In this study, Son et al. used C. elegans to study how HSF-1 and FOXO cooperate to confer insulin/IGF-1 signaling (IIS)-mediated longevity. They show that prefoldin 6 (PFD-6), a component of the molecular chaperone prefoldin-like complex, relays longevity response from HSF-1 to FOXO under reduced IIS, and their findings suggest that the prefoldin-like chaperone complex mediates longevity response from HSF-1 to FOXO to increase the life span in animals with reduced IIS.
Keywords: aging, C. elegans, DAF-16/FOXO, HSF-1/HSF1, PFD-6/PFDN6
Abstract
Heat shock factor 1 (HSF-1) and forkhead box O (FOXO) are key transcription factors that protect cells from various stresses. In Caenorhabditis elegans, HSF-1 and FOXO together promote a long life span when insulin/IGF-1 signaling (IIS) is reduced. However, it remains poorly understood how HSF-1 and FOXO cooperate to confer IIS-mediated longevity. Here, we show that prefoldin 6 (PFD-6), a component of the molecular chaperone prefoldin-like complex, relays longevity response from HSF-1 to FOXO under reduced IIS. We found that PFD-6 was specifically required for reduced IIS-mediated longevity by acting in the intestine and hypodermis. We showed that HSF-1 increased the levels of PFD-6 proteins, which in turn directly bound FOXO and enhanced its transcriptional activity. Our work suggests that the prefoldin-like chaperone complex mediates longevity response from HSF-1 to FOXO to increase the life span in animals with reduced IIS.
The insulin/IGF-1 signaling (IIS) pathway regulates life span in many species, including Caenorhabditis elegans, Drosophila, and mammals (Kenyon 2010; Altintas et al. 2016). In C. elegans, inhibition of daf-2, an insulin/IGF-1 receptor homolog, or its downstream phosphoinositide 3-kinase cascade components substantially increases life span (Kenyon et al. 1993; Antebi 2007; Kenyon 2010). The longevity conferred by reduced IIS is mediated by the activation of transcription factors, including the DAF-16/forkhead box O (FOXO) transcription factor, which results in the induction of its target genes (Kenyon et al. 1993; Kenyon 2010). Heat shock transcription factor 1 (HSF-1) also acts downstream from IIS to regulate longevity (Hsu et al. 2003; Morley and Morimoto 2004; Cohen et al. 2006; Kenyon 2010; Chiang et al. 2012; Seo et al. 2013). Activation of HSF-1 induces genes encoding molecular chaperones, including HSP70 and HSP90 family members and small heat shock proteins, for the maintenance of protein homeostasis. HSF-1 and DAF-16/FOXO act together in part to promote longevity by the transactivation of common targets, most notably small heat shock proteins that contribute to longevity (Hsu et al. 2003; Murphy et al. 2003). In addition, the up-regulation of HSF-1 in neurons increases the activity of DAF-16/FOXO in other tissues in a cell-nonautonomous manner, which is crucial for extending life span (Douglas et al. 2015). Nevertheless, how HSF-1 and DAF-16/FOXO cooperate to exert their longevity response remains poorly understood.
In addition to classical heat shock proteins whose expression is regulated by HSF-1, other chaperones and chaperonins contribute to protein homeostasis. For example, the prefoldin complex functions as a molecular chaperone for various essential proteins (Boulon et al. 2010; Lundin et al. 2010). In eukaryotes, the prefoldin complex is composed of six protein subunits: two α-type subunits (prefoldin 3 [PFD-3] and PFD-5) and four β-type subunits (PFD-1, PFD-2, PFD-4, and PFD-6) (Boulon et al. 2010; Lundin et al. 2010). The prefoldin complex binds to nascent polypeptide chains of cytoskeletal proteins, actins, and tubulins to help transfer these proteins to the group II chaperonin, TRiC (TCP1 ring complex)/CCT (chaperonin-containing TCP1) for proper folding (Boulon et al. 2010; Lundin et al. 2010) in the cytoplasm. Several prefoldin subunits have other functions in addition to those as components of the prefoldin complex. Mammalian PFD-2 and PFD-6 are components of the R2TP (Rvb1, Rvb2, Tah1 [TPR-containing protein], and Pih1 [protein interacting with Hsp90])/prefoldin-like complex (Cloutier et al. 2009). The R2TP/prefoldin-like complex is required for the assembly of noncytoskeletal complexes, such as RNA polymerase II in the cytoplasm (Boulon et al. 2008, 2010; Cloutier et al. 2009). In addition, several components in prefoldin and R2TP/prefoldin-like complexes function as transcription cofactors acting in the nucleus (Millan-Zambrano and Chavez 2014). Despite the important roles of prefoldin and R2TP/prefoldin-like complex subunits in various cellular functions, their roles in aging and life span regulation remain unknown.
In the present study, we functionally characterized the life span-regulating role of C. elegans prefoldin-6/PFD-6, which we identified from a genome-wide hsf-1 modifier RNAi screen. We showed that pfd-6 was required for life span extension conferred by daf-2 mutations. We found that pfd-6 expression in the intestine and hypodermis was required for daf-2(−) longevity. We showed that PFD-6-containing R2TP/prefoldin-like complex contributed to the longevity of daf-2 mutants. Furthermore, PFD-6 was up-regulated in daf-2(−) mutants. We then showed that PFD-6 up-regulated DAF-16/FOXO through physical interaction to induce a subset of DAF-16/FOXO target genes. Thus, PFD-6 appears to extend the life span in animals with reduced IIS by mediating longevity response from HSF-1 to DAF-16/FOXO.
Results
A genome-wide RNAi screen identifies genes that affect longevity conferred by reduced IIS
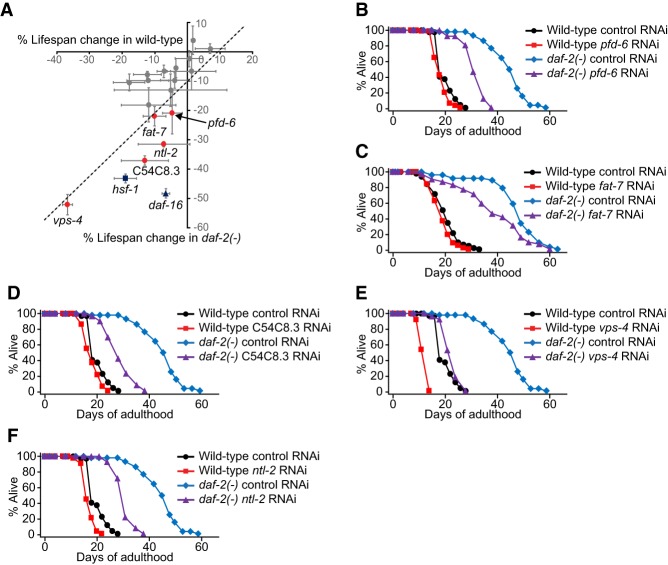
We sought to identify novel longevity genes that interacted with hsf-1. To this end, we performed a genome-wide RNAi screen using reduction-of-function hsf-1(sy441) mutants, which display a temperature-sensitive sterility phenotype (Hajdu-Cronin et al. 2004). We identified 17 RNAi clones that enhanced the sterility of the hsf-1 mutants compared with wild-type animals at 20°C (Supplemental Table S1). We then tested whether any of these RNAi clones affected longevity conferred by daf-2/insulin/IGF-1 receptor mutations, which up-regulate HSF-1 and require HSF-1 for exerting longevity (Hsu et al. 2003; Morley and Morimoto 2004). RNAi targeting each of five genes—pfd-6, fat-7 (fatty acid desaturase 7) (Murphy et al. 2003), C54C8.3/acetylgalactosaminyltransferase, vps-4 (vacuolar protein sorting 4), and ntl-2/CCR4–NOT transcription complex subunit—decreased the life span of the daf-2 mutants at least 10% greater than that of wild type (Fig. 1A–F); these extents are similar to the effects of RNAi targeting hsf-1 or daf-16 (Fig. 1A). Thus, our genetic screen identified novel hsf-1-interacting genes required for longevity caused by reduced IIS.
Figure 1.
Five hsf-1(sy441) sterility enhancer RNAi clones decrease the longevity of daf-2 mutants. (A) The graph shows percentage changes in the average life span of wild-type (X-axis) and daf-2(e1370) [daf-2(−); Y-axis] animals treated with each of 17 RNAi clones obtained from the primary screen. Each circle, square, or triangle indicates an average value of two different sets of life spans. A dashed line indicates an arbitrary cutoff showing RNAi clones that have 10% more life span-decreasing effects on daf-2 mutants than on wild type. Error bars indicate the standard error of the mean (SEM) of two independent experiments. Worms were treated with RNAi only during adulthood to exclude the possibility that RNAi clones may affect the health of the animals during development. hsf-1 RNAi and daf-16 RNAi clones were used as positive controls. (B–F) pfd-6 RNAi (B), fat-7 RNAi (C), C54C8.3 RNAi (D), vps-4 RNAi (E), and ntl-2 RNAi (F) substantially decreased the life spans of daf-2 mutants while having little effect on that of wild-type animals. For pfd-6, we confirmed the increased sterility caused by pfd-6 RNAi in hsf-1 mutants using solid plates (Supplemental Fig. S1E). See Supplemental Table S2 for statistical analysis and additional repeats.
