Significance
Mutations in cystic fibrosis transmembrane conductance regulator (CFTR) cause cystic fibrosis, a lethal genetic disease occurring in people of northern European descent. Decades of study have been directed toward a molecular understanding of this ion channel. The structure presented here enables a direct correlation of structure with function, most of which has been characterized in human CFTR.
Keywords: human CFTR, anion channel, ABC transporter, cryo-EM
Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is an anion channel important in maintaining proper functions of the lung, pancreas, and intestine. The activity of CFTR is regulated by ATP and protein kinase A-dependent phosphorylation. To understand the conformational changes elicited by phosphorylation and ATP binding, we present here the structure of phosphorylated, ATP-bound human CFTR, determined by cryoelectron microscopy to 3.2-Å resolution. This structure reveals the position of the R domain after phosphorylation. By comparing the structures of human CFTR and zebrafish CFTR determined under the same condition, we identified common features essential to channel gating. The differences in their structures indicate plasticity permitted in evolution to achieve the same function. Finally, the structure of CFTR provides a better understanding of why the G178R, R352Q, L927P, and G970R/D mutations would impede conformational changes of CFTR and lead to cystic fibrosis.
The cystic fibrosis transmembrane conductance regulator (CFTR) is a unique ATP-binding cassette (ABC) transporter that functions as an ion channel. Whereas other ABC transporters transport substrates against their chemical gradients, CFTR conducts anions down their electrochemical gradient (1). Loss-of-function mutations of the CFTR gene cause cystic fibrosis, the most common heritable disease in people of northern European descent (2, 3). Even with modern medical care, the average survival age of cystic fibrosis patients is ∼40 y old (4).
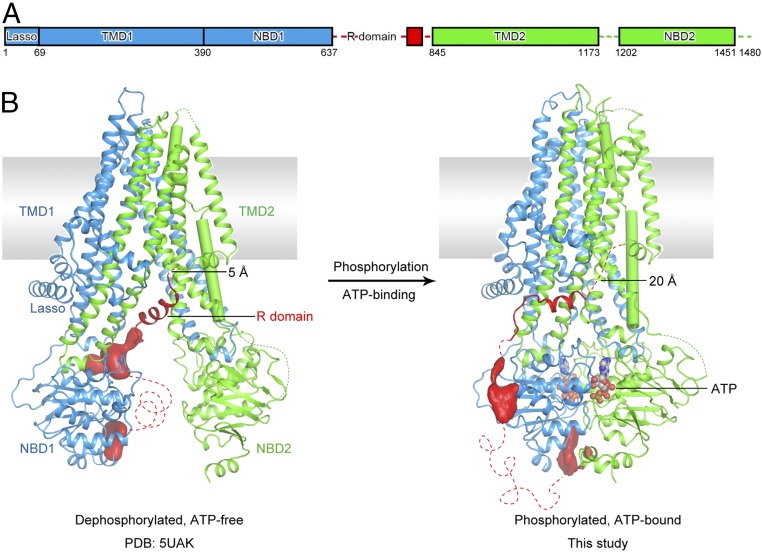
CFTR is a single polypeptide containing an N-terminal lasso motif, two transmembrane domains (TMDs), and two nucleotide-binding domains (NBDs) (Fig. 1A). Distinct from other ABC transporters, CFTR also contains an ∼200-residue cytoplasmic regulatory (R) domain that regulates the activity of CFTR (Fig. 1A). The R domain contains 19 predicted sites for protein kinase A (PKA); up to six have been found phosphorylated in vivo (5–7). Phosphorylation of the R domain increases the open probability of the CFTR channel (8) and also stimulates ATP hydrolysis by CFTR (9).
Fig. 1.
Two functional states of human CFTR. (A) Schematic diagram showing the domain structure of CFTR. The numbers represent the range of residues visible in the cryo-EM map, not the exact boundaries of different domains. (B) Ribbon diagrams of the dephosphorylated, ATP-free conformation (Left; PDB ID code 5UAK) and the phosphorylated, ATP-bound structure (Right). Regions not resolved in the structure are shown as dashed lines. The EM densities, shown in red, correspond to unstructured regions within the R domain. TM 8 is highlighted in cylinder. ATP is shown in ball-and-stick and colored by heteroatom. The distances between the last visible residue in the R domain and the first residue in TMD2 are indicated.
