Abstract
Purpose:
The risk of subsequent breast cancer among female childhood cancer survivors is markedly elevated. We aimed to determine genetic contributions to this risk, focusing on polygenic determinants implicated in breast cancer susceptibility in the general population.
Experimental Design:
Whole-genome sequencing (30X) was performed on survivors in the St Jude Lifetime Cohort, and germline mutations in breast cancer predisposition genes were classified for pathogenicity. A polygenic risk score (PRS) was constructed for each survivor using 170 established common risk variants. Relative rate (RR) and 95% confidence interval (95%CI) of subsequent breast cancer incidence were estimated using multivariable piecewise exponential regression.
Results:
The analysis included 1,133 female survivors of European ancestry (median age at last follow-up=35.4 years, range=8.4–67.4) of whom 47 were diagnosed with one or more subsequent breast cancers (median age at subsequent breast cancer=40.3 years, range=24.5–53.0). Adjusting for attained age, age at primary diagnosis, chest irradiation, doses of alkylating agents and anthracyclines, and genotype eigenvectors, RRs for survivors with PRS in the highest vs. lowest quintiles were 2.7 (95%CI, 1.0–7.3), 3.0 (95%CI, 1.1–8.1) and 2.4 (95%CI, 0.1–81.1) for all survivors and survivors with and without chest irradiation, respectively. Similar associations were observed after excluding carriers of pathogenic/likely pathogenic mutations in breast cancer predisposition genes. Notably, the PRS was associated with the subsequent breast cancer rate under the age of 45 years (RR=3.2; 95%CI, 1.2–8.3).
Conclusions:
Genetic profiles comprised of small-effect common variants and large-effect predisposing mutations can inform personalized breast cancer risk and surveillance/intervention in female childhood cancer survivors.
Keywords: subsequent breast cancer, childhood cancer survivors, polygenic determinants
Introduction
Female survivors of childhood cancer, particularly those treated with chest irradiation, have a substantially elevated risk of subsequent breast cancer. The reported cumulative incidence of 30% by age of 50 years for irradiated survivors is among the highest described for any population(1). For female survivors without a history of chest irradiation, high-dose alkylator and anthracycline chemotherapy increase the risk of subsequent breast cancer(2). Our recent study based on whole-genome sequencing (WGS) with deep coverage on blood DNA samples from 3,006 survivors from St Jude Lifetime Cohort Study (SJLIFE)(3–5) showed survivors carrying a pathogenic/likely pathogenic (P/LP) mutation in one of 60 cancer predisposition genes had a significantly increased rate of various subsequent neoplasms including subsequent breast cancer. However, in addition to the monogenic determinants, consisting of rare P/LP mutations with moderate-to-high penetrance, the full allelic spectrum of the genetic architecture of breast cancer susceptibility also encompasses common variants with low penetrance.
In the general population, genome-wide association studies (GWAS) have thus far identified 172 common variants that confer susceptibility to breast cancer with small effect sizes (per-allele odds ratio range=1.03–1.31)(6). Additionally, a small number of risk modifiers have been identified among carriers of BRCA1 or BRCA2 mutations(7,8). Although each of these variants confers only modest risk individually, the combined effect, assessed in the form of a polygenic risk score (PRS), can be substantial. Incorporation of PRS into risk prediction models has shown significant utility in the development of improved tools for population risk screening and stratification(9–11). Based on the variants identified to date, women in the highest 1% of the PRS distribution have a 3.5-fold greater breast cancer risk than the population average(6).
The WGS data of the SJLIFE survivors(3,4) provided a unique opportunity to examine both rare and common genetic variations and model them simultaneously to fully explore the allelic spectrum of the genetic architecture underlying the risk of developing breast cancer among female survivors of childhood cancer. While early screening and risk reduction strategies are recommended for P/LP mutation carriers, a PRS derived from established common risk variants can potentially identify additional high-risk survivors who may benefit from similar cancer surveillance and risk reduction interventions.
