Significance
SNARE proteins facilitate the synaptic vesicle fusion with the plasma membrane. The structure of the fusion pore and the mechanism of its formation are not clearly understood. Using coarse-grained molecular dynamics simulations of a nanodisc bridged by four SNARE proteins to a bilayer, we identify the importance of the C termini of synaptobrevin 2 and syntaxin 1 in facilitating fusion pore formation. The conductances of the simulated fusion pores agree with experimental values, and SNARE mutants that inhibit fusion experimentally fail to form fusion pores in the simulations. Confinement of SNARE complexes is essential for rapid fusion pore formation, but release of the SNARE complexes from this confinement is required for fusion pore expansion.
Keywords: membrane fusion, exocytosis, transmitter release, molecular dynamics, nanodisc
Abstract
Release of neurotransmitters from synaptic vesicles begins with a narrow fusion pore, the structure of which remains unresolved. To obtain a structural model of the fusion pore, we performed coarse-grained molecular dynamics simulations of fusion between a nanodisc and a planar bilayer bridged by four partially unzipped SNARE complexes. The simulations revealed that zipping of SNARE complexes pulls the polar C-terminal residues of the synaptobrevin 2 and syntaxin 1A transmembrane domains to form a hydrophilic core between the two distal leaflets, inducing fusion pore formation. The estimated conductances of these fusion pores are in good agreement with experimental values. Two SNARE protein mutants inhibiting fusion experimentally produced no fusion pore formation. In simulations in which the nanodisc was replaced by a 40-nm vesicle, an extended hemifusion diaphragm formed but a fusion pore did not, indicating that restricted SNARE mobility is required for rapid fusion pore formation. Accordingly, rapid fusion pore formation also occurred in the 40-nm vesicle system when SNARE mobility was restricted by external forces. Removal of the restriction is required for fusion pore expansion.
Transmitter release from secretory vesicles begins with the formation of a narrow fusion pore (1). The soluble N-ethyl-maleimide-sensitive factor attachment protein receptor (SNARE) proteins form a minimal membrane fusion machinery (2). The v-SNARE synaptobrevin-2 (syb2/VAMP2) and the t-SNARE syntaxin-1 (stx1) are anchored to the vesicle and plasma membrane, respectively, by their helical transmembrane domains (TMDs) (3), while the t-SNARE SNAP-25 is anchored at the plasma membrane by four palmitoylated cysteines (4). The SNARE motifs of syb2, stx1, and SNAP-25 associate via zippering of conserved heptad repeats to form a tight four-helix bundled trans-SNARE complex (5). The zippering of SNAREs from the membrane-distal N termini toward the membrane-proximal C termini is believed to pull the two opposing membranes together to drive fusion (6).
A number of studies in which either SNARE transmembrane domains were replaced by lipid anchors (7–9) or a TMD was partially deleted (10), or in which the C terminus of the syb2 TMD was extended by polar residues (11), indicated that fusion was inhibited or even arrested, pointing to an important function of the TMD in fusion pore formation. The precise nanomechanical mechanism of fusion and the structure of the nascent pore remain unclear. One model proposes a lipid-based fusion pore (12), while other studies have suggested a proteinaceous fusion pore with the TMDs of stx1 and syb2 lining the fusion pore, like an ion channel or gap junction pore (9). Recent experiments using varied-sized nanodiscs (NDs) (13) support the hypothesis that the fusion pore is a hybrid structure composed of both lipids and proteins (14); however, there is still no structural model of the fusion pore.
Conventional in vitro reconstitution experiments use liposome/liposome fusion assays to study SNARE mediated fusion (2, 7, 15). Accordingly, SNARE-mediated fusion has been studied using coarse-grained (CG) molecular dynamics (MD) simulations of 20-nm vesicles and has been shown to require microsecond-long simulations to observe fusion (16). Recently, ND/liposome systems in which various copy numbers of syb2 were incorporated in an ND while the t-SNAREs were present on a liposome have been used experimentally to study SNARE-mediated membrane fusion (13, 17). The small dimensions of the ND compared with a spherical vesicle makes such systems ideally suited for MD simulations without introducing extreme curvature, which is well known to strongly influence the propensity of fusion (18–20).
MARTINI-based CGMD simulations have been used in several studies of membrane fusion (16, 21–23). To elucidate the fusion pore structure and the mechanism of its formation, we performed unbiased CGMD simulations of ND/bilayer fusion using physiological mixed lipid/cholesterol membranes with asymmetric leaflet composition (24) bridged by four trans-SNARE complexes. Our results show that zippering of the SNARE complex exerts a mechanical force on the membranes, pulling the two membranes together. The force transfer occurs via the helical linker and TMDs, leading to changes in membrane curvature and spontaneous fusion pore formation. The early fusion pore has a proteolipidic structure with a few lipid head groups along with polar residues near the C termini of syb2 and stx1 lining the nascent fusion pore. The simulated fusion pores are consistent with experimental fusion pore conductance measurements. Two SNARE mutants that strongly inhibit fusion experimentally did not produce fusion pores in our ND/bilayer simulations.
Performing simulations with a complex mixture of lipids and asymmetric leaflets aims to understand fusion in physiological membranes but at the same time introduces uncertainty in relating the observed fusion mechanisms to specific system properties. The use of asymmetric membranes has been shown to have significant effects on the fusion barriers (25). Thus, the simulations of the complex system were complemented by simulations using a comparatively well-defined POPC-membrane vesicle system with a single SNARE complex.
Restricting the mobility of the SNARE complexes by the ND or by application of suitable forces in a vesicle/bilayer system is critical for rapid fusion pore formation, but removal of this restriction is required to prevent rapid closure and allow fusion pore expansion.
