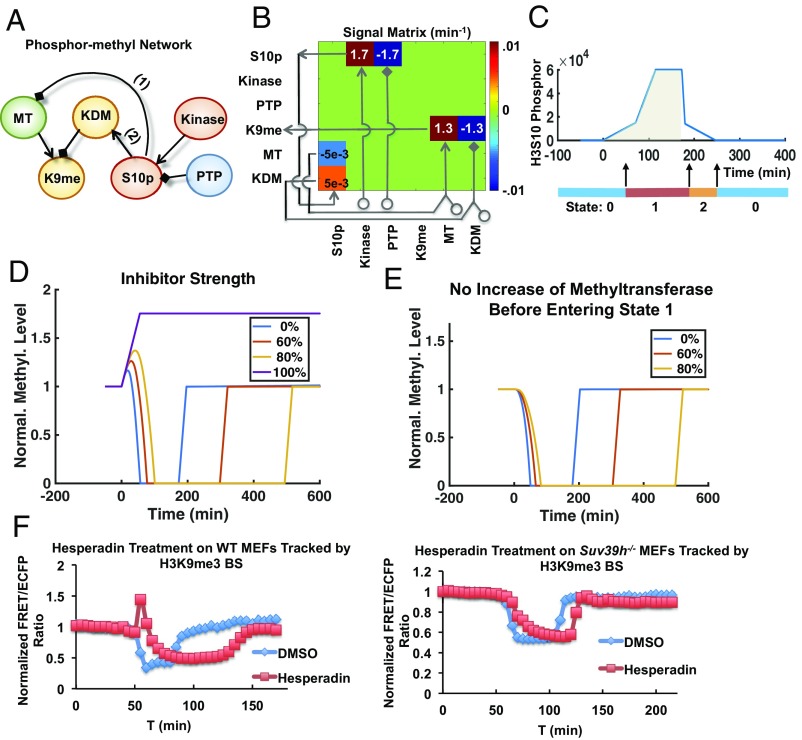
To obtain an in-depth understanding of dynamic H3S10p and H3K9me3 interregulation during mitosis, we constructed a mathematical model based on our experimental results. This model contains a time-dependent piecewise linear differential equation system (38) describing a signaling network containing six variables, H3K9me3, H3S10p, methyltransferases (MTs), demethylases (KDMs), kinases, and phosphatases (Fig. 5A). We implemented the following model assumptions: H3K9me3 is positively and negatively regulated by the active enzymes MTs and KDMs, respectively, whereas H3S10p is regulated positively by active kinases and negatively by phosphatases (Fig. 5 A and B). The model is separated into three main states: 0 for interphase, 1 for mitosis, and 2 for mitosis exit (Fig. 5C). The entrance of a cell into late G2 phase (state 0) triggers a small increase of kinase activities accompanied by an increase of methyltransferase activity (39, 40); meanwhile, H3S10 phosphorylation in the model is programmed to recruit KDMs and repel MTs based on earlier reports (Fig. 5A) (13). When the phosphorylation level rises above a threshold value, a large increase of demethylase follows, which subsequently causes quick demethylation of H3K9. After that, the cell enters mitosis (state 1) and the kinase activity increases sharply (41). When the cumulative phosphorylation level rises above another threshold value, the cell will undergo mitosis exit as suggested by an earlier report (Fig. 5C) (42), hence triggering a sharp decrease of kinase and switching from state 1 to 2. All of the model parameters are based on the literature or obtained by fitting to the experimental results of H3K9me3 dynamics observed in HeLa cells shown in Figs. 2 and 3E (detailed description in SI Appendix, Materials and Methods and Table S1). This model configuration is also supported by our mass spectrometry results revealing that histone H3 during mitosis has an increase in association with Aurora kinase B and demethylase cofactor GLYR1 but a decrease with phosphatase PP1 and H3K9 methyltransferase SUV39H2 (SI Appendix, Fig. S18).
Fig. 5.