Materials and Methods
Study cohort
SJLIFE is a retrospective cohort with prospective clinical follow-up of childhood cancer survivors treated at St. Jude Children’s Research Hospital beginning in 1962(3,4). WGS data were previously generated for 3,006 survivors who consented for a SJLIFE clinical assessment before April 2016 and were followed until December 31st, 2016(5). Female participants in the SJLIFE study were eligible for the current analysis. Ascertainment of subsequent malignancies, including invasive and in situ breast cancer, was carried out using a combination of approaches, including direct clinical care/surveillance, monitoring by the institutional Cancer Registry, systematic searches of the National Death Index, and study questionnaires for self-reporting. All 55 subsequent breast cancer cases (31 ductal/lobular invasive and 24 ductal/lobular in situ) were confirmed by medical records, consisting of pathology reports (n=54) and clinical notes (n=1). The SJLIFE genomic study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board at St. Jude Children’s Research Hospital. Consent for participants under 18 years of age was provided by a parent or legal guardian. All participants aged 14 years and older provided written informed consent; individuals aged between eight and 13 years provided verbal assent.
Population Admixture Analysis
Genotypes for 12,000 loci selected with low linkage disequilibrium (LD) across three continental reference populations(12) were extracted from the WGS data for 3,006 childhood cancer survivors. Similarly, genotype data were extracted from the 1000 Genomes Project Phase 3 version 5 data for CEU (Utah residence with Northern and Western European ancestry, n=99), JPT+CHB (Japanese and Chinese, n=207) and YRI (Yoruban in Nigeria, n=108), and subsequently used as the reference populations in a STRUCTURE analysis which inferred the admixture coefficients for each survivor (https://code.google.com/archive/p/glu-genetics/). Among the females within the SJLIFE cohort, 1133 survivors having ≥ 80% CEU were genetically identified as individuals of European ancestry and retained for further association analysis. In the remaining 278 survivors of non-European ancestry, 236 were African American ancestry (four breast cancers in three survivors), 34 were of Asian/Native-Indian American ancestry (three breast cancers in two survivors), and eight were three-way admixed ancestry (none with breast cancer).
Principal Components Analysis
For the group of female survivors of European ancestry, a principal components analysis was carried out on a genotype matrix as described previously using the GLU software package (https://code.google.com/archive/p/glu-genetics/). The top 10 eigenvectors derived from this analysis were included as adjustment variables in association analyses.
Polygenic risk score of common variants
Odds ratio (OR) for each common variant was calculated using fixed-effects meta-analysis to combine the published individual estimates(6). PRS was calculated as a weighted sum of the number of risk alleles carried by an individual, in which their weights were taken as the natural logarithm of the meta-analysis-estimated ORs of the corresponding loci. Among 172 breast cancer associated loci, one locus (rs71557345) had multiple mappings to GRCh38; another locus (rs140936696) was insufficiently covered (< 10x) in many samples in the WGS data and also lacked a good surrogate variant (pairwise r2 > 0.8). These two loci were excluded, and PRS calculation was based on the remaining 170.
Germline predisposition mutation carriers
The following cancer predisposition genes known to confer a high to moderate risk for developing breast cancer(13) were included for analysis: BRCA1, BRCA2, TP53, PTEN, CDH1, STK11, NF1, PALB2, ATM, CHEK2, and NBN. Survivors with a P/LP mutation (5) were categorized as germline predisposition mutation carriers otherwise as non-carriers.
Standardized incidence ratio, absolute excess risk, and cumulative incidence
Standardized incidence ratio (SIR) was calculated as the ratio of the observed number of females with subsequent breast cancer (clinically confirmed) in the cohort during follow-up to the expected number, which was calculated by applying the general population’s age-, sex-, and calendar year-specific rates of breast cancer in white-race females from the Surveillance, Epidemiology, and End Results (SEER) database(14) to the SJLIFE cohort during the same follow-up. Absolute excess risk (AER) was calculated as the difference between the numbers of observed and expected breast cancer incident cases per 1,000 person-years of follow-up. Cumulative incidence of subsequent breast cancer was calculated by age accounting for left-truncation for non-carriers within the highest (5th) quintile, three intermediate quintiles and the lowest (1st) quintile.