Results
Nanodisc/Bilayer Assembly Bridged by Four SNARE Complexes.
Since a previous experimental study (17) had shown that three or more SNARE complexes are required for successful fusion and that in the crystal structure, four SNARE complexes are associated through an X-shaped assembly of the TMDs (3), we used an assembly of four SNARE complexes. The system setup consisted of a ND placed on top of a bilayer, with the two bridged by four partially (up to layer +5) unzipped SNARE complexes (Fig. 1A). The t-SNAREs were placed in a planar bilayer resembling the synaptosomal plasma membrane lipid composition (26) (SI Appendix, Table S1) and the syb2 TMDs in an ND with lipid composition presented in SI Appendix, Table S2.
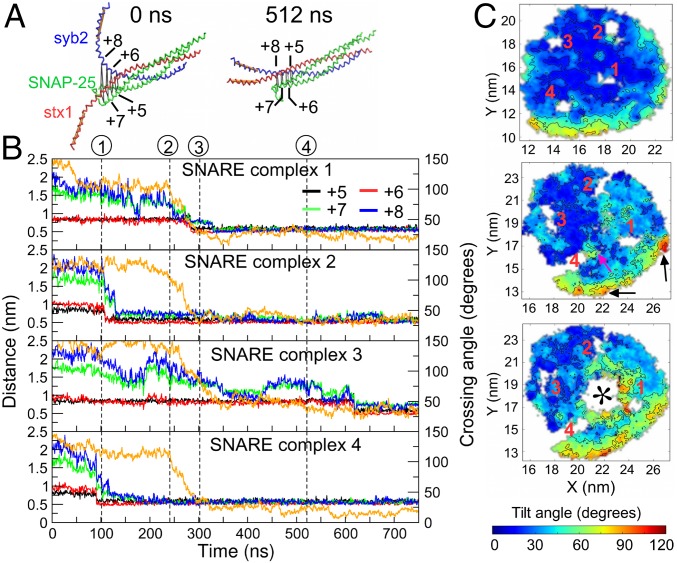
Fig. 1.
Snapshots of ND-bilayer fusion showing key intermediates observed in Sim1. (A) Initial, energy-minimized structure. The ND is shown in orange, and the bilayer is shown in tan. (B) Snapshot after 10 ns of equilibration showing formation of a “dimple” in the bilayer. (C) Zoom-in view (at 239 ns) showing the association of three splayed lipids highlighted in a ball-and-stick representation, two (green and cyan) from the ND and one (magenta) from the bilayer. (D) Expansion of hydrophobic core to from the stalk (at 245 ns). (E) Evolved pore (1 μs). Waters are shown as cyan spheres. (F) Pore closure at 2.1 μs.
Starting from the energy-minimized system (Fig. 1A), a 10-ns equilibration run was performed in which the stx1 TMD tilts spontaneously, pushing the membrane upward, generating a local bending of the plasma membrane bilayer (Fig. 1B), previously referred to as a “dimple” (27). This site later forms the point of contact initiating stalk formation in the ND/bilayer fusion simulations (Fig. 1 C and D).
Evolution of Trans-SNARE Complex Zipping Induces TMD and Lipid Tilting.
During the initial simulation phase, a spontaneous zipping of layers +5 and +6 occurs (Fig. 2A), pulling the ND toward the bilayer. Zipping up to layer +6 of one of the SNARE complexes was sufficient to overcome the repulsive forces between the adjacent membrane surfaces. Thermal fluctuations resulted in initiation of contacts between a few lipid head groups of the juxtaposed monolayers, which occurred close to the SNARE complex(es) zipped up to layer +6 (Fig. 1C). Analysis of the time course (Fig. 2B) shows that two of the SNARE complexes were zipped up to layer +6. In none of the simulations were layers +7 and +8 zipped before the occurrence of proximal lipid head group contacts.
Fig. 2.
SNARE layer zipping and stalk formation. (A) Structures of SNARE complex 4 from Sim1 showing unzipped layers at 0 ns (Left) and zippered layers at 521 ns (Right). (B) Distances between the backbone beads of syb2 residues A74, F77, A81, and L84 and stx1 residues V244, A247, T251, and A254, respectively, corresponding to layers +5 to +8, and the angles (orange) between TMDs of syb2 and stx1 for each SNARE complex (complexes 1–4, from top to bottom). The angle between the TMDs was defined as the angle between a vector connecting the center of mass of residues M95–I98 and I111–F114 of syb2 TMD and a vector connecting the center of mass of residues I266–I269 and I283–I286 of stx1 TMD (orange traces in A). Dashed vertical lines denote the times at which the following events occurred: 1, proximal lipid head group contacts (101 ns); 2, proximal lipid tail contacts (239 ns); 3, distal lipid head group contacts (302 ns); and 4, fusion pore formation. (C) Lipid tilt distribution maps at the start of simulation (5–20 ns; Top), at stalk initiation (230–240 ns; Middle), and at stalk expansion (240–250 ns; Bottom). The site of initial tail contacts is marked with a pink arrow and highly tilted lipids at the ND edge with black arrows (Middle). The color-coding represents the tilt of the lipids in the lower ND leaflet mapped on to the x- and y-coordinates of the corresponding PO4 bead. The positions of the syb2 TMDs from the four SNARE complexes are labeled 1–4. The asterisk marks the formed stalk.