Decades of electrophysiological and biochemical studies have provided detailed functional characterization of human CFTR (10, 11). Recently, the molecular structures of CFTR have been determined for zebrafish and human by cryoelectron microscopy (cryo-EM) (9, 12, 13). In the dephosphorylated, ATP-free conformation, the structures of zebrafish CFTR (zCFTR) and human CFTR (hCFTR) (Fig. 1B) are nearly identical (9, 12). In this conformation, the 12 transmembrane (TM) helices of CFTR pack tightly in the membrane outer leaflet and segregate into two bundles extending into the cytoplasm (Fig. 1B). The two NBDs are separated from each other, with the R domain wedged in between (Fig. 1B). The structure of the phosphorylated, ATP-bound conformation was determined for zCFTR, in which the two NBDs form a closed dimer and the R domain becomes completely disordered (13). Whereas the overall architecture of CFTR is similar to other ABC transporters, it shows several unique features consistent with its function as an ion channel (9, 12, 13). In particular, the helix TM 8 makes two sharp breaks inside the membrane, creating a flexible hinge to facilitate gating (13).
In this work, we report a structure of the phosphorylated, ATP-bound human CFTR channel determined by cryo-EM. The hCFTR structure shares many key features with that of zCFTR determined under the same condition (13). It also reveals several differences that are important to correlate the structure with functional data obtained from hCFTR.
Results
Structure Determination.
Based on previous work showing that eliminating ATP hydrolysis prolongs the lifetime of the open CFTR channel (14), we replaced a catalytic residue in NBD2 (E1371) to abolish its ATPase activity. The mutant construct, E1371Q, was treated with PKA to phosphorylate the R domain. The cryo-EM structure was determined in the presence of saturating ATP-Mg2+ (9 mM). The final reconstruction, with an estimated overall resolution of 3.2 Å (SI Appendix, Fig. S1 and Table S1), shows well-defined densities for the lasso motif, 12 TM helices, 2 NBDs, and 2 ATP-Mg2+ molecules (SI Appendix, Fig. S2). Additional density, likely corresponding to part of the phosphorylated R domain, is also visible (SI Appendix, Fig. S2). With this map, we built a model containing nearly all residues in the TMDs and NBDs, as well as a polyalanine helix of the R domain (Fig. 1B). The structure is refined to excellent geometry and stereochemistry statistics (SI Appendix, Table S1).
With this structure, we can now directly visualize conformational changes within hCFTR upon phosphorylation and ATP binding (Fig. 1). Compared with the dephosphorylated, ATP-free conformation, the two halves of CFTR move toward each other, changing the shape of the molecule from an inverted “V” to an elongated form. The two NBDs form a closed dimer, occluding two ATP molecules at their interface. In CFTR, the phosphorylated R domain relocates to the peripheral surface, away from the NBD dimer interface.
Phosphorylated R Domain.
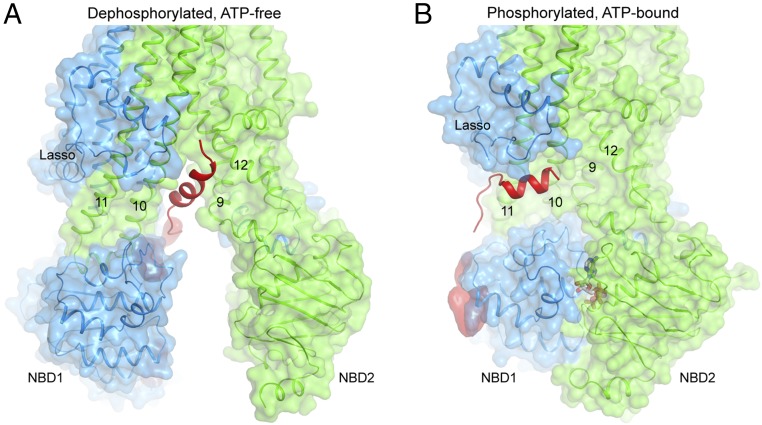
Structural comparison of the R domain before and after phosphorylation provides a plausible explanation of how the R domain regulates channel activity. When dephosphorylated, the R domain is wedged between TM helices 9, 10, and 12 and extends into the cytosol between the two NBDs (Fig. 2A). Dimerization of the NBDs, which leads to channel opening (14), is hindered by the presence of the R domain. In the phosphorylated, ATP-bound conformation, the R domain-binding surface is completely closed off (Fig. 2B). A helical segment is observed parallel to the membrane surface, interacting with TMs 10 and 11 and residues 34–39 of the lasso motif (Fig. 2B). Because the side chain densities in this region are inadequate, we can’t assign its amino acid sequence. NMR studies of the phosphorylated R domain indicated that residues 766–776, 801–810, and 826–835 are likely to be helical (15). The distance between the observed helix and the first visible residue in TMD2 (T845) is 20 Å (Fig. 1B), in line with all three possibilities. In the dephosphorylated hCFTR structure, a helical segment of the R domain was also observed (Fig. 1A). Because this helix is connected to TMD2, it is most likely to correspond to residues 825–843 (Fig. 1A).