Multivariable regression analyses
Initially, non-genetic baseline models were fit with piecewise exponential regression for the rate of subsequent breast cancer with covariates including age at primary diagnosis, radiotherapy (RT, yes/no), cumulative doses of alkylating agents and anthracyclines, genotype eigenvectors and attained age as cubic splines, for overall and by chest-RT, respectively. Subsequently, for the genetic risk model, the mutation-carrier status was added as a binary variable (0/1) and PRS as a categorical variable (quintiles), with the lowest quintile of PRS (1st quintile) as the reference group, comparing adjusted rate of subsequent breast cancer in the highest quintile of PRS (5th quintile) or middle quintiles (2nd-4th quintiles) versus the lowest quintile of PRS (1st quintile). The quintile cutoffs were determined based on PRS distribution among all female survivors of European ancestry. The following stratified analyses were considered: 1) chest-RT (yes/no); 2) attained age (<45 or ≥45); and 3) mutation non-carriers only.
Results
Distribution of polygenic risk score and prevalence of mutation
Among 3,006 survivors with WGS data available, a total of 1,133 females were genetically identified as individuals of European ancestry with European admixture coefficients >80% and considered for further analyses (Table 1 and supplementary Figure S1). Among this group of survivors (median age at last follow-up=35.4 years, range=8.4–67.4), 47 have been diagnosed with one or more subsequent breast cancers (median age at subsequent breast cancer=40.3 years, range=24.5–53.0). The PRS was calculated using well-established breast cancer susceptibility variants (supplementary Table S1) and the values (median=10.1, range=8.3–12.2) are shown in supplementary Table S2. The PRS follows approximately a normal distribution (supplementary Figure 2). The quintile cutoffs were close to the expected numbers based on minor allele frequencies from the OncoArray control population(15) (supplementary Table S3). Thirty-four survivors (3.0%) were carriers of P/LP mutations in one of the 11 breast cancer predisposition genes, with four mutation carriers having developed subsequent breast cancer (supplementary Table S4).
Table 1.
Characteristics of Female Survivors of European Ancestry in the SJLIFE Cohort
| Characteristic | Survivors with subsequent BC |
Survivors without subsequent BC |
|---|---|---|
| N (%) | N (%) | |
| Total | 47 | 1086 |
| Race | ||
| Non-Hispanic White | 47 (100%) | 1069 (98%) |
| Hispanic White | 0 (0%) | 13 (1%) |
| Other | 0 (0%) | 4 (0%) |
| Diagnosis | ||
| Leukemia | ||
| Acute lymphoblastic leukemia | 3 (6%) | 389 (36%) |
| Acute myeloid leukemia | 2 (4%) | 34 (3%) |
| Other leukemia | 0 (0%) | 1 (0%) |
| CNS tumors | ||
| Astrocytoma or glioma | 1 (2%) | 58 (5%) |
| Medulloblastoma or PNET | 0 (0%) | 22 (2%) |
| Ependymoma | 0 (0%) | 12 (1%) |
| Other | 0 (0%) | 21 (2%) |
| Lymphoma | ||
| Hodgkin lymphoma | 30 (64%) | 119 (11%) |
| Non-Hodgkin lymphoma | 4 (9%) | 52 (5%) |
| Sarcoma | ||
| Ewing sarcoma | 0 (0%) | 36 (3%) |
| Osteosarcoma | 1 (2%) | 37 (3%) |
