In this study, Rubio et al. researched how the dynamic genome-wide landscape of m6A-modified mRNAs impacts virus infection and host immune responses. They show that type I interferon (IFN) production triggered by dsDNA or human cytomegalovirus (HCMV) is controlled by the cellular m6A methyltrasferase subunit METTL14 and ALKBH5 demethylase, and their results demonstrate that responses to nonmicrobial dsDNA in uninfected cells are regulated by enzymes controlling m6A epitranscriptomic changes.
Keywords: RNA m6A modification, dsDNA signaling, human cytomegalovirus, innate immunity, virus infection
Abstract
Modification of mRNA by N6-adenosine methylation (m6A) on internal bases influences gene expression in eukaryotes. How the dynamic genome-wide landscape of m6A-modified mRNAs impacts virus infection and host immune responses remains poorly understood. Here, we show that type I interferon (IFN) production triggered by dsDNA or human cytomegalovirus (HCMV) is controlled by the cellular m6A methyltrasferase subunit METTL14 and ALKBH5 demethylase. While METTL14 depletion reduced virus reproduction and stimulated dsDNA- or HCMV-induced IFNB1 mRNA accumulation, ALKBH5 depletion had the opposite effect. Depleting METTL14 increased both nascent IFNB1 mRNA production and stability in response to dsDNA. In contrast, ALKBH5 depletion reduced nascent IFNB1 mRNA production without detectably influencing IFN1B mRNA decay. Genome-wide transcriptome profiling following ALKBH5 depletion identified differentially expressed genes regulating antiviral immune responses, while METTL14 depletion altered pathways impacting metabolic reprogramming, stress responses, and aging. Finally, we determined that IFNB1 mRNA was m6A-modified within both the coding sequence and the 3′ untranslated region (UTR). This establishes that the host m6A modification machinery controls IFNβ production triggered by HCMV or dsDNA. Moreover, it demonstrates that responses to nonmicrobial dsDNA in uninfected cells, which shape host immunity and contribute to autoimmune disease, are regulated by enzymes controlling m6A epitranscriptomic changes.
Differential chemical modification of mRNA in theory provides a powerful means to dynamically alter gene expression. In particular, methylation of adenosine at the N6 position (m6A) constitutes the most widespread internal base modification to mRNA (Yue et al. 2015; Roundtree et al. 2017). A methyltransferase writer core complex composed of the METTL3 catalytic subunit, METTL14, and WTAP installs m6A marks on nascent mRNA cotranscriptionally (Fig. 1A; Liu et al. 2014). Distributed primarily within mRNA-coding sequences and reportedly enriched at the start of 3′ exons and proximal to termination codons, m6A is also found within 3′ untranslated regions (UTRs) and extended 5′ cap structures where the 2′-O-methyl nucleotide immediately adjacent to the 7-methylguanosine (m7G) cap is an adenosine (Dominissini et al. 2012; Meyer et al. 2012). In addition to modifying RNA secondary structure (Liu et al. 2015, 2017; Roost et al. 2015), m6A is specifically recognized by a family of “reader” proteins, including YTH domain-containing proteins that reside primarily in the cytoplasm (YTHDF1 and YTHDF2) or the nucleus (YTHDC1 and YTHDF3) (Fig. 1A; Patil et al. 2018). Identification of demethylases FTO and ALKBH5, capable of erasing m6A marks in vitro, suggested that m6A modifications were potentially dynamic and reversible, sculpted by the opposing actions of writers that install m6A and erasers that remove it (Jia et al. 2011; Zheng et al. 2013). Recently, however, FTO was shown to have limited capacity to demethylate internal m6A residues, with its preferred substrate being N6, 2′-O-dimethyl-adenosine (m6Am), located beside the m7G cap (Mauer et al. 2017). Although m6A methylation occurs to nascent pre-mRNA, it is predominately stable in the cytoplasm until the mRNA decays (Ke et al. 2017). While this argues against methylation removal in the cytoplasm, internal m6A removal by a demethylase such as ALKBH5 within the nucleus could not be excluded and remains possible (Ke et al. 2017). Numerous aspects of RNA biology and metabolism have been reportedly regulated by m6A mRNA modification, including nuclear processing and export (Fustin et al. 2013; Zheng et al. 2013, 2017; Haussmann et al. 2016), translation (Meyer et al. 2015; Coots et al. 2017; Zhou et al. 2018), and mRNA decay (Wang et al. 2014; Du et al. 2016; Chen and Shyu 2017; Edupuganti et al. 2017). In addition, mRNA modification by m6A influences development, differentiation, reprogramming, circadian rhythm, cell cycle, disease pathogenesis, and stress responses (Fustin et al. 2013; Aguilo et al. 2015; Merkestein et al. 2015; Zhou et al. 2015; Vu et al. 2017; Wojtas et al. 2017; Wen et al. 2018).
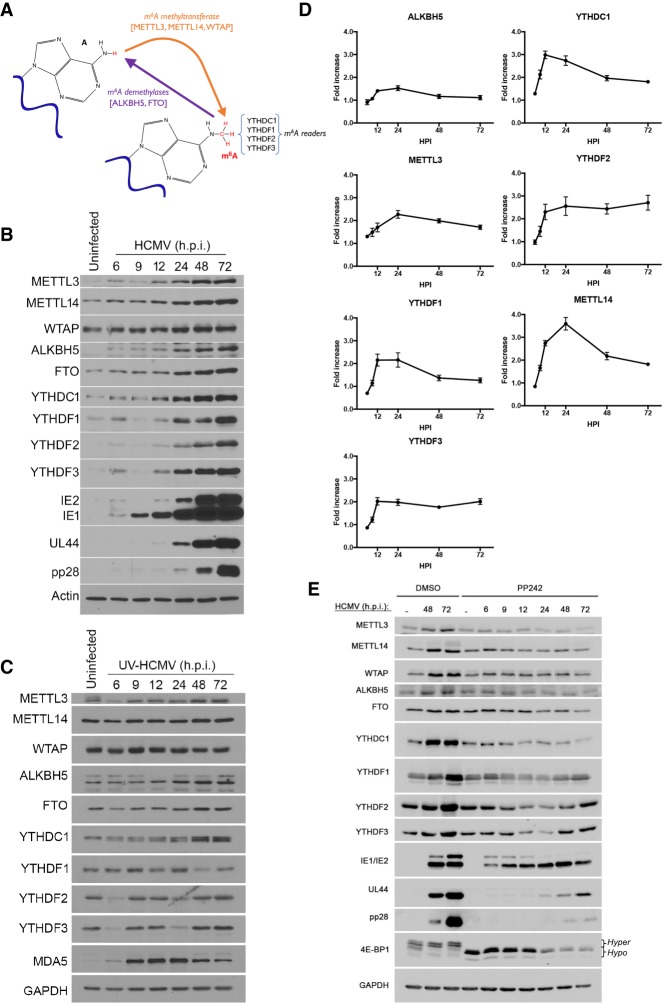
Figure 1.
Accumulation of the cellular m6A machinery is induced by human cytomegalovirus (HCMV). (A) Regulation of m6A levels in mRNA by the opposing action of a multisubunit methyltransferase and demethylases. A subset of reader proteins that recognize m6A-modified residues is depicted. (B) Normal human dermal fibroblasts (NHDFs) were mock-infected (uninfected) or infected with HCMV (multiplicity of infection [MOI] = 3). At the indicated hours post-infection (hpi), total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antibodies. Actin represents a host antigen whose levels remain unchanged during infection. (C) As in B except cells were infected with UV-inactivated HCMV, and GAPDH was used as a control host antigen. (D) Total RNA collected from HCMV-infected cells (MOI = 3) at the indicated times (hours) post-infection. For each of the indicated genes, mRNA was analyzed by real-time quantitative PCR (qPCR) and normalized to PPIA mRNA levels. Three biological replicates were performed. Error bars indicate SEM. (E) Uninfected NHDFs or NHDFs infected with HCMV (MOI = 3) were treated with the mechanistic target of rapamycin (mTOR) active site inhibitor PP242 or DMSO. At the indicated times (hours post-infection), total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera. GAPDH served as a loading control. The change in 4E-BP1 migration in uninfected cells from a slow-migrating hyperphosphorylated species (hyper) to a faster-migrating hypophosphorylated (hypo) form validates the activity of PP242.
