Significance
The phytohormone abscisic acid (ABA) controls root responses to environmental signals such as abiotic stress. ABA signaling in roots depends on the nonredundant role of the PYL8 receptor. This study reveals special features of this ABA receptor. ABA binding triggers hormone-dependent stabilization of PYL8 through reduced ubiquitination and induces nuclear localization of the receptor. ABA-induced stabilization also allows movement of the PYL8 receptor from the root epidermis and stele to adjacent tissues. Hence, like mobile transcription factors that regulate plant development, the PYL8 protein can move between cells. In summary, our study reports a novel non-cell-autonomous mechanism to regulate hormone perception and root growth.
Keywords: ABA, PYL8, non-cell-autonomous, ABA biosensor, root
Abstract
The phytohormone abscisic acid (ABA) plays a key role regulating root growth, root system architecture, and root adaptive responses, such as hydrotropism. The molecular and cellular mechanisms that regulate the action of core ABA signaling components in roots are not fully understood. ABA is perceived through receptors from the PYR/PYL/RCAR family and PP2C coreceptors. PYL8/RCAR3 plays a nonredundant role in regulating primary and lateral root growth. Here we demonstrate that ABA specifically stabilizes PYL8 compared with other ABA receptors and induces accumulation of PYL8 in root nuclei. This requires ABA perception by PYL8 and leads to diminished ubiquitination of PYL8 in roots. The ABA agonist quinabactin, which promotes root ABA signaling through dimeric receptors, fails to stabilize the monomeric receptor PYL8. Moreover, a PYL8 mutant unable to bind ABA and inhibit PP2C is not stabilized by the ligand, whereas a PYL85KR mutant is more stable than PYL8 at endogenous ABA concentrations. The PYL8 transcript was detected in the epidermis and stele of the root meristem; however, the PYL8 protein was also detected in adjacent tissues. Expression of PYL8 driven by tissue-specific promoters revealed movement to adjacent tissues. Hence both inter- and intracellular trafficking of PYL8 appears to occur in the root apical meristem. Our findings reveal a non-cell-autonomous mechanism for hormone receptors and help explain the nonredundant role of PYL8-mediated root ABA signaling.
Responses to environmental conditions in plant roots are coordinated by different hormones. Thus, hormone signaling regulates root growth, root system architecture, and tropic root responses (1–3). Abscisic acid (ABA) mediates root responses to different environmental factors, such as the presence of nitrate in the soil, water deficit, moisture gradients, and salt or nutrient deficiency (4). ABA signaling through the PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR)-Protein phosphatases type 2C (PP2Cs) and ABA-activated SNF1-related protein kinases (SnRK2s) core components is linked to different plant adaptive responses to water deficit and osmotic and salt stress, such as the maintenance of primary root elongation and the repression of lateral root formation (4–10). For example, in maize seedlings under water-deficit stress the primary root growth is maintained through ABA action, which acts partially through ethylene antagonism (5). In Arabidopsis roots exposed to salt stress, ABA also has a growth-promoting role during the recovery phase (11). Additionally, ABA signaling is required for root hydrotropism, an adaptive response that facilitates soil exploration under heterogeneous water availability (3). Regulation of root growth by ABA is closely connected with hydrotropism, as the hydrotropic response involves asymmetric ABA signaling in the root cortex through the PYR/PYL/RCAR-PP2C-SnRK2 core signaling pathway (3, 12, 13). Other environmental cues, such as salinity, induce root adaptations that are mediated by ABA (10, 14, 15). Nutrient-induced root plasticity is also regulated by ABA, for example, the suberization of the endodermis in response to either sodium chloride treatment or sulfur or potassium deficiencies (14). Thus, harmful minerals can be excluded by the endodermis.
Although the role of ABA in root physiology has been well studied, the molecular and cellular mechanisms that operate to coordinate the action of core components are not well known. For example, the expression of the ABA-activated kinase SnRK2.2 is observed in all root tissues, but expression of the ABA receptor promoters is restricted to some of them (3, 13). Therefore, it is not well known how ABA perception is connected with the activation of SnRK2.2 in different root tissues. Additionally, different PYR/PYL/RCAR ABA receptors are expressed at high levels and contribute to the quantitative regulation of ABA sensitivity in the root, but uniquely the pyl8/rcar3 single knockout shows reduced sensitivity to ABA-mediated inhibition of root growth (8, 13). Therefore, PYL8/RCAR3 plays a nonredundant role for ABA signaling in the root, which relies on PYL8-mediated inhibition of at least five clade A PP2Cs, i.e., HAB1, HAB2, ABI1, ABI2, and PP2CA (13). Compared with other ABA receptors, PYL8 shows a unique expression pattern in the root epidermis and lateral root cap (LRC) (13). Recent studies investigating the degradation of ABA receptors in seedlings have revealed that PYL8 is ubiquitinated and degraded by the 26S proteasome in Arabidopsis thaliana (16, 17). In those studies, we found that ABA treatment increased PYL8 protein levels, but had no significant effect on other receptors such as PYR1 and PYL4 (17). Moreover, ABA treatment limited PYL8 degradation in seedlings and reduced PYL8 polyubiquitination (16). It is currently unknown whether such a mechanism operates in root tissues or whether ligand perception by either PYL8 or other receptors is required to stabilize PYL8. Further investigation of ligand-induced effects on PYL8 stability might help explain its nonredundant role for ABA signaling in roots.
Studies to investigate how different hormones control root growth, tropic root responses, or stress adaptation have revealed single tissue layers or discrete spatial domains that are differentially targeted by hormones (1). For example, auxin targets elongating epidermal cells during the gravitropic response whereas ABA targets elongating cortical cells during the hydrotropic response (3, 18). On the other hand, endodermal ABA signaling promotes lateral root quiescence under saline conditions (9). ABA also promotes quiescence of the quiescent center (QC), which may be considered as positive regulation of root growth as it promotes QC maintenance (19). However, ABA also inhibits cell division in the proximal part of the Arabidopsis root meristem, which can explain the inhibitory effect of high ABA concentrations on root growth (19). In contrast, low levels of ABA promote root elongation through an increased rate of cell production and elongation (3). ABA signaling is also important in the mature root, where most of the absorption of minerals and water takes place (8). In agreement with the above-mentioned physiological studies, the expression pattern of SnRK2.2 indicates that ABA signaling is required in all root tissues (3). Altogether, these studies suggest that ABA perception in different root domains is important to regulate root physiology and root growth; however, a detailed molecular and cellular understanding of root ABA perception is still lacking in the different root tissues where ABA acts.
