Summary
Generation of the “epitranscriptome” through post-transcriptional ribonucleoside modification embeds a layer of regulatory complexity into RNA structure and function. Here we describe N4-acetylcytidine (ac4C) as an mRNA modification that is catalyzed by the acetyltransferase NAT10. Transcriptome-wide mapping of ac4C revealed discretely acetylated regions that were enriched within coding sequences. Ablation of NAT10 reduced ac4C detection at the mapped mRNA sites and was globally associated with target mRNA down-regulation. Analysis of mRNA half-lives revealed a NAT10-dependent increase in stability in the cohort of acetylated mRNAs. mRNA acetylation was further demonstrated to enhance substrate translation in vitro and in vivo. Codon content analysis within ac4C peaks uncovered a biased representation of cytidine within wobble sites that was empirically determined to influence mRNA decoding efficiency. These findings expand the repertoire of mRNA modifications to include an acetylated residue and establish a role for ac4C in the regulation of mRNA translation.
In Brief (eTOC blurb)
Post-transcriptional acetylation of cytidines in mammalian mRNAs enhances RNA stability and translation.
Graphical Abstract
INTRODUCTION
Analogous to the widely studied epigenome, generation of the “epitranscriptome” through chemical modification of ribonucleosides expands the regulatory content intrinsic within messenger RNAs (mRNAs) (Roundtree et al., 2017). Occurring in all four nucleobases, over 140 ribonucleoside modifications have been reported in prokarya, archaea and eukarya (Boccaletto et al., 2018). While the multiplicity of modified residues in RNA implies regulatory potential, limited availability of reagents and poor mechanistic knowledge pose substantial obstacles to comprehensive surveys. To date, most studies have focused on detailed examination of abundant transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). In contrast, modifications within mRNAs remain poorly understood. Where examined, mRNA modifications have been demonstrated to influence posttranscriptional metabolism through the regulation of mRNA stability, processing and/or translation (Roundtree et al., 2017).
Focusing solely on cytidine, eleven base modifications have been detected in RNA, three of which are conserved in all domains of life: 5-methylcytidine (m5C), 5-hydroxymethylcytidine (hm5C) and N4-acetylcytidine (ac4C) (Boccaletto et al., 2018). Amongst these, direct analogs of m5C and hm5C are found in DNA and studies into their regulation, distribution and function in RNA have been facilitated through existing knowledge of DNA methylation (Delatte et al., 2016; Squires et al., 2012; Yang et al., 2017). In contrast, ac4C remains relatively unexplored. Initially described in the bacterial tRNAmet anticodon (Stern and Schulman, 1978), ac4C was subsequently detected in eukaryotic serine and leucine tRNAs and 18S rRNA (Boccaletto et al., 2018). In all cases, ac4C production has been catalyzed by the N-acetyltransferase 10 (NAT10) enzyme or its homologs (Chimnaronk et al., 2009; Ito et al., 2014; Sharma et al., 2015). Suggestive of a non-redundant relationship, ac4C is the sole acetylation event to have been described in eukaryotic RNA and NAT10 is the singular human enzyme to have both acetyltransferase and RNA binding activities (Figure S1A) (Ito et al., 2014). Recently, unbiased mass spectrometry (MS) studies raised the possibility that the NAT10/ac4C axis extends to polyadenylated (poly(A)) RNAs: proteomic characterization of the mRNA-interactome revealed NAT10 as a poly(A)-interacting factor, and ac4C was consistently detected in liquid chromatography (LC)-MS/MS of poly(A) RNA isolated from a variety of human cell types at a level comparable to the 5’ 7-methylguanosine (m7G) cap (Castello et al., 2012; Dong et al., 2016). Together, these results suggested that ac4C exists within mRNA at physiologically relevant levels.
Here, we utilize transcriptome-wide approaches to investigate ac4C localization and function in mRNA. We find that ac4C is widely distributed within the human transcriptome with a majority of sites occurring within coding sequences (CDS). Disruption of the NAT10 gene ablated ac4C detection at mapped mRNA sites and revealed a role for acetylation in promoting target gene expression through improved mRNA stability and translation. Bioinformatic analysis of codon composition within ac4C peaks further demonstrated a strong enrichment for cytidine specifically within wobble sites. Notably, ac4C results in increased thermal stability upon Watson-Crick base pairing with guanosine as compared to unmodified cytosine (Kumbhar et al., 2013) and may thus influence interaction with cognate tRNAs during translation. In support of such a role, translation of acetylated reporter mRNAs was robustly stimulated, both in vitro and in vivo, particularly when ac4C was present at wobble positions. In sum, we describe ac4C as a component of the epitranscriptome that functions in the regulation of mRNA expression through enhanced stability and translation, potentially at the level of decoding efficiency.
RESULTS
Ablation of ac4C through NAT10 disruption
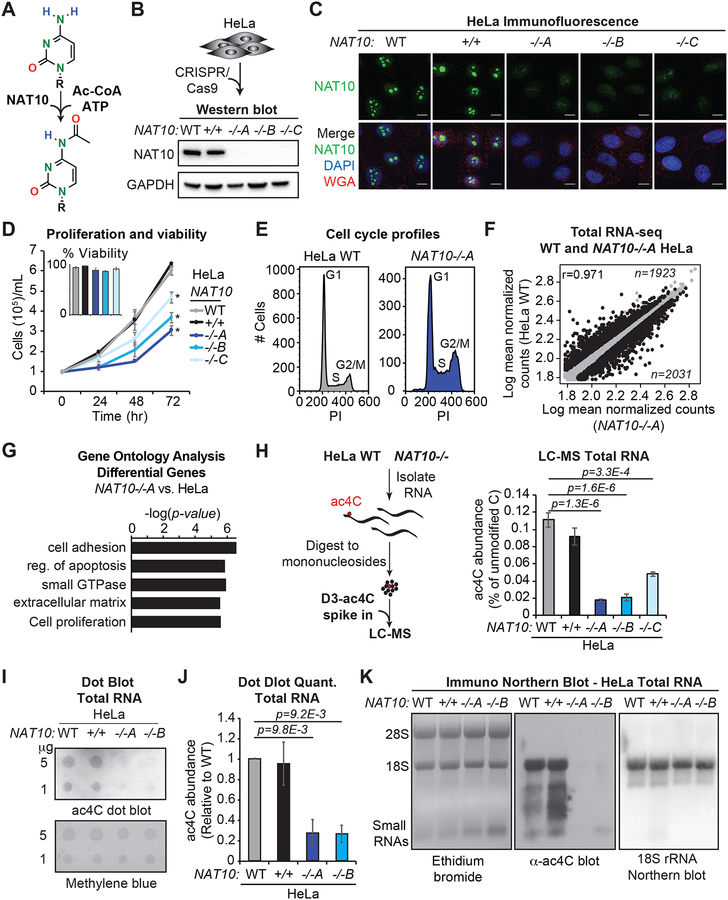
Analysis of RNA modifications can be hampered by poor knowledge or redundancy of the responsible enzyme(s). Based on the unique architecture of the NAT10 enzyme and the singular occurrence of acetylation within eukaryotic RNA, we hypothesized that NAT10 is the main source of ac4C in human cells (Figure 1A). To investigate this possibility, we genetically ablated NAT10 in HeLa cells. Of the 29 exons in NAT10, only exon 5 is utilized in all coding isoforms (Figure S1A). We thus pursued cleavage-directed frame shift mutations against NAT10 exon 5. Genomic sequencing revealed preferential selection for adenine insertion at one allele, coupled to distinct fates at the second allele (out-of-frame deletion, in-frame deletion and adenine insertion for NAT10−/− clones A-C, respectively) (Figure S1B). In addition, a HeLa clone that was transfected with guide RNA, but failed to show NAT10 mutation was selected as a NAT10+/+ control. Western blot and immunofluorescence in the CRISPR-Cas9 generated clones revealed effective loss of NAT10 expression (Figures 1B and 1C). However, consistent with its description as an essential gene in yeast (Sharma et al., 2015), minor residual protein was observed that was attributed to low efficiency skipping of exon 5 (Figures 1C and S1C). Characterization of NAT10−/− cells revealed high viability, but reduced proliferation kinetics and an increase in the fraction of cells in G2/M as compared to wildtype HeLa (Figures 1D, 1E, S1D and S1E). High-throughput sequencing of total RNA from wildtype and NAT10−/− HeLa cells showed strong concordance between replicates and identified 3954 protein coding genes that were differentially expressed upon NAT10 ablation (Figures 1F and S1F, Tables S1 and S2). Gene ontology (GO) analysis of the altered genes revealed a strong enrichment for cell survival and proliferation, providing a rationale for the observed phenotype (Figure 1G, Table S2).
Figure 1. NAT10-catalyzed ac4C in human HeLa cells.
(A) NAT10 catalyzes cytidine acetylation.
(B-C) NAT10−/− cells were generated through CRISPR/Cas9. Western blot and immunofluorescence (IF) for NAT10 in parental (WT) and targeted NAT10−/− cells. DAPI and wheat germ agglutinin (WGA) were used to mark the nucleus and cytoplasm, respectively, in IF.
(D) Cell proliferation was evaluated through trypan blue counting at the indicated times. Insert represents percent viability at 72 hrs. Mean ± SEM, n=4, Two-Way ANOVA.
(E) Representative of propidium iodide (PI) staining and flow cytometry for cell cycle analysis in NAT10−/−A and parental HeLa cells.
(F) Scatter plots for differentially expressed genes (black) in NAT10−/−A vs. parental HeLa cell RNA-seq (adjusted p-value < 0.05).
(G) GO enrichment on the subset of dysregulated genes from (F).
(H) Total RNA was digested to mononucleosides, spiked with D3-ac4C or 15N3-C and analyzed by LC-MS. Mean ± SEM, n=3, One-Way ANOVA with Tukey’s post hoc test.
(I) Representative anti-ac4C dot blot performed on total RNA with methylene blue staining as loading control.
(J) Densitometry quantitation of (I). Mean ± SEM, n=3, One-Way ANOVA with Tukey’s post hoc test.
(K) Anti-ac4C immuno-Northern blot in HeLa total RNA with ethidium bromide staining (left) and hybridization to 18S rRNA-specific probe (right) for general RNA visualization. Representative of biological triplicates.
To explore the involvement of NAT10 in catalyzing RNA acetylation in HeLa cells, we performed LC-MS for ac4C in total RNA. Relative quantification against isotopically labeled internal standards (D3-ac4C, 15N3-C) revealed a substantial loss of ac4C in total RNA from NAT10−/− clones, as compared to parental HeLa and the NAT10+/+ control (Figure 1H). Independently performed LC-MS/MS for quantification at attomole sensitivity (Basanta-Sanchez et al., 2016) indicated a similar fold reduction for NAT10−/− clone A as compared to parental HeLa (Figure S1G), thereby demonstrating the validity of the LC-MS approach. Based on these findings, NAT10−/− clones A and B, which yielded an ~80–90% reduction in ac4C (Figure 1H), were selected for downstream studies. Consistent with the LC-MS results, dot blot using monoclonal antibody against ac4C (Sinclair et al., 2017) (Figure S1H) demonstrated a near complete loss of signal in total RNA from clones A and B relative to control (Figures 1I and 1J). Likewise, anti-ac4C immuno-Northern blot showed abundant signal in regions corresponding to 18S rRNA and tRNA species that was ablated in the NAT10−/− clones (Figure 1K). Immuno-Northern additionally revealed a preponderance of NAT10-dependent acetylated species occurring in the size range expected to contain poly(A) RNAs, thus supporting the premise that NAT10 activity extends beyond rRNA and tRNA (Figure 1K). Importantly, re-expression of NAT10 cDNA in NAT10−/− clone A effectively reconstituted ac4C in total RNA thereby establishing NAT10 as a bona fide RNA acetyltransferase in HeLa cells (Figure S1I).
Of note, NAT10 was previously described as a protein acetyltransferase with demonstrated activities against α-tubulin, histones and p53 (Larrieu et al., 2014; Liu et al., 2016; Lv et al., 2003; Shen et al., 2009). In contrast to the dramatic reduction observed in ac4C levels, immunoblotting with acetyl-specific antibodies showed little change in protein acetylation in NAT10−/− as compared to wildtype HeLa cells (Figure S1J). Likewise, re-expression of NAT10 in clone A did not impact acetylation of established protein substrates (Figure S1K). To reconcile our results with previous reports, we explored generality through CRISPR-Cas9 directed NAT10 inactivation in Flp-In T-Rex 293 cells. As in HeLa, immunoblot revealed a considerable decrease in ac4C upon NAT10 ablation that was efficiently restored upon single copy integration of full-length NAT10 cDNA, but not NAT10 lacking the RNA helicase domain (Figures S1L and S1M). In contrast, acetylated α-tubulin levels remained stable upon NAT10 modulation (Figure S1N). Of relevance, initial investigations into NAT10 protein-acetyltransferase activity largely focused on an isoform that lacks the N-terminal RNA-interacting region (Lv et al., 2003; Shen et al., 2009), whereas HeLa and 293 cells uniformly express full-length NAT10. Together, these findings point to RNA as the preferred substrate for full-length NAT10 and suggest that NAT10 protein acetyltransferase activity is regulated in a cell-type specific manner through the production of isoforms that lack critical RNA binding determinants. Overall, these results solidify NAT10 as the principal source for RNA acetylation in human cells.
Detection of ac4C in poly(A) RNA
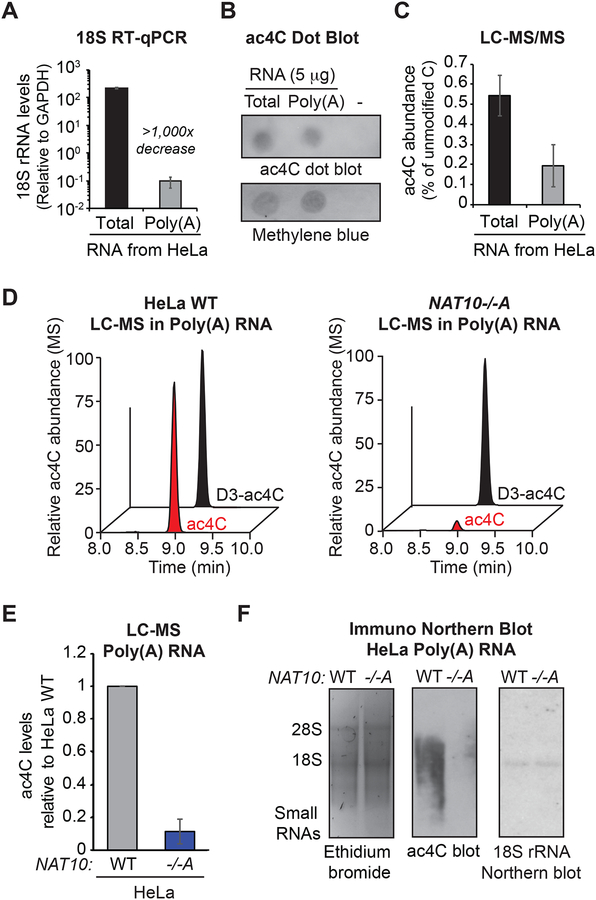
To gain direct evidence for ac4C within poly(A) RNA, oligo(dT)-purified RNA was isolated from parental HeLa cells for determination of ac4C levels. Poly(A) enrichment was confirmed through reduced RT-qPCR detection of 18S rRNA relative to total RNA (Figure 2A) and bioanalyzer analysis (Figure S2A). ac4C levels were examined in the purified poly(A) RNA through dot blot and LC-MS/MS. Both techniques detected substantial ac4C in the poly(A) RNA that was estimated to be ~40% the level in total RNA (Figures 2B, 2C and S2B). Given the near 1000× reduction in 18S rRNA relative to GAPDH mRNA in the poly(A) preparation (Figure 2A), identification of ac4C in HeLa poly(A) RNA is inconsistent with abundant rRNA contamination and instead points to a bona fide presence in poly(A) RNA. To further explore the dependency on NAT10, poly(A) samples from parental and NAT10−/− clone A were subjected to LC/MS with isotopically labeled internal standards (Figure 2D). Consistent with the extent of ablation in total RNA, ac4C levels in poly(A) RNA from clone A were reduced ~90% (Figure 2E). Finally, immuno-Northern blot in poly(A) RNA established that the ‘unknown’ signal observed in total RNA (Figure 1K) derives from polyadenylated species and confirmed NAT10-dependency through loss of signal in the NAT10−/− clones (Figure 2F). Importantly, Northern blot with probe against 18S rRNA shows that the observed ac4C smear does not emanate from contaminating 18S rRNA degradation products (Figure 2F). The sum of these analyses performed in HeLa RNA strongly support the presence of NAT10-dependent ac4C in poly(A) RNA.
Figure 2. ac4C detection in polyadenylated RNA.
(A) Determination of poly(A) RNA purity through RT-qPCR with primers specific to 18S rRNA and GAPDH. Mean ± SEM, n=3.
(B) Representative anti-ac4C dot blot performed on total and poly(A) RNA from (B).
(C) LC-MS/MS of total and poly(A) RNA from (B). Mean ± SEM relative to parental HeLa cells, n=3.
(D) Chromatograms of representative LC-MS performed in poly(A) RNA from NAT10−/−A and parental HeLa cells.
(E) Relative quantification of ac4C detection in poly(A) RNA LC-MS. Mean ± SEM relative to parental HeLa cells, n=3.
(F) Anti-ac4C immuno-Northern as in poly(A) RNA. Representative of biological triplicates.
ac4C mapping in poly(A) RNA
We next pursued transcriptome-wide mapping to solidify the occurrence of ac4C within protein-coding mRNAs (Figure 3A). Since RNA mapping strategies typically rely on conversion to cDNA and select modifications negatively impact this step (Hauenschild et al., 2015), we first assessed the behavior of acetylated RNA in reverse transcription. To generate substrates of known acetylation status, plasmid encoding mouse ß-globin RNA was in vitro transcribed in the presence of unmodified CTP or ac4CTP (Figure 3B). Stable ac4C incorporation was validated through dot blot with anti-ac4C antibody (Figure S1H). Reverse transcription of the in vitro transcribed probes using gene-specific radiolabeled primer showed efficient generation of full-length cDNA from both the acetylated and unmodified probes, thus confirming that ac4C is amenable to cDNA-based sequencing methods (Figure 3B).