Significantly, virus-encoded mRNAs are also chemically modified by m6A, and a role for m6A in infection biology is emerging. While m6A has been detected in viral RNA, precisely how host enzymes that control m6A modification levels impact virus reproduction remains unclear. For two RNA viruses that replicate in the nucleus, cellular m6A functions stimulate virus reproduction. The METTL3/14 m6A writer complex reportedly promotes HIV replication, whereas m6A erasers ALKBH5 and FTO suppress it (Kennedy et al. 2016; Lichinchi et al. 2016a; Tirumuru et al. 2016). However, different mechanisms have been proposed to account for this, including differential nuclear export, translation, and virus genome replication. Similarly, the METTL3 methyltransferase promotes influenza A virus replication and m6A sites in the viral HA segment are required for proper protein expression and pathogenicity in mice (Courtney et al. 2017). Recently, RNA viruses that replicate exclusively in the cytoplasm have been found to contain m6A in their (+) sense RNA genomes. In contrast to nuclear RNA viruses, m6A writers METTL3/14 limited HCV infectious particle production and protein expression without detectably changing RNA replication (Gokhale et al. 2016; Brocard et al. 2017; Gonzales-van Horn and Sarnow 2017). The FTO demethylase, however, stimulated HCV protein expression. In addition, METTL3/14 restricted Zika virus reproduction, whereas FTO and ALKBH5 demethylases enhanced it. Zika virus infection also enriched for m6A modifications of host mRNAs but did not detectably change the abundance of host antiviral transcripts (Lichinchi et al. 2016b). DNA viruses such as SV40, adenovirus, and herpesviruses replicate in the nucleus and produce mRNAs that contain m6A (Lavi and Shatkin 1975; Sommer et al. 1976; Moss et al. 1977; Hesser et al. 2018; Tan et al. 2018; Tsai et al. 2018). Intriguingly, interfering with the host m6A machinery in cells infected with the γ-herpesvirus KSHV resulted in complex proviral and antiviral outcomes that were dependent on the cell type analyzed (Hesser et al. 2018). How cellular functions that control steady-state mRNA m6A levels impact the reproduction of large DNA viruses and the DNA-sensing pathways that impact host innate defenses remain unknown.
Infection with human cytomegalovirus (HCMV), a large DNA virus that replicates within the nucleus, is predominately asymptomatic in healthy individuals (Britt 2008; Boeckh and Geballe 2011). However, HCMV causes life-threatening disease among the immunocompromised, including solid organ or stem cell transplant recipients (Ljungman et al. 2010; Razonable and Humar 2013), and is a significant source of congenital morbidity and mortality among newborn infants in the developed world (Cannon et al. 2010; Manicklal et al. 2013). Unlike many viruses, including α- and γ-herpesvirus subfamily members, the β-herpesvirus HCMV does not globally impair ongoing host protein synthesis. Instead, extensive remodeling of the host translational landscape plays a critical role in regulating HCMV productive replication (McKinney et al. 2014; Tirosh et al. 2015). Significantly, changes in host gene expression induced in response to HCMV can either stimulate or restrict acute virus reproduction. While some HCMV-induced alterations to host gene expression require de novo expression of viral genes, others do not and are in fact triggered by exposure to the viral dsDNA genome. Whether any of these responses to HCMV are impacted by cellular m6A RNA modification enzymes and how this might influence HCMV reproduction are unknown. Furthermore, the unanticipated extent to which HCMV manipulates host gene expression post-transcriptionally, balancing expression of proviral cellular factors and antiviral host responses, makes it a powerful model to investigate how the cellular m6A modification machinery impacts virus reproduction.
Here, we show that the overall abundance of cellular m6A writers METTL3/14, erasers ALKBH5 and FTO, and reader proteins increased in response to HCMV infection. Interfering with METTL3/14 accumulation reduced virus protein accumulation and reproduction, whereas depleting the ALKBH5 demethylase enhanced viral protein expression and stimulated virus reproduction. Unexpectedly, interferon β (IFNB1) mRNA accumulated upon METTL14 depletion. This impaired HCMV reproduction, implicating an underlying response to dsDNA. Significantly, METTL14 depletion stimulated IFNB1 accumulation in response to dsDNA in uninfected primary human fibroblasts, while ALKBH5 depletion restricted it. Codepleting ALKBH5 with m6A readers YTHDF1 or YTHDF2 intensified the reduction in dsDNA-induced IFNB1 mRNA accumulation. Moreover, IFNB1 mRNA was found in an RNA fraction enriched for m6A. While IFNB1 mRNA biogenesis following dsDNA exposure was increased by METTL14 depletion and reduced by ALKBH5 depletion, altered IFNB1 mRNA decay was detected only in response to depleting METTL14. Genome-wide profiling following ALKBH5 depletion identified differentially expressed genes regulating antiviral immune responses, while METTL14 depletion altered pathways involving metabolic reprogramming, stress responses, and aging. This establishes that m6A RNA modification enzymes regulate cellular responses to dsDNA sensing, which shapes host immunity and contributes to autoimmune disease. It further suggests that m6A epitranscriptomic changes play a fundamental role in cell-intrinsic innate immune responses.
Results
Regulation of HCMV replication by host m6A modification enzymes
Since host protein synthesis is not impaired by HCMV infection (McKinney et al. 2014), we could examine how cellular functions required for m6A RNA modification responded to HCMV and influenced virus reproduction. Compared with uninfected primary human fibroblasts (normal human dermal fibroblasts [NHDFs]), HCMV infection resulted in accumulation of m6A methyltransferase subunits METTL3/14; m6A readers YTHDC1 and YTHDF1,2,3; and the demethylases ALKBH5 and FTO between 12 and 72 h after infection (Fig. 1B). This most closely coincided temporally with accumulation of the representative HCMV early (E) protein UL44 (Fig. 1B) and was severely curtailed following infection with UV-inactivated HCMV, indicating that virus-induced accumulation of cellular m6A effector polypeptides was largely dependent on viral gene expression (Fig. 1C). These modest increases in abundance observed 48–72 h after infection with UV-inactivated HCMV are unlikely to result from incomplete UV inactivation, as they were not observed for all m6A effector proteins. It could, however, indicate a secondary level of induction independent of virus gene expression.
In response to viral gene expression, a less than twofold increase in ALKBH5 mRNA was observed over time (Fig. 1D). Whereas METTL3, YTHDF1, and YTHDF3 mRNA increased approximately twofold, a threefold to fourfold increase in mRNA levels was detected for YTHDC1, YTHDF2, and METTTL14 (Fig. 1D). Finally, the HCMV-induced increase in cellular m6A effectors was repressed by treatment with the mechanistic target of rapamycin (mTOR) active site inhibitor PP242 (Fig. 1E). This indicates that the increase in m6A writers, readers, and erasers was dependent on mTOR activation in HCMV-infected cells. The HCMV UL38 protein, which is expressed at early times post-infection, activates mTOR complex 1 (mTORC1) and has been shown to regulate cap-dependent translation of host mRNAs in HCMV-infected cells (McKinney et al. 2012, 2014). Taken together, these data suggest that the increase in cellular m6A effector proteins induced by HCMV is in part regulated by changes in mRNA abundance and cap-dependent mRNA translation. Similar changes in mRNA abundance and mRNA translation inferred from ribosome profiling have been reported by others (Tirosh et al. 2015) in cells infected with a different HCMV strain (Supplemental Fig. S1).
To determine how host m6A writers and erasers influenced virus replication, cultures were treated with siRNA specific for METTL3, METTL14, ALKBH5, or control nonsilencing siRNA. Subsequently, siRNA-treated NHDFs were mock-infected or infected with HCMV at a low multiplicity of infection, which allows for measurement of virus reproduction and spread through the culture. Compared with nonsilencing siRNA-treated cultures, METTL3/14-specific siRNAs inhibited HCMV reproduction up to 11-fold (Fig. 2A) and reduced accumulation of representative virus immediate–early (IE), early, and late (L) proteins (Fig. 2B). Even under these conditions, where approximately one in 20 cells was initially infected, a virus-induced increase in METTL14 and ALKBH5 abundance remained detectable (Supplemental Fig. S2A), albeit modest in comparison with the increase observed at greater viral inoculums, where the majority of cells was infected (Fig. 1B). In addition, a greater reduction in HCMV reproduction was observed with METTL14 siRNA-treated compared with METTL3 siRNA-treated cultures. As both METTL3 and METTL14 are components contained within a multisubunit methyltransferase (Liu et al. 2014), this likely reflects the larger reduction in METTL3 and METTL14 levels achieved using METTL14 siRNA compared with METTL3 siRNA (Fig. 2B). Importantly, METTL3 and METTL14 siRNAs did not detectably change ALKBH5 or GAPDH levels (Fig. 2B). METTL14 depletion similarly reduced accumulation of representative HCMV immediate–early, early, and late proteins over time when high-multiplicity infections were performed (Supplemental Fig. S2B). In contrast, ALKBH5 siRNA enhanced virus protein accumulation and stimulated virus reproduction and spread by nearly 100-fold following infection at low multiplicity (Fig. 2A,B). ALKBH5 depletion did not detectably increase levels of HCMV IE1/2 (immediate–early), UL44 (early), or pp28 (late) proteins in cells infected at high multiplicity (Supplemental Fig. S2B). Depleting the demethylase FTO did not detectably stimulate HCMV reproduction (Supplemental Fig. S3). The selective effect of ALKBH5 on HCMV reproduction (Supplemental Fig. S3) is consistent with the restricted capacity of FTO to demethylate internal m6A residues and its preference for m6Am substrates (Mauer et al. 2017). Finally, depleting METTL3 or ALKBH5 did not measurably impact replication of other DNA viruses, including HSV-1 or vaccinia virus (Supplemental Fig. S3).