Fig. 2.
Conformational changes of the R domain. Close-up view of the intracellular opening in the dephosphorylated, ATP-free (A) and phosphorylated, ATP-bound (B) conformations. The R domain is shown in red. The lasso motif and TM helices 9–12 are indicated.
At a lower contour, amorphous densities are observed along the peripheral surface of NBD1 and at the C-terminal region of NBD1 (Figs. 1B and 2B). These densities most likely correspond to parts of the R domain, consistent with studies showing that the R domain is largely unstructured and interacts with NBD1 at multiple locations (15–17). Interaction of the R domain with the lasso motif was previously reported using isolated peptides (18), but the specific residues identified to mediate the interaction are different from those observed in the cryo-EM structure.
These structural observations indicate that the dephosphorylated R domain inhibits channel activity by preventing NBD dimerization. It seems that phosphorylation by PKA stabilizes the R domain away from the NBD interface, permitting the conformational changes necessary for channel opening. This conclusion was inferred from our previous structural study of the zebrafish CFTR, but here we observe parts of the phosphorylated R domain on the outside surface of NBD1. In fact, the functional effects of phosphorylation can be imitated by simply deleting the R domain. Coexpression of the two halves of CFTR without the R domain results in an ATP-gated channel that opens independent of phosphorylation (19).
The Ion Permeation Pathway.
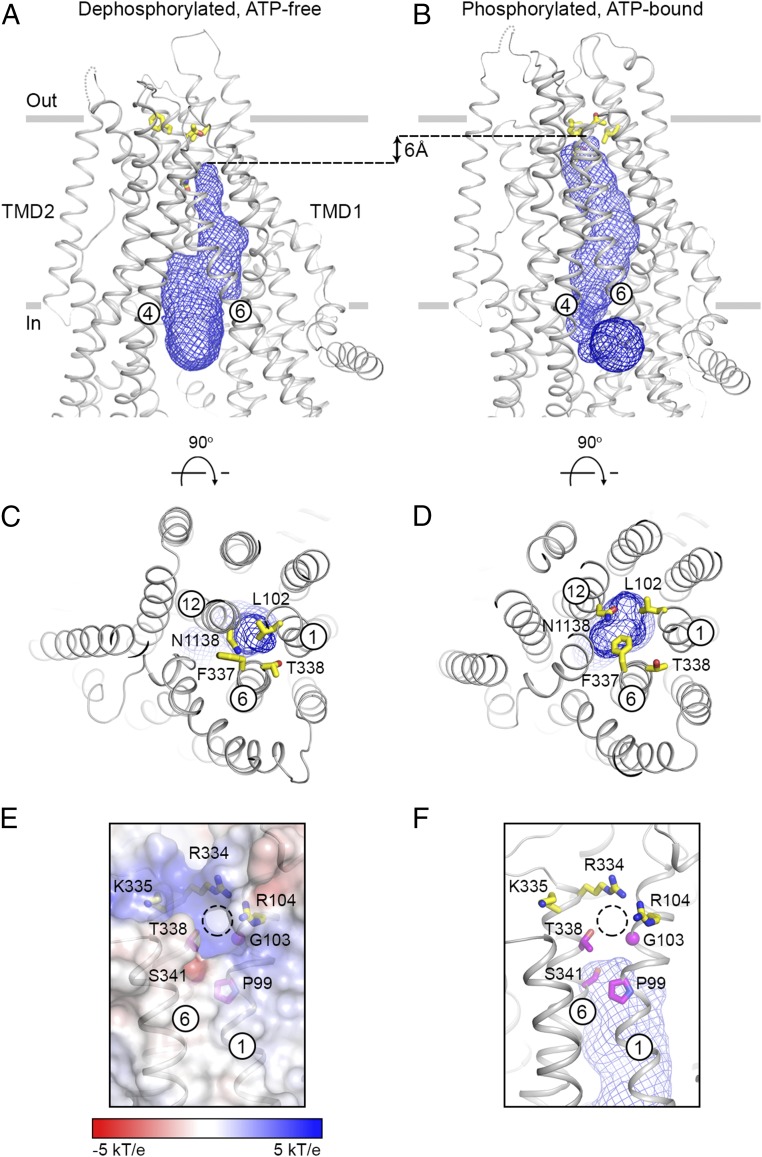
Using a probe equal to the size of a dehydrated chloride ion (1.7-Å radius), a passageway into the heart of the channel can be identified (Fig. 3 A and B). Functional studies indicate that this passageway is the path of ion flow in CFTR (9, 12). Comparison of hCFTR in two conformations show that upon NBD dimerization, the pore penetrates 6 Å further toward the extracellular space, reaching a constriction area comprised of residues L102, F337, T338, and N1138 (Fig. 3 A–D). Consistent with this conformational change, residues L102C, F337C, T338C, and N1138C in cysteine-free constructs are accessible to intracellular cysteine reactive reagents only in the channel-open state (20–22).