| Rhabdomyosarcoma | 0 (0%) | 29 (3%) |
| Non-rhabdomyosarcoma | 2 (4%) | 33 (3%) |
| Embryonal | ||
| Wilms tumor | 2 (4%) | 89 (8%) |
| Neuroblastoma | 0 (0%) | 60 (6%) |
| Germ cell tumor | 1 (2%) | 25 (2%) |
| Other | ||
| Retinoblastoma | 0 (0%) | 28 (3%) |
| Hepatoblastoma | 0 (0%) | 6 (1%) |
| Melanoma | 0 (0%) | 5 (0%) |
| Carcinomas | 1 (2%) | 15 (1%) |
| Other | 0 (0%) | 15 (1%) |
| Radiation (%) | ||
| Any | 40 (85%) | 606 (56%) |
| Cranial radiation therapy | 5 (11%) | 345 (32%) |
| Breast | 36 (77%) | 149 (14%) |
| Pelvis | 10 (21%) | 187 (17%) |
| Chemotherapy | ||
| Alkylating agent | 30 (64%) | 597 (55%) |
| Anthracyclines | 27 (57%) | 636 (59%) |
| Epipodophyllotoxins | 8 (17%) | 408 (38%) |
| Mercaptopurine | 5 (11%) | 436 (40%) |
| Methotrexate | 12 (26%) | 548 (50%) |
| Platinum agents | 2 (4%) | 120 (11%) |
| Vinca alkaloids | 34 (72%) | 795 (73%) |
| Corticosteroids | 17 (36%) | 529 (49%) |
| Median (range) | Median (range) | |
| Age at diagnosis (years) | 15.2 (3.1–21.2) | 6.1 (0.01–22.7) |
| Age at follow-up (years) | 44.5 (24.9–61.1) | 35.0 (8.4–67.4) |
| Length of follow-up (years) | 31.2 (13.4–51.2) | 28.1 (5.6–53.5) |
Standardized incidence ratio and absolute excess risk of subsequent breast cancer
Table 2 shows a SIR of 5.7 (95%CI, 4.3–7.4) comparing the survivor cohort overall with the general population, with subgroup SIRs ranging from 2.9 (95%CI, 1.0–6.3) for survivors with PRS in the lowest quintile to 9.4 (95%CI, 5.7–14.7) in the highest quintile. AER was 1.7 (95%CI, 1.2–2.4) per 1,000 person years of follow-up, ranging from 0.7 (95%CI, 0.0–2.0) for survivors with PRS in the lowest quintile to 3.2 (95%CI, 1.8–5.2) in the highest quintile.
Table 2.
Standardized Incidence Ratio and Absolute Excess Risk of Subsequent Breast Cancer among Female Childhood Cancer Survivors of European Ancestry, Overall and by Polygenic Risk Score Quintile
| PRS Quintiles | Observed | Expected | SIR | AER per 1,000 person years |
|---|---|---|---|---|
| 1 | 6 | 2.1 | 2.9 (1.0 – 6.3) | 0.7 (0.0 – 2.0) |
| 2 | 6 | 1.9 | 3.4 (1.2 – 7.3) | 0.8 (0.1 – 2.1) |
| 3 | 14 | 2.1 | 6.6 (3.6 – 11.0) | 2.3 (1.1 – 4.1) |
| 4 | 10 | 1.7 | 5.9 (2.8 – 10.9) | 1.7 (0.6 – 3.3) |
| 5 | 19 | 2.0 | 9.4 (5.7 – 14.7) | 3.2 (1.8 – 5.2) |
| Overall | 55 | 9.7 | 5.7 (4.3 – 7.4) | 1.7 (1.2 – 2.4) |
Cumulative incidence of subsequent breast cancer by polygenic risk score
After excluding survivors carrying P/LP mutations in breast cancer predisposition genes, the cumulative incidence of subsequent breast cancer increased with PRS (Figure 1 and supplementary Figure S3). By the age of 45 years, the cumulative incidence was 15.5%, 4.2% and 2.8%, respectively, for survivors with PRS in the highest quintile, three intermediate quintiles and the lowest quintile. Among 43 survivors without a predisposing mutation who developed subsequent breast cancer, 15 had a PRS in the highest quintile, 22 in three intermediate quintiles and six in the lowest quintile.
Figure 1. Cumulative Incidence of Subsequent Breast Cancer by Polygenic Risk Score after Excluding Carriers of Breast Cancer Predisposing Mutations.