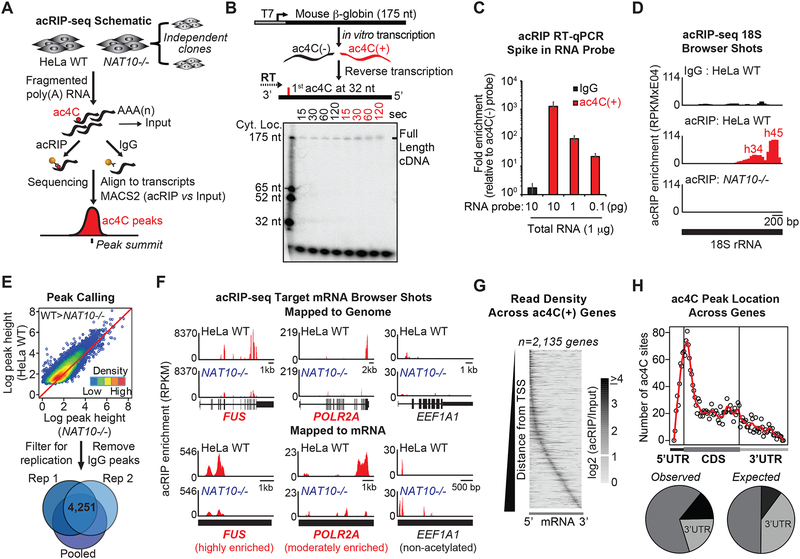
Figure 3. Transcriptome-wide mapping of ac4C in mRNA.
(A) Schematic of acRIP-seq.
(B) ac4C(+) or C-RNA templates were reversed transcribed using 32P-labeled primers. Ladder represents positions of specific cytidines within the probe.
(C) ac4C(+) or C-RNA probes were spiked into total RNA followed by acRIP-RT-qPCR. ac4CRNA levels are represented relative to C-RNA. Mean ± SEM, n=3.
(D) Input-subtracted RPKM browser views of 18S rRNA acRIP-seq reads.
(E) Acetylated regions were defined through acRIP summits displaying higher pileup values in parental (WT) relative to NAT10−/− HeLa, followed by filtering for IgG overlap and experimental replication.
(F) Input-subtracted RPKM browser views of ac4C peaks in highly (FUS) and moderately enriched (POLR2A) ac4C targets, as well as a non-acetylated control (EEF1A1), mapped to the human reference genome or to mRNA sequence, as indicated.
(G) Grayscale heatmap of acRIP-seq positional enrichment within transcripts. Each row represents a gene and columns represent percentiles of gene length. Genes are ordered by increasing distance of the maximum enrichment from the transcription start of the canonical transcript.
(H) Number of ac4C summits parsed by location within CDS or UTRs for all acetylated transcripts (top). Pie charts indicating percentage of summits within CDS or UTRs in the acetylated transcripts (observed) compared to the expected percentage based on the length of each feature (expected) (bottom).
The in vitro transcribed probes further facilitated examination of ac4C IP efficiency. Acetylated and unmodified ß-globin probes were spiked into HeLa total RNA at varying concentrations and RNA immunoprecipitation was performed with anti-ac4C antibody (acRIP). Subsequent RT-qPCR confirmed linear recovery of acetylated ß-globin RNA (Figure 3C). To additionally assess acRIP functionality within the complex modification landscape of cellular RNAs, we investigated 18S rRNA recovery in parental vs. NAT10−/− HeLa cells. Human 18S rRNA possesses two acetylated sites, existing in helices 34 and 45, the latter of which occurs at near 100% stoichiometry (Taoka et al., 2018). Accordingly, acRIP in fragmented total RNA and RT-qPCR directed near helix 45 showed strong ablation of 18S recovery in NAT10−/− cells as compared to control (Figure S3A). In contrast, the abundant non-acetylated 28S and 5S rRNAs were not recovered (Figure S3A). Given that 28S rRNA directly base pairs with 18S rRNA in vivo (Khatter et al., 2015), this demonstration establishes that downstream results are not an artifact of 18S rRNA co-purification. Together, these findings support the technical feasibility of specific recovery of acetylated mRNAs through antibody-based RNA IP and subsequent mapping through cDNA-based methodologies.
To assess for the occurrence of NAT10-regulated ac4C targets within mRNA, we coupled acRIP and next-generation sequencing (acRIP-seq) (Figure 3A, Table S1). IP was performed in duplicate in fragmented poly(A) RNA from parental HeLa cells and two NAT10−/− clones, followed by library construction and sequencing. acRIP-seq reads were mapped to a human reference genome to identify regions of enrichment relative to input and IgG control IP, and to a reference transcriptome to facilitate discrete peak calling across exon junctions (Figure 3A, Table S3). Acetylated ß-globin spike-in recovery validated IP efficiency across samples (Figure S3B). Importantly, residual 18S rRNA was effectively enriched through acRIP in parental, but not in NAT10−/− HeLa cells (Figure 3D). Furthermore, regions of enrichment directly mapped to known 18S acetylation sites in helices 34 and 45 with peak heights roughly reflective of the respective stoichiometries (Taoka et al., 2018). In contrast, specific enrichment of non-acetylated 28S rRNA was not observed when compared to IgG (Figure S3C). Bolstered by these results, we defined acetylated peaks in mRNA based on quantitative enrichment in parental relative to NAT10−/− acRIP, with no evidence of an overlapping peak in the IgG IP. After filtering for replication, a total of 4,250 candidate ac4C peaks were identified (Figure 3E). Examination of peak distributions across transcripts revealed that the majority of acetylated genes possess 1–2 ac4C peaks (Figure S3D). Representative browser shots depicting genome and transcriptome alignments of highly and moderately enriched targets, as well as a non-enriched control (FUS, POLR2A and EEF1A1, respectively) show discrete peaks in the ac4C(+) mRNAs in parental HeLa cells that were substantially ablated in the absence of NAT10 (Figure 3F, Table S3). The attenuated signal in the NAT10−/− clones is consistent with the minor residual ac4C and NAT10 observed during clone validation (Figure 1). RT-qPCR validation of select targets supports acRIP-seq mapping accuracy, wherein enhanced amplification was determined at ac4C-rich vs. -poor regions, that was reduced in NAT10−/− cells (Figure S3E). Additionally, NAT10-RIP followed by RT-qPCR mirrored the ac4C substrate enrichment: defined ac4C(+) targets showed increased interaction with NAT10 as compared to ac4C(−) targets, and association was decreased in response to NAT10 ablation (Figure S3F).
Having established clear ac4C peaks, we next examined for biased localization within target transcripts. Input normalized ac4C read densities were plotted as a function of relative 5’ to 3’ positioning within substrate mRNAs (Figure 3G). The summarized distribution revealed ac4C peaks are not restricted to any particular location across target transcripts but display a general 5’ positional bias (Figure 3G). Moreover, ac4C peak summits were queried for relative location within specific transcript features (Table S4). Here, we observed ac4C sites cluster proximal to translation start sites with the majority of summits occurring within coding sequences (CDS) (Figure 3H). In terms of absolute numbers, ac4C was enriched in 5’ untranslated regions (UTRs) and CDS, and reciprocally depleted in 3’UTRs as compared to the overall percent of mRNA sequence assigned to these features (Figure 3H).
Biased down-regulation of ac4C(+) mRNAs in NAT10−/− cells
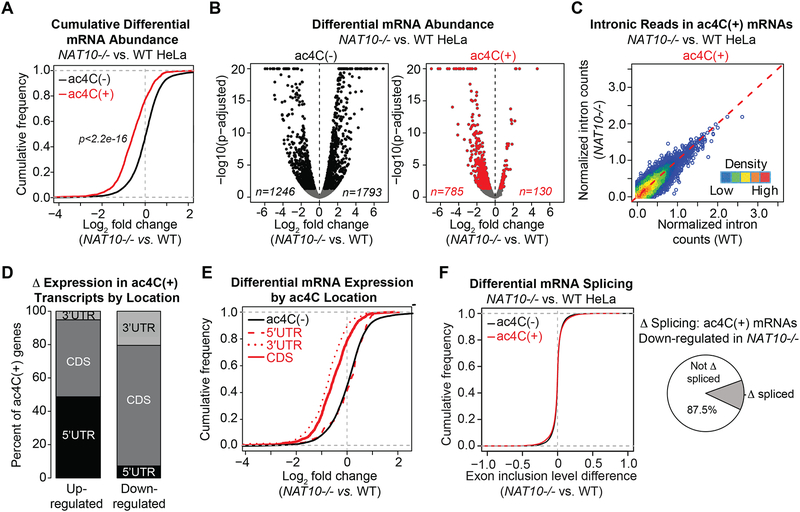
The observed positional bias of ac4C implies a regulatory function in gene expression. In support of this premise, ac4C(+) transcripts were enriched for GO terms related to cell survival and viability, suggestive that reduced proliferation in NAT10−/− cells directly relates to altered expression of acetylated substrates (Figure S4A). We thus investigated the relationship between acetylation status and mRNA abundance. Differential gene expression upon ac4C loss was assessed through RNA-seq performed in NAT10−/−A and parental HeLa cells (Table S2). Examination of the pool of mRNAs with determined ac4C peaks revealed an overall tendency towards decreased expression upon NAT10 loss as compared to transcripts lacking ac4C (Figure 4A). While mRNAs not marked by ac4C showed a balanced response to NAT10 deletion, with comparable numbers of up- and down-regulated genes, acetylated transcripts showed a considerable bias toward decreased expression upon ac4C loss (Figure 4B).
Figure 4. Loss of ac4C is globally associated with target mRNA down-regulation.
(A) Cumulative distribution function (CDF) plot depicting differential expression of ac4C(−) or ac4C(+) transcripts in NAT10−/−A vs. parental HeLa cells (ac4C(−), n=13,202; ac4C(+), n=2,114). p = Kolmogorov-Smirnov (KS) test.
(B) Volcano plots of differentially expressed protein coding genes in NAT10−/−A vs. parental HeLa cells, segregated by acetylation status. Differentially expressed ac4C(−) and ac4C(+) genes are shown in black and red, respectively (adjusted p < 0.05).
(C) Normalized intronic reads from ac4C(+) transcripts in NAT10−/−A vs. parental HeLa cells.
(D) Percentage of ac4C summits occurring within CDS or UTRs in transcripts with differential expression in NAT10−/−A relative to parental HeLa cells from (B).
(E) CDF plot showing expression changes of protein-coding genes in NAT10−/−A vs. parental HeLa cells for ac4C(−) and ac4C transcripts with peaks occurring within the CDS (n=1,131), 5’UTR (n=257) or 3’UTR (n=231). KS test: ac4C(−) vs. 5’UTR, p = 0.15; ac4C(−) vs. 3’UTR, p < 2.2e-16; ac4C(−) vs. CDS, p < 2.2e-16.
(F) CDF plots of exon inclusion differences in NAT10−/−A vs. parental HeLa cells, based on ac4C status (ac4C(−), n=39,876; ac4C(+), n=9,787) (left). Pie chart represents the proportion of down-regulated ac4C(+) transcripts that also showed differential splicing in NAT10−/− A relative to parental HeLa cells (right).
We examined several parameters at the gene expression level to elucidate the mechanism leading to down-regulation of acetylated mRNAs in NAT10−/− cells. Although modifications are deposited to RNA post-transcriptionally, to fully rule out down-regulation through reduced transcription, we compared intronic signal in the defined acetylated targets in NAT10−/− relative to parental HeLa cells. Estimation of depth normalized intronic reads established that overall transcription of ac4C(+) mRNAs was not inhibited in response to NAT10 ablation (Figure 4C). Likewise, pan-H3 acetylation, a marker of active transcription, was equivalently detected at select ac4C(+) mRNAs in NAT10−/− vs. parental HeLa cells (Figure S4B). Next, as UTRs are enriched in target binding sites for regulatory microRNAs, we inspected for biased localization of ac4C within the down-regulated subset. As observed for the total pool of acetylated mRNAs, the majority of down-regulated targets contained ac4C peaks within the CDS (Figure 4D). Reciprocal examination of differential gene expression segregated by ac4C summit location revealed that loss of CDS and 3’UTR acetylation in NAT10−/− cells was globally associated with decreased transcript levels, whereas ac4C within the 5’UTR had little effect on substrate expression (Figure 4E). Given that ac4C was generally depleted within 3’UTRs, we surmise the down-regulation bias is principally driven by loss of CDS acetylation (Figure 4E). Finally, as ac4C in the CDS could influence mRNA levels through the production of less stable isoforms, we queried for changes in splicing relative to altered gene expression. Overall, acetylated mRNAs showed similar changes in splicing in response to NAT10 ablation as compared to the non-acetylated set, and splicing variation was not augmented in transcripts showing the greatest changes in gene expression (Figure 4F). Altogether, these results suggest that the biased reduction in abundance of acetylated mRNAs upon NAT10 deletion likely originates from loss of a modification-associated activity inherent within the mRNA molecule.
ac4C promotes mRNA stability
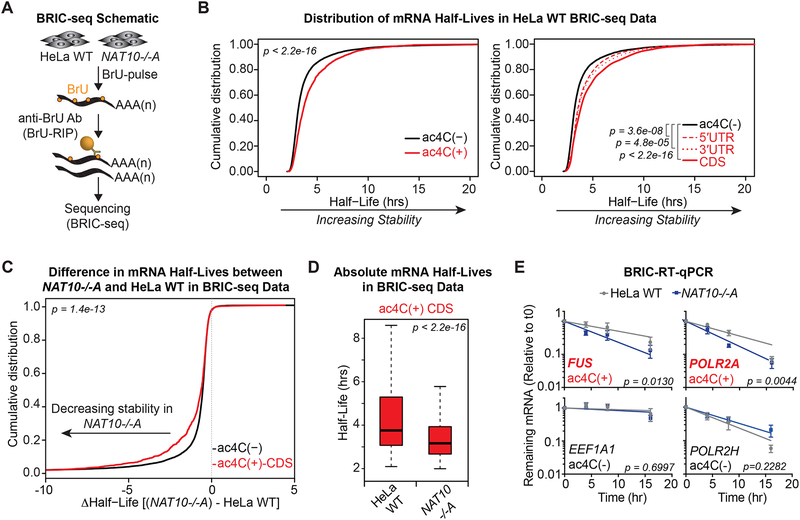
Given the above indications that ac4C regulates mRNA expression post-transcriptionally, we investigated mechanisms that influence mature mRNA abundance. In particular, we explored whether decreased detection of acetylated targets in NAT10−/− cells relates to altered mRNA stability. To this end, we performed BRIC-seq (5’-bromo-uridine [BrU] immunoprecipitation chase-deep sequencing analysis) in parental and NAT10−/− HeLa cells to examine RNA stability genome-wide (Tani et al., 2012). BRIC-seq involves antibody-based enrichment of BrU-pulsed RNAs, followed by sequencing with an internal “spike-in” for normalization (Figure 5A). BRIC-seq performed on biological replicates in parental HeLa cells produced reproducible half-lives that ranged from several minutes to >24 hours. Binning mRNAs by ac4C status revealed a strong correlation to transcript stability: ac4C modified mRNAs were characterized by significantly longer half-lives as compared to all other transcripts (Figure 5B, Table S5). This result was particularly evident for mRNAs with ac4C present within coding sequences, wherein the strongest transcriptome-wide association with enhanced half-life was observed (Figure 5B). Likewise, half-life determination in NAT10−/− cells showed that mRNAs with CDS acetylation were significantly decreased in the absence of NAT10 as compared to ac4C(−) mRNAs (Figures 5C and 5D). Of note, consistent with the reduction in proliferation, BrU uptake was generally diminished in NAT10−/− cells and the spike-in probe constituted a majority of reads at the latter time points. Determined half-lives in BRIC-seq from the NAT10−/− condition are thus generally decreased. Importantly, BRIC-RT-qPCR was not affected by this quantification artifact and a substantial destabilization of targets with CDS acetylation was observed in NAT10−/− relative to parental HeLa cells, while an mRNA with UTR ac4C and ac4C(−) controls were unaffected (Figures 5E and S5A). Finally, to determine whether stabilization of ac4C(+) mRNAs relates to inhibition of exonuclease digestion, we assessed the ability of Xrn1, the major 5’ to 3’ exonuclease activity in cells, to degrade an in vitro transcribed radiolabeled reporter generated in the presence or absence of ac4C. In vitro monitoring of Xrn1 activity revealed no distinction whether the transcript body contained unmodified cytidine or ac4C (Figure S5B). Together, these results indicate that ac4C actively promotes mRNA expression through increased stability via a mechanism uncoupled from exonuclease resistance.
Figure 5. ac4C promotes mRNA stability.
(A) Schematic of 5’-bromo-uridine [BrU] immunoprecipitation chase-deep sequencing (BRIC-seq).
(B) Cumulative distribution plots of mRNA half-lives in parental HeLa cells for ac4C(−) (n=9,821) and all ac4C(+) (n=1,966) transcripts (left), or subdivided by ac4C summits occurring exclusively within 5’UTR (n= 248), 3’UTR (n=219), or CDS (n= 1,048) (right). p = KS test.
(C) CDF plot of differential mRNA half-lives in NAT10−/−A vs. parental HeLa cells for ac4C(−) and ac4C(+) transcripts with summit position within CDS. p = KS test.
(D) Boxplots of median half-lives of ac4C(+) transcripts with CDS summits in parental and NAT10−/−A HeLa cells. Boxes indicate median, 25th, and 75th percentiles, and whiskers extend to 1.5 times the interquartile range (excluding outliers). p = Wilcoxon rank-sum test.