pfd-6 is specifically required for the longevity of daf-2(−) and osm-5(−) mutants
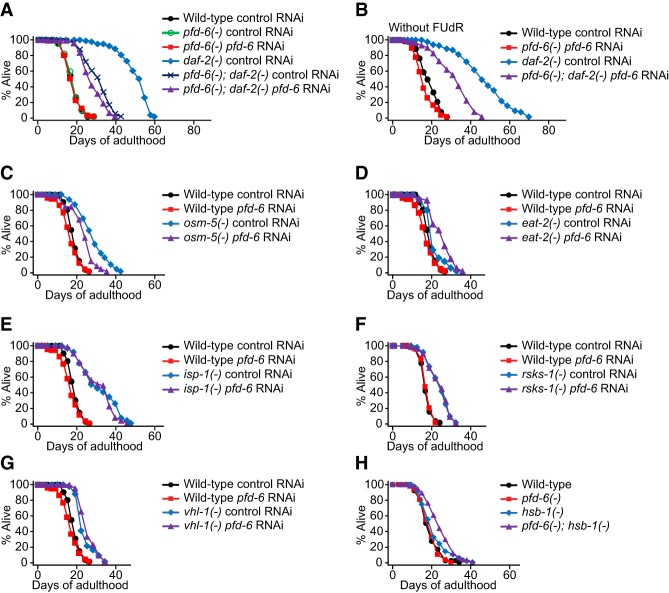
We further determined the requirement of these genes for daf-2(−) longevity using available mutants (Supplemental Fig. S1A–D). Strong loss-of-function alleles of pfd-6 cause semiembryonic lethality and larval arrest phenotypes (Supplemental Fig. S2A). We therefore characterized a point mutant allele of pfd-6(gk493446) that causes a Thr16-to-isoleucine (T16I) change in a highly conserved PFD-6 (Supplemental Fig. S2B–E). Similar to pfd-6 RNAi, pfd-6(gk493446) mutations substantially decreased the life span of daf-2 mutants but had a little effect on that of the wild type (Supplemental Fig. S1A). In addition, pfd-6 mutation and/or RNAi knockdown suppressed the long life span of daf-2 mutants with or without 5-fluoro-2′-deoxyuridine (FUdR) (Fig. 2A,B; Supplemental Fig. S2F,G); FUdR is used for preventing C. elegans reproduction in life span assays (Park et al. 2017). Thus, pfd-6 is required for the longevity of daf-2(−) mutants.
Figure 2.
pfd-6 is required for the longevity of daf-2 mutants. (A,B) pfd-6(gk493446) [pfd-6(−)] point mutation causing a T16I change with pfd-6 RNAi partly suppressed the longevity of daf-2(e1370) [daf-2(−)] mutants with (A) or without (B) FUdR. (A) pfd-6 RNAi during adulthood slightly decreased the short life span of pfd-6; daf-2 double mutants but did not affect that of pfd-6 mutants (two out of four trials). (C) pfd-6 RNAi partially suppressed the longevity conferred by osm-5(p813) [osm-5(−)] mutations. (D–G) pfd-6 RNAi did not decrease the longevity of eat-2(ad1116) [eat-2(−)] (D), isp-1(qm150) [isp-1(−)] (E), rsks-1(tm1714) [rsks-1(−)] (F), and vhl-1(ok161) [vhl-1(−)] (G) animals. (H) pfd-6 mutation did not decrease the life span of hsb-1(cg116) [hsb-1(−)] mutants. Considering our data suggesting that pfd-6 mediates the longevity effects of HSF-1, it is surprising to find that genetic inhibition of pfd-6 did not decrease the longevity of rsks-1 or hsb-1 mutants, which require HSF-1 for a longer life span (Chiang et al. 2012; Seo et al. 2013). We speculate that up-regulation of HSF-1 by rsks-1 and hsb-1 mutations may affect pfd-6 differently than that by daf-2 and osm-5 mutations, which will be interesting to examine in future studies. See Supplemental Table S3 for statistical analysis and additional repeats.
We asked whether pfd-6 was required for the longevity of various other long-lived mutants; these include sensory-defective osm-5 mutants, dietary-restricted eat-2 mutants, mitochondrial respiration-impaired isp-1 mutants, TOR downstream rsks-1/S6 kinase mutants, HIF-1-hyperactive vhl-1 mutants, and HSF-1-hyperactive hsb-1 mutants. We found that pfd-6 RNAi decreased the longevity of osm-5 mutants (Fig. 2C) while not decreasing the life span of the other tested long-lived mutants (Fig. 2D–H; Supplemental Fig. S2H; see figure legends for discussion). Because impaired sensory neuron-mediated longevity also acts through IIS (Jeong et al. 2012), these data suggest that pfd-6 influences life span by acting mainly through IIS.
PFD-6 in the intestine and hypodermis is required for daf-2(−) longevity
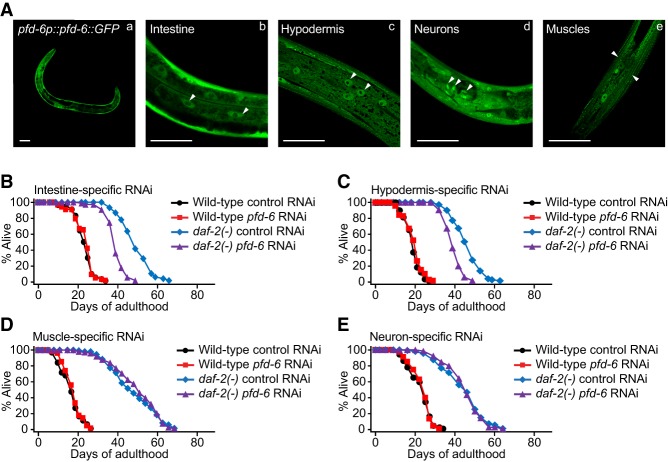
Next, we characterized transgenic animals that expressed pfd-6::GFP under a pfd-6 promoter (pfd-6p::pfd-6::GFP). PFD-6::GFP fusion protein was localized in both the cytosol and nuclei of intestinal, hypodermal, muscle, and neuronal cells throughout the developmental stages (Fig. 3A; Supplemental Fig. S3A). The pfd-6::GFP transgene restored longevity in pfd-6; daf-2 mutants (Supplemental Fig. S3B) while having a marginal effect on the wild-type life span (Supplemental Fig. S3C). We then asked in which tissues pfd-6 was required for the longevity of daf-2(−) mutants using tissue-specific RNAi systems (Qadota et al. 2007; Calixto et al. 2010). As expected, dsRNA of pfd-6 had little effect on the life span of RNAi-defective rde-1(−) or systemic RNAi-defective sid-1(−) mutants (Supplemental Fig. S3D,E). We found that intestine- or hypodermis-specific pfd-6 RNAi significantly decreased the long life span of daf-2(−) mutants but not that of daf-2(+) control animals (Fig. 3B,C). In contrast, muscle- or neuron-specific knockdown of pfd-6 hardly affected the life span in control or daf-2(−) mutant backgrounds (Fig. 3D,E). Thus, together with the expression pattern of pfd-6::GFP (Fig. 3A), pfd-6 in the intestine and hypodermis appears to contribute to the longevity of daf-2 mutants.
Figure 3.
Intestinal and hypodermal PFD-6 contributes to the long life span of daf-2 mutants. (A) pfd-6p::pfd-6::GFP was expressed in various tissues (panel a), including the intestine (panel b), the hypodermis (panel c), neurons (panel d), and muscles (panel e). Arrowheads indicate PFD-6::GFP in the intestine, hypodermis, neurons, or muscles. Bars, 50 µm. (B,C) The longevity of daf-2(e1370) [daf-2(−)] mutants was significantly decreased by intestine {rde-1(ne219); kbls7[nhx-2p::rde-1; rol-6D]}-specific pfd-6 RNAi (B) or hypodermis {rde-1(ne219); kzls9[lin-26p::nls::gfp; lin-26p::rde-1; rol-6D]}-specific pfd-6 RNAi (C). We noticed that life span-decreasing effects of the intestine- or the hypodermis-specific pfd-6 RNAi were greater than the life span-decreasing effects of pfd-6 RNAi on rde-1(−); daf-2(−) animals (Supplemental Fig. S3D). (D,E) Muscle {rde-1(ne219); kzls20[hlh-1p::rde-1, sur-5p::nls::gfp]}-specific (D) or neuron {sid-1(pk3321); uIs69[myo-2p::mCherry; punc-119::sid-1)]}-specific (E) pfd-6 RNAi did not affect the life span of daf-2(+) or daf-2(−) animals. We confirmed the efficiency of neuron- and muscle-specific pfd-6 RNAi knockdown using fluorescence-tagged PFD-6 proteins (Supplemental Fig. S3F–I). See Supplemental Table S4 for statistical analysis and additional repeats.
PFD-6 as a component of the R2TP/prefoldin-like complex contributes to longevity
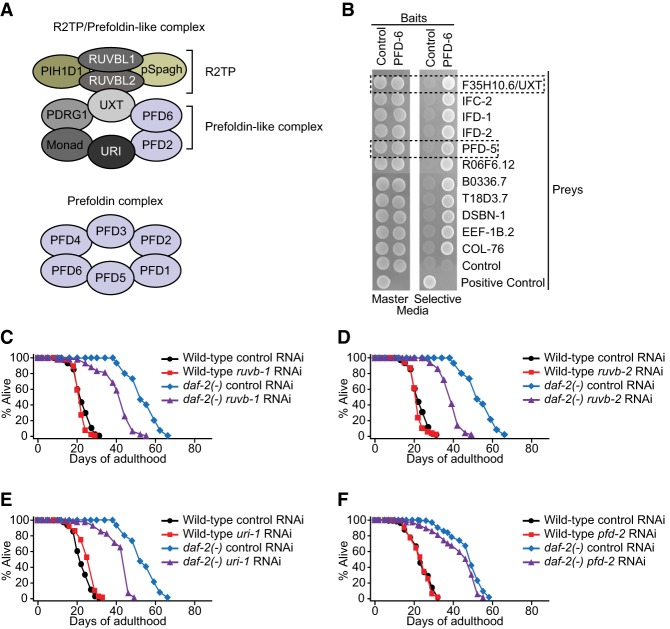
Mammalian PFD-6 acts as a subunit of R2TP/prefoldin-like and prefoldin complexes (Fig. 4A). By performing a large-scale yeast two-hybrid screen, we found that PFD-6 bound various proteins, including F35H10.6/UXT and PFD-5, homologs of R2TP/prefoldin-like and prefoldin complex components, respectively (Fig. 4B; Supplemental Table S7). Thus, C. elegans PFD-6 appears to be a subunit of R2TP/prefoldin-like and prefoldin complexes, similar to that in other species (Lundin et al. 2010; Millan-Zambrano and Chavez 2014). We then examined whether daf-2(−)-induced longevity was affected by RNAi targeting various components of these two complexes (Fig. 4A; Supplemental Fig. S4A,B). We found that the full longevity of daf-2(−) mutants was decreased by RNAi knockdown of ruvb-1, ruvb-2, or uri-1 (Fig. 4C–E). We also found that knockdown of pfd-2 modestly decreased the longevity conferred by daf-2 mutations (Fig. 4F). In addition, the life span-shortening effects of ruvb-1 RNAi, ruvb-2 RNAi, or uri-1 RNAi were not additive to those of pfd-6 mutations (Supplemental Fig. S4D–F). In contrast, RNAi clones targeting prefoldin complex-specific components showed minimal effects on longevity (Supplemental Fig. S4H–K). These data suggest that the R2TP/prefoldin-like complex, rather than the prefoldin complex, contributes to the long life span of daf-2(−) mutants.
