Abstract
FLIRT (Fast Local InfraRed Thermogenetics) is a microscopy-based technology to locally and reversibly manipulate protein function during cellular behaviors while simultaneously monitoring the effects in vivo. FLIRT locally inactivates fast-acting temperature sensitive (ts) mutant proteins, using non-ts mutants as controls. We demonstrate that FLIRT can control ts proteins required for cell division, Delta-Notch cell fate signaling, and germline structure in C. elegans with cell-specific and even subcellular precision.
Molecular control of transient cellular behavior such as cell division involves precise spatiotemporal regulation. Fast-acting (≤20 sec) ts mutants enable studies of temporally regulated protein function by upshifting cells from permissive to restrictive temperature to conditionally inactivate protein function during the cellular process of interest (Figure 1a). Current techniques to inactivate fast-acting ts mutants rely on changing the temperature of the whole cell or organism using temperature-controlled stages or specimen holders1–6. While these techniques have high temporal resolution, they are unable to perturb protein function with spatial resolution.
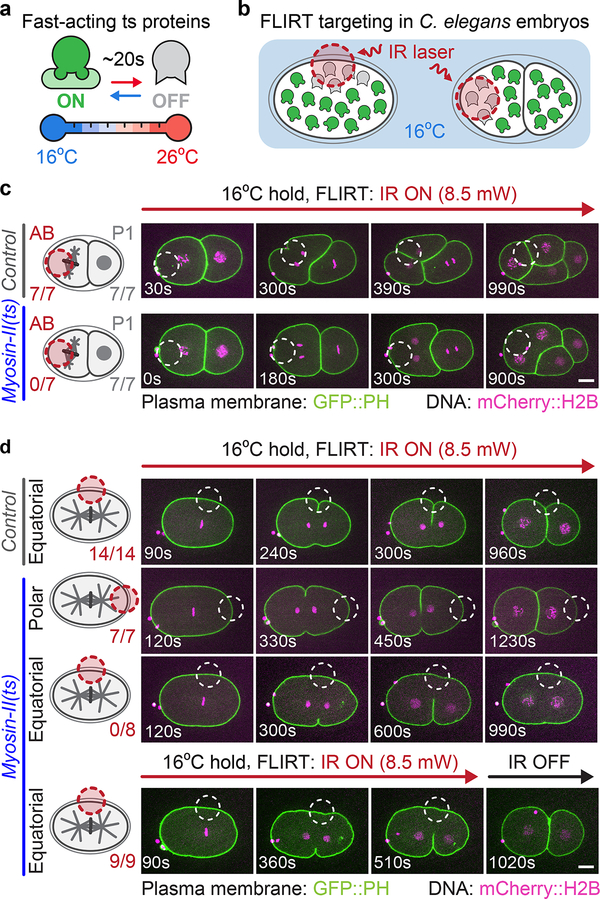
Figure 1:
FLIRT calibration and application for spatiotemporal control of ts protein function in vivo
(a) Fast-acting ts mutant proteins are rapidly inactivated upon temperature upshift3, 6. (b) Schematic of FLIRT targeting in which an IR laser is used to locally inactivate ts mutant protein function. (c) Experimental schematic (left) and representative time lapse images (right) of cell-specific FLIRT targeting in 2-cell C. elegans embryos. See Supplementary Video 1. (d) Experimental schematic (left) and representative time lapse images (right) of subcellular FLIRT targeting either an equatorial or polar region in myosin-II(ts) 1-cell embryos. Embryos were FLIRT-targeted either throughout division (top 3 rows, see Supplementary Video 3) or for ~8 min to test reversibility (bottom row, see Supplementary Video 5). The number of AB and P1 cells (c) or 1-cell embryos (d) from biologically independent embryos that successfully completed cell division is indicated below each experimental schematic (left). Red (schematic) or white (images) dashed circles indicate the FLIRT-targeted regions. Time is in sec after FLIRT initiation. Scale bars=10 μm.
To harness the power of fast-acting ts mutants for high-resolution spatiotemporal studies, we developed FLIRT (Fast Local InfraRed Thermogenetics), which uses infrared (IR) light to rapidly and locally control ts mutant protein function. In brief, an IR laser focused on a distinct subcellular structure or specific cell within an organism locally heat-inactivates ts proteins at precise moments during a cellular behavior while monitoring the effects in vivo using a spinning disc confocal microscope (Figure 1b). Here we describe the FLIRT system, validate its ability to alter local temperature in vivo, and demonstrate its precision and resolution in altering ts mutant protein function at the cellular and subcellular levels in C. elegans.
The FLIRT system is built on an inverted spinning disk confocal microscope equipped with a multimode 1470 nm wavelength laser (Supplementary Figure 1a). 1470 nm was selected, as previously used for IR-induced gene expression7, due to the high absorption of water and low cellular toxicity in that spectral region. The area targeted for IR irradiation is controlled by a wheel containing different mask shapes and sizes; the IR mask is then projected onto the specimen plane. The IR laser is focused at the beginning of each imaging session using photon upconverting nanoparticles (UCPs) (Supplementary Figure 1b), which are excited by IR light but emit visible light at wavelengths similar to commonly used fluorophores (e.g. GFP) via a 3-photon anti-Stokes luminescence mechanism, allowing precise focus of the laser-targeted region onto the specimen plane8. A microfluidic temperature control system maintains specimen temperature and functions as a heat sink.