(E) BrU-labeled RNA was immunoprecipitated as described in (A) followed by RT-qPCR. Decay graphs were generated by applying the One-Phase Decay model. Mean ± SEM, n=4, Sum-of-squares F test.
mRNA acetylation enhances translation
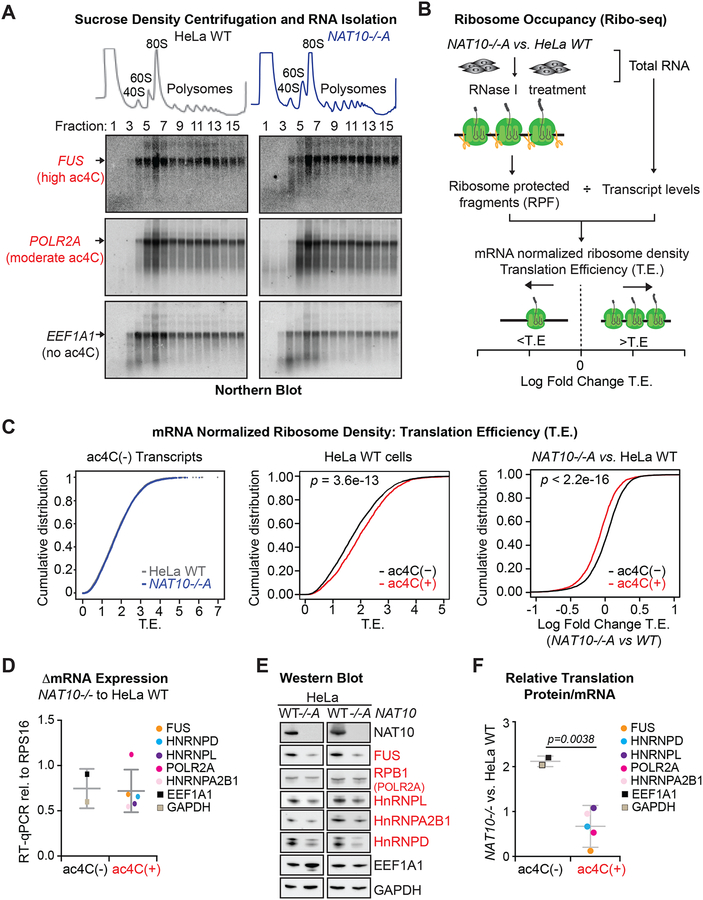
mRNA decay and translation are intricately linked, such that a reduction in mRNA stability manifests in decreased translation, and decreased translation reciprocally reduces mRNA stability (Hanson and Coller, 2018). We thus explored whether the observed influence of ac4C on transcript levels is reflected in enhanced translation. However, as ac4C is also found in 18S rRNA and tRNAser/leu, we first examined for pleiotropic effects that could globally impact translation. Centering on tRNA, a deficiency in tRNAser/leu function would be predicted to have the largest impact on transcripts enriched in these cognate codons. We instead find Ser/Leu content is unrelated to gene expression changes in NAT10−/− compared to parental HeLa cells (Figures S6A and S6B). Likewise, while defective tRNAser/leu function would result in ribosomal stalling at the cognate codons in mRNA, ribosome profiling showed no distinction in A-site occupancy on either serine or leucine codons in parental vs. NAT10−/− HeLa cells (Figure S6C). Turning to 18S rRNA, RNA profiles indicative of intact 40S, 80S and polyribosomes were clearly visible in sucrose density gradients from both parental and NAT10−/− conditions, whereas 45S rRNA, which accumulates when processing of 18S rRNA is compromised (Tafforeau et al., 2013), was not visualized in either condition (Figure 6A, top). In addition, mRNA association with polyribosomes was uncompromised in NAT10−/− vs. parental HeLa cells as determined through Northern blot of RNA purified from the sucrose gradient fractions (Figure 6A, bottom). Thus, the changes in gene expression observed in ac4C modified mRNAs are unrelated to the known trans-acting roles of NAT10 (i.e. 18S rRNA and tRNAser/leu acetylation).
Figure 6. ac4C enhances translation efficiency.
(A) Absorbance at 254 nm in sucrose density gradient fractions from parental and NAT10−/−A HeLa cells (top). Total RNA isolated from each fraction was hybridized to probes specific for two ac4C(+) transcripts, FUS and POLR2A, and an ac4C(−) transcript, EEF1A1 (bottom). Blots are representative of biological triplicates.
(B) Schematic of Ribo-seq.
(C) CDF plots of mRNA-normalized ribosome footprint reads (T.E.) for ac4C(−) transcripts in NAT10−/−A and parental HeLa cells (left); ac4C(−) and ac4C(+) transcripts in HeLa WT cells (middle), or in NAT10−/−A vs. HeLa WT (right). ac4C(−), n=5445; ac4C(+), n=1733. p = KS test.
(D) RT-qPCR for differential expression of determined ac4C(−) and ac4C(+) mRNAs in NAT10−/−A and HeLa WT cells. Dots represent the mean from three biological replicates. Error bars depict the average and SD within ac4C(+) and ac4C(−) transcripts.
(E) Representative Western blots of proteins associated with ac4C(+) and ac4C(−) transcripts from parental and NAT10−/−A HeLa cells.
(F) Relative translation of select ac4C(+) and ac4C(−) transcripts as determined through the change in protein expression compared to the change in mRNA expression in NAT10−/− vs. parental HeLa cells. Dots represent the mean delta T.E. from three biological replicates. Error bars depict the average and SD within ac4C(+) and ac4C(−) transcripts. Two-tailed student’s t-test.
We next examined the influence in cis of ac4C on substrate mRNA translation. As ac4C exhibits a strong 5’ localization bias, we first assessed whether acetylation affects translation initiation. To this end, we measured 48S pre-initiation complex accumulation in vitro and found no distinction whether reporters were generated in the presence of ac4C or unmodified cytidine (Figure S6D). Likewise, detection of full-length message in ribosome free sucrose-density gradient fractions was not influenced by acetylation status (Figure 6A, black arrows, fractions 1–4). Considering that 5’UTR acetylation does not associate with target mRNA expression (Figure 4E), NAT10-dependent variations in mRNA abundance are most consistent with a direct role for ac4C on transcript stability and/or translation downstream of initiation.
To explore the influence of ac4C on mRNA translation, we examined ribosome occupancy across the transcriptome through sequencing of ribosome protected fragments (Riboseq). Translation efficiency (T.E.), as calculated through Ribo-seq, is a direct metric of ribosome density per mRNA molecule (Figure 6B, Table S6). Accordingly, Ribo-seq assesses the impact of cellular perturbations on mRNA translation in vivo. In comparing Ribo-seq performed in parental and NAT10−/− HeLa cells, we found no discernible difference in the translation of ac4C(−) mRNAs (Figure 6C). In contrast, ac4C(+) mRNAs displayed elevated T.E. in parental HeLa that was specifically ablated in response to NAT10 deletion (Figure 6C). As ac4C stabilizes mRNAs (Figure 5B) and pleiotropic effects on translation were not observed in NAT10−/− cells (Figures 6A and S6), this global demonstration of increased ribosome density specific to ac4C(+) mRNAs is indicative that acetylation intrinsically promotes translation. To gain direct support for the positive role of ac4C in translation, we examined several target mRNAs in detail. While steady state mRNA levels were unchanged or only moderately affected in NAT10−/− vs. parental HeLa cells (Figure 6D), Western blotting showed a dramatic reduction in protein expression exclusive to ac4C(+) targets (Figure 6E). Quantification of relative translation (change in protein vs. change in mRNA) depicts a clear defect in protein output associated with ac4C(+) as compared to ac4C(−) mRNAs upon NAT10 loss (Figure 6F). These results documenting a NAT10-dependent increase in translation specific to ac4C(+) mRNAs in HeLa solidifies the involvement of mRNA acetylation in translational regulation in vivo.
mRNA acetylation enhances translation when present within wobble cytidine
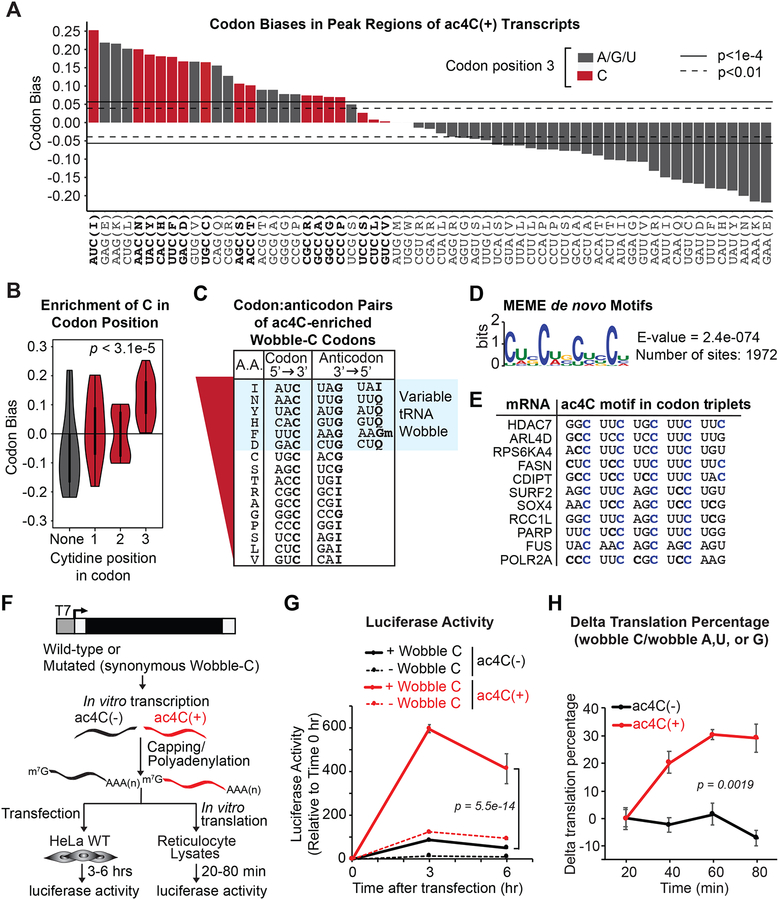
Several lines of evidence suggest that the positive impact of mRNA acetylation on translation and stability occurs at the level of tRNA decoding efficiency. Precedence for such an association exists in prokaryotes, wherein the presence of ac4C in the anticodon wobble site of tRNAmet enforces an amide group conformation that facilitates hydrogen bonding with guanosine and ensures appropriate recognition of AUG sequences in mRNA (Stern and Schulman, 1978; Taniguchi et al., 2018). This result reflects an important aspect of mRNA/tRNA association: wobble site interactions are geometrically distinct from mRNA codon positions 1 and 2, thus allowing for non-standard base-pairing (Agris et al., 2007), ac4C in this sense safeguards efficient tRNAmet decoding in prokaryotes. Our detection of ac4C within coding sequences of human mRNAs raises the possibility that mRNA acetylation reciprocally enhances translation by promoting interaction with cognate tRNAs. Following this logic, ac4C in human mRNAs would be predicted to have the strongest impact when present within codon wobble sites. That is precisely what we observe: all 16 mRNA codons with cytidine in position 3 were enriched within our acetylated transcripts as compared to the transcriptome (Figure S7A). These effects were even more dramatic when focusing exclusively within acetylated peaks (Figure 7A). In contrast, codons with cytidine at positions 1 and 2 were balanced and showed an equal tendency to be enriched or de-enriched in acetylated regions (Figures 7B and S7B). Remarkably, comparison of the most enriched codons within ac4C peaks to the amino acid code revealed a striking relationship to codon degeneracy. Within the genetic code, a codon is considered “degenerate” if substitution at any site does not alter amino acid selection. For the eight least enriched codons in ac4C peaks (Figure 7C), wobble site substitutions are universally tolerated and have no influence on amino acid identity. Reciprocally, these codons are recognized by invariant tRNA anticodon compositions with wobble site G or I (inosine). Improper wobble discrimination would accordingly have no influence on these codons. In contrast, for the eight most enriched codons (Figure 7C), wobble site substitutions induce coding changes that alter amino acid content or introduce stop codons. Amongst these, the top six enrichment scores correspond to codons decoded by multiple tRNAs, such that a single mRNA codon is presented with G vs. Q (queuosine) or G vs. I in the cognate tRNA anticodon (Figure 7C). These results demonstrating a biased prevalence of codon contexts corresponding to multimodal mRNA:tRNA interactions are highly suggestive of a direct role for wobble site ac4C in decoding efficiency.
Figure 7. ac4C statistically and functionally associates with mRNA wobble cytidines.
(A) Codon bias within CDS-localized ac4C peaks relative to the entire transcriptome. Red bars depict codons with C in the wobble position. Horizontal lines indicate the magnitude of codon bias expected by random sampling at the significance level of p = 0.01 or p = 1e-4, as indicated.
(B) Violin plot of aggregated codon bias results from (A).
(C) ac4C-peak enriched codons with wobble C were ranked according to (A). Anticodon sequences of the respective tRNAs are shown with variable decoders highlighted in blue.
(D) Sequence logo of enriched motifs within ac4C peaks determined using MEME. Enrichment p-value (E-value) derived from FDR corrected Fisher’s Exact Test.
(E) Alignment of top scoring motifs in ac4C peaks to substrate mRNAs. Cytidines in blue designate occurrence within the third (wobble) position of each codon.
(F) Firefly luciferase mRNA naturally containing C within wobble sites (wobble C) or with synonymous codon substitutions that removed C from all wobble sites (wobble A, U or G) was generated in the presence of CTP or ac4CTP.
(G) mRNAs from (F) were transfected into HeLa cells. Luciferase activity was monitored through luminescence. Mean ± SEM, n=3. Two-Way ANOVA with Tukey’s post hoc test.
(H) mRNAs from (F) were in vitro translated in reticulocyte lysates. Data represent the % difference in luminescence of wildtype versus mutated luciferase in the presence or absence of ac4C, mean ± SEM, n=3. Two-Way ANOVA.
Building on the selective codon enrichment within ac4C peaks, we examined for biased representation of specific motifs using MEME (Bailey and Elkan, 1994). Constrained at ≤12 nucleotides, MEME revealed one highly enriched cytidine-containing motif present in ~74.0% of examined ac4C peaks: a C-rich sequence characterized by four obligate cytidines separated by two non-obligate nucleotides (CXX) (Figure 7D). Likewise, MEME performed without a length constraint identified a 29-nucleotide repeating CXX motif occurring in ~41.0% of ac4C peaks (Figure S7C). In support of a role in decoding efficiency, mapping of top scoring motifs to their source mRNAs revealed codon phasing that uniformly placed the obligate cytidine in the wobble position (Figure 7E). The broad peak in the validated ac4C target POLR2A contained eight distinct CXX repeats ranging in length from 12–18 nucleotides, resulting in 40 codons with wobble cytidines (Figure S7D). These results suggest overall ac4C detection in human cells may be boosted by highly modified substrates that are characterized by an over-representation of cytidine within wobble sites.
To directly examine the role of ac4C in wobble site decoding, synonymous mutations were incorporated into a luciferase reporter to remove cytidine from all wobble positions without altering amino acid sequence. The resulting wildtype (+ wobble C) and mutated (− wobble C) constructs served as templates for the generation of variably acetylated mRNA through in vitro transcription, m7G-capping and polyadenylation (Figures 7F and S7E). Subsequent transfection into HeLa cells and protein monitoring through relative luminescence revealed a substantial reduction in luciferase translation in response to synonymous codon substitutions associated with unmodified mRNA (Figure 7G). These results are consistent with the introduction of non-optimal codons, as tRNA concentrations are limiting in vivo. Impressively, the presence of ac4C in positions 1 and 2 effectively eliminated the codon penalty associated with mutated luciferase (− wobble C) and produced wildtype protein levels, thereby demonstrating the general stabilizing effect for ac4C on Watson-Crick base-pairing in any context. However, in support of a major role in wobble site discrimination, introduction of ac4C into mRNA codon position 3 dramatically stimulated luciferase translation, resulting in a ten-fold increase in protein as compared to unmodified wildtype luciferase, or mutated luciferase with ac4C exclusive to positions 1 and 2 (Figure 7G). These results were independent of secondary effects related to mRNA transfection: eIF-2α phosphorylation, a known side-effect of RNA transfection, was equally detected in ac4C(+) and (−) reporters (Figure S7F). Likewise, co-transfection of modified Firefly luciferase and unmodified Nano Luciferase showed that increased translation was specific to the acetylated mRNA, and not a general impact on translation (Figure S7G). Finally, Northern blot for luciferase mRNA isolated from transfected HeLa at the indicated time points showed no evidence for mRNA degradation within the experimental time frame (Figure S7H). These mRNA transfection results conclusively demonstrate the cis-acting role of cytidine acetylation in translation dynamics, in the absence of secondary effects related to tRNA and rRNA acetylation.
A potential caveat to the generality of mRNA transfection relates to limited tRNA levels in vivo. Thus, to ensure the cis effect of ac4C on mRNA translation is robust in any context, we performed in vitro translation in rabbit reticulocytes, wherein tRNA concentrations are saturating. As predicted, in stark contrast to the mRNA transfection results, the penalty for suboptimal codons was minimal in reticulocytes and the precise wobble site substitutions that effectively abolished luciferase translation in HeLa only minorly affected unmodified luciferase production in vitro (Figure S7I). Reticulocyte extracts were thus programed with variably acetylated luciferase mRNA and protein output was monitored through luminescence over time. To normalize for the general impact of codon substitutions on translational efficiency, data are presented as the percent change in translation of luciferase mRNA containing C within wobble sites vs. luciferase with synonymous A, U or G substitutions at those locations. Consistent with the mRNA transfection results, while the influence of synonymous codon substitutions remained constant over time for the unmodified luciferase reporters (Figure 7H), the presence of ac4C within wobble sites strongly stimulated translation as compared to acetylated substrates bearing substitutions that uniformly removed ac4C from those locations (Figure 7H). These findings solidify the direct impact of ac4C in translation, in the absence of confounding pleiotropic effects. In sum, bioinformatic analysis of sequence biases within ac4C peaks and the empirically determined positive influence of wobble site ac4C on mRNA translation strongly point to a direct role for mRNA acetylation in tRNA discrimination that manifests in the efficient translation of substrate mRNAs.
DISCUSSION
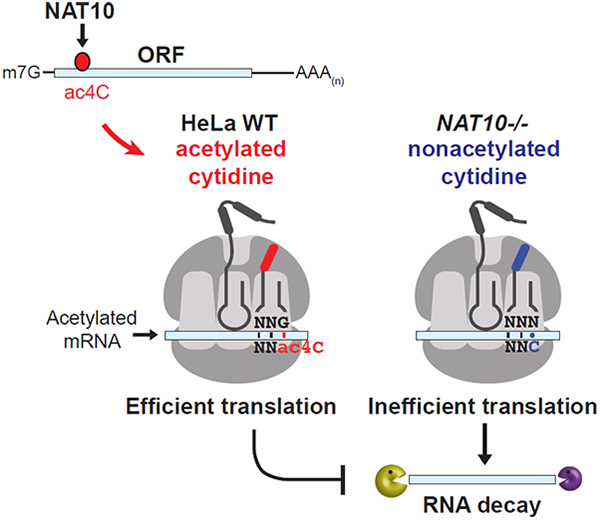
Our data expand the complement of known modifications within mRNAs to include N4-acetylcytidine. ac4C is the first acetylation event to be described in mRNA and we find that its occurrence is regulated by a single enzyme, NAT10. mRNA acetylation was widely distributed within the human transcriptome and overall enriched within coding sequences. Analysis of ac4C function revealed an intrinsic role in promoting mRNA stability and translation. Examination of codon composition within acRIP-seq peaks further exposed a strong enrichment for cytidine within wobble sites, suggesting a direct role for ac4C in the process of ribosomal decoding. Support for this hypothesis came from the demonstration that wobble site ac4C stimulated translation in vitro and in vivo. Together, these data expand the growing list of modifications that impact mRNA regulation. Given the broad number of acetylated targets, ac4C becomes an important component of the epitranscriptome within human cells.