Figure 2.
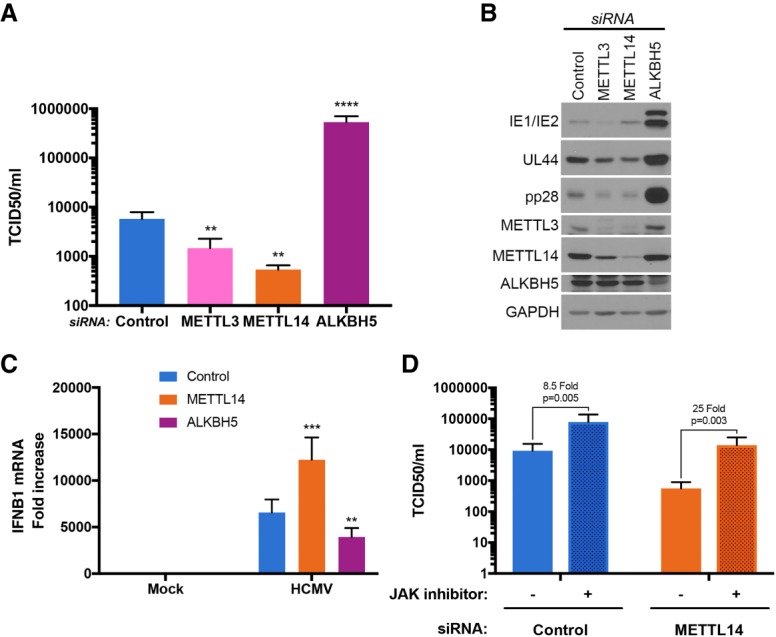
Control of HCMV replication by the host RNA m6A METTL3/14 methyltransferase and ALKBH5 demethylase. (A) NHDFs were treated with control nonsilencing siRNA, siRNA specific for the m6A methyltransferase METTL3 or METTL14 subunits, or siRNA specific for the ALKBH5 demethylase. After 72 h, cultures were infected with HCMV (MOI = 0.05), and the infection allowed to proceed for 7 d. Supernatants were harvested, and virus titer (tissue culture infective dose at which 50% of cultures are infected [TCID/50]) was determined using NHDFs. Error bars indicate SEM. (**) P ≤ 0.01; (****) P ≤ 0.0001 by Student's t-test. (B) As in A except total protein was harvested and fractionated by SDS-PAGE, and the accumulation of viral proteins encoded by representative immediate–early (IE1/2), early (UL44), or late (pp28) genes was measured by immunoblotting with the indicated antisera. GAPDH served as a loading control. (C) NHDFs treated with control nonsilencing siRNA (blue) or siRNA specific for METTL14 (orange) or ALKBH5 (purple) were infected with HCMV (MOI = 3). After 6 h, total RNA was harvested, and IFNB1 mRNA levels were quantified by qRT–PCR. Three biological replicates (n = 3) were performed, and error bars represent SEM. (**) P ≤ 0.01; (***) P ≤ 0.001 by Student's t-test. (D) NHDFs were treated with control nonsilencing siRNA or siRNA specific for METTL14. After 72 h, cultures were treated with either 1 µM Janus kinase (JAK) inhibitor (pyridone 6) or DMSO and infected with HCMV (MOI = 0.05). Supernatants were harvested, and virus titer (TCID/50) was determined after 7 d using NHDFs.
Cellular m6A modification enzymes regulate IFNB1 mRNA accumulation in HCMV-infected cells
To determine whether host functions regulating m6A modification controlled antiviral immune responses, IFNB1 mRNA abundance was measured in HCMV-infected cells treated with nonsilencing siRNA or siRNA specific for METTL14 or ALKBH5. While IFNB1 mRNA levels were significantly elevated in cultures treated with METTL14 siRNA relative to nonsilencing siRNA, ALKBH5 siRNA-treated cultures contained reduced IFNB1 mRNA (Fig. 2C). This suggested that greater HCMV reproduction in ALKBH5-depleted cells might result from less IFNβ production, while reduced virus replication in METTL14-depleted cultures resulted from greater IFNβ production. To test this hypothesis, the involvement of IFN signaling in the inhibition of HCMV replication by METTL14 depletion was examined. NHDFs treated with control nonsilencing siRNA or METTL14 siRNA were infected with HCMV, and virus replication was measured in the presence or absence of a Janus kinase (JAK) inhibitor, which inhibits JAK signaling downstream from the type I IFN receptor. While HCMV replication remained inhibited by METTL14 siRNA compared with control siRNA, JAK inhibitor treatment to a large extent restored HCMV reproduction in METTL14 siRNA-treated cultures to levels observed in cultures treated with control siRNA (Fig. 2D). In contrast, addition of JAK inhibitor to control siRNA-treated cultures resulted in a small, <10-fold increase (Fig. 2D). This indicates that restriction of HCMV reproduction by METTL14 siRNA likely resulted from IFNβ production and subsequent signaling dependent on JAK–STAT.
m6A modification enzymes and reader proteins control cell-intrinsic innate responses to dsDNA in uninfected cells
Since HCMV is a dsDNA virus and since dsDNA alone is sufficient to trigger IFNβ production (Chen et al. 2016), it was conceivable that the control of IFNβ production by m6A-modifying enzymes might not be restricted to virus-infected cells. To test this possibility, the capacity of cellular m6A writers and erasers to regulate innate IFNβ production in uninfected cells was evaluated. Addition of dsDNA effectively stimulated IFNB1 mRNA accumulation in nonsilencing siRNA-treated cultures (Fig. 3A). Unexpectedly, METTL14 depletion significantly enhanced IFNB1 mRNA accumulation, while ALKBH5 depletion effectively limited IFNB1 mRNA accumulation in uninfected cells (Fig. 3A). This suggested that IFNB1 mRNA might be m6A-modified. Indeed, compared with GAPDH and DICER, a cellular mRNA known to be m6A-modified, IFNB1 mRNA was substantially enriched in an RNA fraction immunoprecipitated from dsDNA-treated NHDFs using an m6A-specific antibody (Fig. 3B). Analysis of m6A-enriched RNA by RNA sequencing (RNA-seq) identified m6A peaks mapping over two discrete IFNB1 mRNA segments (Fig. 3C; Supplemental Table S1), each of which contains consensus m6A acceptor sites (Linder et al. 2015). While one cluster (supported by two out of three biological replicates) maps entirely within the IFNB1-coding sequences, the second (supported by three out of three biological replicates) includes the junction between the ORF and 3′ UTR (Fig. 3C). Others have similarly found methylated mRNA residues enriched within UTRs near stop codons (Dominissini et al. 2012; Meyer et al. 2012). Finally, depleting METTL14 or ALKBH5 did not detectably alter the responsiveness of target cells to exogenously added IFNβ, as measured by STAT Y701 phosphorylation, or prevent accumulation of a representative IFN-stimulated gene (ISG) (Supplemental Fig. S4). Instead, the observed changes in IFNB1 mRNA levels reflected differential production of functional IFNβ cytokine (Fig. 3D).
Figure 3.