To measure and calibrate the temperature change induced by FLIRT, we used two circular masks (16 and 27 μm diameter, Supplementary Figure 1b) and two independent assays: 1) a thermochromatic dye that undergoes a temperature-dependent color change from opaque (black) to transparent at 15°C, altering light transmittance through a glass coverslip painted with this dye (Supplementary Figure 2); 2) an mCherry-based bioassay in C. elegans embryos expressing mCherry::HistoneH2B, based on previous work showing that the fluorescence intensity of mCherry emission decreases as sample temperature increases (Supplementary Figure 3)9. We generated calibration curves of the temperature-dependent change in light transmission (thermochromatic dye) or fluorescence intensity (mCherry) across a range of temperatures using the microfluidic temperature control system and compared this to the change in intensity observed with increasing IR laser power (Supplementary Figures 2, 3). Both assays gave similar results; we determined that 0.9 and 1.0 mW of IR laser power shift the sample temperature within the targeted area by 1°C in vivo when using 16 and 27 μm diameter circular masks, respectively (Supplementary Figure 3); we confirmed this result using whole-cell FLIRT in vivo (Supplementary Note 1).
To test if FLIRT can inhibit ts mutant proteins in a cell-specific manner without affecting non-targeted cells, we used a 16 μm diameter mask to specifically target the division plane in either the anterior cell (AB) or the posterior cell (P1) of 2-cell control or myosin-II(ts) (nmy-2(ne3409ts)) embryos. Myosin-II is an actin motor protein required for constriction of the actomyosin contractile ring during cytokinesis, the physical division of one cell into two10–12. In myosin-II(ts) embryos upshifted to 26°C, cytokinesis fails in both AB and P1 cells (Supplementary Note 2). In FLIRT-targeted myosin-II(ts) embryos, division failed only in the targeted cell but completed in the other cell (Figure 1c, Supplementary Figure 4a, Supplementary Video 1). When the IR laser was turned off ~4 min after anaphase onset, both AB and P1 cells divided, indicating that FLIRT-mediated protein inhibition is rapidly reversible (Supplementary Figure 4b, Supplementary Video 2). In control non-ts embryos, neither cell failed in division, even when targeted with FLIRT throughout contractile ring constriction (Figure 1c, Supplementary Figure 4a, Supplementary Video 1). Thus, FLIRT specifically and reversibly inactivates ts proteins only in the targeted cell of a 2-cell embryo.
We next tested if FLIRT can control ts mutant protein function with subcellular precision in the 1-cell C. elegans embryo. During cytokinesis, myosin-II is enriched at the cell equator in the actomyosin contractile ring, where it is thought to drive ring constriction10–12. Myosin-II in the polar regions is not thought to drive ring constriction. Thus, we FLIRT targeted (16 μm diameter mask) either an equatorial or polar region of dividing control and myosin-II(ts) embryos (Figure 1d, Supplementary Figure 5a, Supplementary Videos 3, 4). While FLIRT often caused a transient bleb in control embryos with equatorial targeting, likely due to local thermal gradients13, it did not disrupt the normal polarity or asymmetry of this cell division (in contrast to other IR systems14), embryo viability, or developmental timing (Supplementary Figure 6), and all control embryos divided successfully (Figure 1d, Supplementary Figure 5a). In myosin-II(ts) embryos, FLIRT targeting one side of the equatorial region prevented contractile ring constriction on that side, but not on the non-targeted side. On the non-targeted side, the contractile ring initiated constriction but regressed upon approaching the FLIRT-targeted region (Figure 1d, Supplementary Figure 5a, Supplementary Videos 3, 4). When the laser was turned off ~6 min after anaphase onset, both sides of the equator underwent ring constriction and cytokinesis completed, again demonstrating that FLIRT inhibition of protein function is reversible (Figure 1d, Supplementary Figure 5a, Supplementary Videos 5, 6). Targeting the polar regions of myosin-II(ts) embryos or control embryos did not disrupt ring constriction (Figure 1d, Supplementary Figure 5a). Moreover, targeting the equatorial region in Delta(ts) (apx-1(zu347ts)) embryos, a gene required for Delta-Notch cell fate signaling but not cytokinesis, did not disrupt cell division, demonstrating that the effects of FLIRT are specific to the ts mutant tested (Supplementary Figure 5b). These results suggest that FLIRT locally and reversibly inhibits ts mutant protein function with subcellular precision.
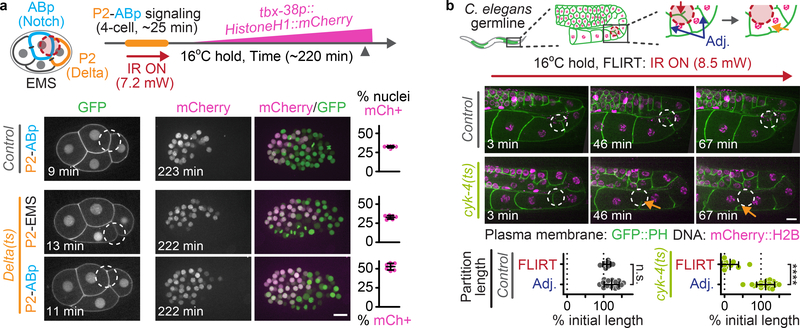
To confirm that FLIRT can disrupt ts mutant proteins with subcellular precision during a different cellular behavior, we used the Delta(ts) fast-acting ts allele to perturb Delta-Notch-mediated cell fate signaling by targeting specific cell-cell contacts within the 4-cell embryo. The 4-cell C. elegans embryo consists of two anterior (ABa, ABp) and two posterior (EMS, P2) cells. Only the P2 cell expresses the transmembrane ligand Delta/APX-1 (hereafter Delta), while both the ABa and ABp cells express its transmembrane receptor, Notch/GLP-1 (hereafter Notch; Supplementary Figure 7a)4, 15. Direct cell-cell contact between P2 and ABp activates Notch in ABp, changing its cell fate and establishing dorsal-ventral embryonic polarity at the 4-cell stage (Supplementary Figure 7a)4, 5, 16. To monitor Delta-Notch signaling, we generated strains expressing a fluorescent Notch transcriptional reporter (tbx-38p::mCherry::HistoneH117, see also18, 19) expressed in ~25% of the cells in late stage embryos when Notch is activated in ABp and ~50% of cells when Notch activation in ABp is blocked (Figure 2a, Supplementary Figure 7b-e). We used FLIRT to target specific cell-cell contacts (16 μm mask) in control and Delta(ts) mutant embryos and monitored the effect on Delta-Notch signaling (Supplementary Figure 7c). FLIRT targeting the P2-ABp contact for ~25 min inhibited Delta-Notch signaling in Delta(ts) mutants, but not in control embryos, whereas FLIRT targeting the P2-EMS contact had no effect (Figure 2a), suggesting that FLIRT locally inhibits Delta-Notch cell fate signaling by targeting P2-ABp cell contacts.