While the precise means by which ac4C promotes mRNA stabilization and translation remain obscure, the prevalence of cytidine-containing wobble sites within acetylated peaks is reminiscent of the mechanism by which tRNAmet acetylation promotes decoding fidelity in prokaryotes (Stern and Schulman, 1978; Taniguchi et al., 2018). ac4C locks cytosine in an unusual ‘proximal’ conformation through adopting a non-standard gauche orientation across the C(4’)-C(5’) bond (Parthasarathy et al., 1978). Acetylation of the wobble cytosine in tRNAmet thus prevents shielding of the Watson-Crick base pairing sites, ensuring strong association with guanosine and consequent proper decoding of methionine in bacteria (Kumbhar et al., 2013). Applying this paradigm to our observations in HeLa mRNA dictates that just as tRNA acetylation supports mRNA codon recognition in E. coli, mRNA acetylation should support tRNA recognition in humans to facilitate decoding efficiency (graphical abstract). The observed enrichment of wobble site cytidines within ac4C peaks, wherein the stabilizing influence on codon:anticodon pairing would be most relevant, bolsters this notion. Indeed, the most enriched wobble cytosine codons within ac4C peaks are those that would maximally benefit from the stabilization of Watson-Crick base pairing; codons where wobble site choice is critical for amino acid identity, all A/U combinations in position 1 and 2, and codons where the corresponding tRNA wobble position can be either G or a nucleotide variant (Q or I). In all cases, ac4C would be predicted to aid in tRNA selection through specific recognition of guanosine, thereby improving cognate tRNA choice. However, the basis by which select wobble C codons are targeted for acetylation whereas others are not remains unclear. Indeed, repeating CXX motifs are also present within UTR ac4C peaks, suggesting that the tripartite spacing directly relates to NAT10 substrate selection. Detailed studies into NAT10 function will be required to resolve the question of targeting specificity.
While our results support the premise that ac4C enrichment within coding sequences improves mRNA decoding efficiency, whether ac4C acts at the level of decoding rate or fidelity remains to be seen. As mRNA stability and translation are tightly coupled, the positive influence of ac4C on both these parameters does not help to discriminate between these possibilities. Importantly, our results reinforce the notion that the specific reduction in stability and translation of determined ac4C(+) transcripts in response to NAT10 ablation is a direct consequence of defective mRNA acetylation, and not an artifact of altered rRNA or tRNA acetylation: general defects in ribosome biogenesis and the translation of Ser/Leu-rich mRNAs were not observed. Most relevantly, the direct role of mRNA acetylation in translation was established through monitoring luciferase production from variably acetylated in vitro transcribed mRNA, wherein ac4C stimulated translation when all else was held constant. Thus, the presence of NAT10-catalyzed ac4C within distinct RNA pools appears to regulate translation at several levels, including through direct modulation of substrate mRNAs.
Although our analysis focuses on CDS acetylation, there are implications associated with finding ac4C in distinct locations within mRNA. Within the CDS, ac4C is enriched towards the 5’ end of target transcripts. The significance of this localization bias remains unclear but may reflect a role in ac4C function or the mechanism by which acetylation is deposited on target transcripts. Notably, ac4C is also enriched within 5’UTRs. While not examined in this study, mRNA acetylation at these locations may signify an analogous function in stabilizing RNA secondary structures. Relatedly, while cytidine was enriched in codon position 3 within CDS ac4C peaks, we cannot rule out a role for ac4C in positions 1 and 2. Strictly based on ac4C thermodynamic properties, occurrence at any location should improve interaction with guanosine in RNA. Indeed, incorporation of ac4C exclusively in positions 1 and 2 of mutated luciferase stimulated translation and rescued protein production associated with non-optimal synonymous codon substitutions. Likewise, it remains possible that ac4C may modulate interactions with other to be determined modified ribonucleotides. Overall, based on the diverse distribution of ac4C, putative roles in controlling translational initiation, mRNA localization, translational repression, deadenylation, etc. should not be unexpected.
Concluding remarks
In summary, we describe, for the first time, the presence of an acetylated base within mRNA. We demonstrate that mRNA acetylation is catalyzed by the NAT10 enzyme and determine that ac4C is globally enriched within coding sequences. The presence of ac4C within target mRNAs conferred enhanced stability and translation efficiency, and occurrence within mRNA wobble sites directly promoted translation in vivo and in vitro. Intriguingly, the distribution and impact of ac4C contrasts with the abundant mRNA modification, N6-methyladenine (m6A). While ac4C is enriched within the 5’ regions of coding sequences and associates with substrate mRNA stability, m6A displays a 3’ localization bias and relates to mRNA destabilization (Roundtree et al., 2017). This dichotomy is reminiscent of the regulation of gene expression through the histone “code.” Whereas histone acetylation is considered an activating mark, histone methylation is generally repressive. Hence, just as histone modifications dynamically regulate gene expression at the chromatin level, an impact on protein expression may occur through altered mRNA modifications. Considering that the regulation of translation emerges as a common theme amongst the documented mRNA modifications (Roundtree et al., 2017), our findings raise the possibility that an ‘epitranslation’ code exists at the mRNA level to post-transcriptionally regulate gene expression. Altogether, these results significantly expand our current concept of the epitranscriptome to include an acetylated residue with a role in the regulation of mRNA expression and translation.
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shalini Oberdoerffer (shalini.oberdoerffer@nih.gov). There are no restrictions on any data or materials presented in this paper.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture and generation of NAT10 mutant cell lines
HeLa (Human cervix carcinoma, female, ATCC) cells were cultured in Dulbeccos’ Modified Eagle Medium (DMEM, ThermoFisher Scientific) containing 25 mM glucose and 1 mM sodium pyruvate and supplemented with 4 mM L-glutamine (ThermoFisher Scientific) and 10% bovine calf serum (BCS, HyClone), in the absence of antibiotics. Flp-In T-Rex 293 (Human embryonic kidney, female, ThermoFisher Scientific) cells were grown in DMEM containing 25 mM glucose, 1 mM sodium pyruvate, 4 mM L-glutamine, 10% BCS and 400 μg/mL zeocin.
CRISPR-Cas9 mediated ablation of the NAT10 gene was achieved with PX458 plasmid (Addgene) containing expression cassettes for pSpCas9–2A-GFP and chimeric guide RNA (Ran et al., 2013). To target exon 5 of the NAT10 gene, a guide RNA sequence of GTGAGTTCATGGTCCGTAGG was selected through the http://crispr.mit.edu website. Plasmid containing the guide RNA sequence was transfected into cells using Lipofectamine 2000 according to the manufacturer’s instructions (ThermoFisher Scientific). Forty-eight hours post-transfection, GFP-positive cells were sorted and collected using the FACSAriaII cell sorter (BD Biosciences). GFP-positive cells (1,000 cells) were seeded in 15-cm dishes in complete DMEM medium. After seven days, single colonies were transferred into 96-well plates. Depletion of NAT10 expression was screened by Western blot. To determine the presence of insertions or deletions (indels) in NAT10 targeted clones, genomic DNA was isolated using a Quick-DNA Miniprep kit (Zymo Research) and NAT10 exon 5 PCR amplification was achieved using 2 U Taq DNA polymerase (NEB), 100 μM dNTPs and 250 nM of primers flanking exon 5; Forward: 5’-TGGCTTTGTGCTCTGAAGTC-3’; Reverse: 5’-GCTCTTAGCCCAGAGGCTGT-3’. PCR product was cloned into pCR2.1-TOPO using the TOPO TA cloning Kit (ThermoFisher Scientific) and transformed into E. coli DH5α competent cells (ThermoFisher Scientific). Plasmids were isolated from 8–10 single colonies and sequenced by Sanger sequencing (Macrogen USA, Rockville, MD). Clones with mutations in both alleles were selected for downstream studies. Cell lines generated using the above strategy include HeLa NAT10+/+, HeLa NAT10−/−A, HeLa NAT10−/−B, HeLa NAT10−/−C and Flp-In T-Rex 293 NAT10−/− cells. All clones were maintained under the same conditions as parental cells.
NAT10 Δhelicase cDNA was generated through PCR amplification of an N-terminal fragment lacking the helicase domain using 5’-ATAGAAGACACCGGGACCGATC-3’ and 5’-tttAAGCTTctagagtcttacagcagtccaccaac-3’ with pICE-FLAG-NAT10-siR-WT (Addgene, Cat#:59365) as template. The C-terminal region of NAT10 was obtained by digestion of pICE-FLAG-NAT10-siR-WT with XbaI and HindIII. The N-terminal and C-terminal regions were assembled into pBluescript to generate a NAT10Δhelicase clone lacking amino acids 259–502. Full-length NAT10 and NAT10Δhelicase were subcloned into the pcDNA5/FRT-chimeric intron vector (pcDNA5/FRT-CI). pcDNA5/FRT-CI vector is a modified version of pcDNA5/FRT (ThermoFisher Scientific, Cat#:V6010–20) which contains a chimeric intron (CI) to aid in high expression levels. The modified vector was produced by replacing the promoter with that of pCI-neo (Promega, Cat#:E1841), via BglII and NheI sites. Final constructs were then confirmed by restriction mapping and sequencing.
For transient expression of NAT10 cDNA, NAT10−/−A HeLa cells were transfected with plasmids encoding full-length NAT10 or empty vector using Lipofectamine 2000. RNA and protein were harvested after 72 hrs. For stable NAT10 expression, single copy integration of full-length or Δhelicase NAT10 was achieved through Lipofectamine 2000 transfection in NAT10−/− Flp-In T-Rex 293 grown in the absence of zeocin for 48 hours, followed by selection with hygromycin (100 ug/mL). Colonies were picked, expanded and confirmed by Western blot. Flp-In T-Rex 293 derived clones were maintained in DMEM containing 25 mM glucose, 1 mM sodium pyruvate, 4 mM L-glutamine, 10% BCS and 100 ug/mL hygromycin. Cell lines generated using the above strategy include Flp-In T-Rex 293 NAT10−/− + full-length NAT10 and Flp-In T-Rex 293 NAT10−/− + −Δhelicase NAT10. All cells lines generated in this study are listed in the Key Resources Table under “Experimental Models: Cell lines.”
METHOD DETAILS
Analysis of protein expression by Western blot
Cells were seeded at a density of 2.5 × 105 cells/mL in 6-well plates and grown for 24 hrs, rinsed with PBS once and detached using 0.05% trypsin for 5 min at 37 °C. Reaction was stopped by 1 volume of complete media and cells were pelleted by centrifugation at 3,000 rpm for 5 min at 4 °C. Cells were rinsed with cold PBS and lysed in NP40 lysis buffer containing 0.5 % (v/v) NP40, 50 mM HEPES [pH 7.5], 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5 mM fresh DTT and 1× EDTA-free protease inhibitor cocktail III (EMD Millipore), followed by Bioruptor sonication for three cycles of 30 sec on/off at low setting (Diagenode). Cell lysates were cleared by centrifugation at 13,000 rpm for 10 min at 4 °C and protein concentration was quantified using the Bradford reagent (BioRad). Equal amounts of protein (25 μg) were loaded on 4–12% Bis-Tris gels, separated using NuPAGE MOPS SDS running buffer (ThermoFisher Scientific) and transferred onto nitrocellulose membranes using Tris-Glycine buffer (250 mM Tris [pH 6.8], 1.92 M Glycine, 20% methanol).
For analysis of cleaved caspase-3, cells were lysed in buffer containing 0.5 % (v/v) NP40, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 2.5 mM EDTA, 1 mM PMSF and 1% Halt™ protease inhibitor cocktail (ThermoFisher Scientific) and sonicated as described above. Equal amounts of protein were separated through 12% SDS-PAGE using SDS-Tris-Glycine running buffer (1% SDS, 250 mM Tris [pH 8.8], 1.92 M Glycine) and transferred onto Immobilon-P polyvinylidene difluoride membranes as described above.
After blocking membranes with 5% milk in 0.05% Tween-20 PBS buffer, immunoblot analysis was performed with primary antibodies as follows: rabbit polyclonal anti-NAT10 (1:2500 dilution, Cat#:13365–1-AP, ProteinTech), rabbit polyclonal anti-hnRNPD (1:2500 dilution Cat#:12770–1-AP, ProteinTech), rabbit polyclonal anti-EEF1A1 (1:2000 dilution, Cat#:11402–1-AP, ProteinTech), mouse monoclonal anti-GAPDH (1:2500 dilution, clone 6C5, Cat#:sc-32233, Santa Cruz Biotechnology), mouse monoclonal anti-p53 (1:1000 dilution, clone DO-1, Cat#:sc-126, Santa Cruz Biotechnology), rabbit polyclonal anti-cleaved caspase-3 (Asp175) antibodies (1:1000 dilution, Cat#:9661, Cell Signaling Technology), rabbit monoclonal anti-α-Tubulin (1:2500 dilution, clone 11H10, Cat#:2125S, Cell Signaling Technology), rabbit monoclonal anti-Histone H3 (1:2500 dilution, clone D2B12, Cat#:4620S, Cell Signaling Technology), mouse monoclonal anti-hnRNPL (1:1000 dilution, clone 4D11, Cat#:ab6106, Abcam), mouse monoclonal anti-hnRNPA2B1 (1:1000 dilution, clone DP3B3, Cat#:ab6102, Abcam), mouse monoclonal anti-RPB1 (POLR2A) (1:150000 dilution, clone CTD4H8, Cat#:05–623, Millipore) and rabbit polyclonal anti-FUS (1:1000 dilution, Cat#:A300–292A, Bethyl Laboratories) were incubated in a solution containing 1% milk in 0.05% Tween-20 PBS buffer overnight at 4 °C. Rabbit monoclonal anti-Acetyl-α-Tubulin (Lys40) (1:1000 dilution, clone D20G3, Cat#:5335S, Cell Signaling Technology), rabbit polyclonal anti-Histone H3 (acetyl K9+K14+K18+K23+K27) (1:1000 dilution, Cat#:ab47915, Abcam) and mouse monoclonal anti-p53 (acetylK120) (1:1000 dilution, clone 10E5, Cat#:ab78316, Abcam) were incubated in a solution containing 2% BSA in 0.05% Tween-20 TBS buffer overnight at 4 °C. After three washes in 0.05% Tween-20 PBS buffer, membranes were incubated with horseradish peroxidase (HRP)-conjugated 2secondary antibodies; anti-mouse IgG (1:10000 dilution, GE Healthcare), or anti-rabbit IgG (1:10000 dilution, Cell Signaling Technology). Western blots were visualized by enhanced chemiluminescence using the ProSignal Pico ECL Reagent (Genesee Scientific). Chemiluminescence was detected using the ChemiDoc Imaging System (BioRad) and quantified by densitometry using ImageLab software (version 6.0.0, BioRad).
Analysis of NAT10 expression and localization by immunofluorescence
Cells were seeded on poly-L-lysine coated coverslips (Sigma-Aldrich) at a density of 2.5 × 105 cells/mL in 12-well microplates and grown overnight in complete DMEM medium. Coverslips were rinsed with PBS once and incubated in a 2% paraformaldehyde solution for 5 min at room temperature, followed by three PBS washes. Cells were stained with 5 μg/mL Alexa594-conjugated WGA (Wheat Germ Agglutinin, ThermoFisher Scientific) for 10 min at room temperature, permeabilized with PBS containing 0.2% Triton X-100, 0.5% BSA and 5% Donkey serum for 30 min at 4°C and blocked in PBS containing 0.5% BSA and 5% Donkey serum for 1 hr at room temperature. Slides were washed three times in PBS solution and incubated with rabbit polyclonal anti-NAT10 antibodies (1:200 dilution, Cat#:PA5–31376, ThermoFisher Scientific) overnight at 4 °C. After washing, coverslips were incubated with Donkey anti-rabbit DyLight® 488 Abcam). Slides were then rinsed three times with PBS and mounted with ProLong Gold antifade reagent with DAPI onto slides (ThermoFisher Scientific). Confocal images were obtained in a Carl Zeiss LSM780 microscope equipped with Plan-Apochromat 63×/1.40 Oil DIC lens and ZEN software, followed by maximum intensity Z-projection using ImageJ software.
Analysis of cell proliferation and viability
Cells were seeded at a density of 105/mL in complete medium and harvested by trypsinization after 24, 48 and 72 hr. Viable cells were counted by Trypan blue exclusion in a Cellometer Auto T4 (Nexcelom Biosciences). For cell cycle analysis, cells were seeded at a density of 105/mL in complete medium and harvested by trypsinization after 72 hr. Cells were washed with PBS prior to fixation in 70% ethanol, followed by two additional PBS washes. Fixed cells were next stained with propidium iodide (50 mg/mL; Roche) containing 0.2 mg/mL DNase-free RNase (Roche) for 30 min at room temperature and immediately analyzed by a BD FACScalibur using the BD Cell Quest Pro software (BD biosciences).
Isolation of total and polyadenylated RNA
Total RNA was purified from cultured cells using Trizol (ThermoFisher Scientific) followed by treatment with Turbo™ DNase I (ThermoFisher Scientific). Enrichment of polyadenylated RNA [poly(A) RNA] was achieved through two rounds of selection with Oligo-(dT)25 Dynabeads (ThermoFisher Scientific) for LC-MS, dot blot or acRIP-seq, or two rounds of poly(A)Purist MAG (ThermoFisher Scientific) for ImmunoNorthern blot, according to the manufacturer’s instructions. Poly(A) RNA precipitations were carried out using 0.3M sodium acetate [pH 5.5], 15 μg/mL linear acrylamide (carrier) and 2.5× ethanol. Purification was estimated through Bioanalyzer picoRNA chips (Agilent Technologies) and by RT-qPCR using specific primers for 18S rRNA and GAPDH (see Table S7 for primer description). Briefly, RNA was reverse transcribed (RT) with random hexamers using the Superscript III system (ThermoFisher Scientific) according to the manufacturer’s suggestions, followed by qPCR using LightCycler 480 SYBR Green I Master Mix (Roche, Basil, Switzerland) in a LightCycler 96 Instrument (Roche).
Synthesis of isotope-labeled ac4C internal standards
To synthesize isotope-labeled D3-ac4C, cytidine (200 mg, 0.82 mmol) and D6-acetic anhydride (78 μL, 0.82 mmol, Sigma-Aldrich) were dissolved in methanol (4 mL) and heated to reflux with stirring. The reaction was monitored by thin layer chromatography (TLC), and additional aliquots of D6-acetic anhydride were added every hour for 3 hours. After 5 hr the reaction was cooled to room temperature and solvent removed under reduced pressure. Silica gel chromatography (CH2Cl2/MeOH) yielded the pure product, D3-ac4C, as a white solid (80 mg, 34%). Electrospray ionization-mass spectrometry (ESI-MS, positive mode): [M+H]+ calculated: 288.3, [M+H]+ found: 288.7. ESI-MS (negative mode): [M]− calculated: 287.1, [M]− found: 287.2. λmax = 249 nm, 300 nm. 1H-NMR (400 MHz, MeOD). δ 8.51 (d, J= 8.0 Hz, 1H), 7.43 (d, J= 8.0 Hz, 1H), 5.90 (d, J= 4.0 Hz, 1H), 4.23 (q, J= 4.0 Hz, 1H), 4.16 (m, 2H), 4.00 (dd, J= 12.0, 4.0 Hz, 1H), 3.84 (dd, J= 8.0, 4.0 Hz, 1H), 13C-NMR (126 MHz,D2O/d8-1,4 dioxane) δ 174.08, 162.71, 156.95, 145.43, 97.93, 91.38, 83.68, 74.45, 68.59, 60.16, 23.62–23.16 (q, J(C-D)= 75 Hz, 1C).