Fig. 3.
Conformational changes of the ion-permeation pathway. Two orthogonal views of the dephosphorylated, ATP-free (A and C) and the phosphorylated, ATP-bound (B and D) structures. The pore, shown as a blue mesh, is defined by a probe with the size of a dehydrated chloride ion (1.7-Å radius). It is connected to the cytosol through an opening between TMs 4 and 6. The side chains of residues L102, F337, T338, and N1138 are shown as yellow sticks. (E and F) The extracellular opening (shown as a dashed circle). CFTR is shown in ribbon along with the electrostatic surface (E) or the pore as blue mesh (F). Residues located around the external region of the opening are shown in stick models. In purple are potential gating residues; in yellow are positively charged residues influencing ion conduction.
In the NBD-dimerized conformation, the pore remains connected to the cytosol through a gap between TMs 4 and 6 (Fig. 3B). At the extracellular surface, a small opening is observed between TM 1 and TM 6 (Fig. 3 E and F). The radius of the opening is about 1.2 Å, too small to permit ion or water permeation. Among the residues surrounding this opening, S341 and T338 have been proposed to gate the pore (20, 21). Three positively charged residues, R104, R334, and K335, cluster on the extracellular side of the opening (Fig. 3 E and F). Modification of these residues influences anion conduction (22–24), suggesting that these residues are involved in attracting extracellular anions through electrostatic interactions. It seems possible that this small opening is the extracellular mouth of the pore. However, we suspect a fully conductive channel would require further dilation.
Structural Comparison Between Zebrafish and Human CFTR.
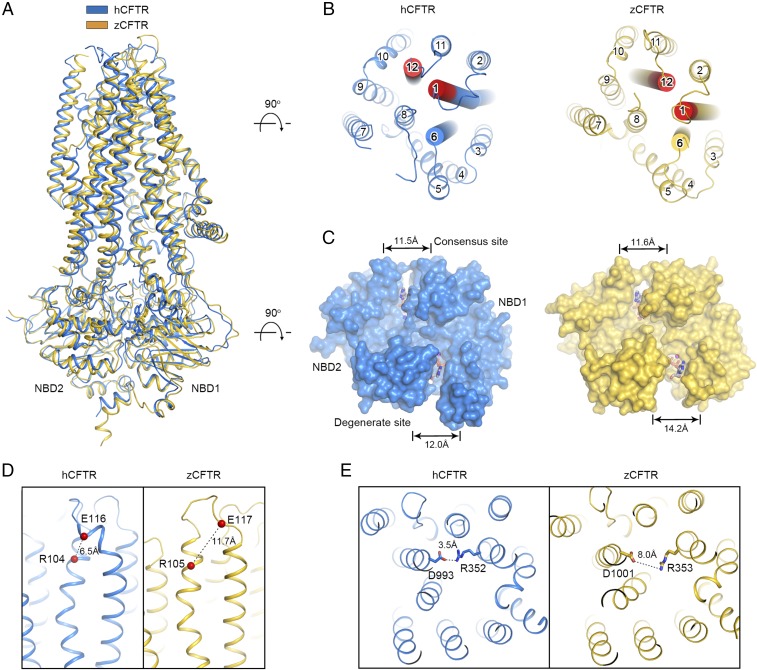
The structure of the phosphorylated, ATP-bound hCFTR share many features with that of zCFTR (13) (Fig. 4). For example, the two NBDs form a “head-to-tail” dimer occluding two ATP molecules, the ion conduction pathway is lined with positively charged residues, and the pore is open to the cytoplasm between TMs 4 and 6. Also similar to zCFTR, TM 8 shows an unusual helix–loop–helix configuration inside the membrane and TM 7 locates outside the ion conduction pathway in a position atypical of ABC transporters. These conserved features emphasize structural aspects central to CFTR function.
Fig. 4.