Results
ABA Specifically Stabilizes the PYL8 Receptor.
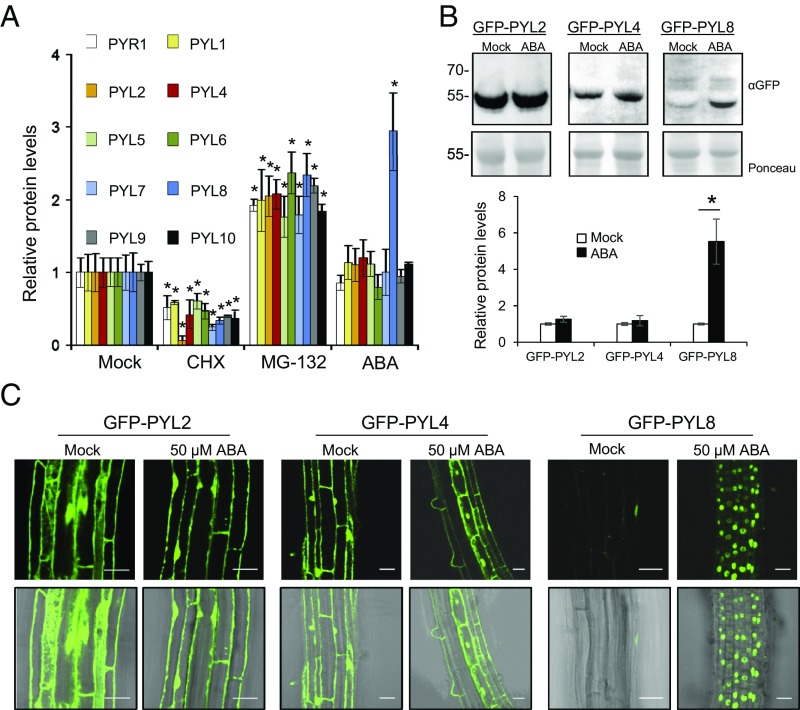
ABA receptor stability and degradation is an emerging topic in ABA signaling (16, 17, 20, 21). To obtain a comprehensive picture of the turnover of ABA receptors, we analyzed protein dynamics of 10 epitope (HA)-tagged receptors (PYR1, PYL1, PYL2, PYL4, PYL5, PYL6, PYL7, PYL8, PYL9, and PYL10, the most highly expressed gene products of the gene family) in 2-wk-old plants. Treatment with the translation inhibitor cycloheximide (CHX) led to diminished protein synthesis of all ABA receptors, whereas treatment with the proteasome inhibitor MG132 led to their accumulation (Fig. 1A). Interestingly, addition of ABA specifically led to the accumulation of PYL8 protein (Fig. 1A). ABA treatment of transgenic lines that express GFP-tagged versions of PYL2, PYL4, and PYL8 also revealed a selective ABA-induced accumulation of PYL8 (Fig. 1 B and C). qRT-PCR analyses corroborated that this effect was not caused by changes in the expression of 35S promoter-driven 3HA- or GFP-tagged transgenes (SI Appendix, Fig. S1). ABA therefore appears to enhance PYL8 accumulation in these lines through a posttranscriptional mechanism. Confocal laser scanning microscopy (CLSM) also revealed that GFP-PYL8 exhibited a predominantly nuclear localization in root cells following ABA treatment, whereas GFP-PYL2 and GFP-PYL4 localized to both the nucleus and cytosol of mock or ABA-treated roots (Fig. 1C).
Fig. 1.
ABA treatment specifically increases PYL8 protein levels in seedlings. (A) Effect of CHX, MG132, or ABA treatment on protein levels of HA-tagged receptors. The 10-d-old seedlings expressing HA-tagged receptors were either mock or chemically treated with 50 μM CHX, MG132, or ABA for 6 h. Immunoblot analysis using anti-HA was performed to quantify protein levels. A single asterisk (*) indicates P < 0.05 (Student’s t test) compared with the corresponding mock-treated sample. (B) Effect of ABA treatment on GFP-PYL2, GFP-PYL4, and GFP-PYL8 protein levels. Seedlings expressing GFP-tagged PYL proteins were either mock- or 50 μM ABA-treated for 6 h. Immunoblot analysis using anti-GFP was performed to quantify protein levels. (C) ABA treatment leads to selective accumulation of GFP-PYL8 in the nucleus. CLSM analysis of the Arabidopsis root differentiation zone in lines expressing GFP-tagged PYL proteins that were either mock- or ABA-treated for 1 h. (Scale bars, 30 μm.)