Figure 4.
Prefoldin-like complex components are required for the long life span of daf-2 mutants. (A) Schematic model of the R2TP/prefoldin-like complex and the prefoldin complex in mammals; the R2TP/prefoldin-like complex interacts with RNA polymerase II in the cytoplasm to help its assembly, and the prefoldin complex is composed of PFD-1, PFD-2, PFD-3, PFD-4, PFD-5, and PFD-6, which bind to nascent actin or tubulin polypeptides. Prefoldin complex-bound polypeptides are transferred to TRiC/CCT chaperonin for proper folding. (B) Yeast two-hybrid screening with full-length PFD-6 as bait was performed using the yeast PBN204 strain. The interaction was validated using nutritional selection (ADE2) for the growth assay. See Supplemental Table S7 for the description of candidate genes. (C–F) Several R2TP/prefoldin-like complex components were required for life span extension caused by reduced IIS. RNAi targeting ruvb-1 (C), ruvb-2 (D), uri-1 (E), or pfd-2 (F) significantly decreased the long life span of daf-2(e1370) [daf-2(−)] mutants. Knockdown of potential UXT homolog F35H10.6 did not affect the life span of wild-type, daf-2(−), pfd-6(−), or pfd-6; daf-2 mutants (Supplemental Fig. S4C,G). See Supplemental Table S5 for statistical analysis and additional repeats.
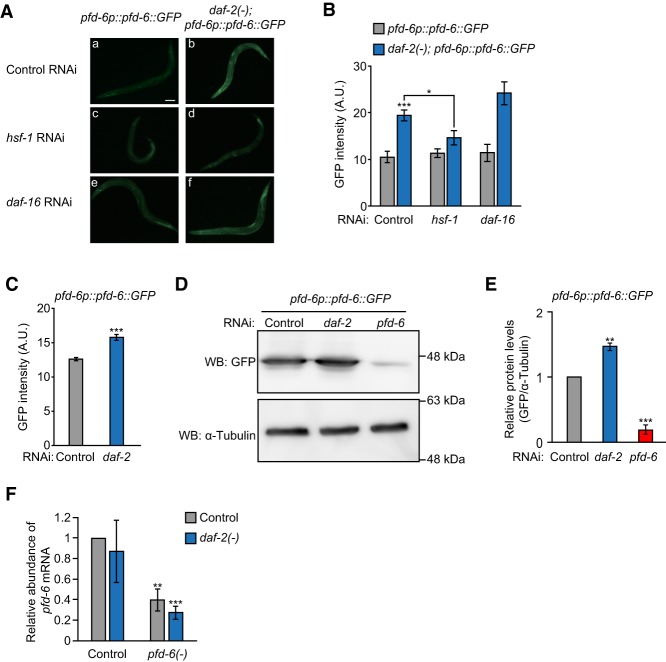
daf-2 mutations increase PFD-6 levels in an HSF-1-dependent manner
We next asked whether reduced IIS affected the level of PFD-6 and found that the PFD-6::GFP level was increased by genetic inhibition of daf-2 using fluorescence imaging and Western blot assays (Fig. 5A–E). Importantly, we showed that this increase was suppressed by hsf-1 RNAi (Fig. 5A,B; Supplemental Fig. S5). In contrast, daf-16 RNAi did not affect the level of PFD-6::GFP (Fig. 5A,B; Supplemental Fig. S5). Different from the PFD-6 protein level, the pfd-6 mRNA level was not changed in daf-2 mutants (Fig. 5F). These data suggest that inhibition of daf-2 up-regulates PFD-6 protein levels via HSF-1.
Figure 5.
HSF-1 increases the levels of PFD-6. (A) Mutations in daf-2 increased PFD-6 levels, which were decreased by hsf-1 RNAi but not by daf-16 RNAi. Bar, 100 µm. The following animals that expressed an extrachromosomal array of pfd-6::GFP were used: IJ570 (yhEx134[pfd-6p::pfd-6::GFP; odr-1p::RFP]) and IJ810 {daf-2(e1370); yhEx134[pfd-6p::pfd-6::GFP, odr-1p::RFP]}. (B) Quantification of A. n > 15 from two independent experiments. (C) GFP levels in integrated pfd-6::GFP transgenic animals (IJ1249: yhIs74[pfd-6p::pfd-6::GFP; odr-1p::RFP]) were examined. n > 32 from three independent experiments. (D) Western blot assays were performed using the IJ1249 strain. A representative blot is shown from three repeats that displayed consistent results. α-Tubulin was used as a loading control. (WB) Western blot. (E) Quantification of D. (F) mRNA levels of pfd-6 in wild-type, daf-2(e1370) [daf-2(−)], pfd-6(gk493446, RNAi) [pfd-6(−)], and pfd-6(−); daf-2(−) animals were measured by using quantitative RT–PCR. n = 3. Error bars represent SEM. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, two-tailed Student's t-test.
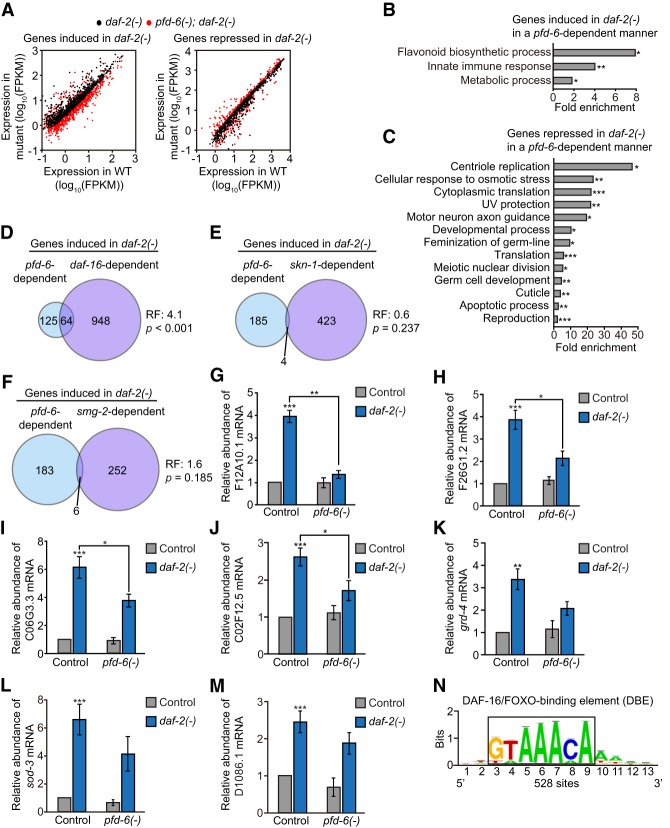
PFD-6 regulates the expression of a large subset of DAF-16/FOXO target genes in daf-2 mutants
Because the R2TP/prefoldin-like complex can regulate transcription (Millan-Zambrano and Chavez 2014), we wondered whether pfd-6 influenced global gene expression in daf-2(−) mutants. By performing mRNA sequencing (mRNA-seq), we identified 189 up-regulated and 162 down-regulated genes in daf-2 mutants whose expression was dependent on pfd-6 (fold change >1.5 and <0.67 for up-regulated and down-regulated genes, respectively; P < 0.05) (Fig. 6A; Supplemental Fig. S6A–D). Gene ontology (GO) analysis using these genes identified several enriched GO terms, including flavonoid biosynthetic process among up-regulated genes and centriole replication among down-regulated genes (Fig. 6B,C). We then examined which longevity factors in IIS acted together with PFD-6. Specifically, we compared the transcriptome regulated by PFD-6 in daf-2(−) in this study with those regulated by DAF-16/FOXO (Riedel et al. 2013; Chen et al. 2015), SKN-1/NRF (Ewald et al. 2015), and SMG-2/UPF1 (Son et al. 2017), which are crucial for longevity conferred by reduced IIS (Kenyon 2010; Blackwell et al. 2015; Son and Lee 2017). Interestingly, genes differentially regulated in daf-2 mutants by PFD-6 displayed a highly significant overlap with those regulated by DAF-16/FOXO but not with those regulated by SKN-1/NRF or SMG-2/UPF1 (Fig. 6D–F; Supplemental Fig. S6E–K). Thus, PFD-6 appears to act together with DAF-16/FOXO to regulate a subset of gene expression in daf-2(−) mutants. We confirmed these results for selected genes using quantitative RT–PCR (qRT–PCR) (Fig. 6G–M; Supplemental Fig. S6L–N). We also found that the consensus sequence of the DAF-16-binding element (DBE) was significantly enriched in the 5′ upstream regions of pfd-6-dependent up-regulated genes in daf-2(−) mutants (Fig. 6N; Supplemental Fig. S6O,P). Together, these data suggest that PFD-6 regulates the expression of a subset of DAF-16/FOXO target genes in daf-2(−) mutants.