Structural comparison of phosphorylated human and zebrafish CFTR. (A) Superposition of the overall structures. hCFTR is shown in blue, and zCFTR (PDB ID code 5W81) is in yellow. (B) An extracellular view showing the different locations of TMs 1, 6, and 12 (highlighted as cylinders) in the two structures. The extracellular region of TMs 1 and 6, where the residues have the largest displacement between these two structures, are colored in red (residues 107–109 and 1129–1134 of hCFTR, and 107–109 and 1137–1142 of zCFTR). (C) The symmetric NBD dimer in hCFTR versus the asymmetric dimer in zCFTR. The distances shown are between the Cα atoms of the conserved lysine in the Walker A motif and the glycine in the signature motif (LSGGQ) at each ATP-binding site. (D) The distance between R104 and E116 in the two structures. (E) The R352/D993 salt bridge is only observed in hCFTR.
One major structural difference between the phosphorylated zCFTR and hCFTR occurs in the TMDs (Fig. 4B). Although the overall configuration of the pore is similar, the positions of many individual residues are not identical. When superimposed, the root-mean-square deviation (rmsd) for all 627 Cα positions in TMD is 2.0 Å. The difference is more prominent in the extracellular region of TMs 1, 6, and 12, where the largest displacement of equivalent residues is 6 Å (residues 107–109 of TM 1 and 1129–1134 of TM 12; Fig. 4B, red region). The small extracellular opening observed in hCFTR (Fig. 3 E and F) is completely sealed in zCFTR.
A second major structural difference is found in the NBD dimer (Fig. 4C). In CFTR, as well as other ABC transporters in the ABCC subfamily, only one ATPase site is catalytically competent. The degenerate site binds but does not hydrolyze ATP (25, 26). The NBD dimer in zCFTR is asymmetric: The degenerate site is more open than that of the consensus site (Fig. 4C). However, in hCFTR, the NBD dimer is symmetric: Both ATPase sites are completely closed off when ATP is bound. Symmetric ATP binding was also observed for the multidrug resistance protein 1, a closely related ABC transporter in the ABCC subfamily (27). Thus, the asymmetric NBD dimer observed in zCFTR does not represent a general feature caused by the degenerate ATPase site.
Because of these structural differences, the hCFTR structure provides a better framework to interpret functional data, most of which have been obtained from hCFTR. For example, cysteines introduced at positions 104 and 116 were reported to form a spontaneous disulfide bond (28). Whereas the Cα atoms of these two residues are 11.7 Å apart in zCFTR, in hCFTR the distance is 6.5 Å, within the range of a disulfide bond (Fig. 4D). Another example is R352, whose functional role has been debated. Based on analysis of relative permeability of Cl− to Na+, it was suggested that R352 functions as a selectivity determinant by providing positive charge in the pore (29–31). However, in a different study, the R352Q mutant was proposed to alter the structure such that an endogenous cysteine becomes accessible to thiol reagents (23). Additional studies show that the charge-swapping mutant R352E/D993R functions similar to the wild-type protein, suggesting that R352 forms a salt bridge with D993 (32). In the structure of phosphorylated hCFTR, this salt bridge is indeed observed (Fig. 4E), thus directly supporting the structural role of R352 in stabilizing the open pore. In zCFTR, the equivalent residues are too far apart to interact with each other.
Disease-Causing Mutations Interfere with Conformational Changes.
More than 300 mutations have been identified to cause cystic fibrosis (https://cftr2.org/). Some mutations lead to defects in protein synthesis and processing, and others diminish channel activity. Understanding the molecular mechanisms underlying these mutations is key to personalized treatment as different therapy is needed to facilitate folding versus improve function. In addition, these mutations provide valuable information to help us understand the molecular details underlying CFTR gating. Previously, we mapped all of the 53 missense mutations onto the structure of zCFTR and categorized the mutants into four groups based on their structural consequences (12). At that time, we could not explain several mutations, including G178R, G970R, and L927P (12). Now with the structure of hCFTR, we can better rationalize why these mutations cause cystic fibrosis.