Chemical acetylation of polycytidylic acid
PolyC (5 μg, Sigma-Aldrich) was resuspended in 0.2 mL water and mixed with 0.4 mL tri-nbutylamine and 0.1 mL of acetic anhydride. The reaction was incubated overnight at 4 °C. Next, 0.1 mL acetic anhydride and 0.25 mL of tri-n-butylamine were added and incubated at room temperature for another 24 h. Finally, the reaction was diluted to 3 mL with water and dialyzed against 0.2 M NaCl, then against water, lyophilized and resuspended at 1 μg/mL in water.
ac4C detection and quantification by mass spectrometry
Digestion of total or poly(A) RNA (2.5–10 μg) was performed as previously described (Sinclair et al., 2017). Briefly, RNA was incubated with 1U/10 μg RNA of nuclease P1 (Sigma-Aldrich) in 100 mM ammonium acetate [pH 5.5] for 16 hr at 37 °C. Five microliter of 1 M ammonium bicarbonate [pH 8.3] and 0.5U/10 μg RNA of Bacterial Alkaline Phosphatase (ThermoFisher Scientific) were added for 2 hrs at 37 °C. Following digestion, sample volumes were adjusted to 150 μL with RNase-free water and spin filtered to remove enzymatic constituents (Amicon Ultra 3K, Cat#:UFC500396). Filtrate and washes (200 μL × 3, RNase-free water) were collected and lyophilized. Lyophilized samples were reconstituted in 250 μL H2O containing internal standards (D3-ac4C, 500 nM; 15N3-C, 5 μM, Cambridge Isotopes). Individual samples (15 μL for ac4C analyses, 5 μL for major bases) were then analyzed via injection onto a C18 reverse phase column coupled to a Thermo Quantum Ultra Triple Quadrupole mass spectrometer in positive electrospray ionization mode (Agilent Technologies). Quantification was performed based on nucleoside-to-base ion transitions using standard curves of pure nucleosides and stable isotope labeled internal standards described above.
For attomole sensitivity of ac4C analysis, 100 ng of total or poly(A) RNA was analyzed by LC-MS/MS at the Mass Spectrometry Center at State University of New York (SUNY)-Albany, using a previously reported method (Basanta-Sanchez et al., 2016).
ac4C detection by HPLC
Detection of ac4C by HPLC was performed as described previously (Sinclair et al., 2017). Briefly, RNA was incubated with 1U/10 μg RNA of nuclease P1 (Sigma-Aldrich) in 100 mM ammonium acetate [pH 5.5] for 16 hr at 37 °C. Five microliter of 1 M ammonium bicarbonate [pH 8.3] and 0.5U/10 μg RNA of Bacterial Alkaline Phosphatase (ThermoFisher Scientific) were added for 2 hrs at 37 °C. Following digestion, samples were lyophilized and reconstituted in 10 μL RNase-free water and injected into an Agilent Technologies 1260 Infinity HPLC equipped with a UV detector (Agilent Technologies). Nucleosides were separated on a Kinetex 2.6u C18 100A 100×2.1 mm column at a flow rate of 0.25 mL/min. UV detector was set at 254 nm with a band width of 4 nm. Buffer A: 0.01% formic acid; buffer B: 50% acetonitrile, 0.01% formic acid [pH 3.5] with the gradient as follows: 0−1 min, 100% A; 1−2.4 min, 99.8% A; 2.4−3.8 min, 99.2% A; 3.8− 5.2 min, 98.2% A; 5.2−6.6 min, 96.8% A; 6.6−10 min, 95% A; 10− 12.5 min, 92% A; 12.5−18 min, 70% A; 18−18.5 min, 0% A; 18.5−20 min, 0% A; 20−21 min, 100% A; 21−30 min, 100% A.
ac4C detection by dot blot
Dot blots were performed using rabbit monoclonal anti-ac4C antibodies as described previously (Sinclair et al., 2017). Briefly, 1–10 μg RNA were denatured at 75 °C for 5 min, immediately placed on ice for 1 min and loaded onto Hybond-N+ membranes. Membranes were crosslinked twice with 150 mJ/cm2 in the UV254nm Stratalinker 2400 (Stratagene), blocked with 5% non-fat milk in 0.1% Tween-20 PBS (PBST) for 30 min at room temperature, and probed overnight with anti-ac4C antibody in 1% non-fat milk (1:1000,) at 4 °C. Membranes were next washed three times with 0.1% PBST, incubated with HRP-conjugated secondary anti-rabbit IgG in 1% non-fat milk (1:10000 dilution, Cell Signaling Technology) at 4 °C overnight, washed four times with 0.1% PBST and developed with the SuperSignal ELISA Femto Maximum Sensitivity Substrate (ThermoScientific).
ac4C detection by ImmunoNorthern blot
ImmunoNorthern blot was performed using the NorthernMax kit (ThermoFisher Scientific). Equal amounts of total (20 μg) or poly(A) RNA (10 μg) were mixed with formaldehyde denaturing loading dye, heated to 65 °C for 15 min and separated on 1% agarose denaturing gel containing 1 μg/L ethidium bromide (Sigma-Aldrich). Loading control was verified by UV imaging before transfer. RNA was transferred onto Amersham Hybond-N+ membranes (GE Healthcare) by capillary transfer using 20× SSC buffer (3 M NaCl, 0.3M Na-citrate, [pH 7.-], following the manufacturer’s instructions (ThermoFisher Scientific). Membranes were rinsed with PBS, crosslinked twice with 150 mJ/cm2 in the UV254nm Stratalinker 2400 (Stratagene), blocked with 5% non-fat milk in 0.1% Tween-20 PBS (PBST) for 30 min at room temperature, and probed overnight with anti-ac4C antibody in 1% non-fat milk (1:1000,) at 4 °C. Membranes were next washed three times with 0.1% PBST, incubated with HRP-conjugated secondary anti-rabbit IgG in 1% non-fat milk (1:10000 dilution, Cell Signaling Technology) at 4 °C overnight, washed four times with 0.1% PBST and developed with the SuperSignal ELISA Femto Maximum Sensitivity Substrate (ThermoScientific). Chemiluminescence was detected on X-ray films. Stripping was achieved by microwaving the blots in solution containing 0.1% SDS and 0.1×6 SSC. Hybridization was performed with an 18S rRNA specific 5’ 32P-end labeled oligo (see Table S7 for sequence) at 42°C overnight in oligo probe hybridization buffer (10× Denhardt’s solution, 6× SSC, 0.1% SDS), followed by exposure to a phosphorimager.
In vitro transcription of ac4C-containing RNA probes
β-globin or luciferase DNA templates (see Table S7 for oligonucleotide sequence) were in vitro transcribed using the MAXIscript T7 Transcription Kit (ThermoFisher Scientific), according to the manufacturer’s instructions. For modified transcripts, ac4CTP (Sinclair et al., 2017), m5CTP (Trilink), or hm5CTP (Trilink) replaced CTP in the reaction mix. Incorporation and stability of ac4C in RNA probes was assessed by dot blots and HPLC.
Analysis of ac4C effects on reverse transcription
To evaluate whether ac4C affects reverse transcription, first strand cDNA synthesis was performed on the C- or ac4C-RNA probes (see in vitro transcription). For this purpose, primer complementary to the probe sequence (5 pmol, 5’-CACATTCTACC-3’) was radiolabeled with 20 U T4 Polynucleotide kinase (NEB) and 10 μCi 32P-γATP (3000 Ci/mmol, PerkinElmer) in a 10 μl reaction. The radiolabeled primer was subsequently annealed to 25 ng of probe by first heating at 65 °C for 5 min followed by 5 min at room temperature. To remove excess of non-incorporated 32P-γATP, reactions were filtered using Illustra™ MicrosSpin™ G-50 Columns (GE Healthcare). RT reactions were initiated by adding one volume of a mixture containing 0.5 mM dNTPs, 50 U SuperScript III (ThermoFisher Scientific), 40 U RNase Out (ThermoFisher Scientific), 1× SuperScript buffer (ThermoFisher Scientific), 5 mM MgCl2 and 0.05 mM DTT in a 20 μl reaction for 15, 30, 60 or 120 sec at 50 °C. Reactions were stopped by heating at 95 °C for 5 min. Template RNA was digested with 2U RNase H (ThermoFisher Scientific) for 30 min at 37 °C and reactions were stopped by adding one volume of 2× loading dye (95% formamide, 0.025% bromophenol blue, 0.025% xylene cyanol, 0.5 mM EDTA) and heating at 95 °C for 5 min. RT products were resolved in 8M Urea/8% PAGE gels and examined through phosphorimager analysis.
Acetylated RNA immunoprecipitation (acRIP)
To confirm anti-ac4C antibody enrichment potential, RNA immunoprecipitation was performed on the in vitro transcribed probes. Briefly, anti-ac4C antibody (1 μg) or rabbit monoclonal IgG Isotype control (1 μg) were pre-coupled to 300 μg Protein G Dynabeads (ThermoFisher Scientific) in PBS for 1 hr at room temperature. DNase-treated total RNA (1 μg) from HeLa cells was spiked with 10 pg (1:10−5), 1 pg (1:10−6) or 0.1 pg (1:10−7) of ac4C- or CRNA and immunoprecipitated for 4 hr at 4 °C in 100 μl of acRIP buffer containing PBS, 0.05% Triton X-100, 0.1 % BSA, 40U murine RNase inhibitor (NEB) and anti-ac4C or IgG pre-coupled Protein G Dynabeads. After immunoprecipitations, beads were washed five times in acRIP buffer and elution of RNA was carried out by RNase-free Proteinase K (50 μg, ThermoFisher Scientific) digestion in 100 μl buffer containing 50 mM Tris-HCl [pH7.5], 75 mM NaCl, 6.25 mM EDTA and 1 % SDS for 1h at 37 °C. RNA was extracted by Phenol:Chloroform [pH 4.5] and ethanol precipitation using 0.3M sodium acetate [pH 5.5] and 15 μg/mL linear acrylamide. RNA in the acRIPs was reversed transcribed using the Superscript III system and a mouse β-globin probe specific reverse primer (Table S7) according to the manufacturer’s suggestions. The level of probe in the acRIPs was evaluated by qPCR using LightCycler 480 SYBR Green I Master (Roche, Basel, Switzerland) in a LightCycler 96 Instrument (Roche). Data are represented as ac4C-RNA levels relative to C-RNA levels.
For immunoprecipitation of acetylated 18S rRNA, total RNA was fragmented using the NEBNext® Magnesium RNA Fragmentation buffer for 4 min at 94 °C. Fragmented RNA (10 μg) was immunoprecipitated with 1 μg anti-ac4C or IgG pre-coupled to Protein G Dynabeads as described above. Immunoprecipitated RNAs and 1% inputs were reverse transcribed using random hexamers and the levels of 18S rRNA, an acetylated RNA, or 28S rRNA and 5S rRNA, non-acetylated RNAs, were analyzed by qPCR and represented as percentage of input. Primers used in qPCR analyses are described in Table S7. Since the ac4C site in helix 45 is located near the 3’ end, proximal to two m62A (N6, N6-dimethyladenosine) sites that impair reverse transcription, qPCR-quality primers to test 18S rRNA enrichment are located 150 nucleotides (nt) 5’ of the acetylation site in helix 45, but 239 nt 3’ of the acetylation site in helix 34.
Acetylated RNA immunoprecipitation and sequencing (acRIP-seq)
Poly(A) RNA from parental and NAT10−/− was used for acRIP-seq analysis. To evaluate clones representing distinct residual ac4C levels, in replicate 1, the NAT10−/− RNA sample corresponded to the pool of NAT10−/−A and NAT10−/−C. In replicate 2, the NAT10−/− sample corresponded only to NAT10−/−A. Poly(A) RNA was isolated by two rounds of oligod(T) selection and fragmented using NEBNext® Magnesium RNA Fragmentation buffer for 5 min at 94 °C. Eight picograms of ac4C-RNA probe was spiked into 8 μg of fragmented poly(A) RNA followed by immunoprecipitation with 1 μg anti-ac4C or IgG pre-coupled to Protein G Dynabeads as described above (see Acetylated RNA immunoprecipitation). Illumina libraries were constructed for inputs (parental vs. NAT10−/−, replicate 1 and 2), acRIPs (parental vs. NAT10−/−, replicate 1 and 2), and IgG (parental, replicate 1) using the NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®. Libraries were multiplexed on an Illumina HiSeq2500 instrument using TruSeq V4.0 chemistry and sequenced for 126 cycles in paired-end mode. See Table S1 for sample details for all sequencing experiments.
For validation of acRIP-seq, poly(A) RNA was immunoprecipitated as above. Immunoprecipitated RNAs and 1% inputs were reverse transcribed using random hexamers and the levels of ac4C-positive and ac4C-negative regions within the same transcript were analyzed by RT-qPCR. Primers used in qPCR analyses are described in Table S7.
NAT10 immunoprecipitation
Cells were grown to reach ~80% confluency in 15 cm dishes, rinsed with cold PBS, placed on ice and detached mechanically using a cell scraper in 5 mL of cold PBS. Cells were centrifuged at 2,000 rpm for 5 min at 4 °C and resuspended in 1 mL NP40 lysis buffer containing 0.5 % (v/v) NP40, 50 mM HEPES [pH 7.5], 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5 mM fresh DTT, 1× EDTA-free protease inhibitor cocktail III (EMD Millipore) and 400 U/mL murine RNase inhibitor. Lysates were treated with 2 U/mL DNase I at 37 °C for 5 min and immediately put on ice followed by clearing at 13,000 rpm for 5 min. Protein concentration was quantified using the Bradford reagent and adjusted to 2 mg/mL. Per each IP, 2.5 μg polyclonal anti-NAT10 antibody (ProteinTech, Cat#:13365–1) or 2.5 μg rabbit polyclonal IgG control (Cell Signaling Technology, Cat#2729S), were pre-coupled to 900 μg Protein G Dynabeads for 1 hr at room temperature.
Cell Lysates (1 mg protein) from HeLa WT or NAT10−/−A cells were immunoprecipitated overnight at 4 °C in 500 μL of NP40 lysis buffer. After immunoprecipitations, supernatants were collected and beads were washed five times in RIP buffer containing 0.05 % NP40, 50 mM HEPES [pH 7.5], 300 mM KCl, 0.5 mM fresh DTT, 1× EDTA-free protease inhibitor cocktail III and 400 U/mL murine RNase inhibitor. Elution was achieved by adding 100 μL of RIP elution buffer (20 mM Tris pH 8.0, 2% SDS) and incubating at 95 °C for 5 min. A fraction of the IPs (5%), inputs and supernatants were reserved for Western blot analysis. RNA was next extracted by Phenol:Chloroform [pH 4.5] and ethanol precipitation using 0.3M sodium acetate [pH 5.5] and 15 μg/mL linear acrylamide. RNA in the NAT10-RIPs and inputs (1%) were spiked in with an in vitro transcribed luciferase RNA and reversed transcribed using the Superscript III system with random hexamers according to the manufacturer’s suggestions. The levels of specific ac4C(+) and ac4C(−) transcripts were assessed by qPCR using LightCycler 480 SYBR Green I Master in a LightCycler 96 Instrument. Data are normalized to the spiked luciferase RNA and represented as percentage of input in HeLa WT and NAT10−/−A. Primers sequences are detailed in Table S7.
Transcriptome analysis by RNA-seq
For expression profiling, total RNA was isolated from two biological replicates of parental and NAT10−/−A HeLa cells. Sequencing libraries were constructed with the Illumina TruSeq Stranded Total RNA Library Prep Kit (RS-122–2201), including RiboZero treatment. Libraries were multiplexed on one lane of an Illumina HiSeq2500 instrument using TruSeq V4.0 chemistry and sequenced for 126 cycles in paired-end mode. See Table S1 for sample details for all sequencing experiments.
Determination of mRNA half-life
Transcriptome-wide analysis of mRNA half-life was determined using the 5’-bromo-uridine (BrU) immunoprecipitation chase-deep sequencing (BRIC-seq) method, as previously described (Tani et al., 2012). For this purpose, parental HeLa or NAT10−/−A cells were incubated in complete DMEM medium containing 150 μM BrU (Sigma-Aldrich) for 24 hr. Cells were washed twice with PBS and medium was replaced with complete DMEM medium containing 150 μM Uridine (Sigma-Aldrich) for 0, 2, 4, 8 and 16 hrs. After each time point, medium was removed and Trizol was added directly to culture dishes. Total RNA was isolated and DNase-treated as described in the section “RNA purification.”
To obtain a BrU-labeled RNA, plasmid DNA encoding a partial sequence of the firefly luciferase mRNA under control of the T7 RNA polymerase promoter (pJC880, sequence detailed in Table S7) was linearized with NotI-HF then purified by phenol/chloroform extraction and ethanol precipitation. Probe was in vitro transcribed using the MAXIscript T7 Transcription by replacing UTP with BrUTP (Sigma-Aldrich).
For immunoprecipitations, mouse anti-BrU antibody (2 μg, Cat#:555627, BD Biosciences) was pre-coupled to 300 μg Protein G Dynabeads in PBS for 1 hr at room temperature. Ten micrograms of DNase-treated total RNA from each time point was spiked with 1 ng BrU-labeled luciferase RNA and immunoprecipitated for 2 hr at 4 °C in 200 μl of BRIC buffer containing 0.5× PBS, 0.025% Triton X-100, 0.05 % BSA, 5 mM Tris-HCl [pH7.0], 0.5 mM EDTA, 40U murine RNase inhibitor (NEB) and anti-BrU pre-coupled Protein G Dynabeads. After immunoprecipitation, beads were washed five times in BRIC buffer and elution of RNA was carried out by adding 500 μl of Trizol directly to the beads. RNA was extracted by the Trizol method.
Illumina libraries were constructed from two biological replicates using the NEBNext® UltraII™ Directional RNA Library Prep Kit for Illumina®, including the NEBNext® rRNA Ribodepletion step (NEB). Libraries were multiplexed on an Illumina HiSeq2500 instrument using TruSeq V4.0 chemistry, and sequenced for 126 cycles in paired-end mode. See Table S1 for sample details for all sequencing experiments.
To study the effect of NAT10 ablation on specific targets, BrU labeling and immunoprecipitation of mRNA was performed in four replicates of parental and NAT10−/−A HeLa cells as described above. Immunoprecipitates were analyzed by RT-qPCR using gene specific primers (See Table S7 for sequence) and normalized to the levels of immunoprecipitated BrU-labeled spiked probe. Normalized mRNA levels at each time point were further normalized to time zero to obtain the fraction of mRNA remaining. Decay graphs were generated using PRISM (Version 7.0a) and applying the One-Phase Decay model and setting the intercept (Time 0h) to 1 and plateau to 0. The statistical test used to determine differences in decay rates was the Extra sum-of-squares F test.