Zipping of the SNARE domains was associated with a decrease in the angle between TMDs of syb2 and stx1 from ∼120–150° to ∼10–30° (Fig. 2B), which facilitated tilting of lipids with respect to the membrane normal. Initially, only the lipids located at the periphery of the ND near the scaffold protein were comparatively more tilted and disordered than those in the more central regions of the ND, as shown previously (28). As the SNARE complex layers zip, lipid tilt increased in the central region of the ND (Fig. 2C). A few tilted lipids in the proximal leaflets of the ND and the planar membrane adopted a splay conformation (three in Fig. 1C), associated with each other, forming a hydrophobic core, which rapidly evolved into a metastable stalk within ∼10 ns (Fig. 1D). Among the lipids in the central region of the ND, the lipids located close to the largely unzipped SNARE complex 3 (Fig. 2B) were more ordered while the lipids located between the zipped SNAREs were more disordered (Fig. 2C). In all 12 simulations, the hydrophobic core was always formed by splaying of the few disordered lipids located between the zipped SNAREs leading to a stalk. Although the time course of specific events varied between different simulations, the stalk was typically formed within 100–600 ns of initial head group contacts. The appearance of lipid tail contacts between the proximal leaflet lipids from the ND and bilayer is shown in SI Appendix, Fig. S1A.
Induction of Distal Leaflet Lipid Interactions and Fusion Without Stable Hemifusion Intermediate.
With the increasing tail contacts between the proximal leaflets, SNARE complex zipping continued. The zippering of layers +7 and +8 was synchronous (Fig. 2B green, blue), as in case of layers +5 and +6. The continued zipping and tilting of the TMDs pulled the phosphate head groups of the distal leaflets toward the hydrophobic membrane/ND cores by their interactions with C-terminal residues of the SNARE TMDs. Simultaneously, the stalk expanded radially, except for three simulations (Sim3, Sim4, and Sim6) in which the stalk expanded into an elongated shape. Nevertheless, in all 12 simulations, stalk expansion was followed by lipid mixing of distal leaflets without the appearance of a stable hemifusion intermediate, as indicated by a rapid drop in distance between the closest phosphates of lipids from the upper ND leaflet and from the lower planar bilayer leaflet to a value <1 nm (SI Appendix, Fig. S1B).
The fusion pathway proceeded through similar states as shown in Fig. 1, except for simulations Sim3, Sim4, and Sim6, in which more gradual distance changes involved transient formation of an inverted HII phase (SI Appendix, Fig. S2). Appearance of such an HII phase is not a specific feature of the ND system, as it has also been observed in simulations of ∼15-nm vesicle-vesicle fusion in the absence of SNAREs (29), as well as a vesicle bilayer system in the presence of SNARES (16). These simulations were done on membranes with simpler lipid composition. The elongated inverted HII phase was initiated at the stalk periphery, as previously reported (16), and later extended into the hydrophobic core, with several TMD residues of syb2 transiently exposed to water. The water contacts of these residues were broken as the hexagonal phase ruptured. The interaction of polar lipid head groups with the penetrating TMD C termini eventually led to fusion pore formation.
C Termini of Syb2 and Stx1 Promote Lipid Mixing and Fusion Pore Formation.
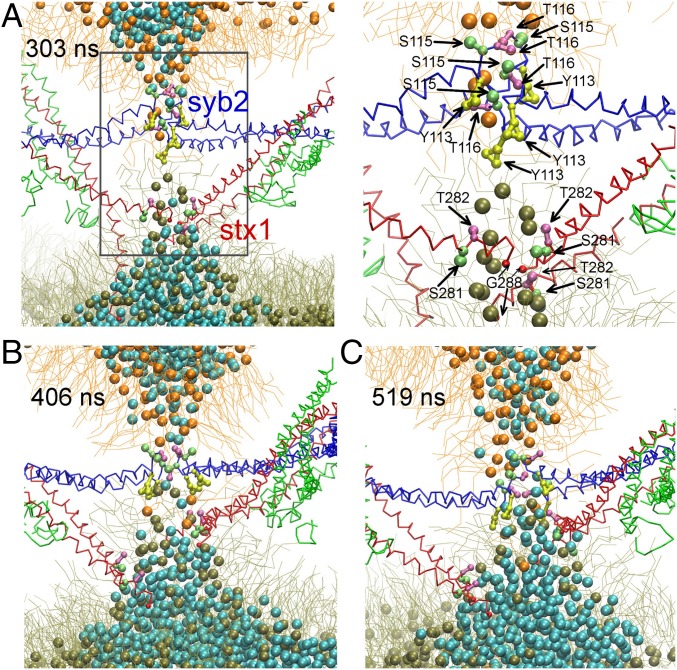
The syb2 C-terminal TMD residues S115 and T116 interacted with lipid head groups in the distal ND leaflet, pulling them into the hydrophobic core (Fig. 3). At 303 ns (Fig. 3A), only SNARE complex 4 was zipped up to layer +8, SNARE complexes 1 and 2 had layers +5 and +6 zipped, while in SNARE complex 3, layers +5 to +8 were still unzipped (Fig. 2B). The inward movement of the head groups was further stabilized by Y113 (Fig. 3A, arrows), reducing the free energy cost of desolvating the head groups in the hydrophobic interior. In the distal leaflet of the bilayer, the lipid head groups were correspondingly pulled in by the C terminus of stx1 and were further stabilized by interactions with stx1 TMD residues S281 and T282 (Fig. 3A, arrows). At 340 ns, three of the SNARE complexes were completely zipped up to layer +8, with the C termini of syb2 and stx1 now positioned close together. At 406 ns (Fig. 3B), the polar TMD residues Y113, S115, and T116 of syb2 and S281, T282, and G288 of stx1 formed a hydrophilic core between the two distal leaflets, facilitating lipid mixing and fusion pore formation (Fig. 3C). In all 12 simulations, the C-terminal polar residues of the SNARE TMDs formed a hydrophilic core, facilitating distal lipid mixing, although the time course of lipid mixing, SNARE complex zipping, and precise geometry of the hydrophilic core varied considerably.