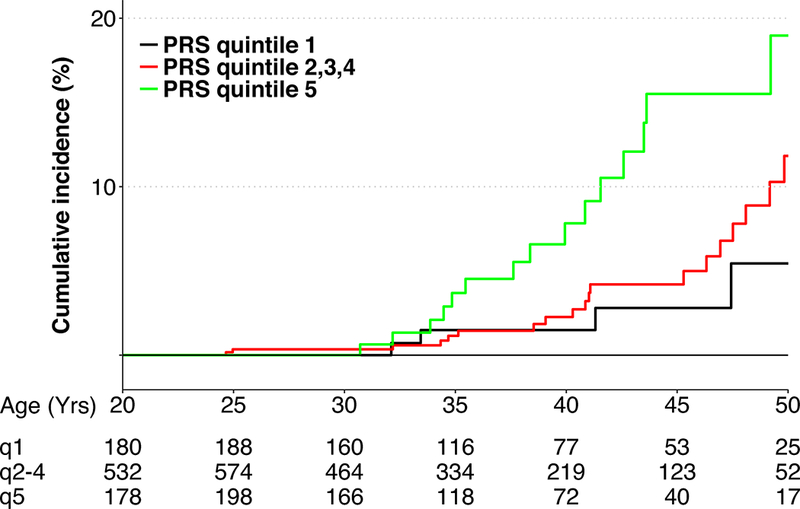
The table below X-axis listed the number of survivors at risk by age and by the polygenic risk scores in the lowest quintile (q1), middle quintiles (q2–4), and the highest quintile (q5).
Monogenic and polygenic risk associations
Clinical risk-factor models were fit for the overall cohort and stratified by chest-RT status (supplementary Table S5). Table 3 shows results after adding both the mutation-carrier status as a binary variable and PRS as a categorical variable. The adjusted relative rates (RR) of subsequent breast cancer were 21.8 (95%CI, 7.1–66.8) for mutation carriers vs. non-carriers, and 2.7 (95%CI, 1.0–7.3) for survivors with PRS in the highest vs. lowest quintiles. Among irradiated survivors, these RRs were 10.3 (95%CI, 3.6–29.8) and 3.0 (95%CI, 1.1–8.1), respectively. Among non-irradiated survivors, the RRs were 42.8 (95%CI, 9.9–183.8) and 2.4 (95%CI, 0.1–81.1), respectively. The RR of PRS in the non-irradiated survivors was comparable to that in the irradiated survivors, but was not statistically significant, likely related to the small number of subsequent breast cancer cases (14 occurrences in 11 survivors) among non-irradiated survivors. In both overall and irradiated survivors, higher PRS was associated with the elevated breast cancer rate (Ptrend<0.02). Similar associations were observed after excluding mutation carriers (supplementary Table S6). Of note, the PRS was associated with the rate of subsequent breast cancer among survivors under the age of 45 years (RR=3.2; 95% CI, 1.2–8.3; Ptrend=0.011) but not clearly over the age of 45 years (Table 4).
Table 3.
Monogenic and Polygenic Associations with Subsequent Breast Cancer among Female Childhood Cancer Survivors of European Ancestry
| Total survivors 55 (47)b |
Chest-RT 41 (36)b |
Chest non-RTa 14 (11)b |
||||
|---|---|---|---|---|---|---|
| Variable | RR (95% CI) | P | RR (95% CI) | P | RR (95% CI) | P |
| PRS quintiles | 0.011c | 0.009c | 0.56c | |||
| 1 | Ref | NA | Ref | NA | Ref | NA |
| 2 | 0.9 (0.3 – 3.0) | 0.89 | 0.8 (0.2 – 2.7) | 0.71 | 2.0 (0.1 – 28.2) | 0.62 |
| 3 | 2.1 (0.7 – 5.9) | 0.18 | 1.9 (0.7 – 5.6) | 0.24 | 5.0 (0.3 – 85.6) | 0.26 |
| 4 | 2.1 (0.8 – 5.7) | 0.16 | 1.6 (0.5 – 5.3) | 0.45 | 5.3 (0.4 – 69.1) | 0.21 |
| 5 | 2.7 (1.0 – 7.3) | 0.05 | 3.0 (1.1 – 8.1) | 0.03 | 2.4 (0.1 – 81.1) | 0.62 |
| Mutation Effect | 21.8 (7.1 – 66.8) | <.001 | 10.3 (3.6 –29.8) | <.001 | 42.8 (9.9 – 183.8) | <.001 |
For chest non-irradiated survivors, tertiles of anthracycline were combined into a binary variable (yes vs. no) so the model would converge.
Number of occurrences of subsequent breast cancer (number of unique survivors)
P value for trend for PRS
Table 4.