Figure 2:
FLIRT for subcellular control of ts protein function
(a) Experimental timeline schematic (top) and representative images (bottom) depicting FLIRT targeting either the P2-ABp or P2-EMS cell-cell contacts in 4-cell control and Delta(ts) embryos. Dot plots (right) show the percent total mCherry+ nuclei at the ~50 cell stage (control P2-ABp N= 7, Delta(ts) P2-EMS N=7, Delta(ts) P2-ABp N=7 biologically independent embryos). Green, GFP::PH (plasma membrane) and Histone(H2B/H3)::GFP (DNA); magenta, tbx-38p::HistoneH1::mCherry (see Methods) (b) Schematic (top) and representative time lapse images (bottom, representative from 7 biologically independent adult worms) depicting FLIRT targeting an individual membrane partition within the cyk-4(ts) adult C. elegans syncytial gonad. Arrows indicate membrane partition retraction. See Supplementary Video 7. Dot plots (bottom) showing the change in FLIRT-targeted and adjacent partition length after FLIRT targeting a single membrane partition in control and cyk-4(ts) worms shown as a percent of initial length (control N=7, cyk-4(ts) N=10 biologically independent worms). Time elapsed is shown in min after FLIRT initiation. Red dashed circles (schematics) and white dashed circles (images) indicate the FLIRT-targeted ROIs. Unpaired two-tailed t-test; n.s., no significance, p>0.05; ****, p≤0.0001. Error bars represent mean ± SD, see Supplementary Table 1 for additional statistical analysis. Scale bars=10 μm.
Finally, we tested if FLIRT can locally inhibit ts mutant protein function in the context of an adult worm tissue. Adult hermaphroditic C. elegans have a syncytial gonad in which germline nuclei undergo incomplete mitosis and are separated by membrane partitions but remain connected to the gonad via an intercellular bridge. The Rho-family GTPase activating protein CYK-4/MgcRacGAP (hereafter CYK-4) localizes to the intercellular bridges between germline membrane partitions and is required for oocyte production20. As previously shown, upshift of cyk-4(or749ts) (hereafter cyk-4(ts)) adult worms to 26°C led to a shortening of the membrane partitions that separate germline nuclei (Supplementary Figure 8)20. FLIRT targeting (16 μm mask) a single germline membrane partition in cyk-4(ts) worms led to a shortening of the FLIRT-targeted membrane partition but did not affect the length of membrane partitions between adjacent germline nuclei (Figure 2b, Supplementary Video 7). FLIRT targeting membrane partitions in control worms did not change partition length (Figure 2b). Thus, FLIRT can locally inhibit ts protein function with spatial resolution in germline tissue within the adult worm.
Together, our results demonstrate that FLIRT is a powerful technique to locally inhibit protein function with cellular and subcellular precision. While we have applied this technology to studies of cell division, cell fate specification, and germline structure in C. elegans, we anticipate FLIRT will be applicable to a myriad of cellular behaviors in model systems accessible to the light microscope with available fast-acting ts mutants.
Online Methods
The FLIRT microscope, imaging, and image analysis
The FLIRT microscope (Supplementary Figure 1a) was built on a Ti inverted microscope stand (Nikon) equipped with a 60 × 1.4 NA oil-immersion Plan Apo objective, a spinning disc confocal unit (CSU-10; Yokogawa) upgraded with Borealis (Spectral Applied Research), two 150 mW excitation lasers at 491 and 561 nm (Cairn), an emission filter wheel (Sutter Instruments) with 2 bandpass filters corresponding to 525/50 nm (Chroma #ET525/50m) and 620/50 nm (Chroma #620/60m), and a cooled charge-coupled device (CCD) camera (Orca-R2; Hamamatsu Photonics). The system was controlled with MetaMorph software (Molecular Devices). A ~3 W multimode 1470 nm laser was used for FLIRT experiments (Rapp Optoelectronics GmbH). A mask wheel made of NBK-7 glass coated with aluminum (Rapp Optoelectronics GmbH) containing masks of distinct shapes and sizes was used to precisely target the IR light on regions of interest (here a 16 or 27 μm diameter mask was used) focused at the specimen plane. The IR system, mask position, and laser power were controlled through Syscon Geo software (Rapp Optoelectronics GmbH). During all FLIRT experiments, focus was maintained relative to the coverslip using the CRISP auto focus system (Applied Scientific Instrumentation) before each image acquisition. Sunstone upconverting nanoparticles (also called Sunstone UCPs or nano545-UCPs, Sigma Aldrich #90992) mounted in H2O between a slide and coverslip were used for IR laser co-alignment with the visible light image path at the beginning of each experimental session as previously described21.