Fig. 3.
Mechanism of fusion pore formation. (A) Approaching distal monolayers at 303 ns; residues Y113 (yellow), S115 (lime), and T116 (pink) of syb2 and residues S281 (lime), T282 (pink), and C-terminal G288 (red) of stx1 are shown in a ball-and-stick representation. On the right is a zoomed-in view of the boxed area in the left panel. For clarity, only lipids from distal leaflets are shown. SNARE proteins and lipids are shown as in Fig. 1. Waters are shown as cyan spheres. (B) Distal leaflet mixing (406 ns). (C) Early stage of aqueous fusion pore (519 ns).
Simulated Fusion Pore Conductances Are in the 150–380 pS Range.
Because the best experimentally characterized fusion pore property is its conductance (1), we estimated the conductances of the simulated pores (Fig. 4). Transient fusion pore flickers, some more stable with extended lifetimes, and lipid mixing of the distal leaflets occurred in all simulations (SI Appendix, Fig. S1B). The MARTINI force field has weaker attractive interactions between the lipid head group and water than those appearing in atomistic simulations, resulting in smaller and less stable water defects in CG membranes (30), which may promote the brief duration of some fusion pore flickers.
Fig. 4.
Fusion pore conductance (in pS). (A) Estimated conductances (black) calculated using SI Appendix, Eq. S1 and diffusion-corrected pore conductances (red) calculated using SI Appendix, Eq. S3 for trajectory segments in which a continuous water density in the pore lasted for at least 25 ns. Conductance traces are shown for two different cases: long-lived pores (Sim1; Left) and flickering pores (Sim6; Right). (B and C) Distributions of uncorrected (B) and diffusion-corrected (C) conductances for Sim1, Sim2, Sim9, and Sim10. The scale at the top shows corrected conductance values based on the correction factor from atomistic simulation.
The conductances of the simulated fusion pores were estimated based on fusion pore geometry assuming a conductivity of 15 mS cm−1 for the solution inside the fusion pore (SI Appendix, Methods and Eqs. S1 and S2), averaging over 1-ns intervals (Fig. 4A and SI Appendix, Fig. S7, black traces). For fusion pores with open times ≥25 ns, the conductance distributions provided mean values of 654 pS for Sim1, 584 pS for Sim2, 428 pS for Sim9, and 460 pS for Sim10 (Fig. 4B). However, for narrow pores, Eq. S1 overestimated the pore conductances owing to a reduced water/ion diffusion coefficient resulting from interactions with the pore wall (31). Accounting for the reduced water diffusion, factor-corrected fusion pore conductances (Fig. 4A and SI Appendix, Fig. S3, red traces) had mean values of 214 pS for Sim1, 122 pS for Sim2, 71 pS for Sim9, and 68 pS for Sim10, threefold to sevenfold lower than the uncorrected values (Fig. 4C). The fusion pore conductance correction factor obtained in CG simulations is likely overestimated owing to the larger size of CG water; therefore, we converted selected fusion pore states of Sim1 to atomistic models and repeated the determination of fusion pore conductance correction. These atomistic simulations yielded an approximately twofold-reduced mean corrected conductance. Thus, we consider mean fusion pore conductance values in the range of 150–380 pS to be reasonable estimates.
Restricted Mobility of the SNARE Complexes Prevents Fusion Pore Expansion.
The ND imposes constraints, keeping the four SNARE complexes in close proximity, which promotes rapid formation of fusion pores but allows the fusion pores to flicker, fail to expand, and reclose (Fig. 1F). To test whether a formed pore is able to expand when these constraints are removed, a stable pore geometry was chosen (Sim1, 650 ns), and the ND region of the fusion pore was transferred into a ∼40-nm vesicle by removing overlapping lipids from the vesicle. After extensive equilibration with the pore geometry restrained, the system (Fig. 5A, Left) was subjected to 170 ns of free simulation, during which the pore rapidly expanded (Fig. 5A, Right).
Fig. 5.
Simulation of the ∼40-nm vesicle-bilayer system. (A) Side view of the ND-bilayer pore geometry, with the ND replaced by a vesicle. Shown are snapshots taken at the start of simulation (Left) and at the end of the free simulation (Right, 170 ns). (B) Side view showing the starting configuration (Left) and the configuration at 1.5 μs (Right) for the vesicle-bilayer fusion setup.
Restricted SNARE Mobility Is Required for Rapid Fusion Pore Formation.
In three 600-ns simulations, in which the ND in the initial setup for ND/bilayer simulations was replaced by a ∼40-nm vesicle (Fig. 5B, Left), lipid mixing of the proximal leaflets and stalk formation occurred, but the distal leaflets failed to fuse, and no fusion pores formed. An HII phase was not evident in the vesicle/bilayer simulations, but for this large system, only three simulations were performed, and HII structures were also rare in the ND simulations (3 out of 12). The formed stalk expanded into an extended hemifusion diaphragm, as has been observed experimentally in liposome fusion assays in which SNARE complex mobility was unrestricted (32). During this phase, the SNARE complexes move radially away from one another (Fig. 5B, Right), presumably driven by entropic forces (33). Thus, the confinement of the SNARE complexes is required for rapid fusion pore formation.
To determine whether fusion pore formation in the ND system is indeed due to the restricted mobility of SNARE complexes and not to other properties of the ND, we carried out two simulations of the ∼40-nm vesicle/bilayer system with the lateral mobility of SNARE complexes constrained using two sets of harmonic restraints (SI Appendix, Fig. S4). These simulations were based on one of the unconstrained simulations described above and were started at the stage when the stalk had formed, while the SNARE complexes were still in close proximity (270 ns). In both simulations, a fusion pore was formed within 500 ns. In contrast, in the simulations without restrictions, in which the SNARE complexes were free to diffuse, no fusion pores were formed, even when the simulation was extended up to 2 µs.