Age-specific Effect for Polygenic Risk Score on the Rate of Subsequent Breast Cancer after Excluding Carriers of Predisposing Mutation
| Total survivorsa | ||||
|---|---|---|---|---|
| <45 years | ≥45 years | |||
| Variable | RR (95% CI) | P | RR (95% CI) | P |
| PRS Quintile | 0.011b | 0.540b | ||
| 1 | Ref | Ref | ||
| 2 | 0.4 (0.1 – 1.8) | 0.22 | 1.3 (0.3 – 6.0) | 0.69 |
| 3 | 1.3 (0.4 – 4.4) | 0.71 | 3.5 (0.9 – 14.4) | 0.08 |
| 4 | 1.7 (0.6 – 4.9) | 0.29 | 2.5 (0.4 – 14.6) | 0.30 |
| 5 | 3.2 (1.2 – 8.3) | 0.02 | 0.7 (0.1 – 7.3) | 0.79 |
Model did not converge for either chest RT or non chest-RT survivors.
P value for trend for PRS
Discussion
To our knowledge, this is the first study to evaluate the joint effects of rare predisposing mutations with moderate-to-high penetrance and common susceptibility variants implicated in the etiology of breast cancer in the general population among childhood cancer survivors for whom subsequent breast cancer risk is known to be elevated by exposures to treatment of childhood cancer(1,2,16). Multivariable regression analysis revealed that survivors with PRS in the highest quintile have a 3-fold increased rate of subsequent breast cancer as compared to survivors with PRS in the lowest quintile. Moreover, our results showed a clear PRS effect on the rate of subsequent breast cancer among younger survivors (<45 years of age). The differential PRS effect by age was also evident as the cumulative incidence curves deviated more dramatically before reaching 45 years of age (Figure 1).
Monogenic predisposing mutations contributed partly to the overall higher incidence of subsequent breast cancer among survivors because their carrier prevalence is 10-fold higher among survivors compared with controls(5). However, germline mutations in TP53 represent the only gene mutation that is etiologically related to both pediatric cancer (e.g. leukemias) and breast cancer, as described in Li-Fraumeni syndrome(17).
The distribution of PRS was similar between survivors of childhood cancer and the general population, suggesting that the PRS does not contribute to the markedly elevated risk of subsequent breast cancer among survivors. Nevertheless, we showed that the PRS constructed from the findings in the general population is also associated with breast cancer risk in this high-risk population of childhood cancer survivors. Within the high-risk survivor population, even a small increase in relative risk by PRS can translate to substantial increase in absolute risk. Survivors in the top range of PRS values would therefore potentially benefit from intervention measures to lessen the risk, and screening with appropriate frequency and modality to improve the likelihood of early detection. In our study, survivors with PRS in the top 5% have 1.9-fold increased rate using the middle PRS-quintile survivors as the baseline (supplementary Table S7), and more than a 10-fold increase if compared with the average of the general population (the overall SIR = 5.7). Based on these data, we would be able to identify more than 50 high-risk survivors beyond the 34 P/LP mutation carriers who would then serve as candidates for screening and possible cancer risk reducing measures in our cohort of 1,133 female survivors.
There are limitations of our study that need to be considered in the interpretation of our findings. First, survivors in the SJLIFE cohort are still relatively young and the effect of rare P/LP mutations and common risk variants on breast cancer risk might change as the members of the cohort age. Hence, we should continue to follow-up and monitor changes in risk with increasing age, particularly in the older age ranges when breast cancer incidence is higher in the general population. It will also be important to investigate the PRS in larger heterogeneous survivor cohorts to further define the potential clinical utility of the PRS within the context of precision preventive medicine and to further inform clinical practice recommendations(18). Lastly, our analysis was restricted to survivors of European ancestry and thus there is a need to assess risk associated with rare P/LP mutations and common variants (i.e. PRS) within other non-European ethnic groups(19–22).
In summary, we demonstrate that the rates of subsequent breast cancer are significantly increased for survivors with a higher PRS and/or carrying a P/LP mutation, independent of prior chest-irradiation and other clinical risk factors. These findings suggest that genetic profiles comprised of rare P/LP mutations and common susceptibility variants improve identification of those survivors at highest risk and may facilitate implementation for personalized breast cancer surveillance and prevention among female childhood cancer survivors. In addition, the study provides a working paradigm for investigation of monogenic and polygenic contributions to other late effects(23), for instance, obesity among survivors of childhood cancer. With further advancement in the genetics and genomics contributing to complex human diseases, practical utilization of the PRS (polygenic) in combination with rare predisposing mutations (monogenic) to guide precision prevention is on the horizon(24–26).