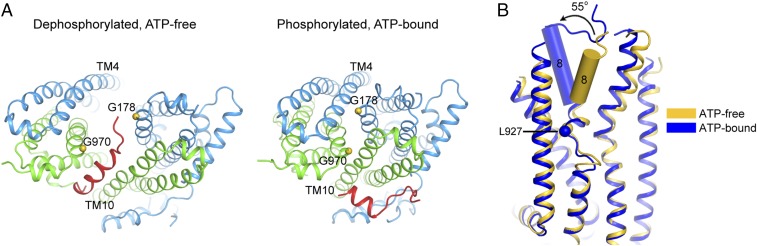
In the dephosphorylated, ATP-free conformation, residues G178 and G970 line the intracellular opening where the R domain resides (Fig. 5A). Upon phosphorylation and NBD dimerization, both glycine residues are engaged in close interhelical packing: G178 packs against TM 4 and G970 against TM 10 (Fig. 5A). Replacing either residue with arginine, as found in some cystic fibrosis patients, would be incompatible with the NBD-dimerized conformation. Consistent with this rationale, although both G178R and G970R mutants are expressed at the cell surface, they show little channel activity in iodide efflux assays (33, 34). Furthermore, the G970R mutant has a lower conductance compared with the G970D mutant, suggesting that the larger side chain of the arginine has a larger effect on NBD dimerization (35).
Fig. 5.
Disease-causing mutations that obstruct conformational changes. (A) The positions of G178 and G970. R domain is shown in red. (B) Superposition of the hCFTR in the ATP-free and ATP-bound conformations, showing the rotation of the outer leaflet segment of TM 8 and the location of L927.
Similarly, L927P may also cause cystic fibrosis by interfering with conformational changes necessary for channel opening (Fig. 5B). The surface expression level of the L927P mutant is 43% that of the wild-type protein, but its channel activity is only 0.1% (36). Comparison of the two structures of CFTR indicates that the extracellular portion of TM 8 rotates about 55° upon phosphorylation and NBD dimerization. L927 is located within the hinge region in TM 8 (Fig. 5B). As proline contains a constrained cyclic side chain, it is unable to adopt many of the main chain conformations that leucine permits. Thus, the L927P mutation would limit the conformational flexibility of the hinge or redirect TM 8 into a nonconductive conformation. The fact that this mutation nearly abolishes the channel activity suggests that the TM 8 hinge is essential for normal gating.
Summary
Here, we have reported the structure of human CFTR in the phosphorylated, ATP-bound conformation. This structure reveals a previously unresolved helix belonging to the phosphorylated R domain, thus offering a more complete understanding of how R domain phosphorylation regulates channel activity. Direct comparison of human CFTR in two functional states establishes conformational changes necessary for channel opening. It also provides a better understanding of several disease-causing mutations.
Methods
Protein Expression and Purification.
The E1371Q mutation was introduced to the wild-type construct with QuikChange Site-Directed Mutagenesis (Stratagene). The E1371Q mutant construct was expressed and purified using a protocol previously established for the wild-type CFTR (12). Briefly, the construct was expressed in HEK293S GnTl− cells and purified using a nanobody resin against the GFP tag attached to the C-terminal of CFTR (37). The membrane was solubilized with 1% 2,2-didecylpropane-1,3-bis-β-d-maltopyranoside (LMNG) and 0.2% cholesteryl hemisuccinate (CHS). The supernatant was applied to the GFP nanobody-coupled affinity resin followed by buffer exchange to 0.06% digitonin. The GFP tag was removed by PreScission protease and the protein was phosphorylated with PKA (NEB P6000L). Phosphorylated CFTR was further purified by gel filtration chromatography using a Superose 6 10/300 column (GE Healthcare) in a buffer containing 20 mM Tris⋅HCl pH 7.5, 200 mM NaCl, 2 mM DTT, 1 mM MgCl2, 1 mM ATP, and 0.06% digitonin.
EM Sample Preparation.
The protein sample was concentrated to 6 mg/mL and supplemented with additional 8 mM ATP, 8 mM MgCl2, and 3 mM fluorinated Fos-Choline-8 before applied to Quantifoil R1.2/1.3 400 mesh Au holey carbon grids (Quantifoil). Grids were frozen with Vitrobot Mark IV (FEI).
Data Collection and Processing.
Data were collected on a 300-kV Titan Krios (FEI) microscope with a K2 summit detector (Gatan) in superresolution mode. Each micrograph was exposed for 10 s at a rate of 8 electrons/pixel per second and fractionated into 50 subframes. The images were normalized with gain reference and binned by 2, resulting in a physical pixel size of 1.03 Å. Contrast transfer function estimation and whole-frame drift correction were carried out using CTFFIND4 (38) and Unblur (39), respectively. Subsequent data processing, including particle picking, 2D, and 3D classification, was carried out in Relion (40), except that drifts of individual particles were corrected using the program alignparts_lmbfgs (41). The ATP-bound zebrafish CFTR (13) (EMD-8782) was used as the initial model for 3D classification. The map generated from Relion 3D refinement was then used as an initial model for further refinements in Frealign (42) using all 677,308 particles. The resolution of the final map is estimated to be 3.2 Å based on the correlation of the two half maps at the 0.143 value (SI Appendix, Fig. S1).
