Abstract
Almost all patients with EGFR-driven lung cancer who are treated with EGFR tyrosine kinase inhibitors (TKI) develop resistance to treatment. A single base (c.2369C>T) transition mutation, EGFR T790M, is the most frequent resistance event after first-generation exposure to EGFR TKI. Whether T790M mutation is acquired or is selected from a pre-existing clone has been a matter of significant debate. In this study, we show that treatment with EGFR TKI leads to activation of the NFᴋB pathway, which in turn induces expression of Activation Induced Cytidine Deaminase (AICDA). In turn, AICDA causes deamination of 5-methylcytosine to thymine at position c.2369 to generate the T790M mutation. Pharmacologic inhibition of the NFᴋB pathway or knockout of AICDA decreased the frequency or prevented the development of T790M mutation, respectively. In addition, patients treated with first line EGFR TKI displayed increased expression of AICDA and detection of the T790M mutation upon progression. These results identify the mechanism of T790M acquisition and present an opportunity to target the process to delay or prevent it.
Keywords: AICDA, Cytosine Deamination, EGFR, Lung Cancer, Methylation
Introduction
Lung cancer is the leading cause of cancer death in the USA with estimated 230,000 new cases and 156,000 deaths this year (1). Multiple driving mutations have been identified in non-small cell lung cancer (NSCLC) including epidermal growth factor receptor (EGFR) mutations, which occur in 10–15% of lung adenocarcinoma patients of Caucasian ethnicity and in 50% of Asian population (2). EGFR mutations mainly affect the EGFR tyrosine kinase domain with the two most common mutations being deletions in exon 19 and missense substitution in exon 21, L858R, where leucine is replaced by arginine (3–5). These mutations lead to constitutive activation of EGFR and NSCLC cellular dependence on this pathway. Treatment with EGFR tyrosine kinase inhibitors (TKIs) decreases the activity of downstream cell survival and proliferation signaling pathways (6, 7). Currently, EGFR TKIs are the standard of care for patients with EGFR mutant NSCLC who has a better response rate and progression free survival as compared to chemotherapy (8, 9). Unfortunately, almost all patients with EGFR-mutant NSCLC treated with first generation EGFR TKIs develop resistance with mean progression free survival (PFS) range of 9.2–13.1 months (9). Mechanisms of resistance include various gene amplifications and mutations as well as epithelial mesenchymal transition and transformation into small cell lung cancer (10–13). However, the main mechanism of developing resistance to first generation TKIs is a single nucleotide transition mutation in EGFR; a cytosine to thymine (C>T) at position 2369 (CAC/CAT) causing threonine to methionine amino acid change at codon 790, T790M (14, 15). This T790M mutation occurs in cis with the original driving mutation, and leads to steric hindrance and increased binding affinity for ATP (16, 17).
T790M mutation is usually not detected at baseline and it has been controversial whether the lack of detection is due to assay sensitivity. Multiple reports have suggested high incidence of T790M mutation (18), however, others have refuted this as artifact due to formalin tissue fixation. T790M mutations were detected in the formalin fixed cancer specimen but not their fresh frozen counterparts; in addition, they were even present in formalin fixed normal lung tissue (19). Although the presence of T790M at baseline cannot be totally ruled out, reports have shown that it can be acquired after TKI treatment (20).
Deamination of cytosine can lead to the generation of either uracil or thymine depending on the methylation status (Supplementary figure 1). Uracil is usually removed by the activity of mismatch repair system through uracil-DNA glycosylase (UDG). This creates an apurinic/apyrimidinic (AP) site, which could lead to mutation at that position. Activation induced cytosine deaminase, AICDA, is usually expressed in germinal center B-lymphocytes upon antigen exposure. AICDA translocates to the nucleus, deaminates cytosine (C) in single-stranded DNA and converts it to uracil (U). The U:G mismatch lesions are converted into point mutations known as somatic hypermutation allowing for a diverse antibody repertoire (21). Furthermore, AICDA causes other genes mutations and is implicated in the development of diffuse large B cell lymphoma (22). Overexpression of AICDA in chronic myeloid leukemia (CML) is associated with increased frequency of C>T mutation and resistance to imatinib exposure (23). In solid tumors, AICDA is expressed in human hepatocellular carcinoma (24), gastric cancer (25), colorectal cancer (26), and lung cancer (27), suggesting a potential role in these tumors too. Since the T790M mutation is a C>T mutation that is acquired upon TKIs exposure, in this report, we explored whether AICDA is activated after EGFR TKIs use in lung adenocarcinoma. We further assessed whether AICDA manipulation affects T790M mutation development frequency and if cytosine at position 2369 is methylated (5-methylcytosine) where its deamination would directly lead to the observed C>T mutation rather than C>G or C>A (Supplementary figure 1).
Materials and Methods
Cell lines
Cell line PC9 was obtained from Sigma-Aldrich (St. Louis, MO). NCI-H3255 was a generous donation from Pasi Janne (Dana Farber Cancer Institute). NCI-H1650, NCI- H1299, NCI-H1975 and NCI-H460 were obtained from ATCC. All cell lines were authenticated by STR profiling and mycoplasma testing was conducted. Cell lines were grown in RPMI-1640 with 10% FBS final concentration per recommendation.