A mathematic model depicting the dynamic coupling of H3K9me3 and H3S10p. (A) The model describes a time-evolving signaling network containing H3K9me3 (K9me), H3S10p (S10p), methyltransferases (MT), demethylase (KDM), serine kinases (Kinase), and phosphatase (PTP). Positive and negative regulations are indicated by the shapes of arrow and diamond, respectively. Numbers 1 and 2 represent the pathways to be perturbed in coupling H3K9me3 and H3S10p. (B) The stoichiometric matrix shown is used to evolve the piecewise linear ordinary differential equation (ODE) system in time, with the values of all nonzero entries indicated. (C) The model is separated into three states: 0 for interphase, 1 for entering mitosis and mitosis, and 2 for exiting mitosis. After the H3S10 phosphorylation reaches an initial threshold (at ∼50 min), the cell switches to state 1 and enters mitosis. The cumulative phosphorylation level is calculated in the shaded area. After the accumulated phosphorylation exceeds a second threshold value, the phosphorylation level decreases, and cell switches from state 1 to 2. (D) The simulated level of H3K9me3 in cells with 0–100% blockages of H3S10p. (E) Simulation of the H3K9me3 dynamics as shown in D but without methyltransferase. (F) Time courses of normalized FRET/ECFP ratio of H3K9me3 biosensor in double thymidine-arrested WT (Left) and Suv39h−/− MEFs (Right) treated with DMSO and 50 nM hesperadin. Hesperadin was added to the culture 1 h before imaging started.
Based on the model and assumptions, we simulated the time course of H3K9me3 before and after mitosis in a HeLa cell under the treatment of kinase inhibitor (Fig. 5D and SI Appendix, Fig. S19). The simulation results indicate that in cells treated with kinase inhibitors (e.g., hesperadin), the H3K9me3 level increases transiently upon entrance into mitosis, due to the reduced phosphorylation level and subsequently less recruitment of KDMs to histones (Fig. 5D). The duration of mitosis (from the beginning of entrance to the end of exit) was also significantly longer due to the slower H3S10p accumulation and hence the longer time needed to reach the threshold value for triggering mitosis exit in these cells (Fig. 5D and SI Appendix, Fig. S19), consistent with the live-cell imaging experiments (Fig. 4A). It is of note that removing MTs in the model eliminated the transient increase of the H3K9me3 at the mitotic entrance in the kinase-inhibited groups, suggesting that H3K9 MTs are responsible for causing more H3K9me3 when H3S10p is inhibited (Fig. 5E). We further assessed the accuracy of our model predictions by experimental approaches. We monitored the effects of inhibiting H3S10p with hesperadin on the dynamics of H3K9me3 during mitosis in Suv39h−/− MEFs, which are deficiency in methyltransferases (Suv39h1/2) (Fig. 5F). The results show that the transient H3K9me3 increase at mitotic entrance in WT MEFs with H3S10p inhibition (Fig. 5F, Left) disappeared in Suv39h−/− MEFs (Fig. 5F, Right), consistent with the modeling results (Fig. 5E), thus verifying the accuracy of our model. Our modeling results hence reveal a simplicity of regulation mechanism underlying H3S10p–H3K9me3 coupling during mitosis, with the two sets of enzymes (kinase/phosphatase and methyltransferase/demethylase) sufficient to provide the kinetic controls.
Discussion
We have developed a highly sensitive and specific FRET biosensor capable of visualizing the dynamic H3K9me3 in single live cells. Together with another FRET biosensor with distinct colors for the simultaneous monitoring H3S10p in the same cell, the results revealed an anticorrelation between the dynamics of H3K9me3 and H3S10p in a cell cycle, with H3S10p facilitating the decrease of H3K9me3 at the entrance of mitosis. This decrease of H3K9me3 is accompanied by a dissolution of heterochromatin-like structures, which may facilitate the access of chromatin by remodeling complexes before the global chromatin reorganization and nuclear envelope breakdown during mitosis. As such, the coordinated regulation of H3S10p and H3K9me3 may facilitate the timely dissolution of heterochromatin-like structures and allow accessibility by chromatin-remodeling complex for an efficient and timely chromatin reorganization during mitosis.