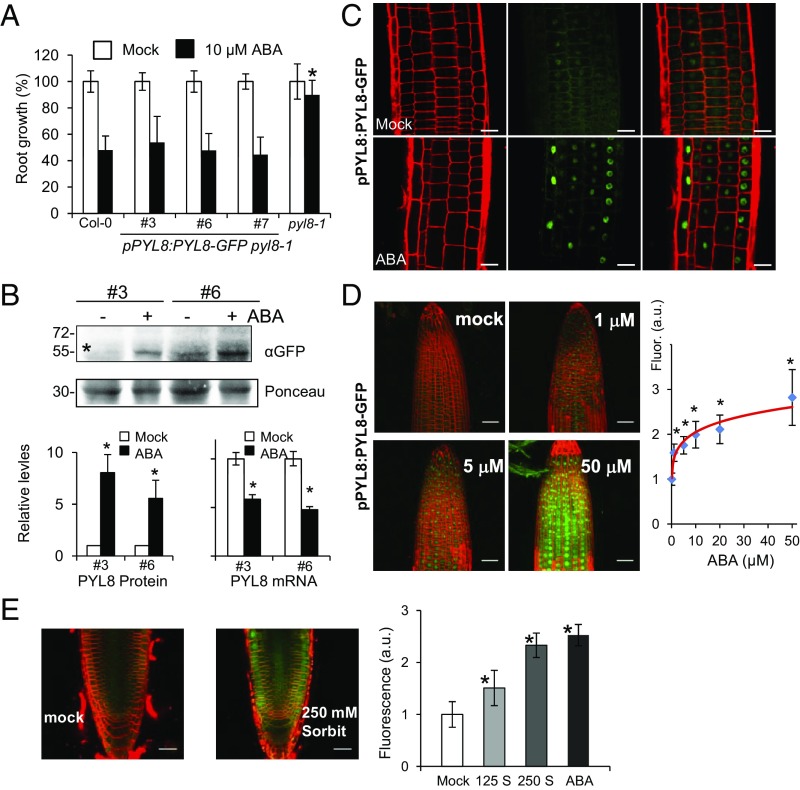
To gain insight on the root localization of PYL8, we expressed PYL8-GFP driven by its own promoter in a pyl8-1 mutant background (ProPYL8:PYL8-GFP pyl8-1). PYL8-GFP complemented the ABA-insensitive pyl8-1 phenotype in a root growth assay, indicating that PYL8-GFP is functional (Fig. 2A). Next, we analyzed PYL8-GFP protein levels in roots by immunoblotting and found that these protein levels increased five- to sevenfold after ABA treatment (Fig. 2B). qRT-PCR analysis showed that ABA treatment does not induce up-regulation of the PYL8 transcript; in fact, ABA down-regulates PYL8 gene expression (Fig. 2B), in agreement with previous reports in seedlings (22, 23). Instead, ABA treatment led to an accumulation of PYL8-GFP in the nucleus, as observed for GFP-PYL8 expressed from a 35S promoter (Figs. 1C and 2C). Next, we investigated whether PYL8-GFP fluorescence may report changes in ABA concentration in the root. A dose–response analysis indicated that PYL8-GFP fluorescence was sensitive to changes in ABA concentration induced by exogenous ABA addition or osmotic stress (Fig. 2 D and E and SI Appendix, Fig. S2 A and B). Finally, a kinetic analysis of PYL8-GFP fluorescence was performed in response to 10 μM ABA treatment, and a gradual increase of the fluorescent signal, which could be detected from 30 min after ABA treatment, was observed (SI Appendix, Fig. S2C). We conclude that ABA specifically stabilizes the PYL8 receptor and leads to its accumulation in root-cell nuclei.
Fig. 2.
ABA increases PYL8 protein levels in roots through a posttranscriptional mechanism. (A) PYL8-GFP complements the ABA-insensitive pyl8-1 phenotype. The 5-d-old seedlings germinated on Murashige and Skoog plates were transferred to new plates lacking or supplemented with 10 μM ABA, and quantification of root growth was performed after 10 d. Data are averages ±SD from three independent experiments (n = 20). A single asterisk (*) indicates P < 0.05 (Student’s t test) compared with Col-0 in the same assay conditions. (B) ABA treatment leads to accumulation of PYL8-GFP protein and down-regulation of PYL8-GFP mRNA in roots. The 10-d-old seedlings expressing PYL8-GFP were either mock- or 50 μM ABA-treated for 3 h and protein or RNA extracted from root tissue. Immunoblot analysis using anti-GFP was performed to quantify protein levels of PYL8-GFP (asterisk) in roots. A major 30-kDa root protein was used to normalize protein loading. qRT-PCR analyses were performed to quantify mRNA expression of PYL8-GFP. A single asterisk (*) indicates P < 0.05 (Student’s t test) compared with mock-treated samples. (C) ABA treatment leads to accumulation of PYL8-GFP in the nucleus. CLSM analysis of Arabidopsis root expressing ProPYL8:PYL8-GFP in the pyl8-1 background after mock or ABA treatment for 6 h. (Scale bars, 25 μm.) (D) Dose–response analysis of PYL8-GFP accumulation in response to treatment with the indicated ABA concentrations for 6 h. Fluorescence was quantified in arbitrary units (a.u.) using images acquired by CLSM. (E) Accumulation of PYL8-GFP after 250 mM sorbitol treatment. Fluorescence was measured after treatment with 125 or 250 mM sorbitol (S) or 50 μM ABA for 3 h. (Scale bars, 30 μm.) A single asterisk indicates P < 0.05 (Student’s t test) compared with mock-treated samples.
PYL8 Stabilization in Roots Is Triggered by Ligand Binding and Requires PP2C Interaction.