Polysome isolation and analysis by Northern blot
Parental and NAT10−/−A cells were grown in complete DMEM medium to 80% confluency. Following quick aspiration of media, plates were placed on top of liquid nitrogen and immediately transferred to ice. Lysis buffer (500 μL, 10 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, 100 μg/mL cycloheximide) was added and cells were scraped into lysis buffer. Cell lysates were passed through a 26-G needle 10 times and clarified by spinning at 20,000 × g for 10 min at 4 °C. Four OD260 units were loaded on a 15%–45% sucrose gradient prepared in gradient buffer (0.5 M Tris-acetate [pH 7.0], 0.5 M NH4Cl, 0.12M MgCl2) using a BioComp Gradient Master (1:48, 81.5°, 17 rpm) and spun at 41,000 rpm in a SW 41 Ti rotor for 2:26 hr at 4°C. Fractionation, recording of 260 nm absorbance and fractions collection were performed using a Brandel Density Gradient Fractionation system.
RNA from each of 16 collected fractions was ethanol precipitated overnight at −80°C. Pelleted RNA was resuspended in LET (25 mM Tris-HCl, pH 8.0, 100 mM LiCl, 20 mM EDTA, pH 8.0) and SDS to 1%, extracted twice with phenol/chloroform/LET, and then ethanol precipitated using NH4OAc and 1 μL Glycoblue (ThermoFisher Scientific AM9515). Following resuspension in LET, samples were run on 1.4% agarose/5.92% formaldehyde gels and transferred onto Hybond-N membrane (GE Healthcare RPN303N). POLR2A and EEF1A1 were detected by Northern blot analysis following incubation with 5’ 32P-end labeled oligo probes oJC3234 and oJC3740, respectively (Table S7), at 42°C overnight in oligo probe hybridization buffer (10× Denhardt’s solution, 6× SSC, 0.1% SDS). FUS mRNA was detected by Northern blot analysis using an asymmetric PCR probe using oJC3732 as template and oJC3733 as reverse oligo. Probing for FUS mRNA was performed overnight at 42°C in asymmetric PCR probe hybridization buffer (50% formamide, 5× SSC, 1× Denhardt’s solution, 0.5 mg/mL fish sperm DNA, 10 mM EDTA, and 0.2% SDS).
Ribosome profiling
Parental and NAT10−/−A cells were grown in complete DMEM medium to 80% confluency in three 10 cm plates per sample per replicate. Media was quickly aspirated, and cells were flash frozen by placing the plates on top of liquid nitrogen and immediately transferred to ice. Cells were scraped on wet ice into 500 μL lysis buffer (10 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM KCl, 1% Triton X-100, 2 mM DTT, and 100 μg/mL cycloheximide), which was transferred from one plate to the next to collect the cells from all three plates for each sample. Cell lysates were next triturated 10 times with a 26-gauge needle and clarified by centrifugation at 20,000 × g for 10 min at 4°C.
For ribosome profiling, 300–450 μL of each lysate was treated with 0.3 U/μL RNase I for 40 min at room temperature, while the remaining lysate was flash frozen in liquid nitrogen for later total RNA isolation. Following the RNase I digestion, 5 μL of Superase-In were added to each sample, and the samples were loaded onto 15–45% sucrose gradients which were prepared, centrifuged, and fractionated as described in the section “Polysome isolation and analysis by Northern blot”. Ribosome footprint RNA was isolated from fractions containing 80S monosomes using two phenol/chloroform/LET (25 mM Tris-HCl, pH 8.0, 100 mM LiCl, 20 mM EDTA, pH 8.0) extractions, and depleted of ribosomal RNA only once, using the human/mouse/rat Ribo-Zero Gold rRNA removal kit (Illumina).
For the total RNA fragmented controls, 200 μL LET was added to each 50 μL aliquot of lysate, and total RNA was extracted once with phenol/LET, once with phenol/chloroform/LET, and once with chloroform. Following ethanol precipitation, the total RNA was treated with DNase I (Sigma-Aldrich). An RNA Spike-In mix (ThermoFisher Scientific) was added to 5 μg of DNase I-treated total RNA according to the manufacturer’s instructions, and ribosomal RNA was depleted using the human/mouse/rat Ribo-Zero Gold rRNA removal kit. Next, the total RNA samples were fragmented in alkaline fragmentation buffer (1 mM EDTA, 50 mM Na2CO3, 50 mM NaHCO3, [pH 9.2] for 40 min at 95°C, as described previously (Ingolia, 2010), and fragments between 26 and 34 nucleotides in size were gel purified and used for library preparation in parallel with the ribosome profiling samples as described above (Ingolia, 2010).
In vitro translation assay
Plasmid DNA encoding firefly luciferase mRNA under the control of the T7 RNA polymerase promoter (pLGENB1) was linearized with BamHI and purified by phenol/chloroform extraction and ethanol precipitation. Luciferase mRNA was transcribed by incubating 2 μg of linearized DNA, 1× transcription buffer (40 mM Tris-HCl [pH 8.0], 6 mM MgCl2, 10 mM DTT, 2 mM spermidine), 1 mM each of ATP, GTP, and UTP with either ac4CTP or CTP, and 80 units of T7 RNA polymerase (Roche) in a final volume of 40 μL at 37°C for 2 hours. mRNA was then precipitated using 2.5 M lithium chloride at −20°C overnight. mRNA pellets were washed with 70% ethanol, air dried, then resuspended in H2O to 1 μg/μL. mRNA integrity was assessed by running 1 μg of each mRNA on a 1% agarose gel.
To evaluate the effect of wobble-ac4C on translation efficiency, a firefly luciferase construct lacking cytidines in wobble positions was generated. Briefly, a luciferase sequence in which all wobble cytidines were replaced through synonymous substitutions was purchased as a gBlock and cloned into pLGENB1 using the Gibson Assembly Cloning Kit (NEB). 127 out of a total of 550 codons were substituted (sequence detailed in Table S7). In vitro transcription was performed as described above.
In vitro translation assays were performed by incubating 20 ng of luciferase mRNA containing ac4C or cytidine with 3.5 μL of rabbit reticulocyte lysate (Promega) in a final volume of 5 μL at 30°C. Reactions were stopped at 20, 40, 60, and 80 minutes by putting reactions on dry ice. For luciferase assays, reactions were diluted with 95 μL of 1 mg/mL BSA, then 2 μL was mixed with 25 μL of luciferase assay reagent (Promega) in polystyrene tubes. Relative light units (RLU) were immediately measured for 10 seconds in a Lumat LB 9507 luminometer. The effect of wobble substitutions in the presence or absence of ac4C was determined as: Δ% translation= 100 * [(RLUwobble C/RLUwobble A,G,U)t1 - (RLUwobble C/RLUwobble A,G,U)t0], where t1= 20–80 min. and t0=20 min.
In vitro capping and polyadenylation of luciferase mRNA
Plasmid DNA encoding firefly luciferase mRNA under control of the T7 RNA polymerase promoter (pLGENB1) was linearized with BamHI and purified by phenol/chloroform extraction and ethanol precipitation. CTP or ac4CTP-containing firefly luciferase mRNAs were transcribed in vitro with the HiScribe T7 High Yield RNA Synthesis Kit (NEB). Transcription reactions were purified by LiCl-precipitation and were subsequently capped with a 7-methylguanosine cap using the Vaccinia Capping System (NEB) according to the manufacturer’s instructions. Capped transcripts were purified by LiCl-precipitation and 4.3 μg of each transcript were 3’-polyadenylated for 15 min at 37 °C using E. coli Poly(A) Polymerase (NEB) in a 15 μL reaction. Polyadenylation reactions were purified by LiCl-precipitation before being used for transfection.
For assessment of poly(A) tail lengths, a PCR-based assay was performed. Briefly, 100 ng of in vitro transcribed RNA was heated with 0.2 pmols of a preadenylated linker (Cat#S1315S, NEB) at 80°C for 2 min and then was cooled to room temperature for 5 min. Reactions to ligate the linker to the RNA were prepared using truncated T4 RNA ligase 2 (NEB) and were incubated for 2 hours at 24°C with gentle agitation. The RNA was then LiCl-precipitated before being isopropanol-precipitated with NaOAc and GlycoBlue (ThermoFisher Scientific). Next, reverse transcription of the RNA was performed with SuperScript III using a primer that binds to the adenylated linker (primer oJC3288). Subsequently, PCR amplification was performed using PfuTurbo DNA Polymerase (Agilent 600252) and primers oJC3291/oJC3841 (primers used are described in Table S7). PCR products were resolved on a 2% agarose gel with size differences indicating differences in poly(A) tail length.
Transfection of HeLa cells with luciferase mRNA
HeLa cells at ~75% confluency were split 1:2 the day before transfection. Immediately prior to transfection, cells were detached by trypsinization, pelleted, resuspended in complete DMEM and then diluted to 275,000 cells/mL. Cells in 900 μL DMEM were transfected in suspension with 150 ng of capped and polyadenylated CTP- or ac4CTP-containing firefly luciferase transcripts per time point. Transfections were scaled up such that cells for all time points were transfected at once. Transfection reactions were prepared using the TransIT-mRNA Transfection Kit (Mirus) according to the manufacturer’s instructions. Immediately following addition of the transfection complexes to the cells, 900 μL of cells were aliquoted into each of three wells on a 12-well plate for each time point (3 and 6 hours) and plates were incubated at 37°C/5% CO2. Once the cells for the 3 and 6 hour time points were plated, 900 μL of cells were aliquoted into 1.5 mL tubes in triplicate for the 0 hour time point. After adding 500 μL PBS, cells were pelleted, resuspended in 500 μL PBS, and then 75 μL was transferred to a new tube to be used for RNA isolation. Cells were re-pelleted, frozen on dry ice, and stored at −80°C until further use. At exactly 3 hours or 6 hours, cells from three wells transfected with CTP- and ac4CTP-containing firefly luciferase mRNA were scraped into the media present in the well and transferred to a 1.5 mL tube. The wells were re-scraped into 500 μL PBS to collect residual cells which were combined with the cells from the first scraping. Cells were then pelleted and washed exactly as was done for the 0 hour time points.
For luminescence detection, HeLa cells transfected with CTP- or ac4CTP-containing firefly luciferase transcripts were lysed in 100 μL 1× Passive Lysis Buffer (Promega) for 15 min at room temperature and 20 μL of lysate were mixed with an equal volume of ONE-Glo EX Reagent (Promega). Luminescence was measured using a Lumat LB 9507 Luminometer (Berthold Technologies).
Translation initiation assay
Plasmid DNA encoding firefly luciferase mRNA under control of the T7 RNA polymerase promoter (pLGENB1) was linearized with BamHI then purified by phenol/chloroform extraction and ethanol precipitation. Luciferase mRNA was transcribed and radiolabeled by incubating 1 μg of linearized DNA, 1× transcription buffer, 1 mM each of ATP, GTP, and UTP with either ac4CTP or CTP, 2 μL UTP [α−32P] (800 Ci/mmol), and 40 units of T7 RNA polymerase (Roche) in a final volume of 20 μL at 37°C for 2 hours. mRNA was then precipitated using 2.5 M lithium chloride at −20°C overnight. mRNA pellets were washed with 70% ethanol, air dried, then resuspended in 20 μL H2O and radioactivity determined by liquid scintillation counting. mRNA integrity was assessed by autoradiography after running 100,000 cpm of each mRNA on a 1.4% agarose-formaldehyde gel and transferring to a nylon membrane.
In vitro translation initiation complexes were assessed by incubating 333,330 cpm of ac4C or C containing luciferase mRNA with 35 μL of rabbit reticulocyte lysate (Promega; L4960) and either 1 mM GMPPNP (5’-Guanylyl imidodiphosphate, Sigma-Aldrich) or 1 mM GTP (Promega) in a final volume of 50 μL at 30°C. Reactions were stopped after 10 minutes by placing on ice and were then layered onto 5 – 30% (w/v) sucrose gradients prepared using a BioComp Gradient Master in 1× gradient buffer (50 mM Tris-acetate pH=7.0, 50 mM NH4Cl, 12 mM MgCl2, 1 mM DTT). Sucrose gradients were centrifuged in a SW 41 Ti rotor at 41,000 rpm for 2 hours and 26 minutes at 4°C, and then fractionated using a Teledyne Isco Foxy R2. Luciferase mRNA in each gradient fraction was determined by counting half of each fraction (300 μL) using a liquid scintillation counter.
Analysis of Xrn-1 digestion
For each individual reaction, 100,000 cpm of luciferase mRNA (with or without ac4C) was incubated with 0.5 U XRN-1 (NEB) in 1× NEBuffer 3 at 37°C. Reactions were stopped at the indicated times by adding an equal volume of 2× gel loading dye (50% formamide, 6.67% formaldehyde, 1× MOPS buffer, 0.8 mg/mL ethidium bromide, 40 mM EDTA). Samples were then heated at 65°C for 10 minutes, loaded on a 1.4% agarose-formaldehyde gel, transferred to nylon membrane, and signal determined by autoradiography.
Chromatin immunoprecipitation
Antibodies (5 μg polyclonal or 5 μg monoclonal) were pre-bound to 200 μL Protein G magnetic beads (Thermo Fisher Scientific) by overnight incubation in 1 mL PBS/5% BSA. Antibodies used included rabbit monoclonal anti-Histone H3 (clone D2B12, Cat#:4620S, Cell Signaling Technologies), rabbit polyclonal anti-Histone H3 (acetyl K9+K14+K18+K23+K27, Cat#:ab47915, Abcam), rabbit monoclonal IgG control (Cat#:3900S, Cell Signaling Technologies) and rabbit polyclonal IgG control (Cat#:2729S, Cell Signaling Technologies). After 3 washes in PBS/5% BSA, beads were resuspended in 100 μL PBS/5% BSA. Cells were dissociated from the plate using 0.05% trypsin-EDTA and crosslinked at room temperature with 1% formaldehyde (Sigma-Aldrich) for 5 min. Crosslinking was quenched with 125 mM glycine (ICN Biomedical) for 10 min. Cell membranes were lysed using cold NP-40 buffer (1% NP40, 150 mM NaCl, 50 mM Tris–HCl; pH 8.0) and nuclei collected by centrifugation at 12 000 × g for 1 min at 4 °C. Nuclear pellets were resuspended to a concentration of 200 million cells/mL in ChIP sonication buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl; pH 8.0), supplemented with Halt protease inhibitors (Thermo Scientific), and chromatin sheared to an average size between 150 and 400 bp by sonication (Bioruptor Twin, Diagenode). Chromatin preparations were cleared by centrifugation at 20 000 × g for 10 min at 4 °C and chromatin was diluted 10-fold in ChIP dilution buffer (1.1% Triton X-100, 0.01% SDS, 167 mM NaCl, 1.2 mM EDTA, 16.7 mM Tris–HCl; pH 8.1). 100 μL antibody-bead slurry was added to 1 mL diluted chromatin containing 20 million cell equivalents and incubated overnight with rotation at 4 °C. Immune complexes were washed 5 times with LiCl wash buffer (250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 100 mM Tris-HCl; pH 7.5) and once with TE (0.1 mM EDTA, 10 mM Tris-HCl; 7.5). Beads were resuspended in IP Elution Buffer (1% SDS, 0.1 M NaHCO3) and crosslinking reversed by overnight incubation at 65 °C. DNA was purified by column purification (QIAGEN) and qPCR was performed using SYBR Green chemistry (Roche) and gene-specific primers. Acetylated H3 enrichment was determined relative to pan-histone H3 [2^(CTpan-H3 – CTAcH3)].
QUANTIFICATION AND STATISTICAL ANALYSES
Identification of ac4C peaks
Raw reads were pre-processed to remove low quality bases and adapter sequences. Reads were mapped to the human genome (hg19) with Tophat2 v.2.1.1 (Trapnell et al., 2009). Parameters used were: reporting at most one alignment per read (-g 1), allowing maximally five mismatches per read (>95% matching), and supplying the Ensembl Release 75 gene annotation. A post-alignment filter removed alignments to mitochondrial DNA (chrM) and non-concordant mate pairs. Separate alignments to the spiked mouse β-globin probe sequence (Table S7) and to the 43kb human ribosomal DNA complete repeating unit (GenBank U13369.1) were performed with Bowtie2 (Langmead and Salzberg, 2012), to specifically analyze reads originating from these features.
Since sites of acetylation in transcripts will be non-contiguous in genomic coordinates, and difficult to assign to alternative isoforms, we sought a representative transcript reference for continuous peak calling. To this end, canonical transcript sequences were downloaded from the UCSC genome browser and used in the generation of a Bowtie2-based index (Langmead and Salzberg, 2012). Reads were aligned in local alignment mode with Bowtie2 (Langmead and Salzberg, 2012). MACS2 was used for peak calling (Zhang et al., 2008), with parameters selected to optimize performance with transcript mapped reads (i.e., turning off the shifting model and local lambda, and using transcript bases as the genome size). Input samples were used as controls for peak calling.
In our MACS2 peak calling, each multi-base-pair peak includes ≥ 1 “summit,” or local peak maxima, defined at a single base position. Each of these summits is a putative “ac4C site.” The acRIP-seq approach is not a base-resolution method, so these ac4C sites are not required to be cytidines. To make a single set of peak and summit definitions that incorporate replicate experiments, we performed peak calling on pooled data. We subjected these peak calls to stringent filtering to remove artifacts such as non-specific binding to immunoglobulin (IgG) and replicability as follows: (i) Peaks were compared to select only those sites with a reduction in signal in NAT10−/− as compared to parental (HeLa), following the expectation that non-artefactual ac4C sites would show diminished signal when the enzyme is reduced. To acquire an enrichment value for comparison, we used the bedtools map function (Quinlan, 2014) to extract the value from the MACS2 pileup output at the position of the summit called in HeLa WT. Peak summits with pileup values higher in HeLa WT than NAT10−/− passed this filtering step. (ii) To remove peaks that result from non-specific IgG binding, we intersected ac4C peaks with peaks called in IgG-IP, and kept only those that had no coordinate overlap (Bedtools, (Quinlan, 2014)). (iii) We next required detection in replicate experiments, by requiring peaks called in the pooled data to overlap with peaks called in each individual replicate. (iv) To investigate the possibility of mapping errors, we spot-checked peak calls against genomically aligned reads to confirm concordance. The list of ac4C(+) targets is presented in Table S3.
For downstream analysis, we analyzed ac4C sites that resided in protein coding genes. When analyzing ac4C localization within UTR and CDS, we counted each summit that was called within the gene, excluding outliers where > 4 sites were called within the gene.