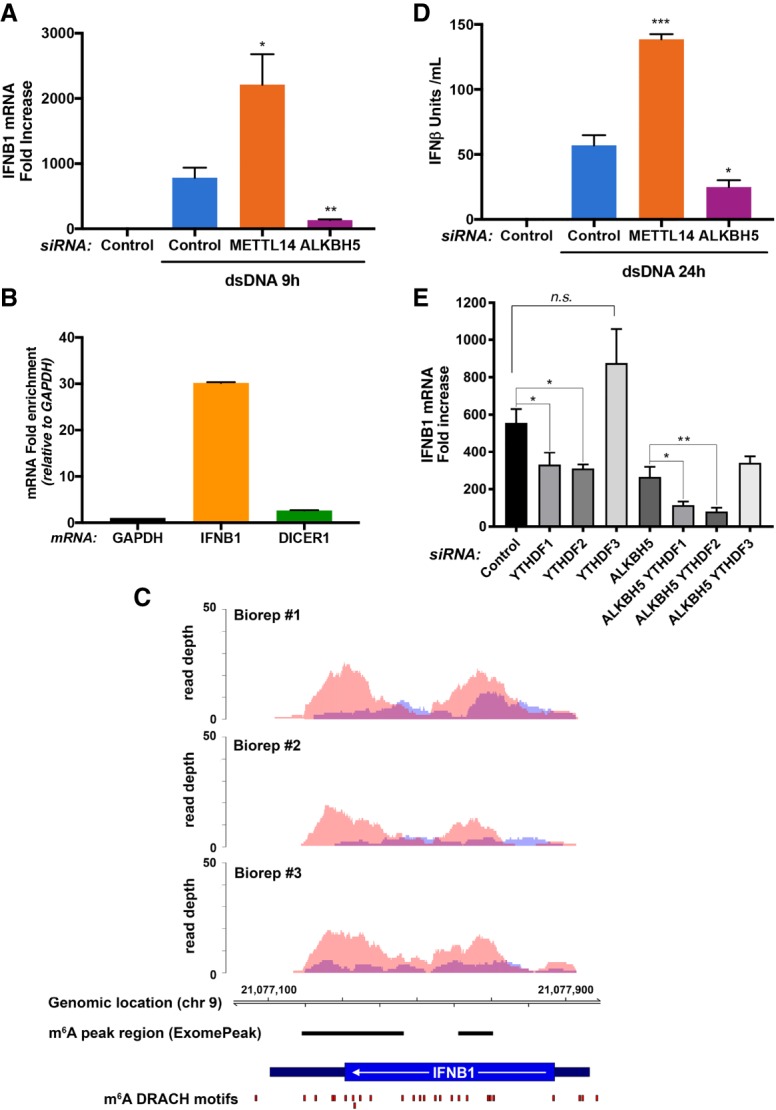
Induction of type I IFN in response to dsDNA is regulated by RNA m6A METTL3/14 methyltransferase and ALKBH5 demethylase. (A) NHDFs treated with control nonsilencing siRNA or siRNAs specific for METTL14 or ALKBH5 were incubated with H2O or dsDNA. After 9 h, total RNA was collected, and IFNB1 mRNA was measured by qRT–PCR. Error bars indicate SEM. (*) P ≤ 0.05; (**) P ≤ 0.01 by Student's t-test. (B) RNA isolated from NHDFs treated with dsDNA for 6 h was immunoprecipitated with anti-m6A antibody. RNA enriched in the immune complex was analyzed by qRT–PCR using primers specific for the indicated genes. (C) Two regions of the IFNB1 transcript are enriched for m6A peaks. Visualization of m6A sequencing (m6A-seq) results shows regions of enrichment for m6A immunoprecipitation (red) over input (blue) for three biological replicates of NHDFs transfected with dsDNA for 12 h. Specific regions enriched for m6A modifications were identified using ExomePeak (see the Supplemental Material) and are shown as horizontal black lines. m6A DRACH motifs (Linder et al. 2015) are shown as red boxes. No IFNB1 mapping reads were detected in control (no dsDNA) immunoprecipitation or input data sets (data not shown), consistent with the minimal background presence of IFNB1 transcripts in untreated cells. The transcript structure of IFNB1 is denoted in dark blue (3′ UTR and 5′ UTR) and blue (CDS). (D) NHDFs treated as in A were incubated with H2O or dsDNA. After 24 h, supernatants or known concentrations of IFNβ standards were placed on indicator cells, and the amount of IFNβ activity was quantified. Error bars indicate SEM. (*) P ≤ 0.05; (***) P ≤ 0.001 by Student's t-test. (E) NHDFs treated with control nonsilencing siRNA or the indicated siRNAs specific for the demethylase ALKBH5 and/or m6A readers YTHDF1, YTHDF2, and YTHDF3 were exposed to dsDNA. After 9 h, total RNA was collected, and IFNB1 mRNA was quantified by qRT–PCR. Error bars indicate SEM. (*) P ≤ 0.05; (**) P ≤ 0.01 by Student's t-test.
In addition to m6A writers and erasers, reader proteins that recognize m6A-modified RNA could potentially impact IFNβ production in response to dsDNA in uninfected cells. Compared with NHDFs treated with control nonsilencing siRNA, depleting m6A readers YTHDF1 or YTHDF2 reduced IFNB1 mRNA induction in response to dsDNA (Fig. 3E). In contrast, IFNB1 mRNA levels were not significantly different in response to dsDNA following m6A reader YTHDF3 depletion (Fig. 3E). This indicated that depleting YTHDF1 or YTHDF2 was similar to depleting the ALKBH5 demethylase, as both suppressed IFNB1 mRNA induction by dsDNA (Fig. 3E). Remarkably, codepletion of ALKBH5 together with YTHDF1 or YTHDF2 exacerbated the reduction in dsDNA-induced IFNB1 mRNA accumulation to levels below that observed when cells were singly depleted for either ALKBH5, YTHDF1, or YTHDF2 (Fig. 3E). This demonstrates a synthetic genetic interaction between ALKBH5 demethylase and the m6A readers YTHDF1 and YTHDF2. It further indicates that these two m6A readers limit the extent to which ALKBH5 depletion suppresses IFNβ production and is consistent with the m6A demethylase and readers acting in the same pathway to regulate IFNB1 mRNA levels.
Regulation of genome-wide responses to dsDNA by METTL14 and ALKBH5
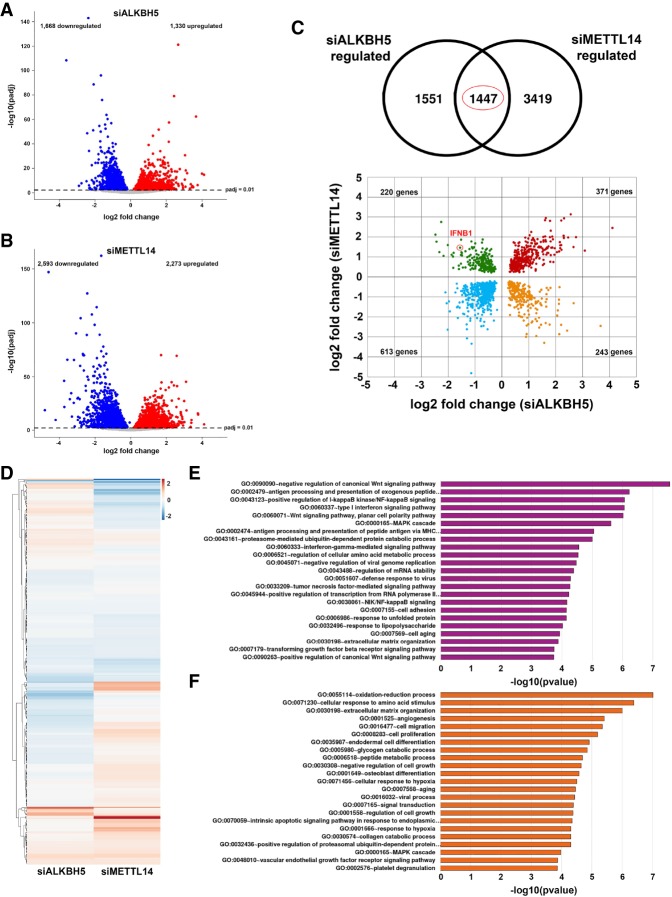
To determine the extent to which cellular m6A functions control genome-wide responses to dsDNA, total RNA isolated from primary NHDFs treated with nonsilencing siRNA or siRNA specific for METTL14 or ALKBH5 and exposed to dsDNA for 6 and 12 h was analyzed by stranded mRNA sequencing (mRNA-seq). Following normalization and classification of differentially regulated genes (adjusted P-value [Padj] of <0.01), 2998 genes proved responsive to ALKBH5 depletion, and 4866 genes were responsive to METTL14 depletion after 12 h, compared with nonsilencing siRNA-treated controls (Fig. 4A,B; Supplemental Fig. S5). A similar number of differentially expressed genes was observed in ALKBH5-depleted NHDFs treated with dsDNA for only 6 h (Supplemental Fig. S6). However, the overall number of differentially expressed genes in METTL14-depleted NHDFs exposed to dsDNA for 6 h was 30%–40% less than that observed after 12 h (Supplemental Fig. S6). The slower or delayed IFNB1 induction observed in METTL14-depleted cells (Supplemental Fig. S6), which conceivably extends to other genome-wide changes when the 6- and 12-h time points are compared, likely accounts for this finding. It further suggests that the differential gene expression phenotypes, including those controlling IFNβ production, occur more rapidly upon m6A eraser depletion than writer depletion.