Figure 6.
PFD-6 up-regulates a large subset of DAF-16 target genes in daf-2 mutants. (A) Scatter plots show the comparison of gene expression between wild-type and daf-2(e1370) [daf-2(−)] or wild-type and pfd-6(gk493446, RNAi); daf-2(e1370) [pfd-6(−); daf-2(−)] animals. Black dots indicate gene expression in daf-2 mutants compared with wild type, whose levels were significantly increased (fold change >1.50; P < 0.05; left) or decreased (fold change <0.67; P < 0.05; right). Red dots indicate expression levels in pfd-6; daf-2 mutants compared with wild type. See Supplemental Table S8 for the list of up-regulated or down-regulated genes in daf-2 mutants compared with wild-type or pfd-6; daf-2 double mutants. (B,C) Overrepresented GO terms of up-regulated (B) or down-regulated (C) genes that are shown in A. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (D–F) Venn diagrams indicate overlapping genes whose expression was increased by daf-2 mutations in a pfd-6- and daf-16-dependent (Riedel et al. 2013) (D), pfd-6- and skn-1-dependent (Ewald et al. 2015) (E), or pfd-6- and smg-2-dependent (Son et al. 2017) (F) manner. Fold change > 1.5. P < 0.05. (RF) Representation factor. P-values for the overlaps of Venn diagrams were calculated using the hypergeometric probability test. (G–M) qRT–PCR results showing mRNA levels of DAF-16 target genes F12A10.1 (G), F26G1.2 (H), C06G3.3 (I), C02F12.5 (J), grd-4 (K), sod-3 (L), and D1086.1 (M). n > 4. Error bars represent SEM. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001, two-tailed Student's t-test. (N) Motif analysis was performed by using 800-base-pair (bp) upstream sequences of pfd-6-dependent genes that were up-regulated in daf-2 mutants. Fold change > 1.5. P < 0.05. See Supplemental Figure S6, O and P, for other enriched motifs.
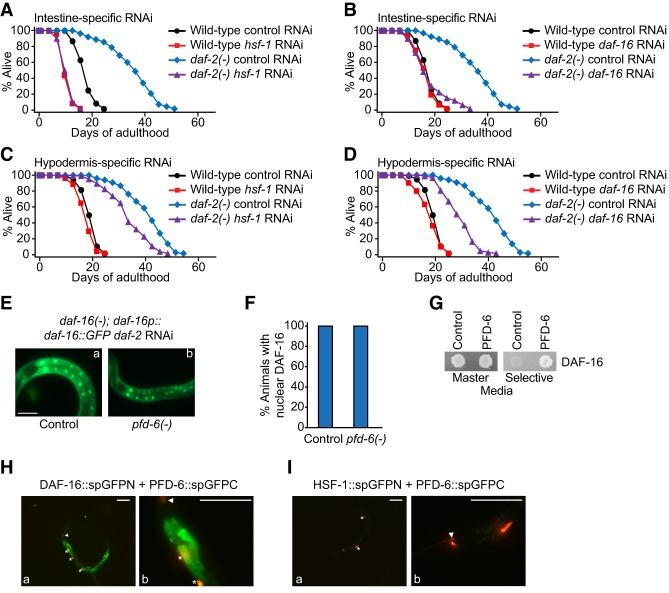
PFD-6 appears to increase the activity of DAF-16/FOXO via physical interaction
Next, we asked how PFD-6 affected the activities of DAF-16/FOXO. First, we established that knocking down hsf-1 or daf-16 in the intestine, hypodermis, or muscles significantly decreased the long life span of daf-2 mutants (Fig. 7A–D; Supplemental Fig. S7A–D). Thus, consistent with published reports regarding the tissue-specific transgenic expression of DAF-16/FOXO (Libina et al. 2003; Zhang et al. 2013) and our tissue-specific pfd-6 RNAi data (Fig. 3B,C), PFD-6 appears to act with HSF-1 and DAF-16 in common tissues, such as the intestine and hypodermis.
Figure 7.
PFD-6 binds and up-regulates DAF-16/FOXO in daf-2(−) mutants. (A–D) Intestine-specific (A,B) or hypodermis-specific (C,D) hsf-1 or daf-16 RNAi decreased the longevity of daf-2(e1370) [daf-2(−)] mutants. See Supplemental Table S6 for statistical analysis and additional repeats. (E) daf-2 RNAi-induced nuclear localization of DAF-16::GFP was not affected by pfd-6 mutations. (F) Quantification of E. n > 18 from three independent experiments. Bars, 100 µm. (G) Yeast two-hybrid assays showed the direct interaction between PFD-6 and DAF-16. (H) GFP N-terminal domain-fused DAF-16 isoform a (DAF-16::spGFPN) bound GFP C-terminal domain-fused PFD-6 (PFD-6::spGFPC). See Supplemental Figure S7J for experimental results using daf-2 RNAi-treated animals. (I) HSF-1::spGFPN did not bind PFD-6::spGFPC. Triangles indicate odr-1p::RFP, a coinjection marker for DAF-16::spGFPN and HSF-1::spGFPN. Asterisks indicate unc-122p::RFP, a coinjection marker for PFD-6::spGFPC.
We next tested whether pfd-6 affected any of several known DAF-16/FOXO activation mechanisms, nuclear translocation, increases in the protein level, and regulation of transcriptional activity via physical binding (Altintas et al. 2016). We found that the genetic inhibition of pfd-6 did not affect the subcellular localization of DAF-16/FOXO (Fig. 7E,F) while having variable effects on the level of DAF-16/FOXO (Supplemental Fig. S7E–H). We then showed that PFD-6 bound to DAF-16/FOXO by using yeast two-hybrid and split GFP system assays (Fig. 7G,H; Supplemental Fig. S7I,J). In contrast, PFD-6 did not bind HSF-1 (Fig. 7I), and this result is consistent with our data suggesting that HSF-1 acts upstream of PFD-6 to regulate its protein level (Fig. 5A,B; Supplemental Fig. S5). Together, these data suggest that HSF-1 up-regulates PFD-6, which increases the activity of DAF-16/FOXO (likely through physical interaction) and promotes the longevity of daf-2 mutants.
Discussion
PFD-6 mediates the cross-talk between HSF-1 and DAF-16/FOXO for longevity
HSF-1 and DAF-16/FOXO are key longevity-promoting transcription factors that act in the IIS pathway. Although the functional relationship between HSF-1 and DAF-16/FOXO for longevity regulation has been established, how HSF-1 and DAF-16/FOXO communicate with each other has remained incompletely understood. In this report, we showed that C. elegans PFD-6 acted as a key life span regulatory protein that mediated cross-talk between HSF-1 and DAF-16/FOXO in the IIS pathway.
Previous studies have indicated that C. elegans HSF-1 and DAF-16/FOXO transcription factors act both separately and together in IIS to elicit physiological responses. HSF-1 is up-regulated in worms with reduced IIS via dissociation from its inhibitory factors, such as DDL-1 (Chiang et al. 2012). Inhibition of IIS promotes the translocation of DAF-16/FOXO from the cytoplasm to the nucleus and increases DAF-16/FOXO activity via diverse regulators (Kenyon 2010; Murphy and Hu 2013). HSF-1 and DAF-16/FOXO also work in parallel in IIS by inducing common targets, including small heat shock proteins (Hsu et al. 2003; Murphy et al. 2003). For example, both DAF-16- and HSF-1-binding motifs exist in the promoter regions of genes encoding several heat shock proteins (GuhaThakurta et al. 2002; Hsu et al. 2003; Murphy et al. 2003; Xu and Kim 2012; Tepper et al. 2013). In addition, HSF-1 overexpression promotes longevity through increasing DAF-16 activity (Hsu et al. 2003; Douglas et al. 2015). However, it remained unclear whether and how a linear process between HSF-1 and FOXO contributes to longevity responses. Here, we show that the up-regulation of PFD-6 via HSF-1 increases the transcriptional activity of DAF-16/FOXO. Therefore, PFD-6 appears to promote longevity by acting in a linear pathway from HSF-1 to DAF-16/FOXO under reduced IIS.
A genome-wide RNAi screen identified novel daf-2(−) longevity-regulating factors
C. elegans is an excellent model organism to identify novel functions of genes through genetic screens. A previous genome-wide RNAi screen identified genes that are required for longevity conferred by daf-2 mutations (Samuelson et al. 2007). Among them, a high fraction of genes is involved in vesicle transport, such as vps-16 (vacuolar protein sorting 16). Despite no direct overlap between the previous study (Samuelson et al. 2007) and the present work, our screen identified a vacuolar protein, vps-4 (Fig. 1E), suggesting that proper vesicle transport is crucial for daf-2(−) longevity. It will be interesting to determine how vesicular transport regulates longevity conferred by reduced IIS in future research.
PFD-6 appears to affect longevity as a component of the R2TP/prefoldin-like complex
The prefoldin complex is composed of PFD-1 through PFD-6 and acts as a chaperone for the proper folding of many essential proteins, including actins and tubulins (Vainberg et al. 1998; Lundin et al. 2010; Millan-Zambrano and Chavez 2014). During aging, intestinal lumens deteriorate and suffer from the loss of microvilli, which contain core actin filaments and are important for cytoskeletal integrity (McGee et al. 2011). ACT-5 (actin 5), a potential target of the prefoldin complex, is required for the formation of microvilli in intestinal cells (MacQueen et al. 2005). Overexpression of a modified HSF-1 extends life span and enhances cytoskeletal integrity (Baird et al. 2014). In addition, the chaperonin activity of the TRiC/CCT complex, which works together with the prefoldin complex for cytoskeletal integrity (Vainberg et al. 1998), contributes to longevity in C. elegans (Noormohammadi et al. 2016). Therefore, we tested whether the genetic inhibition of pfd-6 affected the cytoskeletal integrity in daf-2 mutants during aging. We used filamentous actin (F-actin) reporter Lifeact::gfp-expressing C. elegans (Farooqui et al. 2012) and found that genetic inhibition of pfd-6 did not affect the Lifeact::GFP expression pattern in wild-type or daf-2 mutant worms (Supplemental Fig. S8). These data are consistent with the possibility that pfd-6 regulates the longevity of daf-2 mutants independently of the maintenance of cytoskeletal integrity. However, one caveat of this interpretation is that we used a hypomorphic mutant allele of pfd-6, as pfd-6-null mutations cause semiembryonic lethality and larval arrest. Thus, it will be crucial to further examine the relationship between prefoldins, aging, and cytoskeletal integrity with comprehensive genetic and biochemical tools in future studies.