SNARE Complex Mutations Inhibiting Fusion Do Not Form Fusion Pores in the Simulations.
One critical test to determine the physiological relevance of the simulation of fusion pore formation is to test SNARE protein mutations that strongly inhibit fusion. We tested two such mutations: SNAP-25∆9, which lacks the nine C-terminal residues and corresponds to the Botulinum Toxin A (BoTox) cleavage product, and the syb2-KK construct, which has two lysines added at the syb2 C terminus.
Experimentally, SNAP-25∆9 dramatically reduces the frequency of fusion events, as does cleavage with BoTox (34–36). In three independent simulations of the ND-planar bilayer system, each 3.5-μs long with SNAP-25 replaced by SNAP-25Δ9 in all four SNARE complexes, no fusion pore formation occurred. Interestingly, in all of these simulations, there was no effect on stalk formation (Fig. 6A); however, all of the simulations were arrested at this stage, and none showed mixing of distal leaflets and fusion pore formation, due to the defect in C-terminal SNARE domain zipping (SI Appendix, Fig. S5).
Fig. 6.
SNARE mutants do not lead to fusion pore formation. (A and B) Snapshots at 3 μs for SNAP-25∆9 (A) and syb2-KK mutants (B) showing failure of fusion pore formation. (C) PMF profile along the distance between the centers of mass of C-terminal residues of syb2 and stx1 used as the reaction coordinates. Error bars indicate the errors estimated using the Bayesian bootstrap analysis. (D–F) Final structures from the last umbrella sampling window (reaction coordinate, 3.2 nm) for WT (D), syb2-KK (E), and SNAP-25∆9 (F).
Experimentally, the syb2-KK construct blocks fusion (11), and previous CG MD simulations showed increased activation energy to pull the syb2 C terminus deeper into the membrane (37). We performed three simulations, each 3.5-μs long, with syb2 replaced by syb2-KK in all four SNARE complexes. As for SNAP-25∆9, mixing of proximal leaflets and stalk formation followed a similar time course as wild-type (WT) syb2, while no fusion of distal leaflets was observed (Fig. 6B). The mechanism of inhibition differs from that of SNAP-25∆9, however. In the syb2-KK mutant, zippering was similar to that in WT syb2 (SI Appendix, Fig. S5), but the positively charged lysines at the syb2 C terminus interacted strongly with PO4 groups stabilizing the C terminus at the membrane surface, making the formation of negative curvature unfavorable. Intuitively, the pulling of a lysine residue into the membrane would generate larger defects at the membrane, but this would require much greater energy than that generated by SNARE zipping alone.
To assess the energetics of fusion pore formation in WT and SNARE mutants, we used umbrella sampling simulations, with the distances between the centers of mass defined by S115–T116 of syb2 and F287–G288 of stx1 of all four SNARE complexes as the reaction coordinates. The centers of mass of the TMD C termini were chosen because they spontaneously exert motion toward one another, exerting force that leads to fusion pore formation. By decreasing this distance from an initial value of 6.3 nm to 3.2 nm, a potential of mean force (PMF) was calculated (Fig. 6C). For WT SNARE complexes, the PMF shows very favorable energetics for pore formation. In contrast, the PMF profiles of the mutants that inhibit fusion indicate that pore formation is an energetically unfavorable process. Interestingly, at the early stages along the reaction coordinates, the SNAP-25∆9 mutant is energetically most unfavorable, reflecting the defect in SNARE complex zippering in this mutant, whereas zippering of syb2-KK is very similar to that of WT syb2 (SI Appendix, Fig. S5). Note that no fusion pore forms with syb2-KK (Fig. 6E), which experimentally inhibits fusion completely (11), whereas a fusion pore does ultimately form for SNAP-25∆9 (Fig. 6F), consistent with its ability to support fusion at very low rate (14, 34).
The choice of membrane composition affects the free energy landscape of stalk expansion (25). Moreover, choosing the C termini of the syb2 and stx1 TMDs of all four SNARE complexes as the PMF reaction coordinates results in a lack of control over the fusion pathway and thus might not properly reflect the forces exerted by the SNARE complex and its mutants (SI Appendix, Fig. S7). Therefore, we performed PMF calculations using a well-controlled POPC vesicle-membrane system with a single SNARE complex and choosing as reaction coordinates the distance between two hydrophilic probes representing the hydration shell of C-terminal ends of the TMD of the SNARE complex (SI Appendix, Fig. S8A), as described previously (25, 38). The free energy maximum of the PMF profiles corresponds to the nucleation barrier for pore formation. Once the barrier is overcome, pore formation and expansion proceed along the plateau. The nucleation barrier is ∼10 kBT higher for SNAP-25∆9 and ∼28 kBT higher for the syb2-KK mutant compared with the WT SNARE (SI Appendix, Fig. S8B). Interestingly, pore nucleation in WT and SNAP-25∆9 occurs at nearly the same distance between the probes. In the syb2-KK mutant, the nucleation barrier is shifted along the reaction coordinate to a point at which the TMD C termini are much closer, suggesting that the positively charged C terminus in syb2-KK does not penetrate the membrane and inhibit stalk indentation.
The energetic differences obtained for the different mutants in this simple system confirm the results from the simulations of the complex system described above. In the thermodynamically reversible indentation regime (38, 39), the difference between the energy profiles of WT and SNAP-25∆9 shows a linear range with a slope of ∼20 pN (SI Appendix, Fig. S8B).