Figure 4.
Control of genome-wide responses to dsDNA by m6A demethylase ALKBH5 and m6A methylase subunit METTL14. Volcano plots show differentially expressed genes (adjusted P-value < 0.01) identified from RNA-seq of polyadenylated RNA collected from cells transfected with ALKBH5 siRNA (siALKBH5; n = 3 biological replicates) (A) or METTL14 siRNA (siMETTL14; n = 3 biological replicates) (B) and stimulated with dsDNA for 12 h. Genes up-regulated versus a nonsilencing siRNA control (stimulated with dsDNA for 12 h; n = 3 biological replicates) are shown in red, while down-regulated genes are shown in blue. Nonregulated genes are shown in gray. (C) One-thousand-four-hundred-forty-seven significantly regulated genes (P < 0.01) overlap between data sets generated using siALKBH5 and siMETTL14. A scatter plot of these shows genes that are commonly up-regulated or down-regulated following either METTL14 or ALKBH5 silencing (highlighted in red and blue, respectively). Genes regulated in a reciprocal manner are highlighted in green (up-regulated when METTL14 is silenced and down-regulated when ALKBH5 is silenced) and yellow (down-regulated when METTL14 is silenced and up-regulated when ALKBH5 is silenced). The red circle highlights the IFNB1 mRNA. (D) Heat map depicting 349 ISGs, colored according to log2 fold change in expression versus the nonsilencing siRNA control. (E,F) Pathway analyses (gene ontology direct terms) of significantly differentially expressed genes from A and B were conducted using DAVID and filtered according to a Benjamini-Hochberg procedure (<0.05).
Compared with nonsilencing siRNA-treated cultures, interfering with the m6A writer subunit METTL14 or the eraser ALKBH5 had a complex impact on gene expression in response to dsDNA (Fig. 4A,B). A greater number of genes (3419 genes) was uniquely regulated by METTL14 depletion compared with ALKBH5 depletion (1551 genes) after dsDNA treatment for 12 h (Fig. 4C; Supplemental Table S2). In addition, 984 genes were similarly coregulated in the same direction by depleting either ALKBH5 or METTL14, while 463 genes were reciprocally regulated, including IFNB1 (Fig. 4C). IFN and ISGs figured prominently among those whose induction by dsDNA was impacted by interfering with ALKBH5 or METTL14 (Fig. 4D). Clustered heat maps revealed distinct signatures of IFNB1, IFNL1, and IL6 mRNA accumulation in NHDFs treated with nonsilencing siRNA versus siRNA specific for ALKBH5 or METTL14 in response to dsDNA (Supplemental Fig. S7). Furthermore, 4% of a defined collection of 349 ISGs (Schoggins et al. 2011) together with notable genes controlling innate immunity and inflammation, proliferation, and metabolic control (Supplemental Fig. S7) were up-regulated by METTL14 depletion and down-regulated by ALKBH5 depletion in response to dsDNA.
Top pathways enriched among genes differentially expressed upon ALKBH5 depletion in dsDNA-treated NHDFs included those controlling signaling by IFN, Wnt, NF-κB, TNF, TGFβ, and LPS, many of which impact infection biology and immune responses (Fig. 4E). Notably, ALKBH5 depletion also impacted mRNA biogenesis, including pathways controlling RNA polymerase II transcription and mRNA stability (Fig. 4E). Pathway analysis of 1668 genes down-regulated by ALKBH5 depletion included those controlling NF-κB activation, type I and type II IFN signaling, and antigen processing and presentation, all of which have documented antiviral roles (Supplemental Fig. S8). In contrast, top pathways enriched among genes differentially expressed upon METTL14 depletion in dsDNA-treated NHDFs included those involved in oxidation reduction, responses to amino acid stimulus, catabolic processes, hypoxia, aging, and ER stress (Fig. 4F). Pathway analysis of 2273 genes up-regulated by METTL14 depletion in dsDNA-treated NHDFs included those involved in cilium assembly, catabolic processes, and metabolic alterations (Supplemental Fig. S9), some of which are associated with quiescence or differentiation, senescence, and physiological stress (Favaro et al. 2012; Sánchez and Dynlacht 2016; Takahashi et al. 2018). Analysis of pathways down-regulated by METTL14 depletion included control of RNA polymerase II transcription; circadian regulation of gene expression; Wnt signaling; DNA replication; proliferation; responses to ER stress, LPS, TNF, and hypoxia; and mRNA stability (Supplemental Fig. S9). Overall, this analysis shows that primary human fibroblasts express unique ALKBH5- or METTL14-dependent gene signatures in response to dsDNA. Significantly, while many of the genes differentially expressed following depletion of the ALKBH5 m6A demethylase impact cell-intrinsic antiviral immune responses, depleting the METTL14 methylase subunit impacts pathways involved in metabolic reprogramming, responses to physiological stress, and aging. These changes likely allow rapid metabolic adaptation to a changing environment.
IFNB1 mRNA biogenesis and decay are regulated by m6A modification enzymes
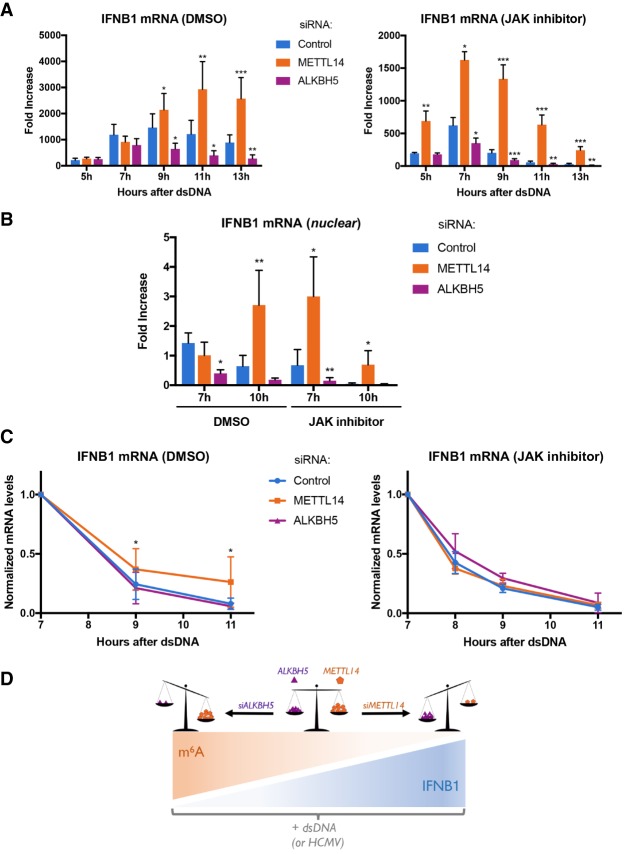
To more precisely understand how METTL14 and ALKBH5 control IFNβ production, we sought to define the underlying regulatory mechanisms. Treating NHDFs with dsDNA for 7 h did not significantly change the subcellular distribution or overall abundance of cellular m6A reader, writer, or eraser proteins (Supplemental Fig. S10A,B). In addition, METTL14 depletion did not appreciably stimulate—and ALKBH5 depletion did not detectably interfere with—NF-κB and IRF3 signaling, as measured by TBK or IRF3 phosphorylation, IκBα accumulation, and IRF3 nuclear accumulation (Supplemental Fig. S10C,D). This suggested that cellular m6A writers and erasers might be controlling IFNB1 mRNA metabolism, in particular mRNA biogenesis or decay of this unspliced cellular mRNA. To prevent secondary effects resulting from IFNβ cytokine signaling following IFNB1 mRNA production, we measured IFNB1 mRNA abundance in response to dsDNA in the presence and absence of JAK inhibitor. Figure 5A shows that compared with nonsilencing siRNA-treated NHDFs, METTL14-depleted cells accumulate greater levels of IFNB1 mRNA from 9 to 13 h after dsDNA exposure. The kinetics of IFNB1 mRNA accumulation were faster in JAK inhibitor-treated cells. In contrast, reduced amounts of IFNB1 mRNA accumulated in ALKBH5-depleted cells (Fig. 5A).
Figure 5.