The R2TP/prefoldin-like complex is expressed in the nucleus as well as cytoplasm (Millan-Zambrano and Chavez 2014), consistent with our results regarding PFD-6 expression (Fig. 3A). The R2TP/prefodin-like complex promotes assembly of RNA polymerase II in the cytoplasm (Boulon et al. 2010). Several molecular functions of the nuclear R2TP/prefodin-like complex have also been discovered (Millan-Zambrano and Chavez 2014). C. elegans uri-1 is required for DNA stability (Parusel et al. 2006). Mammalian UXT is a cofactor of the NF-κB enhanceosome, which promotes the induction of NF-κB target gene expression (Sun et al. 2007). In addition, a large fraction of nuclear proteins that bind the R2TP/prefodlin-like complex are transcription factors (Millan-Zambrano and Chavez 2014). A previous study also reported that PFD-6 physically interacts with DAF-16/FOXO by using mass spectrometry (Riedel et al. 2013). Consistent with this, we showed that PFD-6 bound DAF-16/FOXO in the nucleus as well as cytoplasm using an in vivo split-GFP system assay (Fig. 7H; Supplemental Fig. S7J). Thus, PFD-6 as a component of the R2TP/prefoldin-like complex is crucial for the longevity of daf-2(−) mutants through regulating the transcriptional activity of DAF-16/FOXO.
The tissue-specific requirement of PFD-6 is consistent with that of HSF-1 and DAF-16/FOXO
The activities of HSF-1 and DAF-16/FOXO in several tissues have been shown to contribute to the longevity of daf-2 mutants (Libina et al. 2003; Morley and Morimoto 2004; Zhang et al. 2013; Douglas et al. 2015). Overexpression of HSF-1 in the intestine, neurons, or muscles increases life span in wild-type worms (Morley and Morimoto 2004; Douglas et al. 2015). In addition, a transgene that expresses a dominant-negative HSF-1 in neurons or body wall muscles shortens the long life span of age-1/PI3 kinase mutants (Morley and Morimoto 2004). Transgenic expression of daf-16/FOXO in the intestine, neurons, or hypodermis also partially restores the longevity of daf-2(−) mutants (Libina et al. 2003; Zhang et al. 2013). Here, we performed life span assays with tissue-specific RNAi targeting hsf-1 and daf-16 and found that knockdown of hsf-1 or daf-16 in the intestine, hypodermis, or muscles significantly decreased longevity in daf-2(−) mutants. Overall, these findings are in part consistent with our data using PFD-6, whose activities in the intestine and hypodermis were crucial for the longevity of daf-2 mutants. Moreover, we found that PFD-6 bound DAF-16/FOXO in vivo. Therefore, a life span-regulating pathway from HSF-1 to DAF-16/FOXO through PFD-6 appears to act together at least in these two tissues: the intestine and hypodermis. It will be important to determine whether the communication among HSF-1, PFD-6, and DAF-16/FOXO is strictly tissue-autonomous or has tissue-nonautonomous features.
HSF-1 may up-regulate HSP-70 and HSP-90, which in turn stabilize PFD-6 with reduced IIS
How does up-regulated HSF-1 increase PFD-6 protein levels in daf-2 mutants? In flies and mammals, the R2TP/prefoldin-like complex functions with chaperones HSP70 and HSP90 (Boulon et al. 2010; Benbahouche Nel et al. 2014), both of which are targets of HSF-1 (Vihervaara and Sistonen 2014; Dai 2018). Consistently, we found that both C. elegans HSP-90 and HSP-70 bound PFD-6 (Supplemental Fig. S9). Based on these results, we speculate that HSP-70 and HSP-90, which are up-regulated by HSF-1 in daf-2 mutants, bind and stabilize PFD-6 to contribute to longevity.
The R2TP/prefoldin-like complex may play roles in aging or age-related diseases through heat shock factors and FOXO transcription factors
The physiological roles of heat shock factors and FOXOs in aging and age-related diseases are relatively well established. As master transcription factors for the induction of cytosolic chaperones, heat shock factors are crucial for modulating proteostasis-related and age-associated diseases, including Alzheimer's and Parkinson's diseases (Gomez-Pastor et al. 2018). Multiple studies with human populations have indicated that polymorphisms in FOXO3A are linked to longevity (Kenyon 2010; Tazearslan et al. 2012; Webb and Brunet 2014; Martins et al. 2016). Therefore, it will not be surprising to find that heat shock factors and FOXO transcription factors act together to influence aging and age-associated diseases in mammals, consistent with discoveries made using C. elegans. Prefoldin and R2TP/prefoldin-like complex components, including PFD-6, are evolutionarily conserved from archea to humans. Although the role of the recently discovered R2TP/prefoldin-like complex in diseases remains unexplored, prefoldin complex components are implicated in protecting against Huntington's and Alzheimer's diseases (Sorgjerd et al. 2013; Tashiro et al. 2013). Therefore, it will be interesting to test whether PFD-6 or the R2TP/prefoldin-like complex plays roles in age-related diseases or longevity in mammals through working with heat shock factors or FOXOs.
Materials and methods
Strains
The following strains were used in this study.
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). The strains were N2 wild type, CF1041: daf-2(e1370) III, CF2495: hsf-1(sy441) I, CF1085: daf-16(mu86) I; daf-2(e1370) III, IJ462: pfd-6(gk493446) I, IJ485: pfd-6(gk493446) I; daf-2(e1370) III, IJ547: C54C8.3(gk896531) I, IJ548: C54C8.3(gk896531) I; daf-2(e1370) III, IJ479: ntl-2(gk390728) II, IJ480: ntl-2(gk390728) II; daf-2(e1370) III, IJ373: fat-7(wa36) V, IJ389: daf-2(e1370) III; fat-7(wa36) V, EQ150: hsb-1(cg116) IV, IJ1099: hsb-1(cg116) IV, IJ1105: pfd-6(gk493446) I; hsb-1(cg116) IV, CF1908: eat-2(ad1116) II, CF2172: isp-1(qm150) IV, IJ109: rsks-1(tm1714) III, CF2553: osm-5(p813) X, IJ7: vhl-1(ok161) X, pfd-6(tm3759) I/hT2[bli-4(e937) let-?(q782) qIs48[Pmyo-2::gfp; Ppes-10::gfp; Pges-1::gfp]], pfd-6(tm3510) I/hT2, IJ943: pfd-6(gk493446) I; daf-2(e1370) III; yhEx133[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ944: pfd-6(gk493446) I; daf-2(e1370) III; yhEx134[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ568: yhEx132[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ569: yhEx133[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ570: yhEx134[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ810: daf-2(e1370) III; yhEx134[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ1249: yhIs74[pfd-6p::pfd-6::gfp; odr-1p::rfp], IJ1562: yhEx420[vha-6p::hsp-70::spGFP1-10; odr-1p::rfp]; yhEx406[vha-6p::pfd-6::spGFP11; unc-122p::rfp], IJ1579: yhEx424[vha-6p::hsp-90::spGFP1-10; odr-1p::rfp]; yhEx406[vha-6p::pfd-6::spGFP11; unc-122p::rfp], IJ1732: yhEx413[vha-6p::spGFP1-10; odr-1p::rfp]; yhEx406[vha-6p::pfd-6::spGFP11; unc-122p::rfp], IJ1532: yhEx405[vha-6p::pfd-2::spGFP1-10; odr-1p::rfp]; yhEx406[vha-6p::pfd-6::spGFP11; unc-122p::rfp], IJ1058: daf-16(mu86) I; muIs112[daf-16p::gfp::daf-16cDNA; odr-1p::rfp], IJ1230: pfd-6(gk493446) daf-16(mu86) I; muIs112[daf-16p::gfp::daf-16cDNA; odr-1p::rfp], TJ356: zIs356[daf-16p::daf-16a/b::gfp; rol-6], IJ1125: pfd-6(gk493446) I; zIs356[daf-16p::daf-16::gfp; rol-6D], IJ1530: yhEx402[vha-6p::daf-16a::spGFP1-10; odr-1p::rfp]; yhEx406[vha-6p::pfd-6::spGFP11; unc-122p::rfp], IJ1533: yhEx411[vha-6p::hsf-1::spGFP1-10; odr-1p::rfp]; yhEx406[vha-6p::pfd-6::spGFP11; unc-122p::rfp], IJ1061: yhEx240[rpl-28p::lifeact::egfp; odr-1p::rfp], IJ1063: daf-2(e1370) III; yhEx240[rpl-28p::lifeact::egfp; odr-1p::rfp], WM27: rde-1(ne219) V, NR222: rde-1(ne219) V; kzls9[lin-26p::nls::gfp, lin-26p::rde-1; rol-6D], NR350: rde-1(ne219) V; kzls20[hlh-1p::rde-1; sur-5p::nls::gfp], VP303: rde-1(ne219) V; kbls7[nhx-2p::rde-1; rol-6D], IJ411: daf-2(e1370) III; rde-1(ne219) V, IJ415: daf-2(e1370) III; rde-1(ne219) V; kzls9[lin-26p::nls::gfp; lin-26p::rde-1, rol-6D], IJ416: daf-2(e1370) III; rde-1(ne219) V; kzls20[hlh-1p::rde-1; sur-5p::nls::gfp], IJ417: daf-2(e1370) III; rde-1(ne219) V; kbls7[nhx-2p::rde-1; rol-6D], NL3321: sid-1(pk3321) V, IJ807: daf-2(e1370) III; sid-1(pk3321) V, TU3401: sid-1(pk3321) V; uIs69[myo-2p::mCherry; unc-119p::sid-1], IJ898: daf-2(e1370) III; sid-1(pk3321) V; uIs69[myo-2p::mCherry; unc-119p::sid-1]; sid-1(pk3321) V, IJ1862: uIs69[myo-2p::mCherry; unc-119::sid-1]; yhEx507[unc-119p::pfd-6::gfp; ofm-1p::rfp], and IJ1863: rde-1(ne219) V; kzIs20[pDM#715(hlh-1p::rde-1) + pTG95(sur-5p::nls::GFP)]; yhEx508[hlh-1p::pfd-6::mch; ofm-1p::gfp].