Discussion
The structure of the fusion pore mediating transmitter release and the mechanism of its formation remain unknown. Here we present CGMD simulations starting with an ND and a planar membrane bridged by an assembly of four SNARE complexes, partially unzipped up to layer +5, which lead to spontaneous formation of a proteolipid fusion pore. Our simulations reveal the details of the final zippering dynamics and how the TMD C termini pull lipid head groups in the distal leaflets toward the hydrophobic core of the stalk where, together with the C termini, they form a hydrophilic fusion pore initiation site.
Two mutations that inhibit fusion experimentally failed to form fusion pores in our simulations, supporting the physiological significance of the approach used here. These two mutations inhibit fusion by completely different mechanisms. SNAP-25∆9 prevents proper C-terminal zippering (SI Appendix, Fig. S5), reducing the zippering force by ∼20 pN (SI Appendix, Fig. S8). Experiments have indicated SNARE complex unzipping at 34 pN but not at 11 pN (40) and have shown reversible unzipping-zipping transitions at 18 pN (41). Given the limitations of CG simulations, the ∼20 pN estimate is clearly in the proper range of zippering forces exerted by the SNARE complex.
The syb2-KK mutant zippers normally, but the added lysines stabilize the syb2 TMD in the membrane such that it is unable to translocate lipid head groups as is required to form the hydrophilic fusion pore initiation site. It is conceivable that the addition of uncharged residues at the syb2 or stx1 C terminus would have only a marginal effect on this process, consistent with previous experimental observations on the ability of SNARE to drive fusion when attached to peptide linkers (39) and the inhibition of fusion by the addition of charged residues to the C termini of syb2 (11).
Further evidence for the physiological relevance of the fusion pore structures obtained in the simulations comes from the estimates of fusion pore conductances. The fusion pore structures and conductances obtained in the simulations varied widely, as is the case for experimentally determined conductance values, which range from 35 pS up to ∼1 nS. Patch-clamp studies in different cell types have provided mean fusion pore conductance values within the first 50 μs (42, 43) to a few milliseconds (44, 45) ranging from ∼150 pS in human neutrophils (45) to 200 pS in horse eosinophils (42) to 330 pS in mast cells (43, 46) and chromaffin cells (44). Although the simulations extend only over 2–3 μs, the range of fusion pore conductance estimates is in close agreement with that observed experimentally, suggesting that the fusion pore structures obtained in the simulations closely resemble the structure of fusion pores in living cells.
Experimental evidence indicates that during reconstituted ND/vesicle fusion, syb2 TMD residues I99, V101, C103, I105, and I109 are exposed to the bulk solvent, suggesting that these residues may line the fusion pore (13, 47). The simulations show that only the C terminus of syb2 protrudes into the fusion pore, while the N-terminal residues of the syb2 TMD lie near the stalk periphery. However, in the simulations in which an inverted HII phase was formed at the stalk periphery (SI Appendix, Fig. S1), several hydrophobic syb2 TMD residues were transiently exposed to water, consistent with the experimental data.
The ND/bilayer system used here reveals the detailed molecular mechanics of fusion pore formation by SNARE proteins. The ND constrains the system by keeping the four SNARE complexes in close proximity, promoting rapid formation of fusion pores. When the ND was replaced by a 40-nm vesicle, an extended hemifusion diaphragm formed, as has been observed experimentally with purified SNAREs reconstituted in liposomes (32). Such states may lead to fusion on a time scale of minutes to hours (2). Constraining the SNARE complexes in the vesicle/bilayer system also supported rapid fusion pore formation, demonstrating that it is the constraint, not other features of the ND system, that is key for rapid fusion. Once the fusion pore is formed, the constraints must be released for fusion pore expansion. Brief fusion pore flickers may be due to extended constraint of the SNARE complexes. The formation of a hemifusion state in chromaffin cells that may be followed by fusion pore formation after a long delay (48) may be due to insufficient restriction of the SNARE complexes. Therefore, a switch between rapid fusion pore formation and delayed fusion via a hemifusion intermediate may come from accessory proteins that may restrict the SNARE complexes at a fusion site in the cell.
In the cell, the restriction of SNARE complex mobility may be due to t-SNARE cluster formation (49, 50) and/or interactions with other accessory proteins, such as synaptotagmin or Munc-13 (51). Our results are consistent with the “buttressed ring hypothesis” (52), which proposes that concentric rings of synaptotagmin and Munc-13 have an essential role as organizers of the SNARE complex assembly and fast fusion and then disintegrate at the time of fusion, thereby allowing for fusion pore expansion (50). The ND system simulated here mimics the physiological arrangement with restricted mobility of the SNARE complexes, leads to rapid fusion pore formation, and explains the mechanisms of inhibition of fusion by specific SNARE protein mutations.
Methods
Trans-SNARE Complex Model Generation.
The initial SNARE complex structure was taken from Protein Data Bank ID code 3HD7 (3). The missing C-terminal residues of syb2, stx1, and SNAP-25 were added. The SNAP-25 linker region residues K83–A100 were modeled, assigning a random coil secondary structure. The structures of syb2 and of the stx1/SNAP-25 t-SNARE complex were separately converted to MARTINI CG models and reassembled to the original SNARE complex. The SNARE domains were then partially unzipped by pulling T116 of syb2 away from G288 of stx1 until layer +5 was unzipped (SI Appendix, Methods). The palmitoyl chains were added to residues SNAP-25 C85, C88, C90, and C92.
Trans-SNARE Complex Membrane and ND Insertion.