Position analysis of acetylated sites
To generate a heatmap of ac4C summits within target mRNAs, binned ac4C enrichments over transcripts were transformed into consistent lengths with deepTools (Ramírez et al., 2014) using the computeMatrix scale-regions command. Enrichment is displayed as log2 ratio (acRIP/Input). The resulting matrix was loaded into R, and rows were sorted by the position of the maximum signal as percentage of total transcript length. The heatmap was produced using the non-negative matrix factorization package (http://cran.r-project.org/package=NMF). For other representations of peak location within transcripts or transcript features (CDS and UTR), the relative sizes of CDS and UTR were parsed from annotation BED files, and intersected with summit positions from parsed MACS2 output. Since some target genes may have multiple ac4C peaks, we counted the number of peak summits within each transcript feature (Table S4). When contrasts are made between the relative representation of transcript features, we classify transcripts with summits in >1 feature as “ambiguous,” and exclude them from analysis (Table S4).
Analysis of gene expression and mRNA splicing
Raw reads were processed and aligned to the human genome (hg19), exactly as described for acRIP-seq. Afterwards, genomic alignments were used to quantify transcripts using HTSeq v 0.6.1p1 (Anders et al., 2015) against the Ensembl 75 annotation. Differential expression was calculated using DESeq2 (Love et al., 2014). Overall gene expression levels were filtered based on a summed count of ≥ 1 for all samples. A significance threshold of FDR adjusted p < 0.05 was classified as statistically significant differential expression (Table S2). Only genes with protein coding annotation according to the Ensembl 75 annotation were used for downstream analysis. Prior to classifying transcripts by acetylation status, normalized gene level abundances were generated via variance stabilization normalization (vsn) within DESeq2. These abundances were visualized in scatterplots pairwise for all samples inter se, and between averaged values for HeLa WT and NAT10−/−A. Correlations were calculated with the Pearson correlation coefficient using the cor command in R.
Genes were filtered as acetylated (ac4C+) or non-acetylated (ac4C-), as determined in the acRIP-seq (Table S3), and merged with DEseq2 output for log2 fold expression differences in NAT10 −/−A compared to HeLa WT cells. ac4C(+) transcripts were further separated by the position of summit as 5’UTR, CDS and 3’UTR (Table S4) and merged with DEseq2 output for log2 fold expression differences. Transcripts with summits in two different locations were called “ambiguous” and were excluded from the analysis. The ggplots package in R was used to compute and plot the empirical cumulative distributions. A Kolmogorov-Smirnov (KS) test was used to compare the cumulative distributions of ac4C(+) vs. ac4C(−) transcripts or the cumulative distribution of ac4C(−) vs. 5’UTR, ac4C(−) vs. CDS or ac4C(−) vs. 3’UTR.
To perform the comparison of intronic reads between NAT10−/−A and HeLa WT cells, we calculated intron read-through in the RNA-Seq data using the Spanki tool (Sturgill et al., 2013). Intron read-through for each splice junction was calculated with the spankijunc command and default parameters. To reduce any confounding effects from overlapping alternative isoforms, we restricted our analysis to splice junctions where there were no other isoforms using their donor or acceptor site. A scaling factor was applied to the NAT10−/−A counts to match the total sequencing depth of the HeLa WT sample.
To analyze global splicing differences between NAT10−/−A and HeLa WT, we used rMATS v3.2.5 (Shen et al., 2014) to quantify differences in exon inclusion. After concatenating results together across splicing event types (skipped exons, alternative donors, etc.), we applied a filter on total event abundance (total counts >= 50). Splicing events were then compared within transcripts by acetylation status.
Gene ontology and functional category analyses
Gene ontology (GO) analysis of genes differentially expressed (DE) in response to NAT10−/−was carried out with the Database for Annotation Visualization and Integrated Discovery (DAVID) tool. All DE genes were compared to the transcriptome to identify enrichment of biological processes.
To determine the differential effect of acetylated and non-acetylated transcripts on biological functions, genes were further filtered on the bases of ac4C enrichment in the acRIP-seq (Table S3) as ac4C(+) or ac4C(−). Ingenuity pathway analysis (IPA) software (Qiagen, www.ingenuity.com) was used to compare the statistically significant dysregulated ac4C(−) and ac4C(+) transcripts (adjusted p < 0.05, Table S2) using the option “comparison analysis” and the activation z-core tool with a cutoff p-value < 0.0001. For visualization, only the molecular and cellular functions were displayed. However, redundant categories, including tissue-specific and cancer-type-specific categories, were discarded.
Analysis of Serine/Leucine amino acid bias in differentially expressed transcripts
The R package Biostrings (v2.42.1) was used to calculate amino acid frequencies. Briefly, Ensembl Release 75 peptide sequences were downloaded and the longest transcript was chosen for amino acid frequency calculations. Transcripts were then divided on the basis of the DEseq2 output on whether their expression was unaltered or differential (adjusted p-value of < 0.05) in parental WT versus NAT10−/−A HeLa cells.
Estimation of mRNA half-life
Raw reads were mapped to a modified human genome (hg19 with the luciferase probe sequence added) using Hisat2 v2.0.5 (Kim et al., 2015). Parameters used were: reads that failed to align were not reported (--no-unal), library strandedness was specified (--rna-strandness FR), and a list of known splice sites was generated from the Ensembl 75 annotation (--known-splicesite-infile). Due to skewed sequencing depth, parental HeLa (WT) Rep-1 T2 was down-sampled to a depth representative of the other samples. Afterwards, genomic alignments were used to quantify transcripts using HTSeq v 0.6.1p1 (Anders et al., 2015) against the edited Ensembl 75 annotation with added probe. Library depth normalized gene expression counts were calculated using DESeq2 (Love et al., 2014). Then, all normalized gene count values in a particular time point were divided by the probe value for that time point. Data was further normalized to the first time point and log transformed. Half-lives were calculated by use of a linear model (Table S5). Half-lives for HeLa WT and NAT10−/−A were calculated together within one linear model. A maximum half-life value of 24 hours was applied to calculated values exceeding this value. Genes with R2 values of < 0.8 were excluded from further analysis. Only genes with protein coding annotation according to the Ensembl 75 annotation were used for downstream analysis. Genes were further filtered as acetylated (ac4C+) or non-acetylated (ac4C-) as determined in the acRIP-seq (Table S3), and merged with BRIC-seq data for half-life differences. Additionally, ac4C(+) transcripts were further separated by the position of summit as 5’UTR, CDS and 3’UTR (Table S4). The ecdf function in R was used to compute the empirical cumulative distribution. A Kolmogorov-Smirnov test was used to compare the cumulative distributions of ac4C(+) vs. ac4C(−) transcripts or the cumulative distribution of ac4C(−) vs. 5’UTR, ac4C(−) vs. CDS or ac4C(−) vs. 3’UTR.
Estimation of ribosome density and translation efficiency
Ribosome protected reads were first trimmed of adaptor sequence and reads arising from rRNA were removed from the dataset based on alignment to a set of ribosomal RNA sequences using bowtie2. Remaining reads were then mapped to the UCSC hg19 canonical transcriptome with Tophat (v 2.1.1), with the –prefilter-multihits option enabled to screen out reads that may have arisen from elsewhere in the genome. Uniquely mapped reads were then assigned to the specific codon estimated to be within the A-site of the protecting ribosome, based on identification of the P-site offset from the corresponding peak in read density that occurs up-stream of the start codon (Ingolia, 2010), with the added constraint that each read was mapped to the nearest in-frame codon and under the assumption that all reads will be in frame with the associated gene.
To assess the change in ribosome density over CDS as functions of condition (NAT10−/−A vs. HeLa WT) and the acetylation status of a transcript (ac4C+ vs. ac4C-), we utilized the DESeq2 package within R to normalize ribosome density by mRNA levels for each transcript: T.E. = normalized ribosome protected readstranscript / normalized RNAseq readstranscript (Table S6). These values were then averaged between biological replicates, loaded into python, and plotted as cumulative distribution functions using the ecdf function from numpy.
Analysis of codon biases at A-sites
Codon-level ribosome densities were calculated using various numpy functions in python from arrays of ribosomal A-site counts and codon counts for each transcript. For a given codon, transcript-level A-site density was calculated as: Codon A-site densitytranscript = (Codon A-site countstranscript,/ Total A-site countstranscript) / (Codon counttranscript / Total codon counttranscript). This calculation was repeated for each codon on subsets of transcripts randomly resampled 5000 times to calculate a mean A-site density (y-axis) for each codon and confidence intervals. The human genetic code was analyzed to separate codons for which wobble C substitutions have no influence on amino acid identity from codons for which wobble C substitutions result in coding for distinct amino acids or stop codons. In addition, tRNA sequence diversity and anticodon loop modifications were determined through MODOMICS (Boccaletto et al., 2018).
Sequence motif analysis
The RNA sequence within ac4C peaks was analyzed for the occurrence of over-represented motifs. We performed de-novo motif finding using MEME (Multiple EM for Motif Elicitation, v 4.11.2) in standalone mode (Bailey and Elkan, 1994). We ran MEME with a maximum motif width of 12bp, and also with unrestricted length. Sequence logos presented were produced within MEME output.
Other Statistical Analyses
Number of replicates, statistical tests and p-values are specified in the figures and figure legends.
DATA AND SOFTWARE AVAILABILITY
Generated high-throughput sequencing datasets are publicly available in the Gene Expression Omnibus (GEO) under accession number GSE102113.
Supplementary Material
Table_S1_Summary_of_sequencing_experiments_related_to_Fig1–7.pdf
Table_S2_Differential_gene_expression_related_to_Fig_1and4.xlsx
Table_S3_List_of_acetylated_transcripts_related_to_Fig3.xlsx
Table_S4_Peak_locations_related_to_Fig3.xlsx
Table_S5_mRNA_half_life_related_to_Fig5.xlsx
Table_S6_Translation_Efficiency_related_to_Fig6.xlsx
Table_S7_Oligonucleotides_related_to_Fig1–7.xlsx
Figure S1. Generation and characterization of NAT10−/− cells, related to figure 1.
(A) Schematic of NAT10 protein domains and transcript isoforms. Five different protein coding isoforms are annotated for NAT10, all of which share exon 5 within the DUF1726 domain.
(B) HeLa cells were transfected with guide RNA (sgRNA) directed against exon 5 of NAT10 to target all potential protein coding isoforms. Indels (red) in NAT10 alleles were confirmed by Sanger sequencing in three different clones.
(C) Sashimi plot of NAT10 expression in RNA-seq performed in parental (WT) HeLa cells and NAT10−/− clone A (NAT10−/−A). Minor residual NAT10 expression may be attributed to low level exclusion of exon 5, generating an in-frame protein lacking 41 amino acids of the DUF1726.
(D) Western blot using antibody against cleaved caspase-3, a marker of apoptosis. The positive control corresponds to parental HeLa cells treated with 50 J/m2 and incubated at 37 °C for 7 hr post UV treatment. Representative of n=3.
(E) Cell cycle distribution was determined using propidium iodide staining and flow cytometry in parental, NAT10+/+, NAT10−/−A and NAT10−/−B HeLa cells grown to 80–90% confluency. Data represent percentage of cells in different stages of the cell cycle as indicated. Mean ± SEM, n = 3.
(F) Scatter plots of gene expression estimates from RNA-Seq experiments, all samples compared inter se. Values are depth normalized and variance stabilized. Pearson correlation coefficient (r) inset.
(G) LC-MS/MS evaluation of ac4C levels in total RNA from parental and NAT10−/−A HeLa cells for absolute detection at attomole concentrations (Basanta-Sanchez et al., 2016). Analysis was performed at the Mass Spectrometry facility, SUNY-Albany. Mean ± SEM, n=3. * p < 0.05. Two-tailed student’s t-test.
(H) Anti-ac4C dot blots of serially diluted unmodified or chemically acetylated polycytidylic acid (PolyC) (top) or in vitro transcribed RNA probes containing C, ac4C, m5C or hm5C (bottom).
(I) NAT10−/−A HeLa cells were transfected with empty pcDNA5/FRT vector (q) or vector encoding full-length NAT10 (FL). RNA acetylation was analyzed by Immuno-Northern blot. Representative of biological triplicates.
(J) Western blots of reported substrates of NAT10-catalyzed protein acetylation. Acetyl-specific antibodies to α-tubulin (Ac-α-tub), histone 3 (Ac-H3) and p53 (Ac-p53), as well as antibodies to total protein, were utilized. Representative of biological triplicates.
(K) Representative Western blot of acetylated proteins in NAT10−/−A HeLa cells transfected with empty pcDNA5/FRT (θ) or vector encoding full-length NAT10 (FL).
(L-N) cDNA encoding full-length NAT10 (FL) or NAT10 lacking the RNA helicase domain (Δh, deletion of amino acids 259–502) was integrated in single copy in Flp-In-TRex-293 cells, in which the NAT10 gene was inactivated through CRISPR-Cas9 (NAT10−/−), as in HeLa. RNA and protein acetylation were analyzed by anti-ac4C Immuno-Northern blot (M) or Western blots (N), respectively.
Figure S2. Characterization of ac4C levels in poly(A) RNA, related to figure 2.
(A) Poly(A) RNA was isolated using two rounds of oligo(dT)25 beads and purity verified through size distribution in bioanalyzer profiling.
(B) Chromatograms are representative of LC-MS/MS performed in total and poly(A) RNA from parental HeLa cells. Blank and ac4C standards were included as controls.
Figure S3. acRIP validation and specificity, related to figure 3.
(A) Schematic of the ac4C site in helix 45 of human 18S rRNA (left). Immunoprecipitation with anti-ac4C antibody (acRIP) or IgG control in fragmented total RNA from parental and NAT10-/HeLa cells. Enrichment of acetylated 18S rRNA and the non-acetylated controls 28S rRNA and 5S rRNA were evaluated by RT-qPCR. Mean ± SEM, n=3. * p < 0.001. Two-way ANOVA followed by Tukey’s post hoc test.
(B) Enrichment of in vitro transcribed ac4C-RNA probe in acRIP-seq data. Normalized probe counts in the acRIP or IgG fraction were divided by the normalized probe counts in input to determine probe fold enrichment. As would be expected considering the reduced density of endogenous substrates, the ac4C(+) spike-in was more efficiently recovered in NAT10−/− cells, suggesting that residual ac4C in NAT10−/− cells may in fact be overestimated.
(C) Browser views of 28S rRNA in parental and NAT10−/− acRIP-seq. Data are displayed as input-subtracted IP reads per kilobase per million (RPKM), mapped to the ribosome subunit.
(D) Number of ac4C summits per gene.
(E) RT-qPCR validation of acRIP-seq. Browser views show location of PCR
amplicons (magenta) in the acRIP-seq defined ac4C-positive and ac4C-negative regions (top). Immunoprecipitated RNAs and inputs were reverse transcribed and the levels of ac4C-positive and ac4C-negative regions within the same transcript were analyzed by qPCR (bottom). Data represent transcript levels relative to the determined ac4C-rich region in parental HeLa cells. Mean ± SEM, n=3. * p < 0.05. Two-Way ANOVA followed by Tukey’s post hoc test.
(F) Cell lysates from parental (WT) and NAT10−/−A HeLa cells were immunoprecipitated with anti-NAT10 antibodies or isotypic IgG control. NAT10 levels in the inputs, IPs and flow-through were determined through Western blot. GAPDH blotting confirmed IP specificity (top). RNA was isolated from NAT10 immunoprecipitates and enrichment of defined ac4C(+) and ac4C(−) transcripts evaluated by RT-qPCR. Dots represent mean percent of input from four biological replicates for each specific transcript. Error bars indicate the average and SD within each category. Statistical significance between ac4C(+) and ac4C(−) transcripts in HeLa WT and NAT10−/−A was determined using One-Way ANOVA followed by Tukey’s post hoc test.
Figure S4. ac4C modulates transcript levels post-transcriptionally, related to figure 4.
(A) Functional category analysis of all transcripts that were differentially expressed in NAT10−/−relative to parental HeLa cells, or specifically in the subset of acetylated (ac4C+) or non-acetylated (ac4C-) mRNAs. Activation z-score was calculated and visualized using Ingenuity Pathway Analysis. Blue indicates predicted inhibition while orange indicates predicted activation of specified functional categories.
(B) Pan-acetyl H3 and total H3 ChIP-qPCR were performed in parental and NAT10−/−A HeLa cells. Primers were directed to predicted regions of high [Ac-H3 region (+)] and low histone acetylation [Ac-H3 region (−)] (HeLa-S3 ChIP-seq, UCSC genome browser) in two ac4C(+) transcripts (FUS and POLR2A) and two ac4C(−) transcripts (GAPDH and RPS16). Acetyl H3 signals are normalized to total H3 signals to account for potential changes in nucleosome density. Data are displayed as Mean ± SD from two independent biological replicates.
Figure S5. NAT10 regulates mRNA stability of acetylated transcripts, related to figure 5.
(A) BrU-labeled RNA was prepared and immunoprecipitated from parental and NAT10−/−A HeLa cells at different time points, as described in Fig. 5A, followed by RT-qPCR using specific primers for defined ac4C(+) and ac4C(−) mRNAs. Transcript levels at each time point were normalized to recovery of an acetylated spike-in. Data are represented as the percentage of mRNA remaining relative to time 0 hr from four biological replicates. Decay graphs were generated by applying the One-Phase Decay model and the Extra sum-of-squares F test was used to determine the statistical significance of differences in decay rates. CDS indicates genes with ac4C peaks within the CDS while 3’UTR indicates genes with ac4C peaks within the 3’UTR.
(B) In vitro transcribed mRNA containing either ac4C or unmodified cytosine was treated in the presence of purified XRN1 for the indicated times and resolved on a denaturing agarose gel. RNA was detected by phosphorimaging.
Figure S6. Examination of pleiotropic effects associated with NAT10 ablation, related to figure 6.
(A) Scatter plots depicting the log2 fold change (NAT10−/−A vs. HeLa WT) in gene expression as a function of serine (left) and leucine (right) percentage for all protein coding genes analyzed in the RNA-seq (Figures 4A and 4B).
(B) Boxplots illustrating serine (left) or leucine (right) percentage in genes that were unaltered in NAT10−/−A compared to parental (WT) HeLa (unchanged), versus downregulated ac4C(−) or ac4C(+) genes. The boxes indicate the range between first and third quartiles and whiskers represent the highest and lowest values within 1.5 multiples of the inter-quartile range. Outliers from the inter-quartile range are plotted as individual dots.
(C) Bar plots illustrating the absolute value of average differential ribosome protected fragments (RPF) density [log2 (NAT10−/−A vs. HeLa WT)] at all codons per amino acid.
(D) In vitro reporters containing or lacking ac4C were programed into an in vitro translation system in the presence of either GTP or GMP-PNP. Ribonucleoprotein complexes were separated by sucrose density gradient sedimentation. Graphs indicate the percent of radiolabeled reporter in each fraction. Positions of 48S and 80S complexes are indicated.
Figure S7. Analysis of codon biases within ac4C peaks, related to figure 7.