Genetically encoded biosensors based on FRET, by allowing the visualization and quantification of biochemical signals in single live cells with high spatiotemporal resolutions, have revolutionized research in cell biology. Several FRET and bioluminescence biosensors have been reported to detect histone methylation, acetylation, and phosphorylation (15, 16, 27, 32, 43, 44). However, the limited sensitivity of these genetically encoded FRET biosensors, particularly for histone methylations, has hindered their broad application. In fact, despite the importance of histone methylations in regulating chromatin and genome organizations, there are limited studies on dynamic histone methylations in live cells utilizing these biosensors (11, 45, 46). Our current H3K9me3 FRET biosensor has a significantly improved sensitivity, with a sixfold higher dynamic range comparing to a previously published H3K9 methylation reporter (SI Appendix, Fig. S6). This large dynamic range has allowed us to observe a clear decrease of H3K9me3 during mitosis in different types of mouse and human cells. Our result of a global H3K9me3 decrease in mitosis is consistent with previous proteomic measurements (47), the accumulation of H3K9 demethylase KDM4C at mitotic chromatin (13), and the release of SUV39H1 from chromatin during mitosis (SI Appendix, Fig. S16) (39). In fact, it has been well established that HP1, which can recruit SUV39H1 for the propagation of H3K9me3 marks in chromatin, dissociates from histone tails during mitosis (20, 34, 48, 49). As such, our tool provides convincing evidence on H3K9me3 dynamics during cell cycles at single-cell levels. This tool also allowed the revelation that H3K9me3 reduction occurs at the end of G2 phase before global chromatin reorganization and nuclear envelope dissolution, advancing our understanding on the precise timing of these critical events during cell cycles in live cells.
It is interesting to note that H3K9 is precisely positioned adjacent to H3S10. Given the sheer number of H3 tails in each single cell, i.e., 60 million copies after DNA duplication (30), the posttranslational modifications of H3 require well-coordinated actions of enzymes. Our dual FRET imaging in single live cells indeed revealed an anticorrelation in dynamics between H3S10p and H3K9me3 during cell mitosis, with a clear H3S10p increase accompanied by a decrease of H3K9me3 (Fig. 3 E and F). This observation adds insights on the current understanding of phospho/methyl switch mechanism in the temporal governance of the nuclear chromatin function (24), with H3S10p affecting H3K9me3 and eventually leading to the dissociation of HP1 from H3 (21, 22). Indeed, Aurora kinase B, which controls H3S10p, increases markedly during mitosis (50). The abundant Aurora kinase B and hence rapid H3S10 phosphorylation during mitosis may create docking sites to recruit adaptors and catalyzers to facilitate the dynamics of H3K9me3 demethylations, thus overcoming the potentially slow speed due to the limited copies of demethylases [RNAseq data show that the ratio of aurora kinase B over KDM4C is ∼5.9:1 (51)] (SI Appendix, Table S1). Our simulation models verified and quantitatively mimicked the H3S10p level in regulating the H3K9 demethylation dynamics during mitosis (Fig. 5D). Both our live cell experiments and simulation models showed that the speed of H3K9 demethylation at the entrance of mitosis can be precisely tuned by the levels of H3S10p, leading to alteration of the duration of cell mitosis (Figs. 4 A and B and 5D). In contrast, the manipulation of H3K9me3 does not have significant effect on H3S10p levels (SI Appendix, Fig. S17), consistent with the existence of more copies of Aurora kinase B responsible for H3S10p than those of demethylases that control H3K9me3 demethylation.
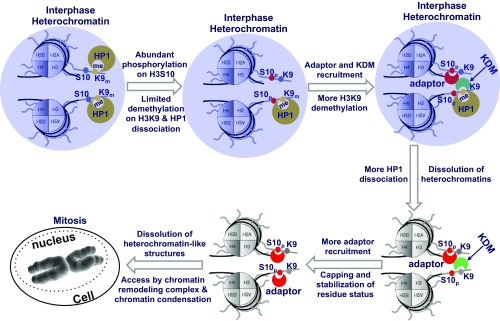
Because H3K9me3 is a hallmark of heterochromatin (7, 34, 35, 52), the demethylation of H3K9me3 at the mitosis entrance may represent a dissolution of heterochromatin-like structures in allowing the access of chromatin remodelers for the preparation of global chromatin reorganization during mitosis. Indeed, our experimental data and clustering analysis showed that there is a clear demethylation associated with a reduction in heterochromatin-like structures, followed by a global chromatin condensation (Fig. 4 C and E). The inhibition of H3S10p and hence H3K9me3 demethylation prevents the dissolution of heterochromatin-like structures before the global reorganization of chromatin structures (Fig. 4 D–H), resulting in a slowdown of the mitosis process. This is consistent with previous studies showing that H3S10p and its dynamic distribution are essential for chromatin condensation during cell mitosis (53). In fact, H3S10p is initially localizing at pericentromeric heterochromatin but gradually spreading along the chromosomal arms to the whole chromosome when entering mitosis (54, 55). However, H3S10p per se may not be sufficient to cause the dissociation of HP1 from H3 in live cells for chromatin remodeling. In fact, the dissociation of HP1 from nucleosomes has been shown to depend on H3K9 demethylation (56), consistent with the observation that the phosphorylation of H3S10 alone by Aurora Kinase B did not disrupt the HP1–H3K9me3 interaction at the interphase (Fig. 3G and SI Appendix, Fig. S15). Therefore, the increase of H3S10p when entering mitosis can facilitate the H3K9me3 demethylation and consequent dissolution of heterochromatin-like structures, allowing the sufficient recruitment of chromatin remodelers for the global chromatin reorganization and condensation (Fig. 6).