Model Construction and Refinement.
We divided the dataset into two halves: one used for model building (the working set) and the other for validation (the free set). To better visualize the side chains for model building, the map calculated from the working set was sharpened using BFACTOR.EXE (written by Nikolaus Grigorieff) with a resolution limit of 3.2 Å and a b-factor of −50 Å2. The model was built in Chimera (43) and Coot (44). The final structure contains residues 1–390 of TMD1; 391–409, 435–637 of NBD1; 845–889, 900–1173 of TMD2; 1202–1451 of NBD2; and 17 residues of R domain (built as alanines).
Phenix (45) and Refmac (46, 47) were used to refine the model in real space and reciprocal space, respectively. Fourier shell correlations (FSCs) were calculated between the model versus the working map, the free map, or the full map, and between the two half maps. Local resolutions were calculated using Blocres (48). The geometry of the model was validated using MolProbity (49, 50). Figures were generated with Pymol (https://pymol.org/2/), Chimera (43), and HOLE (51).
Supplementary Material
Acknowledgments
We thank Mark Ebrahim and Johanna Sotiris of The Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center at The Rockefeller University for assistance in data collection. This work was supported by Cystic Fibrosis Foundation Therapeutics (to J.C.) and the Charles H. Revson Senior Fellowship in Biomedical Science (to Z.Z.). J.C. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6MSM). The Cryo-EM density map has been deposited in the Electron Microscopy Data Bank (accession no. EMD-9230).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815287115/-/DCSupplemental.
References
- 1.Gadsby DC, Vergani P, Csanády L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boat TF, Welsh MJ, Beaudet AL. In: The Metabolic Basis of Inherited Disease. 6th Ed. Scriver CR, Beaudet Al, Sly WS, Valle D, editors. McGraw-Hill; New York: 1989. pp. 2649–2680. [Google Scholar]
- 3.Rommens JM, et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 4.Elborn JS. Cystic fibrosis. Lancet. 2016;388:2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 5.Cheng SH, et al. Phosphorylation of the R domain by cAMP-dependent protein kinase regulates the CFTR chloride channel. Cell. 1991;66:1027–1036. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- 6.Picciotto MR, Cohn JA, Bertuzzi G, Greengard P, Nairn AC. Phosphorylation of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1992;267:12742–12752. [PubMed] [Google Scholar]
- 7.Csanády L, et al. Preferential phosphorylation of R-domain serine 768 dampens activation of CFTR channels by PKA. J Gen Physiol. 2005;125:171–186. doi: 10.1085/jgp.200409076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csanády L, Chan KW, Nairn AC, Gadsby DC. Functional roles of nonconserved structural segments in CFTR’s NH2-terminal nucleotide binding domain. J Gen Physiol. 2005;125:43–55. doi: 10.1085/jgp.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu F, Zhang Z, Csanady L, Gadsby DC, Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–92. doi: 10.1016/j.cell.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 10.Linsdell P. Architecture and functional properties of the CFTR channel pore. Cell Mol Life Sci. 2017;74:67–83. doi: 10.1007/s00018-016-2389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang TC, et al. Structural mechanisms of CFTR function and dysfunction. J Gen Physiol. 2018;150:539–570. doi: 10.1085/jgp.201711946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Chen J. Atomic structure of the cystic fibrosis transmembrane conductance regulator. Cell. 2016;167:1586–1597. doi: 10.1016/j.cell.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Liu F, Chen J. Conformational changes of CFTR upon phosphorylation and ATP binding. Cell. 2017;170:483–491. doi: 10.1016/j.cell.2017.06.041. [DOI] [PubMed] [Google Scholar]
- 14.Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker JM, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulhanty AM, Riordan JR. Phosphorylation by cAMP-dependent protein kinase causes a conformational change in the R domain of the cystic fibrosis transmembrane conductance regulator. Biochemistry. 1994;33:4072–4079. doi: 10.1021/bi00179a036. [DOI] [PubMed] [Google Scholar]
- 17.Bozoky Z, et al. Regulatory R region of the CFTR chloride channel is a dynamic integrator of phospho-dependent intra- and intermolecular interactions. Proc Natl Acad Sci USA. 2013;110:E4427–E4436. doi: 10.1073/pnas.1315104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naren AP, et al. CFTR chloride channel regulation by an interdomain interaction. Science. 1999;286:544–548. doi: 10.1126/science.286.5439.544. [DOI] [PubMed] [Google Scholar]