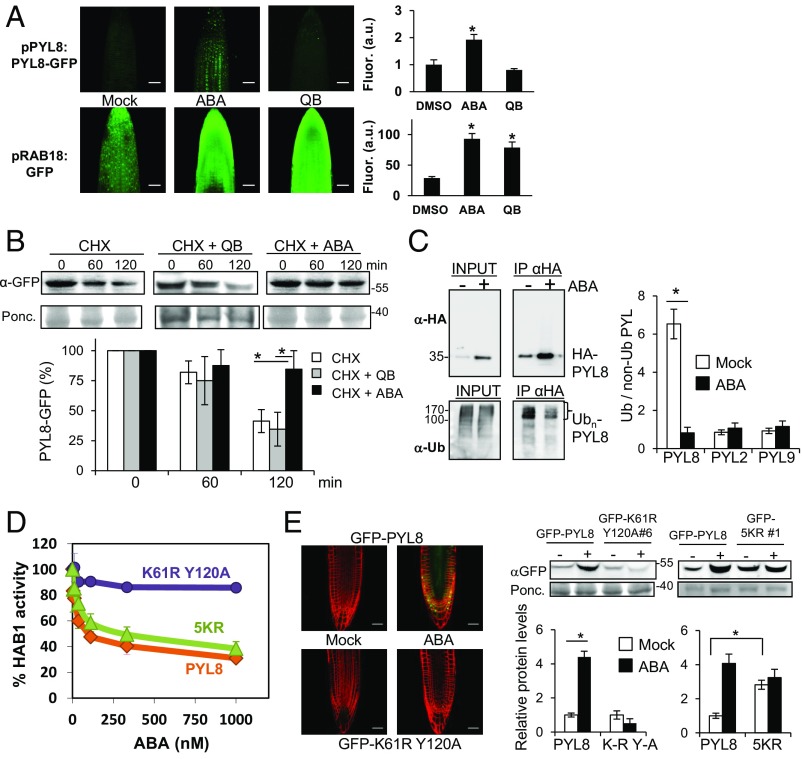
To investigate whether the enhanced PYL8-GFP fluorescence observed after ABA treatment requires ligand perception by PYL8 or simply reflects hormone signaling through other ABA receptors that also operate in the root (13), we switched on ABA signaling through quinabactin (QB) treatment. QB is an ABA agonist that does not activate the PYL8 receptor (SI Appendix, Fig. S3A) but instead activates ABA signaling primarily through dimeric ABA receptors such as PYR1, PYL1, and PYL2, which are expressed at a high level in the root and contribute to the quantitative regulation of ABA signaling (13, 24, 25). QB treatment was able to up-regulate the expression of the ABA-responsive ProRAB18:GFP reporter, but, in contrast to ABA, QB did not enhance PYL8-GFP fluorescence (Fig. 3A). Therefore, the PYL8-GFP accumulation appears to require ABA perception by PYL8.
Fig. 3.
ABA perception by PYL8 is required to trigger its stabilization via reduced receptor ubiquitination. (A) QB treatment does not lead to accumulation of PYL8-GFP. QB induces ABA signaling in the root as revealed by the pRAB18:GFP reporter. CLSM analysis of Arabidopsis root apex expressing either ProPYL8:PYL8-GFP in the pyl8-1 background or ProRAB18:GFP in wild type after mock, 20 μM ABA, or 50 μM QB treatment for 1 h. A single asterisk indicates P < 0.05 (Student’s t test) compared with mock (DMSO)-treated sample. (B) ABA prevents degradation of PYL8-GFP in roots whereas QB does not. The 10-d-old seedlings expressing PYL8-GFP were treated with 50 μM ABA for 6 h to induce accumulation of PYL8. After washing out ABA, a CHX treatment in the absence or presence of 50 μM ABA or QB was performed for 60 or 120 min. Protein extracts of roots were analyzed using an anti-GFP antibody (α-GFP). The histogram shows the quantification of the PYL8-GFP protein during the CHX treatment. A single asterisk indicates P < 0.05 (Student’s t test) when CHX + ABA treatment was compared with CHX or CHX + QB treatments, respectively. (C) ABA treatment increases total HA-PYL8 protein levels in root but reduces polyubiquitinated PYL8 forms. Protein extracts were prepared from mock or ABA-treated root samples and submitted to immunoprecipitation using anti-HA antibodies. Immunoprecipitated PYL8 (IP αHA) was analyzed by immunoblotting using anti-HA and anti-Ub (P4D1) antibodies. The ratio of polyubiquitinated to non-Ub PYL8, PYL2, and PYL9 protein was quantified in mock- and ABA-treated samples. A single asterisk indicates P < 0.05 (Student’s t test) compared with the ABA-treated sample. (D) The PYL8K61R Y120A mutant is unable to inhibit PP2C HAB1, whereas activity of PYL85KR is similar to PYL8 wild type. Phosphatase activity of HAB1 was measured in the presence of PYL8 wild type, PYL8K61R Y120A, or PYL85KR mutants and different ABA concentrations. (E) CLSM of Arabidopsis root apex (Left) and immunoblot analysis of root protein extracts reveal that the PYL8K61R Y120A mutant is not stabilized by ABA. Transgenic seedlings expressing GFP-PYL8, GFP-PYL8K61R Y120A, or GFP-PYL85KR were either mock- or 50 μM ABA-treated for 3 h, root protein extracts were prepared, and immunoblot analysis was performed using anti-GFP to quantify protein levels (Right). A single asterisk (*) indicates P < 0.05 (Student’s t test) when the indicated samples were compared. (Scale bars, 30 μm.)
To ascertain whether ABA treatment leads to decreased degradation of PYL8 in roots, we performed a CHX ± ABA experiment using the ProPYL8:PYL8-GFP pyl8-1 line and analyzed PYL8 protein levels in roots (Fig. 3B). While CHX treatment in the absence of ABA led to a 60% reduction of PYL8 after 120 min, the simultaneous presence of ABA slowed PYL8 degradation to a reduction of only 20%. QB was not effective in slowing PYL8 degradation (Fig. 3B). ABA may prevent degradation of PYL8 in roots through reduced polyubiquitination of the receptor (16). To investigate this possibility, we performed immunoprecipitation of HA-PYL8 in mock and ABA-treated root samples. Immunoprecipitated HA-PYL8 was analyzed using anti-HA and anti-Ub (P4D1) antibodies and the ratio of Ub(n)-PYL8 to PYL8 was found to be approximately sixfold higher in mock compared with ABA-treated roots (Fig. 3C). Thus, even though the total PYL8 protein level was increased about fourfold after ABA treatment, the amount of polyubiquitinated PYL8 was diminished in ABA-treated roots compared with mock conditions (Fig. 3C). In contrast, ABA treatment did not affect the ubiquitination ratio of PYL2 and PYL9 compared with mock conditions (Fig. 3C).