Supplementary Material
Translational Relevance.
Breast cancer is the most common subsequent malignant neoplasm among female childhood cancer survivors. The markedly increased risk of subsequent breast cancer has largely been considered therapy-related. The current clinical screening of this high-risk population, therefore, relies primarily on the radiation dose and volume to the chest/breast tissue used to treat prior childhood cancer. In this study, we assessed the genetic contributions to subsequent breast cancer risk, considering small-effect common variants as a polygenic risk score as well as large-effect rare predisposing mutations implicated in the etiology of breast cancer in the general population. Our findings suggest that small-effect common variants together with large-effect rare mutations can inform personalized breast cancer surveillance in female survivors. The polygenic risk score can be utilized with the rare mutations in clinical settings to enhance the identification of high-risk survivors for screening to enable the early detection and prevention of subsequent breast cancer.
Acknowledgements
This research was supported by funding from the American Lebanese Syrian Associated Charities to St. Jude Children’s Research Hospital and by grants (CA021765, CA216354, and CA195547) from the National Institutes of Health to St. Jude Children’s Research Hospital.
The funders of the study had no role in the design and conduct of the study, and were not involved in collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
The authors thank all the individuals who participated in this study; Clay McLeod, M.Sc., and Liang Ding, Ph.D., from the Department of Computational Biology, St. Jude Children’s Research Hospital, for preparation and submission of the sequencing data to the European Genome-phenome Archive; and Charis Eng, M.D., Ph.D., FACP, Hardis/ACS Professor and Chair, Genomic Medicine Institute, Cleveland Clinic, for her help with pathogenicity review of PTEN promoter mutations. Mr. McLeod and Drs. Ding and Eng did not receive financial compensation. Drs. Robison and Zhang had full access to the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Declaration: The authors declare no potential conflicts of interest
Data Sharing
Aligned BAM files for 3,006 survivors are accessible through the St. Jude Cloud (https://stjude.cloud) and European Genome-phenome Archive (EGA) under accession EGAS00001002499.
References
- 1.Moskowitz CS, Chou JF, Wolden SL, Bernstein JL, Malhotra J, Novetsky Friedman D, et al. Breast cancer after chest radiation therapy for childhood cancer. J Clin Oncol 2014:32(21):2217–23 doi 10.1200/JCO.2013.54.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson TO, Moskowitz CS, Chou JF, Bradbury AR, Neglia JP, Dang CT, et al. Breast Cancer Risk in Childhood Cancer Survivors Without a History of Chest Radiotherapy: A Report From the Childhood Cancer Survivor Study. J Clin Oncol 2016:34(9):910–8 doi 10.1200/JCO.2015.62.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson MM, Ness KK, Gurney JG, Mulrooney DA, Chemaitilly W, Krull KR, et al. Clinical ascertainment of health outcomes among adults treated for childhood cancer. Jama 2013:309(22):2371–81 doi 10.1001/jama.2013.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson MM, Ehrhardt MJ, Bhakta N, Baassiri M, Eissa H, Chemaitilly W, et al. Approach for Classification and Severity Grading of Long-term and Late-Onset Health Events among Childhood Cancer Survivors in the St. Jude Lifetime Cohort. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2017:26(5):666–74 doi 10.1158/1055-9965.EPI-16-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Wilson CL, Easton J, Thrasher A, Mulder H, Liu Q, et al. Genetic Risk for Subsequent Neoplasms Among Long-Term Survivors of Childhood Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018:JCO2018778589 doi 10.1200/JCO.2018.77.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017:551(7678):92–4 doi 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudet MM, Kuchenbaecker KB, Vijai J, Klein RJ, Kirchhoff T, McGuffog L, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS genetics 2013:9(3):e1003173 doi 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS genetics 2013:9(3):e1003212 doi 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Feng B, Miron A, Chen X, Beesley J, Bimeh E, et al. Breast cancer risk prediction using a polygenic risk score in the familial setting: a prospective study from the Breast Cancer Family Registry and kConFab. Genetics in medicine : official journal of the American College of Medical Genetics 2017:19(1):30–5 doi 10.1038/gim.