All image analysis (including intensity measurements and line scan analysis) was performed using FIJI software22 and graphs were generated in Prism 7 (GraphPad Software, Inc.) and imported into Adobe Illustrator CC (Adobe) to assemble the Figures. Images were pseudo-colored and contrast adjusted for display using Adobe Photoshop CC.
Laser power measurements
The power of the 1470 nm IR laser was measured using an XLP12–3S-H2-DO detector and an accompanying Maestro power monitor (Gentec-EO), positioned near the imaging plane of the microscope. For each mask setting (16 μm or 27 μm diameter), the IR laser power was varied and the mW values for each mask were monitored and averaged over a 2 min period from 3 independent measurements at each laser power setting.
Specimen temperature control and calibration
For all experiments, the specimens were mounted in a Peltier-controlled microfluidic specimen holder and temperature control device (CherryTemp, Cherry Biotech) set to the indicated temperatures. Cherry software (Cherry Biotech) was used to control the temperatures of the heat exchangers and to designate the source for the flow chamber. To monitor the stability and accuracy of the set point temperature in the specimen holder, a microthermistor probe (Bioptechs #1917) was mounted with a drop of M9 buffer23, 24 (85 mM NaCl, 42 mM Na2HPO4, 22 mM KH2PO4, 1 mM MgSO4-7H2O), on the coverslip (No. 1.5, VWR #16004–312) with a round 2% agarose pad (Omnipur, CalbioChem) (~280 μm thick and 1 cm in diameter) placed between the probe and the microfluidic specimen holder to mirror the setup for FLIRT experiments with C. elegans embryos (see below). The resistance at each set temperature was measured at the coverslip by a multi-meter (Mastech #MS8220R) across a range of temperatures (8–30°C) and an accurate estimation of temperature was performed by using a temperature versus resistance conversion chart specific for the thermistor used. For FLIRT experiments, samples were mounted on a coverslip in M9 buffer (for all C. elegans experiments, Figure 1, 2, Supplementary Figures 3–7) or H2O (for thermochromatic dye experiments, Supplementary Figure 2) and a round 2% agarose pad (see above) was placed on top of the sample between the coverslip and the flow chamber of the microfluidic specimen holder.
Thermochromatic dye-based temperature measurement
Thermochromatic dyes that undergo a chromatic transition from opaque (black) to transparent at 15°C (LCR Hallcrest LLC #TS-COLDKIT) were mixed with a binder (LCR Hallcrest LLC #TS-COLDKIT) and water in a 1:1:3 ratio respectively. A thin film of the mixture was spread on a coverslip and allowed to air dry for ~30 min. Prior to imaging, ~5 μL of water was added onto the coverslip and a round 2% agarose pad was placed between the water drop and the specimen holder. To calibrate the change in transmitted light versus temperature, the whole thermochromatic dye-coated coverslip was upshifted to specific temperatures at 0.5°C or 1.0°C increments across a thermal-range from 8–20°C, and images were collected at three different stage positions. For FLIRT calibration using the thermochromatic dye, either a 16 μm or 27 μm diameter mask was focused onto the dye at defined IR laser powers (0–15 mW) while holding the background temperature at 9°C, 10°C, or 11°C with the microfluidic specimen holder. Images were acquired every 60 sec for temperature upshift experiments or every 30 sec for FLIRT calibration with the thermochromatic dye, with 2 × 2 binning and 15 × 2.5 μm z-sections. z-sectioning was continually performed during the intervals between image acquisition, and the IR laser was kept on for the duration of the experiment (to mirror FLIRT experiments with C. elegans embryos; see below). Mean intensity values within the FLIRT Region of Interest (ROI) were measured on the maximum intensity projected image at each laser power. For temperature upshift experiments, mean intensity values were measured inside an ROI of the same size as that used for the corresponding FLIRT experiments. Mean intensity values from the initial frame were subtracted and values were normalized to the maximum intensity for the whole sample temperature upshift experiment for each coverslip (Supplementary Figure 2d,e). The temperature to IR laser power ratio (mW/°C) was determined by calculating the laser power required to reach the mean intensity values just before and just after the chromatic transition point (14°C, 14.5°C, 15°C, and 15.5°C) from each of the holding temperatures (9°C, 10°C, or 11°C) (Supplementary Figure 2e).
mCherry::HistoneH2B biosensor-based temperature measurement
A C. elegans strain expressing mCherry::HistoneH2B (OD95)25 was used as a temperature biosensor to calibrate the FLIRT microscope system. For these experiments, embryos at the ~100–200 cell stage were mounted in the microfluidic temperature control device as described above. To plot a calibration curve, the temperature of the sample was varied across a thermal range between 16–26°C and images were acquired at each temperature. For FLIRT experiments, embryos were held at 16°C, subjected to FLIRT with defined laser powers (0–10 mW) and images were acquired every 30 sec with an exposure time of 250 msec. For mCherry calibration experiments, images were acquired with a binning of 8 × 8 and 15 × 2.5 μm z-sections. For all FLIRT experiments, z-sectioning was continually performed between image acquisition. For image analysis, mean intensity values were measured on the sum projected raw image near the center of the FLIRT-targeted region of the embryo (4 × 4 μm square). Two similar sized ROIs were measured outside the embryo to determine the background intensity. The background subtracted intensity values were corrected for bleaching by measuring the average rate of photo-bleaching for each replicate and then applying a bleach correction factor [(bleach rate)*(frame number)*(measured intensity)] for each frame to calculate its bleach-corrected intensity value. The intensity values for each frame were normalized to the average intensity value at 16°C. The relationship between mean intensity value and the change in temperature (ΔT) was measured by monitoring the change in fluorescence intensity after defined temperature shifts and using linear regression modeling on these data in GraphPad Prism 7. Mean y-values (relative fluorescence intensity) were considered for all x values (ΔT relative to 16°C) between 0°C and 10°C, giving the relationship y = −1.438*x + 99.61, R2=0.99 (Supplementary Figure 3b, left panel). By performing similar image and linear regression analysis on embryos held at 16°C and subjected to FLIRT with defined laser powers (3.84 mW-9.75 mW), a linear relationship between fluorescence intensity and laser power was measured for both the 16 μm mask (y = −1.641*x + 100, R2=0.98) and the 27 μm mask (y = −1.595*x + 101, R2=0.99) (Supplementary Figure 3b, right panel). Using linear regression analysis, we derived the relationship between ΔT (°C) and IR laser power (mW) for the 16 μm mask (y = 1.088*x) and the 27 μm mask (y = 1.03*x).