A genome-wide RNAi screen
A genome-wide RNAi screen was conducted as described previously with modifications (Lee et al. 2010). Escherichia coli (HT115) bacteria expressing dsRNA targeting C. elegans genes were cultured in 96-well plates for 16 h at 37°C. The plates were then duplicated into 200 µL of LB that contained 50 µg/mL ampicillin (Usb Corp.) and cultured for 16 h at 37°C. dsRNA expression was induced by the addition of 4 mM isopropyl β-d-thiogalactoside (IPTG) (Gold Biotechnology) for 2 h at 37°C. After pelleting E. coli by centrifugation at 2000 rpm for 10 min to remove LB liquid, the bacteria were resuspended in 100 µL of NG liquid containing 50 µg/mL ampicillin and 4 mM IPTG. Synchronized L1 larvae of N2 and hsf-1(sy441) animals were prepared by performing bleaching (Stiernagle 2006) and subsequent overnight starvation in liquid M9 buffer. Approximately 15–20 N2 or hsf-1(sy441) L1 larvae were deposited into each well of 96-well plates containing dsRNA-expressing E. coli. After 3 d of incubation at 20°C, the sterility of N2 and hsf-1(sy441) was monitored by examining the existence of larval progeny. RNAi clones that enhanced the sterility of hsf-1(sy441) mutants but did not cause sterility in N2 animals were scored as positive hits. The positive hits from the initial RNAi screen were confirmed by repeating four times using liquid medium. To test the sterility on solid plates, gravid N2 and hsf-1(sy441) worms were placed on control RNAi-seeded plates. After 1 d, one L1 worm was transferred to a new plate seeded with control RNAi or pfd-6 RNAi bacteria, and a total of 50 individual L1 worms was transferred onto 50 plates seeded with respective RNAi bacteria. The number of plates that contained egg-laying adults was then measured after 4 d of culture.
Life span assays
Life span assays were performed as described previously with modifications (Son et al. 2017). Worms were maintained at 20°C for at least two generations before life span assays, which were performed at 20°C except for rsks-1(tm1714) mutants. The rsks-1 mutant life span assay was performed as described previously at 22.5°C (Seo et al. 2013). To synchronize the other worm strains, gravid adults were allowed to lay eggs for 12 h on RNAi bacteria-seeded plates. The synchronized progeny were allowed to reach L4 stage, and ∼25–35 worms were transferred to plates seeded with gene-specific RNAi bacteria. After 1 d, worms were transferred onto new RNAi plates treated with 5 µM FUdR (Sigma). Worms were transferred to new RNAi plates every 1 or 2 d until they stopped laying eggs for the life span assay without FUdR. During life span experiments, worms that ruptured, burrowed, crawled off the plates, or had internal hatching of progeny were censored but were included for statistical analysis, which was performed by using OASIS 2 (online application of survival analysis; http://sbi.postech.ac.kr/oasis2) (Yang et al. 2011; Han et al. 2016).
Sequence alignment for PFD-6
Amino acid sequence alignments of PFD-6 among species were conducted by using ClustalW2 format (McWilliam et al. 2013; http://www.ebi.ac.uk/Tools/msa/clustalw2) and ClustalX (Jordan and Piel 2008; http://www.clustal.org/clustal2).
Schematic domain structure of PFD-6
The exon–intron pattern of genomic pfd-6 was made by using Exon–Intron Graphic Maker (http://wormweb.org/exonintron).
Plasmid construction and generation of transgenic worms
The 832-base-pair (bp) region of the pfd-6 promoter and pfd-6 genomic DNA were amplified and cloned into pPD95.75 by using the In-Fusion cloning kit (CloneTech). Plasmids for split GFP assays were generated as described previously (Jeong et al. 2017). The intestine-specific promoter region of vha-6 (893 bp) was inserted into pPD95.75 using the In-Fusion cloning kit. The GFP N-terminal (GFP1-10) or GFP C-terminal (GFP11) region was then inserted into a linearized vha-6 promoter-containing pPD95.75 vector using the In-Fusion cloning kit. daf-16 isoform a (1526 bp), hsf-1 cDNA (2015 bp), pfd-2 cDNA (425 bp), hsp-90 cDNA (2108 bp), or hsp-70 (C12C8.1; 2069 bp) genomic DNA (gDNA) was then inserted into the linearized vha-6p::GFPN vector using the In-Fusion cloning kit. pfd-6 cDNA (380 bp) was inserted into the linearized vha-6p::GFPC vector using the In-Fusion kit. To generate a neuron-specific pfd-6 expression plasmid, a promoter region of unc-119 (1964 bp) and genomic pfd-6 DNA (608 bp) were inserted into pPD95.75 with the In-Fusion cloning kit. For muscle-specific pfd-6 expression, an hlh-1 promoter (2000 bp), genomic pfd-6 DNA (608 bp), and mCherry (711 bp) sequences were cloned into pPD95.75 using the In-Fusion cloning kit. For rpl-28p::lifeact::eGFP, the rpl-28 promoter was inserted into pPD95.75, and the GFP region of pPD95.75 was substituted by lifeact::eGFP obtained from PCR amplification using pTK92-lifeact-GFP as a template (Addgene). The F35H10.6/UXT RNAi construct was made by the insertion of the F35H10.6/UXT (596 bp)-coding sequence from ATG into L4440 using the In-Fusion cloning kit. For generation of transgenic animals, 25 µg/µL DNA construct was injected into the gonad of day 1 adult worms with 75 µg/µL coinjection marker, odr-1p::rfp, unc-122p::rfp, or pRF4 [rol-6(su1006)], except for 6.25 µg/µL rpl-28p::lifeact::eGFP.
Yeast two-hybrid screening
Yeast two-hybrid screen was performed as described previously with modifications (Kim et al. 2008) by Panbionet (http://panbionet.com). The full length of pfd-6 cDNA (381 bp) was amplified by PCR. The PCR product was cloned into pGBKL vector, which contains the DNA-binding domain (BD) of GAL4. Screening was conducted with cotransformation of GAL4 DNA-BD-fused PFD-6 and a C. elegans cDNA activation domain (AD) library in pPC86 vector. Yeast strain PBN204 (Panbionet) was used, and two different reporter genes—URA3 and ADE2—were used as selection markers. Yeast transformants of the pfd-6 bait and the C. elegans cDNA AD library were spread on selection medium lacking leucine and tryptophan (SD−LW). From 8.2 × 106 colonies, 640 colonies grew on medium lacking leucine, tryptophan, and uracil (SD−LWU). Among them, 147 colonies grew on medium lacking leucine, tryptophan, and adenine (SD−LWA). In order to confirm the interaction, the prey genes of 147 candidates were amplified by PCR or transformation of E. coli and retransformed into PBN204 with the PFD-6 bait plasmid. Subsequently, 75 clones were confirmed as real positives.
Construction of plasmids for a yeast two-hybrid assay
cDNAs used in this assay were amplified by PCR using a C. elegans cDNA library as a template. The 381-bp region of pfd-6 and 1593 bp of daf-16 were amplified and cloned into pGBKT7 or pGADT7 (Addgene) by using the In-Fusion cloning system. Transformant yeasts containing GAL-4 DNA-BD-fused PFD-6 and GAL-4 AD-fused DAF-16 were spread on a master plate (SD−LW). After 3 d, colonies were replicated on selective plates (SD−LWA). Four micrograms of DNA were used. Because Gal4 AD-DAF-16 transformed yeast displayed growth defects, the interaction between Gal4 DNA-BD-DAF-16 and AD-PFD-6 was tested for yeast two-hybrid assay.
Imaging transgenic animals
Microscope imaging was performed as described previously with some modifications (Artan et al. 2016). Animals were prepared at mixed stages and transferred onto a 2% agarose pad on glass with 100 mM sodium azide (Daejung). The images of worms were captured by using an AxioCam HRc CCD digital camera (Zeiss Corporation) with a Zeiss Axio Scope A1 compound microscope (Zeiss Corporation). ImageJ (Java-based image processing program; National Institutes of Health, http://rsbweb.nih.gov/ij) was used for the quantification of fluorescent images. The double-RNAi method for Supplemental Figure S5 was used as described previously with some modifications (Rea et al. 2007). HT115 bacteria for control RNAi, daf-2 RNAi, hsf-1 RNAi, or daf-16 RNAi were cultured in LB medium to optical density (OD590) 0.9∼1.0. Bacteria expressing daf-2 dsRNA were mixed at a 1:1 ratio with those expressing control dsRNA, hsf-1 dsRNA, or daf-16 dsRNA. Mixed or single dsRNA-expressing bacteria (100 µL) were seeded onto 50 µg/mL ampicillin-containing NGM plates. IPTG (1 mM) was added and incubated overnight at room temperature. Transgenic worms for the fluorescent imaging (Supplemental Fig. S8) were prepared with the same method that was used for the RNAi-mediated life span assays. The confocal images in Figure 3A were acquired by using Nikon confocal microscopy (A1Rsi) at the Brain Research Core Facilities of the Korea Brain Research Institute (KBRI).
mRNA extraction and quantitative qRT–PCR assays
mRNA extraction and qRT–PCR were performed as described previously with some modifications (Hwang et al. 2015). N2, pfd-6(gk493446), daf-2(e1370), and pfd-6(gk493446); daf-2(e1370) animals were grown on control RNAi-seeded plates until L4 stage and transferred to control RNAi-seeded or pfd-6 RNAi-seeded plates containing FUdR. Synchronized day 1 adult worms were harvested using M9 buffer after two washes and frozen in liquid nitrogen. Total RNA was extracted using RNAiso plus (Takara), and cDNA libraries were obtained from reverse transcription (Promega) with random primers. qPCR was conducted using a StepOne real-time PCR system (Applied Biosystems) with SYBR Green master mix (Applied Biosystems). Two technical repeats were averaged for one biological sample and analyzed for all data sets. The transcripts of samples were normalized to ama-1 mRNA levels.