The t-SNARE TMDs of all four copies of the unzipped SNARE complexes were inserted into the plasma membrane using a self-assembly protocol (24) based on the synaptosomal plasma membrane lipid (26) (SI Appendix, Methods). Beginning with this arrangement of the four SNAREs, the lipid-filled ND was placed in three different orientations with the syb2 T116 residues positioned close to the phosphate groups of the “intravesicular” leaflet and W89 close to the phosphate groups of the “cytoplasmic” leaflet of the ND (37). The system box was resized along the z-axis to 23.0 nm, filled with more water particles and ions to attain a salt concentration of 0.15 M, and subjected to standard CGMD energy minimization and equilibration without any restraints on SNARE proteins. Each of the three systems was subjected to four independent production runs, generating a total of 12 production trajectories (Sim1–Sim12).
Simulation and Analysis.
Simulations were carried out using GROMACS 4.5.x (53) using standard MARTINI MD parameters. Details of simulation methods, PMF calculations, and further analyses are provided in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Dr. Erwin Neher for his critical reading and comments on the manuscript. This work was supported by European Research Council Grant ADG 322699, National Institutes of Health Grant R01 GM121787, and computational resources of the Max Planck Institute’s Computing and Data Facility.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816495115/-/DCSupplemental.
References
- 1.Lindau M, Alvarez de Toledo G. The fusion pore. Biochim Biophys Acta. 2003;1641:167–173. doi: 10.1016/s0167-4889(03)00085-5. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 3.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veit M, Söllner TH, Rothman JE. Multiple palmitoylation of synaptotagmin and the t-SNARE SNAP-25. FEBS Lett. 1996;385:119–123. doi: 10.1016/0014-5793(96)00362-6. [DOI] [PubMed] [Google Scholar]
- 5.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4-Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 6.Jahn R, Scheller RH. SNAREs: Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 7.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieren M, Desfougères Y, Michaillat L, Schmidt A, Mayer A. Vacuolar SNARE protein transmembrane domains serve as nonspecific membrane anchors with unequal roles in lipid mixing. J Biol Chem. 2015;290:12821–12832. doi: 10.1074/jbc.M115.647776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang CW, Chiang CW, Gaffaney JD, Chapman ER, Jackson MB. Lipid-anchored synaptobrevin provides little or no support for exocytosis or liposome fusion. J Biol Chem. 2016;291:2848–2857. doi: 10.1074/jbc.M115.701169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fdez E, Martínez-Salvador M, Beard M, Woodman P, Hilfiker S. Transmembrane-domain determinants for SNARE-mediated membrane fusion. J Cell Sci. 2010;123:2473–2480. doi: 10.1242/jcs.061325. [DOI] [PubMed] [Google Scholar]
- 11.Ngatchou AN, et al. Role of the synaptobrevin C terminus in fusion pore formation. Proc Natl Acad Sci USA. 2010;107:18463–18468. doi: 10.1073/pnas.1006727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao H, et al. Exocytotic fusion pores are composed of both lipids and proteins. Nat Struct Mol Biol. 2016;23:67–73. doi: 10.1038/nsmb.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Q, et al. The role of the C terminus of the SNARE protein SNAP-25 in fusion pore opening and a model for fusion pore mechanics. Proc Natl Acad Sci USA. 2008;105:15388–15392. doi: 10.1073/pnas.0805377105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parlati F, et al. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc Natl Acad Sci USA. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risselada HJ, Kutzner C, Grubmüller H. Caught in the act: Visualization of SNARE-mediated fusion events in molecular detail. ChemBioChem. 2011;12:1049–1055. doi: 10.1002/cbic.201100020. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, et al. SNARE proteins: One to fuse and three to keep the nascent fusion pore open. Science. 2012;335:1355–1359. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nir S, Wilschut J, Bentz J. The rate of fusion of phospholipid vesicles and the role of bilayer curvature. Biochim Biophys Acta. 1982;688:275–278. doi: 10.1016/0005-2736(82)90604-6. [DOI] [PubMed] [Google Scholar]
- 19.Siegel DP. Energetics of intermediates in membrane fusion: Comparison of stalk and inverted micellar intermediate mechanisms. Biophys J. 1993;65:2124–2140. doi: 10.1016/S0006-3495(93)81256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilschut J, Düzgüneş N, Fraley R, Papahadjopoulos D. Studies on the mechanism of membrane fusion: Kinetics of calcium ion induced fusion of phosphatidylserine vesicles followed by a new assay for mixing of aqueous vesicle contents. Biochemistry. 1980;19:6011–6021. doi: 10.1021/bi00567a011. [DOI] [PubMed] [Google Scholar]
- 21.Baoukina S, Tieleman DP. Direct simulation of protein-mediated vesicle fusion: Lung surfactant protein B. Biophys J. 2010;99:2134–2142. doi: 10.1016/j.bpj.2010.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risselada HJ, Grubmüller H. How SNARE molecules mediate membrane fusion: Recent insights from molecular simulations. Curr Opin Struct Biol. 2012;22:187–196. doi: 10.1016/j.sbi.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Yoo J, Jackson MB, Cui Q. A comparison of coarse-grained and continuum models for membrane bending in lipid bilayer fusion pores. Biophys J. 2013;104:841–852. doi: 10.1016/j.bpj.2012.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma S, Kim BN, Stansfeld PJ, Sansom MS, Lindau M. A coarse-grained model for a lipid membrane with physiological composition and leaflet asymmetry. PLoS One. 2015;10:e0144814. doi: 10.1371/journal.pone.0144814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risselada HJ, Bubnis G, Grubmüller H. Expansion of the fusion stalk and its implication for biological membrane fusion. Proc Natl Acad Sci USA. 2014;111:11043–11048. doi: 10.1073/pnas.1323221111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breckenridge WC, Gombos G, Morgan IG. The lipid composition of adult rat brain synaptosomal plasma membranes. Biochim Biophys Acta. 1972;266:695–707. doi: 10.1016/0006-3002(72)90012-1. [DOI] [PubMed] [Google Scholar]