H2B::mCherry biosensor-based thermal gradient measurement
For measurement of the thermal gradient induced by FLIRT, multicellular embryos (~100–200 cells) expressing mCherry::HistoneH2B (OD95) were mounted as described for biosensor temperature measurement. Specimens were held at a permissive temperature (16°C in Supplementary Figure 3d and Supplementary Figure 7d, 10–16°C in Supplementary Figure 3e) and the ROI was targeted to one end of the embryo using either a 27 (Supplementary Figure 3d) or 16 μm (Supplementary Figures 3d, 3e, 7d) diameter mask. Images were acquired with acquisition settings as described above for the mCherry::HistoneH2B biosensor-based temperature measurements, alternating between 5 frames acquired with the FLIRT laser off and 5 frames acquired with the FLIRT laser on with laser powers indicated for each experiment. For Supplementary Figures 3d-e, the FLIRT laser was used to heat the region to ~25.5°C with either the 27 μm mask (9.75 mW and 16°C hold) or the 16 μm mask (8.5 mW and 16°C hold; 10.1 mW and 14°C hold; 12.3 mW and 12°C hold; or 13.9 mW and 10°C hold). For Supplementary Figure 7d, the 16 μm mask was used to heat the region to ~24°C (7.2 mW and 16°C hold). For whole embryo upshifts (Supplementary Figure 3d), the same experimental setup was used and the temperature of the microfluidic temperature device was switched between 16°C and 26°C for 5 frames each.
For image analysis, FIJI22 was used to generate a sum projection of all z-sections and a 50 μm line scan across the entire embryo with a line thickness corresponding to the width of the FLIRT mask region (27 μm for Supplementary Figure 3d, 16 μm for Supplementary Figures 3d, 3e, 7d). A 50 μm long line scan of the same thickness (27 or 16 μm) was used for background measurement and line scans of the embryo were measured for the frames before and after turning the FLIRT laser on or off (4 measurements per embryo). The background measurement was averaged and subtracted from each value across the line scan and the intensity values across the embryo for the frame with the laser turned on were normalized to the frames with the laser off (permissive temperature, 10–16°C). A rolling average of 5 intensity values was calculated across the embryo. These intensity values were converted to calculated ΔT (°C) using the relationship between temperature and intensity found above (Y = −1.438*x + 99.61), and the hold temperature was added to find calculated T (°C). Line scans displayed in Supplementary Figures 3d, 3e, and 7d only include x-values between 2–45 μm to exclude the bright and variable signal from the extra-cellular polar bodies located at one end of the embryo.
Worm husbandry and embryo dissection
All worm strains used in this manuscript were grown on Normal Growth Media (NGM) plates using standard C. elegans husbandry techniques23, 24 and maintained in incubators (Binder) set to 16°C. Immediately prior to imaging, embryos of the appropriate stage were dissected from adult hermaphrodites into chilled M9 buffer (16°C) and transferred onto the microfluidic specimen holder and temperature control device (16°C) using a pulled needle mouth pipette for imaging as described26.
Worm strains used in this study
OD95: unc-119(ed3)* ltIs38[pAA1: pie-1p::GFP::PH(PLC1delta1); unc-119(+)III]; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119(+)]IV]
JCC637: nmy-2(ne3409ts)I; unc-119(ed3)* ltIs38[pAA1: pie-1p::GFP::PH(PLC1∂1); unc-119(+)III]; ltIs37[pAA64: pie-1p::mCHERRY::his-58; unc-119(+)]IV]
JCC744: unc-119(ed3)III; ddIs25 [GFP::par-2;unc-119(+)]; ddIs26 [mCherry::par-6;unc-119(+)]; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV.
JCC596: unc-119(ed3)* ltIs38[pAA1: pie-1p::GFP::PH(PLC1delta1); unc-119(+)III]; zuIs178[his-72(1kb 5’ UTR)::his-72::SRPVAT::GFP::his-72 (1KB 3’ UTR)# + 5.7 kb XbaI - HindIII unc-119(+)]; stIs10024[pie-1p::H2B::GFP; unc-119(+)]#; stIs10138[tbx-38p::HistoneH1-mCherry; unc-119(+)]
JCC623: unc-119(ed3)* ltIs38[pAA1; pie-1p::GFP::PH(PLC1delta1); unc-119(+)III]; apx-1(zu347ts)V; zuIs178[his-72(1kb 5’ UTR)::his-72::SRPVAT::GFP::his-72 (1KB 3’ UTR) # + 5.7 kb XbaI - HindIII unc-119(+)]. stIs10024[pie-1p::H2B::GFP; unc-119(+)]#; stIs10138[tbx-38p::HistoneH1-mCherry; unc-119(+)]
OD239: cyk-4(or749ts) ltIs38 [pAA1; pie-1::GFP::PH(PLC1delta1) unc-119 (+)]III; ltIs37 [pAA64; pie-1::mCHERRY::his-58; unc-119 (+)]IV
*NOTE: The unc-119(ed3) mutation was present in the parental strains but has not been directly sequenced in these strains to determine if the unc-119 gene is mutated.