The results described above using QB treatment suggest that PYL8 stabilization and/or accumulation requires ABA perception by the receptor. To obtain further evidence, we analyzed the PYL8K61R Y120A mutant, which is predicted to be unable to bind ABA because it is impaired in the salt bridge formed between the highly conserved K61 residue and the ABA carboxylate, and the hydrogen bond formed by Y120 with the ABA carboxylate group through an internal water molecule (13). Accordingly, the recombinant PYL8K61R Y120A protein was unable to inhibit PP2C HAB1 (Fig. 3D). We generated GFP-PYL8K61R Y120A transgenic lines and analyzed them following ABA treatment (Fig. 3E). Both CLSM and immunoblot analysis of root protein extracts revealed that GFP-PYL8K61R Y120A fails to accumulate after ABA treatment, in contrast to wild-type PYL8 (Fig. 3E). We conclude that either ABA perception and/or PP2C interaction are required to trigger PYL8 stabilization. Recent proteomic studies led to the identification of ubiquitinated residues in PYL8 (26). In contrast to the K61 residue that affects ABA binding, other N-terminal Lys residues of PYL8 are not predicted to be involved in ABA binding (26). We therefore decided to mutate those Lys residues that are potential ubiquitination sites (26) but presumably do not impair PYL8 function—i.e., Lys24, Lys38, Lys59, Lys70, and Lys84—and generated the quintuple Lys-Arg PYL8 mutant (abbreviated as 5KR). We found that in vitro activity of the 5KR mutant was comparable to wild-type PYL8 (Fig. 3D). Interestingly, GFP- and HA-tagged lines of PYL85KR showed a two- to threefold increase of protein levels compared with wild type in the absence of exogenous ABA treatment (Fig. 3E and SI Appendix, Fig. S3B). We analyzed seedling establishment in the presence of 0.5 μM ABA and ABA-mediated inhibition of root growth in response to 10 μM ABA (SI Appendix, Fig. S3 C and D). In both cases, HA-tagged lines of PYL85KR showed enhanced sensitivity to ABA compared with lines expressing wild-type PYL8. These results suggest that ubiquitination and regulation of PYL8 protein stability is crucial for proper response to ABA.
To investigate whether intracellular movement of PYL8 affects plant sensitivity to ABA, we compared subcellular localization and ABA sensitivity of GFP-PYL8, GFP-PYL85KR, and GFP-PYL8K61R Y120A lines (SI Appendix, Fig. S4). The line expressing GFP-PYL8, which accumulates GFP-PYL8 in the nucleus after ABA treatment, shows enhanced sensitivity in ABA-mediated inhibition of root growth and repression of lateral root formation compared with wild-type Col-0 (SI Appendix, Fig. S4). Some nuclear accumulation of GFP-PYL85KR was observed at endogenous ABA levels (SI Appendix, Fig. S4A), which correlated with enhanced sensitivity to ABA compared with GFP-PYL8 line (SI Appendix, Fig. S4 B and C; see also SI Appendix, Fig. S3 C and D). In contrast, lines expressing GFP-PYL8K61R Y120A, which does not accumulate in the nucleus after ABA treatment, behave as wild-type Col-0 in root growth assays (SI Appendix, Fig. S4). Therefore, nuclear accumulation of GFP-PYL8 induced by ABA is required for ABA response.
PYL8 Behaves Non-Cell-Autonomously in Root Tissues.
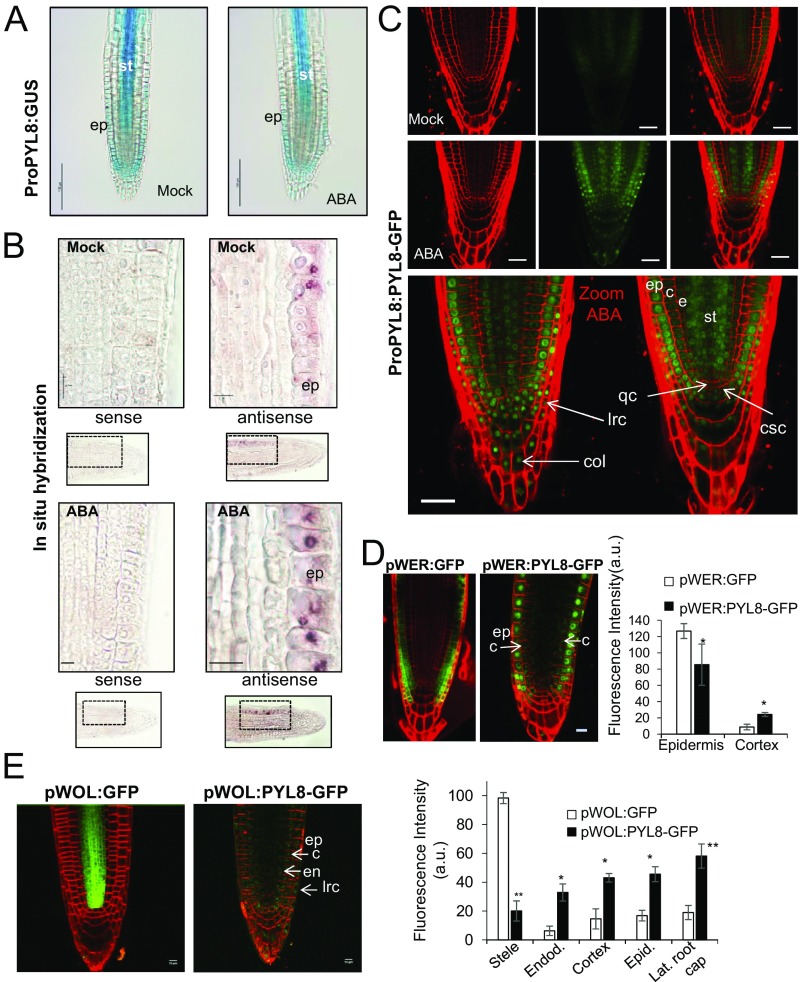
To study the effect of ABA treatment on PYL8 expression in the root apex, we performed β-glucuronidase (GUS) staining after mock or 50 μM ABA treatment for 3 h (Fig. 4A). In control root apices, GUS expression driven by the PYL8 promoter was detected in LRC), root epidermis, and stele cells (SI Appendix, Fig. S5 A and B), as previously reported (13). This PYL8 expression pattern was similar in ABA-treated plants, although some attenuation of PYL8 expression was apparent (Fig. 4A). To gain further insight about the expression of PYL8 in the root apex, we performed in situ hybridization using a digoxigenin-labeled antisense RNA probe to detect the PYL8 mRNA in Col-0 (Fig. 4B). Under both mock and ABA-treatment conditions, the PYL8 transcript was detected mainly in the epidermis. Weak staining of stele cells using the antisense RNA probe was also detected, but was attenuated after ABA treatment (Fig. 4B).