- 27.Kozlov MM, Chernomordik LV. A mechanism of protein-mediated fusion: Coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys J. 1998;75:1384–1396. doi: 10.1016/S0006-3495(98)74056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siuda I, Tieleman DP. Molecular models of nanodiscs. J Chem Theory Comput. 2015;11:4923–4932. doi: 10.1021/acs.jctc.5b00668. [DOI] [PubMed] [Google Scholar]
- 29.Marrink SJ, Mark AE. The mechanism of vesicle fusion as revealed by molecular dynamics simulations. J Am Chem Soc. 2003;125:11144–11145. doi: 10.1021/ja036138+. [DOI] [PubMed] [Google Scholar]
- 30.Bennett WF, Tieleman DP. Water defect and pore formation in atomistic and coarse-grained lipid membranes: Pushing the limits of coarse graining. J Chem Theory Comput. 2011;7:2981–2988. doi: 10.1021/ct200291v. [DOI] [PubMed] [Google Scholar]
- 31.Smart OS, Coates GM, Sansom MS, Alder GM, Bashford CL. Structure-based prediction of the conductance properties of ion channels. Faraday Discuss. 1998;111:185–199, discussion 225–246. doi: 10.1039/a806771f. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez JM, et al. Membrane fusion intermediates via directional and full assembly of the SNARE complex. Science. 2012;336:1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mostafavi H, et al. Entropic forces drive self-organization and membrane fusion by SNARE proteins. Proc Natl Acad Sci USA. 2017;114:5455–5460. doi: 10.1073/pnas.1611506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei S, et al. Exocytotic mechanism studied by truncated and zero-layer mutants of the C-terminus of SNAP-25. EMBO J. 2000;19:1279–1289. doi: 10.1093/emboj/19.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu T, Binz T, Niemann H, Neher E. Multiple kinetic components of exocytosis distinguished by neurotoxin sensitivity. Nat Neurosci. 1998;1:192–200. doi: 10.1038/642. [DOI] [PubMed] [Google Scholar]
- 36.Criado M, Gil A, Viniegra S, Gutiérrez LM. A single amino acid near the C terminus of the synaptosome-associated protein of 25 kDa (SNAP-25) is essential for exocytosis in chromaffin cells. Proc Natl Acad Sci USA. 1999;96:7256–7261. doi: 10.1073/pnas.96.13.7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindau M, Hall BA, Chetwynd A, Beckstein O, Sansom MSP. Coarse-grain simulations reveal movement of the synaptobrevin C-terminus in response to piconewton forces. Biophys J. 2012;103:959–969. doi: 10.1016/j.bpj.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Agostino M, Risselada HJ, Lürick A, Ungermann C, Mayer A. A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature. 2017;551:634–638. doi: 10.1038/nature24469. [DOI] [PubMed] [Google Scholar]
- 39.D’Agostino M, Risselada HJ, Mayer A. Steric hindrance of SNARE transmembrane domain organization impairs the hemifusion-to-fusion transition. EMBO Rep. 2016;17:1590–1608. doi: 10.15252/embr.201642209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min D, et al. Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism. Nat Commun. 2013;4:1705. doi: 10.1038/ncomms2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartmann J, Lindau M. A novel Ca(2+)-dependent step in exocytosis subsequent to vesicle fusion. FEBS Lett. 1995;363:217–220. doi: 10.1016/0014-5793(95)00318-4. [DOI] [PubMed] [Google Scholar]
- 43.Breckenridge LJ, Almers W. Currents through the fusion pore that forms during exocytosis of a secretory vesicle. Nature. 1987;328:814–817. doi: 10.1038/328814a0. [DOI] [PubMed] [Google Scholar]
- 44.Dernick G, Alvarez de Toledo G, Lindau M. Exocytosis of single chromaffin granules in cell-free inside-out membrane patches. Nat Cell Biol. 2003;5:358–362. doi: 10.1038/ncb956. [DOI] [PubMed] [Google Scholar]
- 45.Lollike K, Borregaard N, Lindau M. The exocytotic fusion pore of small granules has a conductance similar to an ion channel. J Cell Biol. 1995;129:99–104. doi: 10.1083/jcb.129.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spruce AE, Breckenridge LJ, Lee AK, Almers W. Properties of the fusion pore that forms during exocytosis of a mast cell secretory vesicle. Neuron. 1990;4:643–654. doi: 10.1016/0896-6273(90)90192-i. [DOI] [PubMed] [Google Scholar]
- 47.Chang CW, et al. A structural role for the synaptobrevin 2 transmembrane domain in dense-core vesicle fusion pores. J Neurosci. 2015;35:5772–5780. doi: 10.1523/JNEUROSCI.3983-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao WD, et al. Hemi-fused structure mediates and controls fusion and fission in live cells. Nature. 2016;534:548–552. doi: 10.1038/nature18598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bademosi AT, et al. In vivo single-molecule imaging of syntaxin1A reveals polyphosphoinositide- and activity-dependent trapping in presynaptic nanoclusters. Nat Commun. 2017;8:13660. doi: 10.1038/ncomms13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gandasi NR, Barg S. Contact-induced clustering of syntaxin and munc18 docks secretory granules at the exocytosis site. Nat Commun. 2014;5:3914. doi: 10.1038/ncomms4914. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, et al. Functional synergy between the Munc13 C-terminal C1 and C2 domains. eLife. 2016;5:e13696. doi: 10.7554/eLife.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothman JE, Krishnakumar SS, Grushin K, Pincet F. Hypothesis: Buttressed rings assemble, clamp, and release SNAREpins for synaptic transmission. FEBS Lett. 2017;591:3459–3480. doi: 10.1002/1873-3468.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.