- 19.Csanády L, et al. Severed channels probe regulation of gating of cystic fibrosis transmembrane conductance regulator by its cytoplasmic domains. J Gen Physiol. 2000;116:477–500. doi: 10.1085/jgp.116.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, Hwang TC. Localizing a gate in CFTR. Proc Natl Acad Sci USA. 2015;112:2461–2466. doi: 10.1073/pnas.1420676112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Hiani Y, Linsdell P. Conformational changes opening and closing the CFTR chloride channel: Insights from cysteine scanning mutagenesis. Biochem Cell Biol. 2014;92:481–488. doi: 10.1139/bcb-2014-0038. [DOI] [PubMed] [Google Scholar]
- 22.Gong X, Linsdell P. Mutation-induced blocker permeability and multiion block of the CFTR chloride channel pore. J Gen Physiol. 2003;122:673–687. doi: 10.1085/jgp.200308889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith SS, et al. CFTR: Covalent and noncovalent modification suggests a role for fixed charges in anion conduction. J Gen Physiol. 2001;118:407–431. doi: 10.1085/jgp.118.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou JJ, Fatehi M, Linsdell P. Identification of positive charges situated at the outer mouth of the CFTR chloride channel pore. Pflugers Arch. 2008;457:351–360. doi: 10.1007/s00424-008-0521-6. [DOI] [PubMed] [Google Scholar]
- 25.Aleksandrov L, Aleksandrov AA, Chang XB, Riordan JR. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J Biol Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- 26.Basso C, Vergani P, Nairn AC, Gadsby DC. Prolonged nonhydrolytic interaction of nucleotide with CFTR’s NH2-terminal nucleotide binding domain and its role in channel gating. J Gen Physiol. 2003;122:333–348. doi: 10.1085/jgp.200308798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson ZL, Chen J. ATP binding enables substrate release from multidrug resistance protein 1. Cell. 2018;172:81–89.e10. doi: 10.1016/j.cell.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Cui G, et al. Three charged amino acids in extracellular loop 1 are involved in maintaining the outer pore architecture of CFTR. J Gen Physiol. 2014;144:159–179. doi: 10.1085/jgp.201311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung M, Akabas MH. Locating the anion-selectivity filter of the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel. J Gen Physiol. 1997;109:289–299. doi: 10.1085/jgp.109.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guinamard R, Akabas MH. Arg352 is a major determinant of charge selectivity in the cystic fibrosis transmembrane conductance regulator chloride channel. Biochemistry. 1999;38:5528–5537. doi: 10.1021/bi990155n. [DOI] [PubMed] [Google Scholar]
- 31.Aubin CN, Linsdell P. Positive charges at the intracellular mouth of the pore regulate anion conduction in the CFTR chloride channel. J Gen Physiol. 2006;128:535–545. doi: 10.1085/jgp.200609516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui G, Zhang ZR, O’Brien AR, Song B, McCarty NA. Mutations at arginine 352 alter the pore architecture of CFTR. J Membr Biol. 2008;222:91–106. doi: 10.1007/s00232-008-9105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seibert FS, et al. Disease-associated mutations in cytoplasmic loops 1 and 2 of cystic fibrosis transmembrane conductance regulator impede processing or opening of the channel. Biochemistry. 1997;36:11966–11974. doi: 10.1021/bi9712652. [DOI] [PubMed] [Google Scholar]
- 34.Seibert FS, et al. Cytoplasmic loop three of cystic fibrosis transmembrane conductance regulator contributes to regulation of chloride channel activity. J Biol Chem. 1996;271:27493–27499. doi: 10.1074/jbc.271.44.27493. [DOI] [PubMed] [Google Scholar]
- 35.Wagner JA, et al. Two novel mutations in a cystic fibrosis patient of Chinese origin. Hum Genet. 1999;104:511–515. doi: 10.1007/s004390050996. [DOI] [PubMed] [Google Scholar]
- 36.Van Goor F, Yu H, Burton B, Hoffman BJ. Effect of ivacaftor on CFTR forms with missense mutations associated with defects in protein processing or function. J Cyst Fibros. 2014;13:29–36. doi: 10.1016/j.jcf.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubinstein JL, Brubaker MA. Alignment of cryo-EM movies of individual particles by optimization of image translations. J Struct Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Grigorieff N. Frealign: An exploratory tool for single-particle cryo-EM. Methods Enzymol. 2016;579:191–226. doi: 10.1016/bs.mie.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis IW, et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.