Fig. 4.
Expression of PYL8 transcript and protein in roots. (A) GUS expression driven by the ProPYL8:GUS gene in the root apex. GUS staining after mock or 50 μM ABA treatment. (Scale bars, 100 μm.) (B) Localization of PYL8 mRNA in the root apex. In situ hybridization was performed on longitudinal sections of the root apex of mock- or 50 μM ABA-treated seedlings using PYL8 antisense or sense probes. The PYL8 transcript was visualized using anti-digoxigenin–AP antibody and nitro-blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate staining. (Scale bars, 10 μm.) (C) CSLM visualization of PYL8-GFP driven by the PYL8 promoter after mock or ABA treatment. Localization of PYL8-GFP after ABA treatment was detected in the root apical meristem, columella, and LRC. (Scale bars, 25 μm.) c, cortex; col, columella; csc, columella stem cells; e, endodermis; ep, epidermis; lrc, LRC; qc, quiescent center; st, stele. (D and E) CSLM visualization of GFP or PYL8-GFP proteins expressed under the control of the pWER and pWOL promoters in pyl8-2 background. To stabilize PYL8, seedlings were treated with 50 μM MG132 and ABA for 6 h. (Scale bars, 10 μm.) Histograms indicate tissue-scale measurements of CLSM images using CellSeT software. *P < 0.05 and **P < 0.01 (Student’s t test) compared with GFP control.
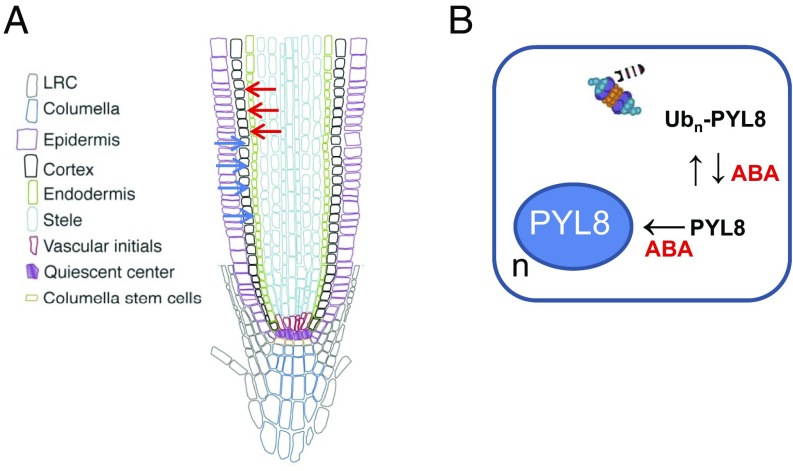
Since ABA signaling has been reported to regulate distinct processes in different root tissues (3, 9, 19) and PYL8 plays a key role in root ABA perception, we investigated the localization of the PYL8 protein in the root apex. Using CLSM, we analyzed the ProPYL8:PYL8-GFP lines after either mock or ABA treatments. Interestingly, expression of PYL8-GFP in mock-treated roots was weakly detected in the epidermis and stele, whereas following ABA treatment it was clearly visible in LRC, epidermis, cortex, endodermis, stele, QC, and columella cells (Fig. 4C). PYL8-GFP expression in the epidermis was markedly enhanced by ABA treatment (Movie S1). Since the PYL8 transcript was not detected in the cortex or endodermis, we conclude that the PYL8 protein is translocated to adjacent tissues from cells where it is initially synthesized. This movement is reminiscent of mobile transcription factors (TFs) such as SHORT ROOT (SHR) that regulate root development (27). To further investigate the movement of PYL8 between different root layers, we expressed PYL8-GFP driven by WER and WOL tissue-specific promoters (3) (SI Appendix) and examined the localization of the fluorescent protein by CLSM (Fig. 4 D and E). Quantification of CLSM images was performed with an updated version of CellSeT software (Fig. 4 D and E), which performs tissue-scale measurements from confocal microscope images (SI Appendix). When PYL8-GFP was expressed under control of the WER promoter, which drives expression in the epidermis and LRC (as confirmed by a pWER:GFP control), we could also detect PYL8-GFP at least in cortex cells (Fig. 4D). In the case of WOL-driven expression, whereas the GFP control was detected in the root vascular cylinder and pericycle as expected, we could detect PYL8-GFP additionally in the LRC, epidermis, cortex, and endodermis (Fig. 4E). Taken together, these results suggest that the PYL8 protein can move from the cells where its transcript is produced.
The ABA-insensitive phenotype of pyl8-2 in root growth assays was not complemented when PYL8 was expressed driven by either WER or WOL promoters (SI Appendix, Fig. S6). PYL8 mRNA expression driven by its endogenous promoter is mostly localized in the LRC, epidermis, and the vascular bundle and is able to complement the ABA insensitivity of pyl8 in root growth assays (Fig. 2A). Driven by the WER promoter, PYL8 protein was markedly detected in LRC and epidermis and weakly in cortex, suggesting that complementation requires intercellular movement of the protein to other root layers. Driven by the WOL promoter, PYL8 protein was detected in most of root tissues; however, expression levels were markedly lower than those obtained using the endogenous PYL8 promoter (Fig. 4 C and E). Therefore, although intercellular movement of the protein seems to be necessary, it is not sufficient unless suitable levels of the protein are achieved. Taken together, these results suggest that combined expression in the epidermis and vascular bundle is required for complementation of the pyl8 phenotype (Fig. 2A).
Finally, given the importance of ABA signaling in the mature root, expression of PYL8-GFP was examined in the root differentiation zone after ABA treatment and was localized in the epidermis, cortex, endodermis, pericycle, and vascular tissue (SI Appendix, Fig. S5 C and D). In particular, a 3D reconstruction after CSLM imaging revealed the presence of PYL8 in procambial cells of the vascular tissue (Movie S2), which suggests a possible role of PYL8 in vascular development.