Sequences of primers used for qRT–PCR
The primers used for qRT–PCR were ama-1-Forward (TGGAACTCTGGAGTCACACC), ama-1-Reverse (CATCCTCCTTCATTGAACGG) pfd-6-Forward (GAAACGGCTTGAGTTTATCG), pfd-6-Reverse (CACCTTGTCTCGCTGTTCCG), pfd-1-Forward (CTGAAGCTAAACAGTCAAAAGAG), pfd-1-Reverse (CGTCTACTTTGAATCAGCTCG), pfd-2-Forward (GCAGAATAATATCGCTAATTTGAC), pfd-2-Reverse (GTGAGCAGACGAATGTTGTGTG), pfd-3-Forward (CAGAAGCCCGAGAAGGTATC), pfd-3-Reverse (CTCGTCGACCACTTTTTGAAC), pfd-4-Forward (CAAACACAAACCCAGTTAAAAGCAG), pfd-4-Reverse (GCAGCTTCCAATTCTGCATGG), pfd-5-Forward (GGAACATATTACAAAGCAGGTTG), pfd-5-Reverse (GTGGCGAGTTGAGACTGAAC), ruvb-1-Forward (CCTAAAAATAAGAAGTGATATCCATTC), rvub-1-Reverse (GGAATGAGATAAGTTGTAGAAGAAG), ruvb-2-Forward (CAAGTTGATAAGGCTTCTGGACG), rvub-2-Reverse (GTCTCTTTTGAATTTCTCCATCTGG), F35H10.6/UXT-Forward (GAGTTTTTATGAATATTGAAGTGAGAG), F35H10.6/UXT-Reverse (CTGTCCATCTTCCGATCACATG), uri-1-Forward (GTTGGAAGCTCCTATTTTGGCG), uri-1-Reverse (CTGTATGAATGCTGCATTCTCCG), F12A10.1-Forward (ATGAATTCGCAAACCTACTTTTGG), F12A10.1-Reverse (GGCGTTTTTCACGAACATTTATATTTG), F26G1.2-Forward (GATGTGGTTTCTTATACAGAAGACGG), F26G1.2-Reverse (GTGGAGCTCCAGTAATCGTACGG), C06G3.3-Forward (GAAACCTGATCAAAGCCAACACG), C06G3.3-Reverse (GAAATGGTATATTAAATGGAAGCCAC), C02F12.5- Forward (CTTGAATCTTGTAAACCAGCTGAC), C02F12.5-Reverse (CTCCCATCACACACACCGTTC), grd-4-Forward (GCTGTAGCTGTTCAAGGCC), grd-4-Reverse (CATCACCAAAATAGCTCAACGTTC), sod-3-Forward (CTATCTTCTGGACCAACTTGG), sod-3-Reverse (GCAAGTTATCCAGGGAACCG), D1086.1-Forward (GGTATTTCAGTGACAAATCAAGAATG), D1086.1-Reverse (CGTGGAATCTCCACACGCC), nlp-35-Forward (CAAGTTCTCGCACCAAGATTCCGA), nlp-35-Reverse (GGTTTTGGAGCAAATGTTGGAGG), M02D8.6-Forward (GCTTCTGTTATATCAGCAAG), M02D8.6-Reverse (CAAGCATCATCTCGAAGAAC), nas-5-Forward (GTGGTCAATGGACGGGGTC), and nas-5-Reverse (CTTTCCCGTATTGGTGAACTATAAAC).
mRNA library preparation and mRNA-seq
mRNA extraction for RNA-seq was performed as described previously with some modifications (Lee et al. 2015). N2, pfd-6(gk493446), daf-2(e1370), and pfd-6(gk493446); daf-2(e1370) were synchronized until the worms reached L4 stage on control RNAi plates and transferred to control or pfd-6 RNAi plates containing FUdR. After 1 d, worms were harvested with M9, washed to remove bacteria, and frozen in liquid nitrogen. Total RNAs were isolated from frozen worms using RNAiso plus (Takara) and ethanol precipitation. The paired-end mRNA-seq library was prepared and sequenced by a HiSeq 2500 platform at Chunlab (Chunlab, Inc.). A total of three biological repeats of samples was analyzed for all conditions. RNA-seq experiments for two biological repeats were performed first, and the last set was added later to increase the robustness of the data.
mRNA-seq analysis
RNA-seq data analysis was performed as described previously (Pertea et al. 2016). Reads were aligned to the C. elegans genome ce11 and analyzed by HISAT2 software (version 2.0.5) (Pertea et al. 2016) with default parameters. Transcript assembly and quantification were performed by using StringTie (version 1.3.3) (Pertea et al. 2015) and Ballgown (version 2.0.0) (Frazee et al. 2015). Differentially expressed genes were analyzed by limma (version 3.24.15) (Phipson et al. 2016) with count values. Batch effects among biological repeats were removed with lmFit, contrasts.fit, and eBayes functions in limma (version 3.24.15) (Law et al. 2014). P-values were calculated by using limma (version 3.24.15) (Phipson et al. 2016) with R version 3.1.3 (http://www.r-project.org). Scatter plot (Riedel et al. 2013), Venn diagram, and heat map analyses (Ewald et al. 2015) were performed as described previously. Heat maps were generated by clustering expression profiles using Cluster 3.0 (Eisen et al. 1998) and visualized by Java Treeview (Saldanha 2004). Venn diagrams were made by using Venn diagram plotter (http://omics.pnl.gov/software/venn-diagram-plotter), and P-values of the overlaps between two different sets of genes were calculated by using hypergeometric probability test (http://nemates.org/MA/progs/overlap_stats.html).
GO biological process
GO analysis for pfd-6-dependent up-regulated genes in daf-2(−) animals was performed by using DAVID (Huang da et al. 2009; https://david.ncifcrf.gov/home.jsp). The enriched GO terms were selected by using the criterion of P-value < 0.05.
Analysis of DNA motifs
DNA motif analysis was performed as described previously with modifications (Seo et al. 2015). Enriched DNA sites located from 800 bp upstream of pfd-6-dependent up-regulated genes and down-regulated genes in daf-2 mutants were searched by using RSAT (RNA sequence analysis tools; http://www.rsat.eu). DNA motifs were predicted by using a default parameter, oligomer lengths of 6 and 7, pseudo-frequency of <0.01, and flanking residue of 3.
Subcellular localization of DAF-16::GFP
Microscope imaging for GFP subcellular localization was performed as described previously with modifications (Seo et al. 2015). Gravid adult worms were transferred to RNAi bacteria-seeded plates and allowed to lay eggs. After 2 or 3 d, day 1 adult progeny were mounted on 2% agar pads with 2 mM levamisole (tetramisole) (Sigma). The number of worms with nuclear-localized DAF-16::GFP in the anterior intestinal cells was counted. ImageJ was used for the quantification of fluorescent images.
Coimmunoprecipitation and Western blot assays using cultured mammalian cells
Coimmunoprecipitation was performed as described previously with some modifications (Seo et al. 2015). HA-tagged HSP-90 and Flag-tagged PFD-6 were expressed using HEK293T cells. The cells were maintained with DMEM (HyClone) supplemented with penicillin/streptomycin (HyClone) and 10% (v/v) FBS (HyClone) in a 5.0% (v/v) CO2-provided incubator at 37°C. Transfection was performed with Lipofectamine LTX and Plus reagent (Invitrogen). Three micrograms of plasmids expressing Flag-HSP-90 and/or HA-PFD-6 was used for transfection. HEK293T cell lysates were collected using PBS (Welgene; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 at pH 7.4) and then centrifuged at 800g for 1 min. The supernatant was removed, and the pellet was resolved with a lysis buffer (20 mM Tris⋅HCl at pH 7.4, 10 mM KCl, 10 mM MgCl2, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 2.5 mM β-glycerophosphate, 1 mM NaF, 1 mM DTT, 1 mM PMSF, protease inhibitor mixture [Biovison]). The cell lysates were sonicated (power output 30 W), and 210 mM NaCl was added. The lysates were incubated for 1 h at 4°C on an end-over-end rotator and subsequently centrifuged at 13,000g for 20 min at 4°C. A total of 2 mg of protein was incubated for 1 h∼overnight with Flag antibody (Sigma). Protein G agarose was added (GE Healthcare) and then incubated for 1 h at 4°C on an end-over-end rotator. The mixture was washed three times with a wash buffer (20 mM Tris⋅HCl at pH 7.4, 500 mM NaCl, 10 mM MgCl2, 2 mM EDTA, 1% Triton X-100) with centrifugation at 800g for 1 min. The mixture was denatured by using 2× SDS sample buffer (Bio-Rad) at 95°C, and proteins were detected by using standard Western blot assays. Primary antibodies against HA (1:2000; Sigma) and Flag (1:5000; Sigma) were used with anti-rabbit (1:5000; Thermo) and anti-mouse (1:5000; Thermo) secondary antibodies, respectively.