Control of IFNB1 mRNA biogenesis and decay by ALKBH5 and METTL14. NHDFs were treated with control nonsilencing siRNA or siRNA specific for the METTL14 m6A methyltransferase subunit or the ALKBH5 demethylase for 72 h. (A) NHDFs treated with the indicated siRNAs were transfected with dsDNA in the presence of DMSO or the JAK inhibitor (pyridone 6). At the indicated times, total RNA was harvested, and the abundance of IFNB1 mRNA was quantified by qRT–PCR. Error bars indicate SEM. (*) P ≤ 0.05; (**) P ≤ 0.01; (***) P ≤ 0.001 by Student's t-test. (B) As in A except cultures were pulse-labeled for 30 min with 5-ethyluridine (EU) at either 7 or 10 h after exposure to dsDNA. Immediately following the EU pulse, nuclear RNA was collected, nascent EU-containing RNA was isolated, and the abundance of IFNB1 mRNA was quantified by qRT–PCR. Error bars indicate SEM. (*) P ≤ 0.05; (**) P ≤ 0.01 by Student's t-test. (C, left panel) NHDFs treated with the indicated siRNAs were exposed to dsDNA for 3 h and pulse-labeled for 2 h with EU. Free EU was washed out, and total RNA was harvested at the indicated time points. Following isolation of nascent EU-containing RNA, overall IFNB1 mRNA levels were measured by qRT–PCR and normalized to GAPDH. Error bars indicate SEM. (*) P < 0.04 by Student's t-test. (Right panel) As in the left panel, except cultures were also treated with the JAK inhibitor (pyridone 6). (D) Model illustrating the relationship between IFNB1 mRNA levels, induced in response to dsDNA in uninfected cells or HCMV infection, and m6A levels in mRNA. The balance between methyltransferase METTL14 and demethylase ALKBH5 activities is depicted as a scale. METTL14 depletion (siMETTL14) tips the scale in favor of the ALKBH5 demethylase, which increases IFNB1 mRNA levels and is predicted to reduce m6A levels. In contrast, ALKBH5 depletion (siALKBH5) disrupts the balance in favor of methyltransferase activity, which decreases IFNB1 mRNA and is predicted to increase m6A levels.
To investigate how cellular m6A writers and erasers influenced mRNA biogenesis, nascent RNA metabolically pulse-labeled with 5-ethyluridine (EU) for 30 min was isolated from nuclei of dsDNA-treated cells to limit the contribution of cytoplasmic mRNA decay. Figure 5B shows that EU-labeled IFNB1 mRNA levels in METTL14-depleted NHDFs increased compared with nonsilencing siRNA-treated controls between 7 and 10 h after dsDNA treatment. In NHDFs treated with JAK inhibitor, a greater increase in EU-labeled IFNB1 mRNA in METTL14-depleted cultures compared with nonsilencing siRNA-treated controls was observed earlier, after only 7 h of dsDNA exposure. This indicated that the kinetics of IFNB1 mRNA production respond to IFN receptor signaling. Conversely, nuclei isolated from ALKBH5-depleted cultures contained less nascent EU-labeled IFNB1 mRNA compared with nonsilencing siRNA-treated cultures (Fig. 5B).
To determine whether METTL14 or ALKBH5 also influenced IFNB1 mRNA decay, total EU pulse-labeled RNA was isolated from NHDFs treated with dsDNA, and IFNB1 mRNA levels were measured by quantitative RT–PCR (qRT–PCR). While decay of EU-labeled nascent IFNB1 mRNA was indistinguishable in nonsilencing siRNA or ALKBH5-depleted cultures, EU-labeled IFNB1 mRNA persisted longer in METTL14-depleted cultures (Fig. 5C). By 11 h after dsDNA treatment, <10% nascent IFNB1 mRNA remained in nonsilencing or ALKBH5-depleted cultures, whereas nearly 30% was detected in METTL14-depleted NHDFs. Unexpectedly, the greater persistence of IFNB1 mRNA in METTL14-depleted NHDFs was dependent on JAK signaling (Fig. 5C), consistent with an ISG selectively influencing the decay of the hypo-m6A methylated IFNB1 mRNA population. Thus, the burst of IFNB1 mRNA biogenesis and decay in response to dsDNA is regulated by cellular functions that control m6A modification of mRNA. In addition, ALKBH5 and METTL14 had a greater impact on IFNB1 mRNA production than decay.
Discussion
Despite its pervasiveness as an internal base modification in cellular and viral mRNAs, how m6A modification of mRNAs impacts infection biology and host responses is incompletely understood. Here, we show host enzymes that install and remove m6A increase in abundance in response to HCMV infection and regulate virus replication. While depleting the ALKBH5 demethylase stimulated viral protein accumulation and HCMV reproduction, interfering with the METTL3/14 methylase had the opposite effect. The impact of the cellular m6A modification machinery on HCMV growth resulted from differential IFNβ production, as IFNB1 mRNA accumulation was stimulated by METTL14 depletion and restricted by ALKBH5 depletion (Fig. 5D). Moreover, IFNB1 production in uninfected cells treated with dsDNA was likewise regulated by METTL14 and ALKBH5, and IFNB1 mRNA was enriched in an m6A RNA-containing fraction. Finally, genome-wide profiling of cells exposed to dsDNA following ALKBH5 depletion identified differentially expressed genes regulating antiviral immune responses, while METTL14 depletion altered pathways involving metabolic reprogramming, stress responses, and aging. Thus, by investigating how the METTL14 m6A methylase subunit and the ALKBH5 demethylase control HCMV replication, we uncovered a fundamental mechanism whereby host cell-intrinsic immune responses to dsDNA in uninfected cells are epitranscriptomically controlled by m6A RNA modification.
While cellular m6A modification enzymes control productive replication of the β-herpesvirus HCMV by influencing IFNβ production, effects on HSV-1, an α-herpesvirus subfamily member, and the prototypical poxvirus vaccinia were not detected. Conceivably, this might reflect differences in cell-intrinsic antiviral responses. Notably, cellular protein synthesis proceeds in HCMV-infected cells, in marked contrast to the strong suppression of ongoing host protein synthesis observed in cells infected with HSV-1 or vaccinia that effectively limit ISG expression (Zhu et al. 1997; Ishikawa et al. 2009). Replication of other viruses, including HIV and influenza A virus, is stimulated by the METTL3/14 m6A writer complex and may also in part reflect reduced IFNβ induction (Courtney et al. 2017).
Our data indicate that IFNB1 mRNA is enriched in an m6A-containing mRNA fraction and that its accumulation is controlled by the methyltransferase subunit METTL14 and the demethylase ALKBH5. Depleting METTL14 increased IFNB1 mRNA stability, which is known to be controlled by ELAVL1/HuR binding to AU-rich sequences in the 3′ UTR. Nucleic acid-sensing pathways are thought to control ELAVL1/HuR activity (Herdy et al. 2015; Takeuchi 2015), potentially explaining why IFNB1 mRNA stability was enhanced by METTL14 depletion in a manner dependent on IFN signaling (Fig. 5C). The impact of METTL14 on IFNB1 mRNA decay is consistent with the reported role for m6A in genome-wide mRNA stability (Wang et al. 2014; Ke et al. 2017). One important difference is that, unlike the majority of cellular mRNAs, which are spliced, IFNB1 mRNA does not contain an intron.
Unexpectedly, METTL14 depletion also stimulated EU incorporation into nascent nuclear IFNB1 RNA following dsDNA treatment, whereas ALKBH5 depletion repressed it (Fig. 5B). In addition, the overall magnitude of differences in EU incorporation into nascent RNA greatly exceeded any differences detected in IFNB1 mRNA stability (Fig. 5, cf. B and C). This suggests that ALKBH5 and METTL14 have a greater impact on IFNB1 mRNA production than decay. As differential IRF3 or NF-κB activation or the subcellular distribution of the cellular m6A machinery could not account for the observed changes in IFNB1 mRNA abundance, limiting m6A installation might stimulate mRNA biogenesis at the level of transcription. This could occur upstream of IFNB1 mRNA transcription. Indeed, pathway analysis of ALKBH5- and METTL14-depleted NHDFs treated with dsDNA implicates biological processes, including mRNA biogenesis and decay, as potential effectors shaping genome-wide responses that impact subsequent IFNB1 mRNA biogenesis. (Fig. 4; Supplemental Figs. S4, S6, S7). Alternatively, cotranscriptional deposition of m6A onto nascent IFNB1 mRNA might directly influence transcription initiation or elongation. Whether one or both of these possibilities prevails and whether effects of METTL14 and ALKBH5 on transcription are limited to rare m6A-containing mRNAs lacking introns such as IFNB1 require further investigation.
Genome-wide profiling data from uninfected NHDFs revealed that responses to dsDNA exposure are regulated by METTL14 and ALKBH5. In particular, opposing reciprocal effects on expression of 463 genes, including IFNB1, were observed and accounted for ∼32% of genes regulated by both METTL14 and ALKBH5 depletion in response to dsDNA. In contrast, the effects of ALKBH5 or METTL14 depletion were not entirely reciprocal genome-wide. Sixty-eight percent of genes responsive to both ALKBH5 and METTL14 depletion were coregulated in the same direction. In addition to targeting an identical cohort of coregulated and reciprocally regulated genes upon dsDNA treatment, METTL14 and ALKBH5 also target discrete gene sets with little overlap. This indicates that while ALKBH5 and METTL14 regulate innate immune responses via IFNB1 mRNA induction, they largely control different, nonoverlapping biological processes. How m6A modification enzymes might impact these diverse processes that respond to dsDNA sensing is in need of additional study.