#NOTE: These strains are expressing a Histone::GFP but they have not been directly sequenced to determine which of the his-72 and/or H2B transgenes are present; the GFP signal was used only as a marker for all nuclei.
Assaying cell division at permissive temperatures
For experiments to assess cell division defects at low background hold temperatures (Supplementary Figure 3f), control 1-cell embryos were dissected and mounted in the microfluidic specimen holder at a permissive temperature (between 10–16°C) and maintained at that temperature throughout the experiment. Images were acquired from prometaphase through anaphase onset of the next cell division. Images were acquired every 30 sec with 200 msec exposure for DIC and 150 msec exposure times for the 491 nm and the 561 nm channels, with 2 × 2 binning and 15 × 2.5 μm z-sectioning. Cell division outcomes were scored and divided into three phenotypic categories: 1) embryos in which cell division completes without any visible defects, 2) embryos in which cytokinesis fails, or 3) embryos in which cell division completes but have minor defects during cell division. Minor division defects occur at low temperatures, likely due to the inherent cold sensitivity of spindle microtubules27, that lead to errors in mitotic spindle positioning, movement, and/or orientation.
FLIRT experiments targeting cell division in myosin-II(ts) and Delta(ts) mutants
For experiments to test the ability of FLIRT to locally inhibit myosin-II (NMY-2) activity with cellular (Figure 1c, Supplementary Figure 4) and subcellular (Figure 1d, Supplementary Figures 5a,b, 6b) precision, embryos from C. elegans strains OD95 (control), JCC637 (myosin-II(ts)), or JCC623 (Delta(ts)), were held at either 16°C (Figure 1c-d, Supplementary Figures 4, 5b, 6b) or 14°C (Supplementary Figure 5a, 6b) using the microfluidic specimen holder and subjected to FLIRT with the 16 μm or 27 μm diameter masks. FLIRT experiments with the 27 μm mask were performed at ~6.1 mW (~22°C) or at 9.8 mW (~26°C) of IR laser power (Supplementary Figures 2, 3; Supplementary Note 1). FLIRT experiments with the 16 μm mask (Figure 1c-d, Supplementary Figures 4, 5, 6b) were performed with 8.5 mW (16°C hold) or 10.1 mW (14°C hold) of IR laser power. FLIRT was turned on during prometaphase for all 1-cell experiments (Figure 1d, Supplementary Figures 5, 6b) and prometaphase of the AB cell for 2-cell experiments (Figure 1c, Supplementary Figures 4) and was maintained on for the duration of the experiment through anaphase onset of the next cell division. During cytokinesis, the contractile ring in the AB cell mostly invaginates from the anterior of the cell and only slightly invaginates from the side of AB in direct cell-contact with P1; thus in 2-cell FLIRT experiments we targeted the division plane on the anterior side of the AB cell. Cytokinesis in P1 initiates from both sides of the cell, but to be consistent with the AB cell experiments, we targeted a posterior region of the P1 cell. For experiments to demonstrate the reversibility of FLIRT, embryos were subjected to an interval of FLIRT with the 16 μm mask for from prometaphase until 6 min after anaphase onset (Figure 1d, Supplementary Figure 5a) or 3–4 min after anaphase onset (Supplementary Figure 4b). Images were acquired every 30 sec with 200 msec exposure for DIC and 150 msec exposure times for the 491 nm and the 561 nm channels, with 2 × 2 binning and 15 × 2.5 μm z-sectioning. z-sectioning with the IR laser was continually performed during the time interval between image acquisition. Completion or failure of cell division was assessed upon anaphase onset of the next cell division.
Effect of FLIRT on anterior-posterior cell polarity
For experiments to test the effect of FLIRT on anterior-posterior cell polarity and daughter cell asymmetry during cell division (Supplementary Figure 6a), C. elegans strain JCC744 (GFP::PAR-2; mCherry::PAR-6; mCherry::HistoneH2B) was held at either 16°C or 14°C in the microfluidic specimen holder. Embryos were monitored using DIC imaging until just prior to anaphase onset. Images were acquired every 3 min with 200 msec exposure for DIC, 150 msec exposure for the 491 nm channel, and 200 msec exposure for the 561 nm channel, with 2 × 2 binning and 15 × 2.5 μm z-sectioning. Experiments were either done with no FLIRT (0 mW) or with the 16 μm mask targeting the equatorial region with either 8.5 mW of laser (16°C hold temperature) or 10.1 mW (14°C hold temperature). When IR was used, FLIRT was initiated prior to anaphase onset and z-sectioning with the IR laser was continually performed during the time interval between image acquisition to maintain local control of temperature. For analysis of cell polarity and daughter cell asymmetry33, measurement of the size of the AB and P1 cells were performed in FIJI22. The GFP and mCherry signals were measured separately to determine cell size both during cell division and at the 2-cell stage and cell polarity and daughter cell asymmetry were measured33 as the difference between AB and P1 size in μm.