Western blot assays with C. elegans samples
Western blot assay using C. elegans was performed as described previously (Jeong et al. 2017).
Supplementary Material
Acknowledgments
Some of the C. elegans strains used in this study were provided by Dr. Cynthia Kenyon and the Caenorhabditis Genetics Center. We thank all Lee laboratory members for help and discussion, Dr. Hong Gil Nam for technical help, and Dr. Kyuhyung Kim and Dr. Christian Riedel for valuable comments. This work was supported by the Korean government (Ministry of Science, ICT [Information And Communication Technology], and Future Planning) through the National Research Foundation of Korea (NRF) (NRF-2016R1E1A1A01941152) to S.-J.V.L., and Korea Brain Research Institute (KBRI) basic research program through the KBRI funded by the Ministry of Science and ICT (18-BR-03-03) to C.M.H.
Author contributions: H.G.S., K.S., M.S., and S.-J.V.L. designed the experiments. H.G.S., K.S., M.S., S.P., S.W.A.A., E.-S.C., Y.L., H.B., E.K., Y.R., C.M.H., and S.-J.V.L. performed experiments. H.G.S., K.S., S.H., A.-L.H., T.-Y.R., S.K.J., and S.-J.V.L. analyzed the data. H.G.S., K.S., and S.-J.V.L. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.317362.118.
References
- Altintas O, Park S, Lee SJ. 2016. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep 49: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A. 2007. Genetics of aging in Caenorhabditis elegans. PLoS Genet 3: 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artan M, Jeong DE, Lee D, Kim YI, Son HG, Husain Z, Kim J, Altintas O, Kim K, Alcedo J, et al. 2016. Food-derived sensory cues modulate longevity via distinct neuroendocrine insulin-like peptides. Genes Dev 30: 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR III, Manning G, Dillin A. 2014. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science 346: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbahouche Nel H, Iliopoulos I, Torok I, Marhold J, Henri J, Kajava AV, Farkas R, Kempf T, Schnolzer M, Meyer P, et al. 2014. Drosophila Spag is the homolog of RNA polymerase II-associated protein 3 (RPAP3) and recruits the heat shock proteins 70 and 90 (Hsp70 and Hsp90) during the assembly of cellular machineries. J Biol Chem 289: 6236–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. 2015. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med 88: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jady BE, Rothe B, Pescia C, Robert MC, Kiss T, et al. 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol 180: 579–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. 2010. HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39: 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. 2010. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods 7: 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AT, Guo C, Itani OA, Budaitis BG, Williams TW, Hopkins CE, McEachin RC, Pande M, Grant AR, Yoshina S, et al. 2015. Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics 201: 613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. 2012. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148: 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Al-Khoury R, Lavallee-Adam M, Faubert D, Jiang H, Poitras C, Bouchard A, Forget D, Blanchette M, Coulombe B. 2009. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods 48: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. 2006. Opposing activities protect against age-onset proteotoxicity. Science 313: 1604–1610. [DOI] [PubMed] [Google Scholar]
- Dai C. 2018. The heat-shock, or HSF1-mediated proteotoxic stress, response in cancer: from proteomic stability to oncogenesis. Philos Trans R Soc Lond B Biol Sci 373: 20160525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Baird NA, Simic MS, Uhlein S, McCormick MA, Wolff SC, Kennedy BK, Dillin A. 2015. Heterotypic signals from neural HSF-1 separate thermotolerance from longevity. Cell Rep 12: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald CY, Landis JN, Porter Abate J, Murphy CT, Blackwell TK. 2015. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui S, Pellegrino MW, Rimann I, Morf MK, Muller L, Frohli E, Hajnal A. 2012. Coordinated lumen contraction and expansion during vulval tube morphogenesis in Caenorhabditis elegans. Dev Cell 23: 494–506. [DOI] [PubMed] [Google Scholar]
- Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL, Leek JT. 2015. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol 33: 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. 2018. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19: 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuhaThakurta D, Palomar L, Stormo GD, Tedesco P, Johnson TE, Walker DW, Lithgow G, Kim S, Link CD. 2002. Identification of a novel cis-regulatory element involved in the heat shock response in Caenorhabditis elegans using microarray gene expression and computational methods. Genome Res 12: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW. 2004. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168: 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SV, Kim S. 2016. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7: 56147–56152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Hwang W, Artan M, Seo M, Lee D, Nam HG, Lee SV. 2015. Inhibition of elongin C promotes longevity and protein homeostasis via HIF-1 in C. elegans. Aging Cell 14: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DE, Artan M, Seo K, Lee SJ. 2012. Regulation of lifespan by chemosensory and thermosensory systems: findings in invertebrates and their implications in mammalian aging. Front Genet 3: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong DE, Lee D, Hwang SY, Lee Y, Lee JE, Seo M, Hwang W, Seo K, Hwang AB, Artan M, et al. 2017. Mitochondrial chaperone HSP-60 regulates anti-bacterial immunity via p38 MAP kinase signaling. EMBO J 36: 1046–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan GE, Piel WH. 2008. PhyloWidget: Web-based visualizations for the tree of life. Bioinformatics 24: 1641–1642. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. 2010. The genetics of ageing. Nature 464: 504–512. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kim JE, Ryu I, Kim WJ, Song OK, Ryu J, Kwon MY, Kim JH, Jang SK. 2008. Proline-rich transcript in brain protein induces stress granule formation. Mol Cell Biol 28: 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hwang AB, Kenyon C. 2010. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol 20: 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Jeong DE, Son HG, Yamaoka Y, Kim H, Seo K, Khan AA, Roh TY, Moon DW, Lee Y, et al. 2015. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev 29: 2490–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. 2003. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502. [DOI] [PubMed] [Google Scholar]
- Lundin VF, Leroux MR, Stirling PC. 2010. Quality control of cytoskeletal proteins and human disease. Trends Biochem Sci 35: 288–297. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Baggett JJ, Perumov N, Bauer RA, Januszewski T, Schriefer L, Waddle JA. 2005. ACT-5 is an essential Caenorhabditis elegans actin required for intestinal microvilli formation. Mol Biol Cell 16: 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R, Lithgow GJ, Link W. 2016. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Weber D, Day N, Vitelli C, Crippen D, Herndon LA, Hall DH, Melov S. 2011. Loss of intestinal nuclei and intestinal integrity in aging C. elegans. Aging Cell 10: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res 41: W597–W600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Zambrano G, Chavez S. 2014. Nuclear functions of prefoldin. Open Biol 4: 140085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. 2004. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Hu PJ. 2013. Insulin/insulin-like growth factor signaling in C. elegans. In WormBook (ed. The C. elegans Research Community), WormBook; 10.1895/wormbook.1.164.1 http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. [DOI] [PubMed] [Google Scholar]
- Noormohammadi A, Khodakarami A, Gutierrez-Garcia R, Lee HJ, Koyuncu S, Konig T, Schindler C, Saez I, Fatima A, Dieterich C, et al. 2016. Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat Commun 7: 13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Jung Y, Lee SV. 2017. Survival assays using Caenorhabditis elegans. Mol Cells 40: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. 2006. URI-1 is required for DNA stability in C. elegans. Development 133: 621–629. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. 2016. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 10: 946–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Inoue M, Hikita T, Koppen M, Hardin JD, Amano M, Moerman DG, Kaibuchi K. 2007. Establishment of a tissue-specific RNAi system in C. elegans. Gene 400: 166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. 2007. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol 5: e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 15: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Samuelson AV, Carr CE, Ruvkun G. 2007. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev 21: 2976–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K, Choi E, Lee D, Jeong DE, Jang SK, Lee SJ. 2013. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell 12: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Seo M, Seo K, Hwang W, Koo HJ, Hahm JH, Yang JS, Han SK, Hwang D, Kim S, Jang SK, et al. 2015. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc Natl Acad Sci 112: E4246–E4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son HG, Lee SV. 2017. Longevity regulation by NMD-mediated mRNA quality control. BMB Rep 50: 160–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son HG, Seo M, Ham S, Hwang W, Lee D, An SW, Artan M, Seo K, Kaletsky R, Arey RN, et al. 2017. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat Commun 8: 14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgjerd KM, Zako T, Sakono M, Stirling PC, Leroux MR, Saito T, Nilsson P, Sekimoto M, Saido TC, Maeda M. 2013. Human prefoldin inhibits amyloid-β (Aβ) fibrillation and contributes to formation of nontoxic Aβ aggregates. Biochemistry 52: 3532–3542. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. Maintenance of C. elegans. In WormBook (ed. The C. elegans Research Community), WormBook; 10.1895/wormbook.1.101.1 http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Tang Y, Lou X, Zhu L, Yang K, Zhang B, Shi H, Wang C. 2007. UXT is a novel and essential cofactor in the NF-κB transcriptional enhanceosome. J Cell Biol 178: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro E, Zako T, Muto H, Itoo Y, Sorgjerd K, Terada N, Abe A, Miyazawa M, Kitamura A, Kitaura H, et al. 2013. Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. J Biol Chem 288: 19958–19972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C, Cho M, Suh Y. 2012. Discovery of functional gene variants associated with human longevity: opportunities and challenges. J Gerontol A Biol Sci Med Sci 67: 376–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154: 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93: 863–873. [DOI] [PubMed] [Google Scholar]
- Vihervaara A, Sistonen L. 2014. HSF1 at a glance. J Cell Sci 127: 261–266. [DOI] [PubMed] [Google Scholar]
- Webb AE, Brunet A. 2014. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci 39: 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kim SK. 2012. The GATA transcription factor egl-27 delays aging by promoting stress resistance in Caenorhabditis elegans. PLoS Genet 8: e1003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6: e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Judy M, Lee SJ, Kenyon C. 2013. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab 17: 85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.