In addition to infection biology, roles for cytoplasmic dsDNA signaling from nonmicrobial sources have emerged. The cytoplasmic DNA sensor cGAS controls senescence, and senescent cells express greater levels of IFNβ and proinflammatory cytokines as part of a senescence-associated secretory phenotype (SASP) (Coppé et al. 2008; Li and Chen 2018; Takahashi et al. 2018). By controlling IFNβ, cellular m6A modification enzymes might also impact aging, senescence, and the pathogenic consequences of overcoming senescence associated with cancer. As cytoplasmic dsDNA signaling is also a trigger for inflammatory diseases, epitranscriptomic regulation of responses triggered by dsDNA could broadly impact pathophysiology associated with systemic lupus erythematosus, rheumatoid arthritis, chronic inflammation, and interferonopathies (Gao et al. 2015; Chen et al. 2016; Crowl et al. 2017; Yan 2017; Li et al. 2018). With this in mind, developing specific inhibitors of the ALKBH5 demethylase could provide a new strategy to limit activation of the type I IFN pathway and a potential therapeutic opportunity to treat autoimmune diseases caused by self DNA.
Materials and methods
Cells, viruses, and chemicals
NHDFs (Lonza, CC-2509) were cultured in Dulbecco's modified Eagle's medium (DMEM; Corning, 10-013-CV) supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, and 5% (v/v) fetal bovine serum (FBS; Invitrogen). Vero cells (American Type Culture Collection) were propagated in DMEM supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, and 5% (v/v) calf serum (FBS; Invitrogen). BSC40 cells were maintained in DMEM supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, and 10% (v/v) FBS. HCMV AD169GFP was kindly provided by Dong Yu and was propagated in NHDFs as described (Bianco and Mohr 2017). Vaccinia virus (Western Reserve strain) was propagated in BSC40 cells as described (Burgess and Mohr 2015). Wild-type HSV-1 (Patton strain) expressing an EGFP-Us11 fusion protein (Benboudjema et al. 2003) was propagated in Vero cells. PP242 (P0037) was purchased from Sigma and used at a concentration of 25 µM.
DNA transfections
DNA was transfected using TransIT-X2 (Mirus, MIR6000) according to the manufacturer's protocol. VACV 70-mer (dsDNA) oligonucleotides were described previously (Unterholzner et al. 2010). Annealing buffer (10 mM Tris-HCl at pH 7.5, 100 mM NaCl, 1 mM EDTA) without nucleic acids was used as negative control.
m6A RNA immunoprecipitation (RIP)
Following poly-A+ selection, ∼1 μg of purified polyadenylated RNA resuspended in IPP buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl at pH 7.5) was denatured for 2 min at 70°C and subsequently placed on ice. Twenty-five microliters of protein-G magnetic beads (Life Technologies, 10004D) washed and suspended in IPP buffer containing RNase inhibitor (Invitrogen, 10777019) were incubated with 1 μg of monoclonal anti-m6A antibody for 15 min at room temperature with end-over-end rotation. RNA was added to the antibody-bound beads and incubated for 2 h at 4°C with end-over-end rotation. RNA-bound beads were washed twice in 200 μL of IPP buffer, twice in low-salt IPP buffer (50 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl at pH 7.5), twice in high-salt IPP buffer (500 mM NaCl, 0.1% NP-40, 10 mM Tris-HCl at pH 7.5), and twice again in 200 μL of IPP buffer and eluted in 30 μL of RLT (Qiagen, 79216). To purify the RNA, 20 μL of MyOne Silane Dynabeads (Life Technologies, 37002D) was washed with 100 μL of RLT, resuspended in 30 μL of RLT, and added to the eluted RNA. After addition of 60 μL of 100% ethanol, the beads were collected magnetically, and the supernatant was discarded. The beads were washed twice in 100 μL of 70% ethanol, and the RNA was eluted in 160 μL of IPP buffer. A second round of immunoprecipitation was performed by incubating the RNA with protein-A magnetic beads (Life Technologies, 10002D) coupled to anti-m6A antibody followed by washes, elution from the protein-A beads, and purification as described above. The final RNA elution from the MyOne Silane Dynabeads was performed in 16 μL of H2O followed immediately by cDNA synthesis.
Isolation of nuclear RNA
RNA from a nuclear fraction was isolated according to a protocol adapted from a published procedure (Wuarin and Schibler 1994). Briefly, NHDFs were washed with PBS, scraped into cold lysis buffer (10 mM Tris-HCl at pH 7.5, 0.15% NP40, 150 mM NaCl), and incubated for 5 min on ice. The lysate was then transferred onto 2.5 vol of a chilled sucrose cushion (ice-cold sucrose buffer [10 mM Tris-HCl at pH 7.5, 150 mM NaCl, 24% sucrose]) and centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant (cytoplasmic fraction) was collected for RNA extraction by TRIzol. The pellet was resuspended in cytoplasmic lysis buffer without NP40 and passed through a fresh sucrose cushion for a second time. The washed nuclear fraction was then dissolved in TRIzol for RNA extraction.
Analysis of nascent RNA synthesis and decay
Transcription and stability of newly synthesized RNA were analyzed using the Click-iT nascent RNA capture kit (Life Technologies, C10365). To measure RNA decay, siRNA-treated NHDFs were stimulated with dsDNA. After 3 h, 0.2 mM EU was added. Following a 2-h incubation with EU, the cells were washed with PBS, and EU-free medium was added. Cells were harvested, and RNA was isolated with TRIzol at the indicated time points after the addition of EU-free medium. The EU-labeled RNAs were biotinylated and captured by using the Click-iT nascent RNA capture kit, according to the manufacturer's instructions. To measure EU incorporation into newly synthesized nuclear RNA, siRNA-treated NHDFs stimulated with dsDNA were exposed to a 30-min EU pulse at the indicated times after DNA addition. Following EU exposure, cells were washed with PBS, and nuclear RNA was isolated and processed as described above.
Library preparation and sequencing
Illumina TruSeq stranded RNA libraries were prepared from 500 ng of poly(A)-selected total RNA (RNA integrity [RIN] score of 10) by staff at the New York University Genome Technology Center (https://med.nyu.edu/research/scientific-cores-shared-resources/genome-technology-center) and sequenced across two PE150 cycle lanes of a HiSeq 4000, yielding between 20,227,308 and 28,768,351 paired-end reads per sample (Supplemental Table S3).
For m6A RNA-seq, RNA fragments with m6A modifications were captured by RIP as described above except that a polyclonal m6A antibody was used (Dominissini et al. 2013). m6A sequencing (m6A-seq) immunoprecipitation and input libraries were prepared from 5–10 ng of RNA using the NEBNext Ultra II RNA library preparation kit (New England Biolabs) following the FFPE entry protocol. Twelve libraries were multiplexed and sequenced on an Illumina NextSeq 500 using a single 75-cycle high-output kit version 2 (single-end read mode), yielding 27,000,000–42,000,000 single-end reads per sample (Supplemental Table S4). Analysis of m6A-seq is described in the Supplemental Material.
RNA-seq analysis
Sequence reads were pseudoaligned against a Homo sapiens transcriptome database comprising the latest cDNA and noncoding RNA databases from Ensembl (https://www.ensembl.org/info/data/ftp/index.html) using Kallisto (Bray et al. 2016) under default parameters. Raw transcript counts were parsed to generate raw gene counts and analyzed using the DESeq2 version 1.18.1 (Love et al. 2014) package in R. Two samples (siAlk-12h-2 and siL14-6h-dsDNA-2) were excluded from analyses based on PCA clustering and low total read counts, respectively (Supplemental Fig. S3). Pairwise contrasts were performed between sample sets for each condition (12 h, no dsDNA/6 h + dsDNA/12 h + dsDNA) and filtered to retain only differentially expressed genes with Padj of <0.01. Pathway analyses for resulting gene lists were performed using the “Functional Annotation Tool” hosted by the David Bioinformatics Research 6.8 platform (Huang et al. 2009a,b).
Data availability
All sequencing data generated during this study are available from the sequence read archive (SRA) under the BioProject ID PRJNA451188.