Fig. 6.
A hypothetical model of the dynamic coupling between H3S10p and H3K9me3 that regulates the transition from interphase to mitosis during a cell cycle. We hypothesize that heterochromatin-like structures can be facilitated by liquid phase separation via HP1 accumulation in association with H3K9me3 at H3 tails (36, 37). At the start of the transition from G2 to M phase, the abundant Aurora kinase B can cause rapid phosphorylation of H3S10 that can recruit adaptors such as 14-3-3 (57, 58). The consequently facilitated recruitment of demethylases and repulsion of methyltransferases can promote the demethylation of H3K9me3 and subsequent dissociation of HP1 from H3 tails. The resulted dissolution of heterochromatin-like structures should allow the enhanced access of chromatin remodeling complex that can help the timely reorganization and condensation of chromatin for mitosis. Purple circle represents heterochromatin-like structures and regions.
Materials and Methods
For information on plasmid construction, cell culture, synchronization, transfection and lentivirus production, imaging, dual FRET imaging, histone protein extraction, immunoblotting analysis, HeLa stable cell line generation by CRISPR, nuclear complexes coimmunoprecipitation, mass spectroscopy, measurement of 2D heterochromatin cluster, construction of 3D heterochromatin cluster, and mathematic modeling, please refer to SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Jenuwein (Max Planck Institute of Immunobiology and Epigenetics) for kindly providing the WT MEFs and Suv39h−/− sister cells. We also thank Dr. Michiyuki Matsuda (Kyoto University, Japan) for kindly providing the pCAGGS vector construct and Dr. Ya-Hui Chi (National Health Research Institute, Taiwan) for kindly providing the SUV39H1 construct. This work is supported by NIH Grants HL121365, GM125379 (to S.C. and Yingxiao Wang), CA204704, and CA209629 (to Yingxiao Wang); NSF Grants CBET1360341 and DMS1361421 (to Yingxiao Wang and S.L.); and the Beckman Laser Institute Foundation and is also funded by China Scholarship Council (to Q.P.). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest statement: Yingxiao Wang and S.L. are scientific cofounders of Cell E&G Inc. However, these financial interests do not affect the design, conduct, or reporting of this research.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811818115/-/DCSupplemental.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Boettiger AN, et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonin W, Neumann H. Chromosome condensation and decondensation during mitosis. Curr Opin Cell Biol. 2016;40:15–22. doi: 10.1016/j.ceb.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Trojer P, Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Duan Q, Chen H, Costa M, Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem. 2008;283:33585–33590. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkins BJ, et al. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343:77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- 7.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 8.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McManus KJ, Biron VL, Heit R, Underhill DA, Hendzel MJ. Dynamic changes in histone H3 lysine 9 methylations: Identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J Biol Chem. 2006;281:8888–8897, and erratum (2006) 281:38966. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- 11.Chu L, et al. SUV39H1 orchestrates temporal dynamics of centromeric methylation essential for faithful chromosome segregation in mitosis. J Mol Cell Biol. 2012;4:331–340. doi: 10.1093/jmcb/mjs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupershmit I, Khoury-Haddad H, Awwad SW, Guttmann-Raviv N, Ayoub N. KDM4C (GASC1) lysine demethylase is associated with mitotic chromatin and regulates chromosome segregation during mitosis. Nucleic Acids Res. 2014;42:6168–6182. doi: 10.1093/nar/gku253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JA, et al. Deacetylation and methylation at histone H3 lysine 9 (H3K9) coordinate chromosome condensation during cell cycle progression. Mol Cells. 2011;31:343–349. doi: 10.1007/s10059-011-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekar TV, Foygel K, Devulapally R, Paulmurugan R. Degron protease blockade sensor to image epigenetic histone protein methylation in cells and living animals. ACS Chem Biol. 2015;10:165–174. doi: 10.1021/cb5008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekar TV, Foygel K, Gelovani JG, Paulmurugan R. Genetically encoded molecular biosensors to image histone methylation in living animals. Anal Chem. 2015;87:892–899. doi: 10.1021/ac502629r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosio C, et al. Mitotic phosphorylation of histone H3: Spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauvé DM, Anderson HJ, Ray JM, James WM, Roberge M. Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J Cell Biol. 1999;145:225–235. doi: 10.1083/jcb.145.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 20.Hathaway NA, et al. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 22.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 23.Mateescu B, England P, Halgand F, Yaniv M, Muchardt C. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 2004;5:490–496. doi: 10.1038/sj.embor.7400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sawicka A, Seiser C. Sensing core histone phosphorylation–A matter of perfect timing. Biochim Biophys Acta. 2014;1839:711–718. doi: 10.1016/j.bbagrm.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang M, Sun J, Chien S, Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc Natl Acad Sci USA. 2008;105:14353–14358. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 27.Lin CW, Jao CY, Ting AY. Genetically encoded fluorescent reporters of histone methylation in living cells. J Am Chem Soc. 2004;126:5982–5983. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- 28.Jayaram H, et al. S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci USA. 2016;113:6182–6187. doi: 10.1073/pnas.1605523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shemiakina II, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 30.Alberts B, et al. Molecular Biology of the Cell. 4th Ed Garland Science; New York: 2003. [Google Scholar]
- 31.Bajar BT, et al. Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat Methods. 2016;13:993–996. doi: 10.1038/nmeth.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuller BG, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shcherbakova DM, Hink MA, Joosen L, Gadella TW, Verkhusha VV. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J Am Chem Soc. 2012;134:7913–7923. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 35.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson AG, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singhania R, Sramkoski RM, Jacobberger JW, Tyson JJ. A hybrid model of mammalian cell cycle regulation. PLOS Comput Biol. 2011;7:e1001077. doi: 10.1371/journal.pcbi.1001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- 40.Dominguez D, et al. An extensive program of periodic alternative splicing linked to cell cycle progression. eLife. 2016;5:e10288. doi: 10.7554/eLife.10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watrin E, Legagneux V. Contribution of hCAP-D2, a non-SMC subunit of condensin I, to chromosome and chromosomal protein dynamics during mitosis. Mol Cell Biol. 2005;25:740–750. doi: 10.1128/MCB.25.2.740-750.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santamaría D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci USA. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin CW, Ting AY. A genetically encoded fluorescent reporter of histone phosphorylation in living cells. Angew Chem Int Ed Engl. 2004;43:2940–2943. doi: 10.1002/anie.200353375. [DOI] [PubMed] [Google Scholar]
- 45.Tan Y, et al. Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat Commun. 2014;5:4619. doi: 10.1038/ncomms5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakaoka S, Sasaki K, Ito A, Nakao Y, Yoshida M. A genetically encoded FRET probe to detect intranucleosomal histone H3K9 or H3K14 acetylation using BRD4, a BET family member. ACS Chem Biol. 2016;11:729–733. doi: 10.1021/cb501046t. [DOI] [PubMed] [Google Scholar]
- 47.Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol. 2012;32:2503–2514. doi: 10.1128/MCB.06673-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto K, Sonoda M. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem Biophys Res Commun. 2003;301:287–292. doi: 10.1016/s0006-291x(02)03021-8. [DOI] [PubMed] [Google Scholar]
- 50.Goto H, Yasui Y, Nigg EA, Inagaki M. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002;7:11–17. doi: 10.1046/j.1356-9597.2001.00498.x. [DOI] [PubMed] [Google Scholar]
- 51.Petryszak R, et al. Expression atlas update–An integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- 54.Hendzel MJ, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 55.Prigent C, Dimitrov S. Phosphorylation of serine 10 in histone H3, what for? J Cell Sci. 2003;116:3677–3685. doi: 10.1242/jcs.00735. [DOI] [PubMed] [Google Scholar]
- 56.Fass E, et al. Phosphorylation of histone h3 at serine 10 cannot account directly for the detachment of human heterochromatin protein 1gamma from mitotic chromosomes in plant cells. J Biol Chem. 2002;277:30921–30927. doi: 10.1074/jbc.M112250200. [DOI] [PubMed] [Google Scholar]
- 57.Winter S, Fischle W, Seiser C. Modulation of 14-3-3 interaction with phosphorylated histone H3 by combinatorial modification patterns. Cell Cycle. 2008;7:1336–1342. doi: 10.4161/cc.7.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winter S, et al. 14-3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J. 2008;27:88–99. doi: 10.1038/sj.emboj.7601954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.