Discussion
We report that, unlike other ABA receptors, PYL8 exhibits distinct regulatory properties. For example, PYL8 is transcribed in the epidermis and stele of the root apex, but the PYL8 protein is also present in the cortex and endodermis. Hence, comparison of the PYL8 transcript expression with the localization of the PYL8 protein reveals translocation of the ABA receptor from epidermis or stele to adjacent tissues, including the cortex and endodermis (Fig. 5A). Moreover, when PYL8-GFP was expressed driven by the WOL promoter, which is specifically expressed in the root vascular cylinder, the protein moved as far as the LRC (Fig. 4E). Hence, PYL8 appears to regulate ABA signaling in the root apex through a non-cell-autonomous mechanism. However, it seems that both intercellular movement and appropriated protein levels are required for complementation of the ABA-insensitive phenotype of pyl8 in root growth assays (Figs. 2A and 4C). The regulatory behavior exhibited by PYL8 is reminiscent of mobile TFs involved in plant development that perform non-cell-autonomous actions by trafficking from cell to cell through plasmodesmata (28, 29). For example, SHORT ROOT (SHR), which is synthesized in all stele cells except the phloem, moves to adjacent cells including the endodermis and phloem (28, 29). The movement of SHR relies on the endomembrane system, interaction with the SHR interacting embryonic lethal (SIEL) protein, and association with endosomes in a SIEL-dependent manner (27). Interestingly, ABA receptors traffic through endosomes (20, 21). In addition to degradation in the vacuole, endosome trafficking can promote signaling functions in both plants and animals by facilitating the movement of proteins between cells (30).
Fig. 5.
Proposed model for ABA-dependent stabilization and movement of the non-cell-autonomous ABA receptor PYL8. (A) PYL8 translocation from epidermis (blue arrows) and stele (red arrows) to adjacent tissues. Translocation could be promoted by increased ABA levels or follow a default mechanism that is reinforced by ABA-induced accumulation of PYL8. The intercellular movement of PYL8 is accompanied by intracellular trafficking and increased nuclear accumulation in response to ABA. (B) ABA reduces polyubiquitination of PYL8 through an unknown mechanism, which stabilizes and increases PYL8 protein levels. ABA also enhances PYL8 localization in the nucleus (n), which prevents vacuolar degradation and might represent an additional mechanism to increase PYL8 levels.
We also report that ABA specifically stabilizes PYL8 compared with other ABA receptors and induces its accumulation in root nuclei. We demonstrate that this requires ABA perception by PYL8 and leads to diminished ubiquitination of PYL8 in roots. Thus, the inactive PYL8K61R Y120A protein is not stabilized by ABA in roots, and activation of root ABA signaling in the absence of ligand binding by PYL8 (as occurs after QB treatment) fails to stabilize PYL8. Different reporters for direct visualization of ABA concentration changes have been described (31–34). However, reporters based on the overexpression of PP2Cs or ABA receptors affect ABA sensitivity per se; for example, ABAleon lines show reduced sensitivity to ABA (33) whereas ABACUS lines show enhanced ABA-mediated inhibition of root growth compared with wild type (34). The ProPYL8:PYL8-GFP pyl8-1 line here described showed wild-type sensitivity to ABA-mediated inhibition of root growth (Fig. 2A), which is a requisite for a root ABA biosensor. Additionally, both kinetic and dose–response analyses indicate that PYL8-GFP might be used as an ABA biosensor in roots to detect exogenous ABA (SI Appendix, Fig. S2). Moreover, the PYL8-GFP biosensor harbors the potential to specifically identify PYL8 agonists through in vivo screening, which is required to confirm the bioactivity of molecules identified by in vitro or in silico screening. However, the current version of the PYL8-GFP biosensor needs to be improved to a ratiometric version to achieve a wider dynamic range of sensitivity to ABA changes (33, 34).
Although the main function of PYL8 is the ABA-dependent inhibition of clade A PP2Cs such as ABI1 and PP2CA (13), an additional role independent of the core ABA-signaling pathway has also been reported (35). Thus, PYL8 promotes lateral root growth by interacting with the TFs MYB77, MYB44, and MYB73 to augment auxin signaling, which points to an additional nuclear role of PYL8 (35). ABA perception by PYL8 in the nucleus is required to inhibit PP2C activity and hence relieve repression of ABA signaling by the SWI/SNF chromatin remodeling ATPase BRAHMA (36). Failure to inhibit nuclear PP2C activity, as exemplified by the phosphatase abi1Gly180Asp that is refractory to inhibition by ABA receptors, blocks ABA signaling and inhibits ABA response element-binding bZIPs that mediate transcriptional response to ABA (37, 38). Interestingly, we observed enhanced nuclear localization of PYL8 in root cells after ABA treatment, which suggests that both intra- and intercellular movement of PYL8 occurs (Fig. 5B). Further insight into the cellular mechanisms involved in movement of ABA receptors is a key issue for future research. We note that the enhanced localization of PYL8 in the nucleus, together with diminished polyubiquitination induced by ABA, can reduce degradation of the receptor through the vacuolar pathway (20, 21) and contributes to stabilization of the receptor (Fig. 5B). The mechanism whereby ABA specifically reduces polyubiquitination of PYL8 is another challenging issue to be investigated.
Materials and Methods
Detailed description is provided in SI Appendix for plant material and growth conditions; generation of HA-tagged and GFP-tagged lines and ProPYL8:PYL8-GFP pyl8-1 lines; expression of PYL8-GFP in root driven by tissue-specific promoters; generation of PYL8 mutations; in vivo protein analysis and degradation assays; root growth assays; RT-qPCR analysis; GUS staining; PP2C inhibition assays; RNA in situ hybridization; CLSM; and measurements and statistical analysis.