Supplementary Material
Acknowledgments
We thank members of the Mohr laboratory, Joel Belasco, and Angus Wilson for helpful discussions. This work was supported by National Institutes of Health grants GM056927 and AI073898 to I.M. L.T. was supported in part by Public Health Service Institutional Research Training Award AI07647.
Authors contributions: R.M.R., D.P.D., C.B., L.T., and I.M. conceived experiments and interpreted data. R.M.R., D.P.D., C.B., and L.T. performed the experiments. I.M., R.M.R., D.P.D., and C.B. wrote and edited the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.319475.118.
References
- Aguilo F, Zhang F, Sancho A, Fidalgo M, Di Cecilia S, Vashisht A, Lee DF, Chen CH, Rengasamy M, Andino B, et al. 2015. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell 17: 689–704. 10.1016/j.stem.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benboudjema L, Mulvey M, Gao Y, Pimplikar SW, Mohr I. 2003. Association of the herpes simplex virus type 1 Us11 gene product with the cellular kinesin light-chain related protein PAT1 results in the redistribution of both polypeptides. J Virol 77: 9192–9203. 10.1128/JVI.77.17.9192-9203.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Mohr I. 2017. Restriction of human cytomegalovirus replication by ISG15, a host effector regulated by cGAS–STING double-stranded-DNA sensing. J Virol 91: e02483-16 10.1128/JVI.02483-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckh M, Geballe AP. 2011. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest 121: 1673–1680. 10.1172/JCI45449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. In Human cytomegalovirus (ed. Shenk TE, Stinski MF), pp. 417–470. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Brocard M, Ruggieri A, Locker N. 2017. m6A RNA methylation, a new hallmark in virus-host interactions. J Gen Virol 98: 2207–2214. 10.1099/jgv.0.000910 [DOI] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. 2015. Cellular 5′–3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17: 332–344. 10.1016/j.chom.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon MJ, Schmid DS, Hyde TB. 2010. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20: 202–213. 10.1002/rmv.655 [DOI] [PubMed] [Google Scholar]
- Chen CA, Shyu AB. 2017. Emerging themes in regulation of global mRNA turnover in cis. Trends Biochem Sci 42: 16–27. 10.1016/j.tibs.2016.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol 17: 1142–1149. 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- Coots RA, Liu XM, Mao Y, Dong L, Zhou J, Wan J, Zhang X, Qian SB. 2017. m6A facilitates eIF4F-independent mRNA translation. Mol Cell 68: 504–514.e7. 10.1016/j.molcel.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J-P, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez P-Y, Campisi J. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: e301 10.1371/journal.pbio.0060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR. 2017. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22: 377–386.e5. 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. 2017. Intracellular nucleic acid detection in autoimmunity. Annu Rev Immunol 35: 313–336. 10.1146/annurev-immunol-051116-052331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. 2013. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8: 176–189. 10.1038/nprot.2012.148 [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L. 2016. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat Commun 7: 12626 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen PWTC, Rossa M, et al. 2017. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 24: 870–878. 10.1038/nsmb.3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro E, Bensaad K, Chong MG, Tennant DA, Ferguson DJP, Snell C, Steers G, Turley H, Li J-L, Gunther UL, et al. 2012. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab 16: 751–764. 10.1016/j.cmet.2012.10.017 [DOI] [PubMed] [Google Scholar]
- Fustin J.-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155: 793–806. 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, Chen ZJ. 2015. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci 112: E5699– E5705. 10.1073/pnas.1516465112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre AB, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. 2016. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20: 654–665. 10.1016/j.chom.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-van Horn SR, Sarnow P. 2017. Making the mark: the role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21: 661–669. 10.1016/j.chom.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M. 2016. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540: 301–304. 10.1038/nature20577 [DOI] [PubMed] [Google Scholar]
- Herdy BKT, Vladimer GI, Tan CSH, Stukalov A, Trefzer C, Bigenzahn J. 2015. The RNA-binding protein HuR/ELAVL1 regulates IFN-β mRNA abundance and the type I IFN response. Eur J Immunol 45: 1500–1511. 10.1002/eji.201444979 [DOI] [PubMed] [Google Scholar]
- Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. 2018. N6-methyl-adenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog 14: e1006995 10.1371/journal.ppat.1006995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792. 10.1038/nature08476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones SY, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr, Darnell RB. 2017. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006. 10.1101/gad.301036.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AVR, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. 2016. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19: 675–685. 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavi S, Shatkin AJ. 1975. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc Natl Acad Sci 72: 2012–2016. 10.1073/pnas.72.6.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen ZJ. 2018. The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215: 1287–1299. 10.1084/jem.20180139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LJ, Fan YG, Leng RX, Pan HF, Ye DQ. 2018. Potential link between m6A modification and systemic lupus erythematosus. Mol Immunol 93: 55–63. 10.1016/j.molimm.2017.11.009 [DOI] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016a. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1: 16011 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. 2016b. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20: 666–673. 10.1016/j.chom.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. 2015. N6-methyl-adenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518: 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. 2017. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45: 6051–6063. 10.1093/nar/gkx141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman P, Hakki M, Boeckh M. 2010. Cytomegalovirus in hematopoietic stem cell transplant recipients. Infect Dis Clin North Am 24: 319–337. 10.1016/j.idc.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The ‘silent’ global burden of congenital cytomegalovirus. Clin Microbiol Rev 26: 86–102. 10.1128/CMR.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. 2017. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541: 371–375. 10.1038/nature21022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Perez C, Mohr I. 2012. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci 109: 5627–5632. 10.1073/pnas.1202829109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, Mohr I. 2014. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep 6: 9–17. 10.1016/j.celrep.2013.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, Sanderson J, Li M, Usher S, Sellayah D, Ashcroft FM, et al. 2015. FTO influences adipogenesis by regulating mitotic clonal expansion. Nat Commun 6: 6792 10.1038/ncomms7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B, Gershowitz A, Stringer JR, Holland LE, Wagner EK. 1977. 5′-terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J Virol 23: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Pickering BF, Jaffrey SR. 2018. Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol 28: 113–127. 10.1016/j.tcb.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable RR, Humar A; AST Infectious Diseases Community of Practice. 2013. Cytomegalovirus in solid organ transplantation. Am J Transplant 13: 93–106. 10.1111/ajt.12103 [DOI] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. 2015. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc 137: 2107–2115. 10.1021/ja513080v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez I, Dynlacht BD. 2016. Cilium assembly and disassembly. Nat Cell Biol 18: 711–717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472: 481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Salditt-Georgieff M, Bachenheimer S, Darnell JE, Furuichi Y, Morgan M, Shatkin AJ. 1976. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res 3: 749–765. 10.1093/nar/3.3.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Loo TM, Okada R, Kamachi F, Watanabe Y, Wakita M, Watanabe S, Kawamoto S, Miyata K, Barber GN, et al. 2018. Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat Commun 9: 1249 10.1038/s41467-018-03555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O. 2015. HuR keeps interferon-β mRNA stable. Eur J Immunol 45: 1296–1299. 10.1002/eji.201545616 [DOI] [PubMed] [Google Scholar]
- Tan B, Liu H, Zhang S, da Silva SR, Zhang L, Meng J, Cui X, Yuan H, Sorel O, Zhang SW, et al. 2018. Viral and cellular N6-methyladenosine and N6,2′-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat Microbiol 3: 108–120. 10.1038/s41564-017-0056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh O, Cohen Y, Shitrit A, Shani O, Le-Trilling VTK, Trilling M, Friedlander G, Tanenbaum M, Stern-Ginossar N. 2015. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog 11: e1005288 10.1371/journal.ppat.1005288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5: e15528 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Cullen BR. 2018. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog 14: e1006919 10.1371/journal.ppat.1006919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11: 997–1004. 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. 2017. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23: 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, et al. 2018. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell 69: 1028–1038.e6. 10.1016/j.molcel.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. 2017. Regulation of m6A transcripts by the 3′ → 5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell 68: 374–387.e12. 10.1016/j.molcel.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol 14: 7219–7225. 10.1128/MCB.14.11.7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N. 2017. Immune diseases associated with TREX1 and STING dysfunction. J Interferon Cytokine Res 37: 198–206. 10.1089/jir.2016.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C. 2015. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29: 1343–1355. 10.1101/gad.262766.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng GQ, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Hou J, Zhou Y, Li Z, Cao X. 2017. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat Immunol 18: 1094–1103. 10.1038/ni.3830 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X, Zhang X, Hess ME, Brüning JC, Qian SB. 2018. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell 69: 636–647.e7. 10.1016/j.molcel.2018.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Cong JP, Shenk T. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci 94: 13985–13990. 10.1073/pnas.94.25.13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated during this study are available from the sequence read archive (SRA) under the BioProject ID PRJNA451188.