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuchenbaecker KB, McGuffog L, Barrowdale D, Lee A, Soucy P, Dennis J, et al. Evaluation of Polygenic Risk Scores for Breast and Ovarian Cancer Risk Prediction in BRCA1 and BRCA2 Mutation Carriers. J Natl Cancer Inst 2017:109(7) doi 10.1093/jnci/djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK, et al. Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 2015:107(5) doi 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K, Wang Z, Li Q, Wacholder S, Hunter DJ, Hoover RN, et al. Population substructure and control selection in genome-wide association studies. PloS one 2008:3(7):e2551 doi 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. The New England journal of medicine 2015:372(23):2243–57 doi 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cronin KA, Ries LA, Edwards BK. The Surveillance, Epidemiology, and End Results (SEER) Program of the National Cancer Institute. Cancer 2014:120 Suppl 23:3755–7 doi 10.1002/cncr.29049. [DOI] [PubMed] [Google Scholar]
- 15.Michailidou K, Lindstrom S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017:551(7678):92–4 doi 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teepen JC, van Leeuwen FE, Tissing WJ, van Dulmen-den Broeder E, van den Heuvel-Eibrink MM, van der Pal HJ, et al. Long-Term Risk of Subsequent Malignant Neoplasms After Treatment of Childhood Cancer in the DCOG LATER Study Cohort: Role of Chemotherapy. J Clin Oncol 2017:35(20):2288–98 doi 10.1200/JCO.2016.71.6902. [DOI] [PubMed] [Google Scholar]
- 17.Kamihara J, Rana HQ, Garber JE. Germline TP53 mutations and the changing landscape of Li-Fraumeni syndrome. Human mutation 2014:35(6):654–62 doi 10.1002/humu.22559. [DOI] [PubMed] [Google Scholar]
- 18.Derman YE. Clinical Practice Recommendations Based on an Updated Review of Breast Cancer Risk Among Women Treated for Childhood Cancer. Journal of pediatric oncology nursing : official journal of the Association of Pediatric Oncology Nurses 2018:35(1):65–78 doi 10.1177/1043454217727515. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh YC, Tu SH, Su CT, Cho EC, Wu CH, Hsieh MC, et al. A polygenic risk score for breast cancer risk in a Taiwanese population. Breast cancer research and treatment 2017:163(1):131–8 doi 10.1007/s10549-017-4144-5. [DOI] [PubMed] [Google Scholar]
- 20.Wen W, Shu XO, Guo X, Cai Q, Long J, Bolla MK, et al. Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast cancer research : BCR 2016:18(1):124 doi 10.1186/s13058-016-0786-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Qian F, Zheng Y, Ogundiran T, Ojengbede O, Zheng W, et al. Genetic variants demonstrating flip-flop phenomenon and breast cancer risk prediction among women of African ancestry. Breast cancer research and treatment 2018:168(3):703–12 doi 10.1007/s10549-017-4638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan CHT, Munusamy P, Loke SY, Koh GL, Yang AZY, Law HY, et al. Evaluation of three polygenic risk score models for the prediction of breast cancer risk in Singapore Chinese. Oncotarget 2018:9(16):12796–804 doi 10.18632/oncotarget.24374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhakta N, Liu Q, Ness KK, Baassiri M, Eissa H, Yeo F, et al. The cumulative burden of surviving childhood cancer: an initial report from the St Jude Lifetime Cohort Study (SJLIFE). Lancet 2017:390(10112):2569–82 doi 10.1016/S0140-6736(17)31610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis CM, Vassos E. Prospects for using risk scores in polygenic medicine. Genome medicine 2017:9(1):96 doi 10.1186/s13073-017-0489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maher BS. Polygenic Scores in Epidemiology: Risk Prediction, Etiology, and Clinical Utility. Current epidemiology reports 2015:2(4):239–44 doi 10.1007/s40471-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurgelun MB, Chenevix-Trench G, Lippman SM. Translating Germline Cancer Risk into Precision Prevention. Cell 2017:168(4):566–70 doi 10.1016/j.cell.2017.01.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