Post-FLIRT embryo viability analysis
Following an individual FLIRT experiment targeting the equatorial region of a dividing control embryo (16 μm mask; either holding at 16°C with 8.5 mW or 14°C with 10.1 mW, see FLIRT experiments targeting cell division in myosin-II(ts) and Delta(ts) mutants section above for details) at the 1-cell stage, the microfluidic temperature device was removed and the 2% agar pad containing the embryo was lifted from its bottom surface using a scalpel. The agar pad was then placed embryo-side down onto NGM worm plates (see also Worm husbandry and embryo dissection section above) containing OP50 bacteria and maintained at 16°C to allow embryo development and hatching. For non-FLIRT controls, 1-cell stage embryos were dissected in M9 buffer (see above) and individually placed onto a separate NGM plate using a pulled needle mouth pipette and maintained at 16°C. Images of the FLIRT and non-FLIRT control isolated embryos were acquired every 16–24 hr from hatching and into adulthood using a Micropublisher 3.3 RTV camera (Q-Imaging) mounted on the C-mount of a dissecting microscope (Olympus SZX16) with a 120 msec exposure time using QCapture Suite Plus software (Q-Imaging) to ensure development was timely and adult reproduction was not grossly affected. Viability was measured as the percentage of embryos that hatched within 24 hr of dissection.
Delta-Notch activity reporter characterization and FLIRT assay
Late 2-cell stage embryos were dissected from control (JCC596) and Delta(ts)34, 35 (JCC623) adult worms and mounted in the imaging chamber of the temperature control device (16°C). For whole cell upshift experiments (Supplementary Figure 7e), embryos were either held at 16°C or were upshifted to 22°C, 24°C, or 26°C for the duration of the experiment. For the 4-cell brief time window temperature upshift experiments, embryos were mounted at 16°C at the late 2-cell stage. The temperature was increased to 24°C or 26°C at the early 4-cell stage, then shifted back down to 16°C following anaphase onset of the P2 cell and maintained at 16°C for the duration of the experiment. For whole cell upshift experiments (Supplementary Figure 7e), images in the 491 and 561 nm channels were acquired every 20 min for 3 hr, from the late 2-cell stage through the ~200 cell stage. For FLIRT experiments (Figure 2a), the embryos were maintained at 16°C. Images were acquired as described above with 200 msec exposure for DIC, 150 msec exposure time for the 491 nm channel, and 250 msec for the 561 nm channel with 2 × 2 binning and 15 × 2.5 μm z-sectioning for all experiments. Following the initial time point at late anaphase of the P1 cell, the P2 cell was targeted with 7.2 mW of IR laser power (16 μm mask) at either the cell-cell contact between P2 and the ABp cell (control and Delta(ts)) or the EMS cell (Delta(ts)). The IR laser remained on and images were acquired every 60 sec with continuous 15 × 2.5 μm z-sectioning between image acquisition through anaphase onset of the P2 cell (~20 min), when the IR laser was turned off. After turning off the IR laser, embryos were maintained at 16°C and images were acquired every 20 min for another 3 hr until the ~200 cell stage.
For ABp cell fate experiments, image analysis was done in FIJI22 using the Cell Counter plugin (https://imagej.nih.gov/ij/plugins/cell-counter.html). At the ~50 cell stage (160 min post-FLIRT), Cell Counter was used to manually identify the total number of GFP+ and mCherry+ nuclei. The percentage of Notch activated cells was determined by quantifying the percentage of all GFP+ nuclei that are also mCherry+. The percentage of GFP+, mCherry+ nuclei were quantified for control and Delta(ts) embryos from whole cell upshifts and FLIRT experiments (for statistical analysis see also Supplementary Table 1).
FLIRT experiments with cyk-4(ts)
Control (OD95) and cyk-4(ts) (OD239) early adult worms were picked into 5 μL of M9 buffer and 0.25–0.5 μL of 0.1 μm diameter polystyrene microspheres (Polysciences 00876–15, 2.5% w/v suspension) then covered with a pad made of 10% agarose in M9 for immobilization36 and mounted in the microfluidic specimen holder set to 16°C. For upshift experiments (Supplementary Figure 8), the temperature control device was shifted to 26°C ~5 min after mounting. Images of the gonad were acquired every 60 sec for 75 min with 200 msec exposure for DIC and 150 msec exposure times for the 491 nm and the 561 nm channels, with 2 × 2 binning and 15 × 2.5 μm z-sectioning. For FLIRT experiments (Figure 2b), the 16 μm mask (8.5 mW) was positioned to target a single membrane partition on the proximal side of the gonad after the bend of the gonad arm and moved to follow the same membrane partition within the worm. Images were acquired every 60 sec and z-sectioning with the IR laser was continually performed during the time interval between image acquisition to maintain local control.
Analysis of compartment partition length was done in FIJI22 by measuring the length of individual compartment partitions on the frame just after FLIRT was turned on and 70 min after upshift or FLIRT targeting. Membrane partition length was measured from the site of contact with the edge of the gonad to the end of the membrane partition at the central germline rachis using a segmented line on the z-section in which the partition was longest. The percent initial length was measured as a percentage of membrane partition length at t=70 min divided by the length at t=0 min. For upshift experiments (Supplementary Figure 8), four individual compartment partitions were analyzed for each worm. For FLIRT experiments (Figure 2b), the FLIRT-targeted partition and the two adjacent non-targeted partitions were analyzed.