Supplementary Material
Acknowledgments
Work in the P.L.R. and F.M. laboratories was supported by the Ministerio de Ciencia e Innovacion, Fondo Europeo de Desarrollo Regional and Consejo Superior de Investigaciones Cientificas Grants BIO2014-52537-R and BIO2017-82503-R (to P.L.R.) and BIO2015-64307-R (to F.M.). J.L.-J. was supported by a Juan de la Cierva contract from Ministerio de Economia y Competitividad (MINECO) and by the Marie Skłodowska-Curie Action H2020-MSCA-IF-2015-707477. B.B.-P. was funded by Programa VALi+d GVA APOSTD/2017/039. J.J. was supported by a FPI contract from MINECO and M.A.F. by a Formacion de Profesorado Universitario contract from MINECO. D.D. and M.J.B. were supported by Biotechnology and Biological Sciences Research Council Grant BB/M002136/1 and Leverhulme Trust Grant RPG-2016-409.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815410115/-/DCSupplemental.
References
- 1.Ubeda-Tomás S, Beemster GT, Bennett MJ. Hormonal regulation of root growth: Integrating local activities into global behaviour. Trends Plant Sci. 2012;17:326–331. doi: 10.1016/j.tplants.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Bao Y, et al. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc Natl Acad Sci USA. 2014;111:9319–9324. doi: 10.1073/pnas.1400966111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietrich D, et al. Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat Plants. 2017;3:17057. doi: 10.1038/nplants.2017.57. [DOI] [PubMed] [Google Scholar]
- 4.Harris JM. Abscisic acid: Hidden architect of root system structure. Plants (Basel) 2015;4:548–572. doi: 10.3390/plants4030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spollen WG, LeNoble ME, Samuels TD, Bernstein N, Sharp RE. Abscisic acid accumulation maintains maize primary root elongation at low water potentials by restricting ethylene production. Plant Physiol. 2000;122:967–976. doi: 10.1104/pp.122.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp RE, et al. Root growth maintenance during water deficits: Physiology to functional genomics. J Exp Bot. 2004;55:2343–2351. doi: 10.1093/jxb/erh276. [DOI] [PubMed] [Google Scholar]
- 7.Deak KI, Malamy J. Osmotic regulation of root system architecture. Plant J. 2005;43:17–28. doi: 10.1111/j.1365-313X.2005.02425.x. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Guzman M, et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan L, et al. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 2013;25:324–341. doi: 10.1105/tpc.112.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng W, Lindner H, Robbins NE, II, Dinneny JR. Growing out of stress: The role of cell- and organ-scale growth control in plant water-stress responses. Plant Cell. 2016;28:1769–1782. doi: 10.1105/tpc.16.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng Y, et al. A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell. 2013;25:2132–2154. doi: 10.1105/tpc.113.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi N, Goto N, Okada K, Takahashi H. Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana. Planta. 2002;216:203–211. doi: 10.1007/s00425-002-0840-3. [DOI] [PubMed] [Google Scholar]
- 13.Antoni R, et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013;161:931–941. doi: 10.1104/pp.112.208678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barberon M, et al. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell. 2016;164:447–459. doi: 10.1016/j.cell.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Ondzighi-Assoume CA, Chakraborty S, Harris JM. Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. Plant Cell. 2016;28:729–745. doi: 10.1105/tpc.15.00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irigoyen ML, et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell. 2014;26:712–728. doi: 10.1105/tpc.113.122234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bueso E, et al. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014;80:1057–1071. doi: 10.1111/tpj.12708. [DOI] [PubMed] [Google Scholar]
- 18.Swarup R, et al. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol. 2005;7:1057–1065. doi: 10.1038/ncb1316. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, et al. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010;64:764–774. doi: 10.1111/j.1365-313X.2010.04367.x. [DOI] [PubMed] [Google Scholar]
- 20.Belda-Palazon B, et al. FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. Plant Cell. 2016;28:2291–2311. doi: 10.1105/tpc.16.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F, et al. ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Mol Plant. 2016;9:1570–1582. doi: 10.1016/j.molp.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 22.Santiago J, et al. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- 23.Szostkiewicz I, et al. Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J. 2010;61:25–35. doi: 10.1111/j.1365-313X.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto M, et al. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA. 2013;110:12132–12137. doi: 10.1073/pnas.1305919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao M, et al. An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Res. 2013;23:1043–1054. doi: 10.1038/cr.2013.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo MC, et al. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci Signal. 2015;8:ra89. doi: 10.1126/scisignal.aaa7981. [DOI] [PubMed] [Google Scholar]
- 27.Wu S, Gallagher KL. The movement of the non-cell-autonomous transcription factor, SHORT-ROOT relies on the endomembrane system. Plant J. 2014;80:396–409. doi: 10.1111/tpj.12640. [DOI] [PubMed] [Google Scholar]
- 28.Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- 29.Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol. 2004;14:1847–1851. doi: 10.1016/j.cub.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 30.Pálfy M, Reményi A, Korcsmáros T. Endosomal crosstalk: Meeting points for signaling pathways. Trends Cell Biol. 2012;22:447–456. doi: 10.1016/j.tcb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christmann A, Hoffmann T, Teplova I, Grill E, Müller A. Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol. 2005;137:209–219. doi: 10.1104/pp.104.053082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TH, et al. Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol. 2011;21:990–997. doi: 10.1016/j.cub.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waadt R, et al. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife. 2014;3:e01739. doi: 10.7554/eLife.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones AM, et al. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife. 2014;3:e01741. doi: 10.7554/eLife.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, et al. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal. 2014;7:ra53. doi: 10.1126/scisignal.2005051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peirats-Llobet M, et al. A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol Plant. 2016;9:136–147. doi: 10.1016/j.molp.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Moes D, Himmelbach A, Korte A, Haberer G, Grill E. Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 2008;54:806–819. doi: 10.1111/j.1365-313X.2008.03454.x. [DOI] [PubMed] [Google Scholar]
- 38.Lynch T, Erickson BJ, Finkelstein RR. Direct interactions of ABA-insensitive(ABI)-clade protein phosphatase(PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol Biol. 2012;80:647–658. doi: 10.1007/s11103-012-9973-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.