(A) Codon bias within ac4C(+) transcripts relative to the transcriptome. Red bars highlight codons with C in the wobble position. Horizontal lines indicate the magnitude of codon bias expected by random sampling, at the significance level of p = 0.01 or p = 1e-4, as indicated.
(B) Codon bias within CDS-localized ac4C peaks, relative to the transcriptome, as described in (A).
(C) Sequence logo of enriched motifs within ac4C peaks determined using MEME. Enrichment p-value (E-value) derived from FDR corrected Fisher’s Exact Test.
(D) Numerous ac4C motifs containing wobble site cytidines were identified within the broad POLR2A acRIP-seq peak. Specific locations are indicated through the red boxes.
(E) Efficiency of polyadenylation in ac4C(+) and ac4C(−) firefly luciferase mRNA was estimated using a PCR-based tailing assay.
(F) HeLa cells were transfected with in vitro transcribed ac4C(−) or ac4C(+) luciferase mRNA and phosphorylation of eIF2α, an indicator of translation inhibition, was evaluated by Western blot. Osmotic stress (200 mM NaCl) and untransfected cells were used as a positive and a negative control, respectively.
(G) HeLa cells were transfected with in vitro transcribed ac4C(+) or ac4C(−) firefly luciferase mRNA along with unmodified Nanoluciferase as an external control. Cells were harvested and assayed for firefly or Nanoluciferase activity at the indicated times. Relative light units were normalized by mRNA levels at each time point and represented as fold enrichment relative to time 20 min.
(H) ac4C(−) or ac4C(+) luciferase mRNAs were radiolabeled during in vitro transcription and transfected into HeLa cells. mRNA was isolated at the indicated time points and the integrity and abundance of luciferase mRNA was evaluated by denaturing agarose gel electrophoresis and phosphorimaging.
(I) Firefly luciferase mRNA naturally containing C within wobble sites (+wobble C) or with synonymous codon substitutions that removed C from all wobble sites (-wobble C) was generated in the presence of CTP, followed by in vitro translation in rabbit reticulocyte lysates. Luciferase activity was monitored at the indicated time points, and the effect of wobble site substitutions (-wobble C) was determined through comparison to + wobble C values. Mean ± SEM, n=3.
Highlights.
- NAT10 catalyzes N4-acetylcytidine (ac4C) modification of a broad range of mRNAs
- mRNA acetylation within coding sequences promotes translation and mRNA stability
- ac4C in wobble sites stimulates translation efficiency
Acknowledgements:
We thank the members of the Center for Cancer Research Sequencing Facility at the National Cancer Institute (Frederick, MD) for providing Illumina sequencing services. We thank Dr. Lin Qishan from the Mass Spectrometry Center at the RNA Institute SUNY-Albany (Albany, NY) for providing LC-MS/MS services. This study utilized the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). This work is supported by the Intramural Research Program of NIH, the National Cancer Institute, The Center for Cancer Research. Support for J.C. was provided by NIH (GM118018, GM125086).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
REFERENCES
- Agris PF, Vendeix FA, and Graham WD (2007). tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366, 1–13. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, and Elkan C (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2, 28–36. [PubMed] [Google Scholar]
- Basanta-Sanchez M, Temple S, Ansari SA, D’Amico A, and Agris PF (2016). Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res 44, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. (2018). MODOMICS: a database of RNA modification pathways. Nucleic Acids Res 46, D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. (2012). Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406. [DOI] [PubMed] [Google Scholar]
- Chimnaronk S, Suzuki T, Manita T, Ikeuchi Y, Yao M, Suzuki T, and Tanaka I (2009). RNA helicase module in an acetyltransferase that modifies a specific tRNA anticodon. EMBO J 28, 1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. (2016). Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285. [DOI] [PubMed] [Google Scholar]
- Dong C, Niu L, Song W, Xiong X, Zhang X, Zhang Z, Yang Y, Yi F, Zhan J, Zhang H, et al. (2016). tRNA modification profiles of the fast-proliferating cancer cells. Biochem Biophys Res Commun 476, 340–345. [DOI] [PubMed] [Google Scholar]
- Hanson G, and Coller J (2018). Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19, 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenschild R, Tserovski L, Schmid K, Thuring K, Winz ML, Sharma S, Entian KD, Wacheul L, Lafontaine DL, Anderson J, et al. (2015). The reverse transcription signature of N-1-methyladenosine in RNA-Seq is sequence dependent. Nucleic Acids Res 43, 9950–9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT (2010). Genome-wide translational profiling by ribosome footprinting. Methods Enzymol 470, 119–142. [DOI] [PubMed] [Google Scholar]
- Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, and Suzuki T (2014). Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18S ribosomal RNA. J Biol Chem 289, 35724–35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, and Klaholz BP (2015). Structure of the human 80S ribosome. Nature 520, 640–645. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhar BV, Kamble AD, and Sonawane KD (2013). Conformational preferences of modified nucleoside N4-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem Biophys 66, 797–816. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu D, Britton S, Demir M, Rodriguez R, and Jackson SP (2014). Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, Deng H, Luo J, Ke Y, and Du X (2016). NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep 17, 349–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Liu H, Wang Q, Tang Z, Hou L, and Zhang B (2003). Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem Biophys Res Commun 311, 506–513. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R, Ginell SL, De NC, and Chheda GB (1978). Conformation of N4-acetylcytidine, a modified nucleoside of tRNA, and stereochemistry of codon-anticodon interaction. Biochem Biophys Res Commun 83, 657–663. [DOI] [PubMed] [Google Scholar]
- Quinlan AR (2014). BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Current Protocols in Bioinformatics 47, 11.12.11–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, and Manke T (2014). deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Research 42, W187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Langhendries JL, Watzinger P, Kotter P, Entian KD, and Lafontaine DL (2015). Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res 43, 2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Zheng X, McNutt MA, Guang L, Sun Y, Wang J, Gong Y, Hou L, and Zhang B (2009). NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res 315, 1653–1667. [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, and Xing Y (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 111, E5593–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair WR, Arango D, Shrimp JH, Zengeya TT, Thomas JM, Montgomery DC, Fox SD, Andresson T, Oberdoerffer S, and Meier JL (2017). Profiling Cytidine Acetylation with Specific Affinity and Reactivity. ACS Chem Biol 12, 2922–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, and Preiss T (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40, 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L, and Schulman LH (1978). The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J Biol Chem 253, 6132–6139. [PubMed] [Google Scholar]
- Sturgill D, Malone JH, Sun X, Smith HE, Rabinow L, Samson ML, and Oliver B (2013). Design of RNA splicing analysis null models for post hoc filtering of Drosophila head RNA-Seq data with the splicing analysis kit (Spanki). BMC bioinformatics 14, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, and Lafontaine DL (2013). The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 51, 539–551. [DOI] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, and Akimitsu N (2012). Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res 22, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Miyauchi K, Sakaguchi Y, Yamashita S, Soma A, Tomita K, and Suzuki T (2018). Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat Chem Biol. [DOI] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Sato K, Ishikawa H, Izumikawa K, Yamauchi Y, Hirota K, Nakayama H, Takahashi N, et al. (2018). Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY, Li A, Wang X, Bhattarai DP, Xiao W, et al. (2017). 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res 27, 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome biology 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table_S1_Summary_of_sequencing_experiments_related_to_Fig1–7.pdf
Table_S2_Differential_gene_expression_related_to_Fig_1and4.xlsx
Table_S3_List_of_acetylated_transcripts_related_to_Fig3.xlsx
Table_S4_Peak_locations_related_to_Fig3.xlsx
Table_S5_mRNA_half_life_related_to_Fig5.xlsx
Table_S6_Translation_Efficiency_related_to_Fig6.xlsx
Table_S7_Oligonucleotides_related_to_Fig1–7.xlsx
Figure S1. Generation and characterization of NAT10−/− cells, related to figure 1.
(A) Schematic of NAT10 protein domains and transcript isoforms. Five different protein coding isoforms are annotated for NAT10, all of which share exon 5 within the DUF1726 domain.
(B) HeLa cells were transfected with guide RNA (sgRNA) directed against exon 5 of NAT10 to target all potential protein coding isoforms. Indels (red) in NAT10 alleles were confirmed by Sanger sequencing in three different clones.
(C) Sashimi plot of NAT10 expression in RNA-seq performed in parental (WT) HeLa cells and NAT10−/− clone A (NAT10−/−A). Minor residual NAT10 expression may be attributed to low level exclusion of exon 5, generating an in-frame protein lacking 41 amino acids of the DUF1726.
(D) Western blot using antibody against cleaved caspase-3, a marker of apoptosis. The positive control corresponds to parental HeLa cells treated with 50 J/m2 and incubated at 37 °C for 7 hr post UV treatment. Representative of n=3.
(E) Cell cycle distribution was determined using propidium iodide staining and flow cytometry in parental, NAT10+/+, NAT10−/−A and NAT10−/−B HeLa cells grown to 80–90% confluency. Data represent percentage of cells in different stages of the cell cycle as indicated. Mean ± SEM, n = 3.
(F) Scatter plots of gene expression estimates from RNA-Seq experiments, all samples compared inter se. Values are depth normalized and variance stabilized. Pearson correlation coefficient (r) inset.
(G) LC-MS/MS evaluation of ac4C levels in total RNA from parental and NAT10−/−A HeLa cells for absolute detection at attomole concentrations (Basanta-Sanchez et al., 2016). Analysis was performed at the Mass Spectrometry facility, SUNY-Albany. Mean ± SEM, n=3. * p < 0.05. Two-tailed student’s t-test.
(H) Anti-ac4C dot blots of serially diluted unmodified or chemically acetylated polycytidylic acid (PolyC) (top) or in vitro transcribed RNA probes containing C, ac4C, m5C or hm5C (bottom).
(I) NAT10−/−A HeLa cells were transfected with empty pcDNA5/FRT vector (q) or vector encoding full-length NAT10 (FL). RNA acetylation was analyzed by Immuno-Northern blot. Representative of biological triplicates.
(J) Western blots of reported substrates of NAT10-catalyzed protein acetylation. Acetyl-specific antibodies to α-tubulin (Ac-α-tub), histone 3 (Ac-H3) and p53 (Ac-p53), as well as antibodies to total protein, were utilized. Representative of biological triplicates.
(K) Representative Western blot of acetylated proteins in NAT10−/−A HeLa cells transfected with empty pcDNA5/FRT (θ) or vector encoding full-length NAT10 (FL).
(L-N) cDNA encoding full-length NAT10 (FL) or NAT10 lacking the RNA helicase domain (Δh, deletion of amino acids 259–502) was integrated in single copy in Flp-In-TRex-293 cells, in which the NAT10 gene was inactivated through CRISPR-Cas9 (NAT10−/−), as in HeLa. RNA and protein acetylation were analyzed by anti-ac4C Immuno-Northern blot (M) or Western blots (N), respectively.
Figure S2. Characterization of ac4C levels in poly(A) RNA, related to figure 2.
(A) Poly(A) RNA was isolated using two rounds of oligo(dT)25 beads and purity verified through size distribution in bioanalyzer profiling.
(B) Chromatograms are representative of LC-MS/MS performed in total and poly(A) RNA from parental HeLa cells. Blank and ac4C standards were included as controls.
Figure S3. acRIP validation and specificity, related to figure 3.
(A) Schematic of the ac4C site in helix 45 of human 18S rRNA (left). Immunoprecipitation with anti-ac4C antibody (acRIP) or IgG control in fragmented total RNA from parental and NAT10-/HeLa cells. Enrichment of acetylated 18S rRNA and the non-acetylated controls 28S rRNA and 5S rRNA were evaluated by RT-qPCR. Mean ± SEM, n=3. * p < 0.001. Two-way ANOVA followed by Tukey’s post hoc test.
(B) Enrichment of in vitro transcribed ac4C-RNA probe in acRIP-seq data. Normalized probe counts in the acRIP or IgG fraction were divided by the normalized probe counts in input to determine probe fold enrichment. As would be expected considering the reduced density of endogenous substrates, the ac4C(+) spike-in was more efficiently recovered in NAT10−/− cells, suggesting that residual ac4C in NAT10−/− cells may in fact be overestimated.
(C) Browser views of 28S rRNA in parental and NAT10−/− acRIP-seq. Data are displayed as input-subtracted IP reads per kilobase per million (RPKM), mapped to the ribosome subunit.
(D) Number of ac4C summits per gene.
(E) RT-qPCR validation of acRIP-seq. Browser views show location of PCR
amplicons (magenta) in the acRIP-seq defined ac4C-positive and ac4C-negative regions (top). Immunoprecipitated RNAs and inputs were reverse transcribed and the levels of ac4C-positive and ac4C-negative regions within the same transcript were analyzed by qPCR (bottom). Data represent transcript levels relative to the determined ac4C-rich region in parental HeLa cells. Mean ± SEM, n=3. * p < 0.05. Two-Way ANOVA followed by Tukey’s post hoc test.
(F) Cell lysates from parental (WT) and NAT10−/−A HeLa cells were immunoprecipitated with anti-NAT10 antibodies or isotypic IgG control. NAT10 levels in the inputs, IPs and flow-through were determined through Western blot. GAPDH blotting confirmed IP specificity (top). RNA was isolated from NAT10 immunoprecipitates and enrichment of defined ac4C(+) and ac4C(−) transcripts evaluated by RT-qPCR. Dots represent mean percent of input from four biological replicates for each specific transcript. Error bars indicate the average and SD within each category. Statistical significance between ac4C(+) and ac4C(−) transcripts in HeLa WT and NAT10−/−A was determined using One-Way ANOVA followed by Tukey’s post hoc test.
Figure S4. ac4C modulates transcript levels post-transcriptionally, related to figure 4.
(A) Functional category analysis of all transcripts that were differentially expressed in NAT10−/−relative to parental HeLa cells, or specifically in the subset of acetylated (ac4C+) or non-acetylated (ac4C-) mRNAs. Activation z-score was calculated and visualized using Ingenuity Pathway Analysis. Blue indicates predicted inhibition while orange indicates predicted activation of specified functional categories.
(B) Pan-acetyl H3 and total H3 ChIP-qPCR were performed in parental and NAT10−/−A HeLa cells. Primers were directed to predicted regions of high [Ac-H3 region (+)] and low histone acetylation [Ac-H3 region (−)] (HeLa-S3 ChIP-seq, UCSC genome browser) in two ac4C(+) transcripts (FUS and POLR2A) and two ac4C(−) transcripts (GAPDH and RPS16). Acetyl H3 signals are normalized to total H3 signals to account for potential changes in nucleosome density. Data are displayed as Mean ± SD from two independent biological replicates.
Figure S5. NAT10 regulates mRNA stability of acetylated transcripts, related to figure 5.
(A) BrU-labeled RNA was prepared and immunoprecipitated from parental and NAT10−/−A HeLa cells at different time points, as described in Fig. 5A, followed by RT-qPCR using specific primers for defined ac4C(+) and ac4C(−) mRNAs. Transcript levels at each time point were normalized to recovery of an acetylated spike-in. Data are represented as the percentage of mRNA remaining relative to time 0 hr from four biological replicates. Decay graphs were generated by applying the One-Phase Decay model and the Extra sum-of-squares F test was used to determine the statistical significance of differences in decay rates. CDS indicates genes with ac4C peaks within the CDS while 3’UTR indicates genes with ac4C peaks within the 3’UTR.
(B) In vitro transcribed mRNA containing either ac4C or unmodified cytosine was treated in the presence of purified XRN1 for the indicated times and resolved on a denaturing agarose gel. RNA was detected by phosphorimaging.
Figure S6. Examination of pleiotropic effects associated with NAT10 ablation, related to figure 6.
(A) Scatter plots depicting the log2 fold change (NAT10−/−A vs. HeLa WT) in gene expression as a function of serine (left) and leucine (right) percentage for all protein coding genes analyzed in the RNA-seq (Figures 4A and 4B).
(B) Boxplots illustrating serine (left) or leucine (right) percentage in genes that were unaltered in NAT10−/−A compared to parental (WT) HeLa (unchanged), versus downregulated ac4C(−) or ac4C(+) genes. The boxes indicate the range between first and third quartiles and whiskers represent the highest and lowest values within 1.5 multiples of the inter-quartile range. Outliers from the inter-quartile range are plotted as individual dots.
(C) Bar plots illustrating the absolute value of average differential ribosome protected fragments (RPF) density [log2 (NAT10−/−A vs. HeLa WT)] at all codons per amino acid.
(D) In vitro reporters containing or lacking ac4C were programed into an in vitro translation system in the presence of either GTP or GMP-PNP. Ribonucleoprotein complexes were separated by sucrose density gradient sedimentation. Graphs indicate the percent of radiolabeled reporter in each fraction. Positions of 48S and 80S complexes are indicated.
Figure S7. Analysis of codon biases within ac4C peaks, related to figure 7.
(A) Codon bias within ac4C(+) transcripts relative to the transcriptome. Red bars highlight codons with C in the wobble position. Horizontal lines indicate the magnitude of codon bias expected by random sampling, at the significance level of p = 0.01 or p = 1e-4, as indicated.
(B) Codon bias within CDS-localized ac4C peaks, relative to the transcriptome, as described in (A).
(C) Sequence logo of enriched motifs within ac4C peaks determined using MEME. Enrichment p-value (E-value) derived from FDR corrected Fisher’s Exact Test.
(D) Numerous ac4C motifs containing wobble site cytidines were identified within the broad POLR2A acRIP-seq peak. Specific locations are indicated through the red boxes.
(E) Efficiency of polyadenylation in ac4C(+) and ac4C(−) firefly luciferase mRNA was estimated using a PCR-based tailing assay.
(F) HeLa cells were transfected with in vitro transcribed ac4C(−) or ac4C(+) luciferase mRNA and phosphorylation of eIF2α, an indicator of translation inhibition, was evaluated by Western blot. Osmotic stress (200 mM NaCl) and untransfected cells were used as a positive and a negative control, respectively.
(G) HeLa cells were transfected with in vitro transcribed ac4C(+) or ac4C(−) firefly luciferase mRNA along with unmodified Nanoluciferase as an external control. Cells were harvested and assayed for firefly or Nanoluciferase activity at the indicated times. Relative light units were normalized by mRNA levels at each time point and represented as fold enrichment relative to time 20 min.
(H) ac4C(−) or ac4C(+) luciferase mRNAs were radiolabeled during in vitro transcription and transfected into HeLa cells. mRNA was isolated at the indicated time points and the integrity and abundance of luciferase mRNA was evaluated by denaturing agarose gel electrophoresis and phosphorimaging.
(I) Firefly luciferase mRNA naturally containing C within wobble sites (+wobble C) or with synonymous codon substitutions that removed C from all wobble sites (-wobble C) was generated in the presence of CTP, followed by in vitro translation in rabbit reticulocyte lysates. Luciferase activity was monitored at the indicated time points, and the effect of wobble site substitutions (-wobble C) was determined through comparison to + wobble C values. Mean ± SEM, n=3.