Statistical analysis
Unpaired two-tailed student’s t-tests were conducted using Microsoft Excel. n.s. = p≥0.05, * = p<0.05, ** = p<0.01, *** = p<0.001, and **** = p<0.0001. In Supplementary Figure 7e, Tukey’s test for multiple comparisons was conducted using GraphPad Prism software, α = 0.05. Error bars represent the SD except for in Supplementary Figure 2d, 2e and the gradient analysis in Supplementary Figures 3d, 3e, and 7d, where error bars represent the SEM. See also Supplementary Table 1 and the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgements
We thank all members of the Canman, Shirasu-Hiza, and Dumont labs for their support; H Kim, I Thomas, C Walsh, B Lesea-Pringle, K Rimu, F Jung, and E Blake for lab assistance; C Connors for comments on this manuscript; I Greenwald, B Bowerman, and B Goldstein for helpful discussions; and the Caenorhabditis Genomics Center for worm strains. We thank S Wildfang, A Ratz, and C Anderson for technical assistance; A Kummel and A Garcia Badaracco for assistance on the thermochromatic dye analysis; and B O’Shaughnessy, S Wang, and S Thiyagarajan for advice on thermal distribution. This work was funded by: a Charles H. Revson Senior Fellowship in Biomedical Science (TD), FRM-DEQ20160334869 (JD); NIH-R01-GM105775 (MSH); NIH-R01-AG045842 (MSH); NIH-DP2-OD008773 (JCC); and NIH-R01GM117407 (JCC).
Abbreviations
- Ts
temperature sensitive
- IR
infrared
- FLIRT
fast local infrared thermogenetics
- ROI
region of interest
- SD
standard deviation
- SEM
standard error of the mean
- Fluor
fluorescence
- Norm
normalized
- Calc
calculated
- mCh
mCherry
- Em
emission
- Ex
excitation
- T or Temp
temperature
Footnotes
Competing Financial Interests Statement
The authors declare no competing interests.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Liu J, Maduzia LL, Shirayama M & Mello CC NMY-2 maintains cellular asymmetry and cell boundaries, and promotes a SRC-dependent asymmetric cell division. Dev Biol 339, 366–373 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Severson AF, Hamill DR, Carter JC, Schumacher J & Bowerman B The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr Biol 10, 1162–1171 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Davies T et al. High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev Cell 30, 209–223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mickey KM, Mello CC, Montgomery MK, Fire A & Priess JR An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signalling cell. Development 122, 1791–1798 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Mello CC, Draper BW & Priess JR The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell 77, 95–106 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Davies T et al. Using fast-acting temperature-sensitive mutants to study cell division in Caenorhabditis elegans. Methods Cell Biol 137, 283–306 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Kamei Y et al. Infrared laser-mediated gene induction in targeted single cells in vivo. Nat Methods 6, 79–81 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Sundaramoorthy S et al. Low Efficiency Upconversion Nanoparticles for High-Resolution Coalignment of Near-Infrared and Visible Light Paths on a Light Microscope. ACS Appl Mater Interfaces 9, 7929–7940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singhal A & Shaham S Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat Commun 8, 14100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Avino PP, Giansanti MG & Petronczki M Cytokinesis in animal cells. Cold Spring Harb Perspect Biol 7, a015834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollard TD Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol 22, 50–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green RA, Paluch E & Oegema K Cytokinesis in animal cells. Annu Rev Cell Dev Biol 28, 29–58 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Oyama K et al. Directional bleb formation in spherical cells under temperature gradient. Biophys J 109, 355–364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittasch M et al. Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nat Cell Biol 20, 344–351 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Crittenden SL, Rudel D, Binder J, Evans TC & Kimble J Genes required for GLP-1 asymmetry in the early Caenorhabditis elegans embryo. Dev Biol 181, 36–46 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Shelton CA & Bowerman B Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122, 2043–2050 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Murray JI et al. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res 22, 1282–1294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good K et al. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development 131, 1967–1978 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Neves A & Priess JR The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell 8, 867–879 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Lee KY et al. CYK-4 functions independently of its centralspindlin partner ZEN-4 to cellularize oocytes in germline syncytia. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sundaramoorthy S et al. Low Efficiency Upconversion Nanoparticles for High-Resolution Coalignment of Near-Infrared and Visible Light Paths on a Light Microscope. ACS Appl Mater Interfaces 9, 7929–7940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner S The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiernagle T Maintenance of C. elegans. WormBook, 1–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Audhya A et al. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol 171, 267–279 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies T et al. Using fast-acting temperature-sensitive mutants to study cell division in Caenorhabditis elegans. Methods Cell Biol 137, 283–306 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Widlund PO et al. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol Biol Cell 23, 4393–4401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Maduzia LL, Shirayama M & Mello CC NMY-2 maintains cellular asymmetry and cell boundaries, and promotes a SRC-dependent asymmetric cell division. Dev Biol 339, 366–373 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies T et al. High-resolution temporal analysis reveals a functional timeline for the molecular regulation of cytokinesis. Dev Cell 30, 209–223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Avino PP, Giansanti MG & Petronczki M Cytokinesis in animal cells. Cold Spring Harb Perspect Biol 7, a015834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard TD Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol 22, 50–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green RA, Paluch E & Oegema K Cytokinesis in animal cells. Annu Rev Cell Dev Biol 28, 29–58 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Bossinger O & Cowan CR Methods in cell biology: analysis of cell polarity in C. elegans embryos. Methods Cell Biol 107, 207–238 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Mickey KM, Mello CC, Montgomery MK, Fire A & Priess JR An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signalling cell. Development 122, 1791–1798 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Mello CC, Draper BW & Priess JR The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell 77, 95–106 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Kim E, Sun L, Gabel CV & Fang-Yen C Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS One 8, e53419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singhal A & Shaham S Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat Commun 8, 14100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray JI et al. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res 22, 1282–1294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.