Droplet Digital PCR
Allele-specific fluorescent primer/probes for wild type (dHsaCP2000020) and T790M (dHsaCP2000019) mutated EGFR were obtained from Bio-Rad (Hercules, California). ddPCR was then performed on DNA extracted from cell lines. Each ddPCR run included a positive control derived from NCI-H1975 sample harboring the T790M mutation and was performed as such: a master mix containing 2X ddPCR Supermix, 20X Primer/Probe Mix, 25X Droplet Stabilizer (RainDance Technologies), and 2–5U Restriction Endonuclease (HindIII; New England BioLabs) was added to separate wells containing 5–500 ng gDNA, for a total reaction volume of 20 μl, and then incubated at room temperature for 15 minutes. Samples were then pipetted into a RainDrop Source Chip (RainDance Technologies), and 3.5–4.5 million droplets were generated with the RainDrop Source machine. The resulting emulsion was amplified using the following settings: 95°C for 10 minutes, and then 40 cycles at 94°C for 30 seconds and 55°C for 60 seconds, followed by 98°C for 10 minutes. Endpoint fluorescence was then measured with a RainDrop Sense machine and analyzed with RainDrop Analyst software. The wild type and mutant signals of each control sample were used to draw positive gates to then analyze events in samples.
To determine sensitivity of ddPCR for T790M mutation, the T790M assay was carried out using 10 ng of H1975 genomic DNA serially diluted by factor of 10 in 50,000 copies of genomic DNA. The mean lowest copy number detected at 0.01ng was three, and thus the limit of detection was 0.012% (Supplementary figure 2). We also detected 1–2 events of T790M in normal genomic DNA, thus, using the mean number of events plus 2 standard deviations as the cut off we determined the specificity of the T790M mutation to be five in 50,000 copies.
Cell line treatment
For continuous long term gefitinib treatment, approximately 1 million cells were plated in T75 flasks overnight and treated the second day with 1/10th the TKI IC50. After 48hrs, cells were harvested and split into 3 lots. One part was used for genomic DNA extraction and the other part for RNA extraction. The third part was re-plated overnight and treated with double the previous dose. This process was repeated, with a DMSO treated control, until a final dose of 100X the IC50 was reached. For combination treatment, Bay11–7082 was added at a constant dose of 0.5uM through-out the increasing dose of gefitinib exposure. For short term EGFR TKIs treatment, cell lines were treated with various indicated doses for 24hrs to assess for AICDA or 6hrs to assess for NFĸB pathway components.
Real-time PCR
Total RNA was isolated using RNeasy Mini Kit (QIAGEN, Germantown, MD) and real-time PCR assays were done in a ABI PRISM 7900HT sequence detection system. Taqman primer/probes were selected from ThermoFisher (Waltham, MA); AICDA Hs00757808_m1 and control RPS13 Hs01945436_u1. The relative expression mRNA level of AICDA was calculated with respect to internal standard RPS13 gene to normalize for variations in RNA quality and the amount of cDNA. The results are a representation of at least two biological repeats and 3 technical replicates. Unpaired t-test was used where p<0.05 was considered statistically significant.
AICDA CRISPR knock out
We used an all-in-one RFP labelled Cas9 and guide RNA (gRNA) expression plasmids from Sigma-Aldrich (St. Louis, MO) targeting AICDA exon 2. Briefly, PC9 cells were transfected with the CRISPR/Cas9 AICDA plasmid using standard transfection methods. RFP positive clones were selected and single cell clones were generated in 96 well plates. Each clone was expanded, the area around the expected cutting site was amplified and sanger sequencing performed. Using the TIDE web-tool software (28), sequencing results were assessed and AICDA mutated clones were selected.
UDG Hydrolysis Assay
We have synthesized an EGFR oligomer, which allows detection of cytosine deamination through the hydrolysis activity of uracil-DNA glycosylase (UDG) in alkaline environment. This oligomer is 79nt in length; the first 5’ end 28nt represent EGFR sequence 2355–2383. The remaining 51nt represent the 2355–2406 nucleotide sequence of EGFR. This will provide a repeat sequence for the deamination process to occur. AICDA protein from a baculovirus system was a generous gift from Dr. Myron Goodman (USC, California) and AICDA was purified by Center for Structural Biology. In summary, AICDA is incubated with 0.5uM probe in a buffer containing, 50mM HEPES, pH 7.5, 10mM MgCl2, 1mM dithiothreitol for 30 min at 37°C. The reaction will be terminated by an extraction with phenol/chloroform/isoamyl alcohol (25:24:1) and incubated with UDG at 37°C for 1hr. Hydrolysis is achieved by heating at 95°C for 10mins in 0.2M NaOH. Electrophoresis was done using a 4–20% TBE gel.
Patient Samples
Patient samples were collected through the UM molecular pathology laboratory. The clinical variables are provided in supplementary table 1. Evaluation of patient samples was conducted using surplus pathology materials with a waiver of informed consent granted by the Institution Review Board (IRB) in accordance with recognized ethical guidelines of Declaration of Helsinki, Belmont Report and US Common Rule. Samples used for this research Patients’ tumors were sequenced for original EGFR driving mutation and upon progression, biopsies were obtained and sequenced for the T790M mutation. RNA was isolated using RNeasy Mini Kit and real-time PCR was done as mentioned above.
Mass Spectrometry
Genomic DNA (500ng) was bisulfite treated and cleaned using EZ DNA Methylation kit (Zymo Research, Irvine, CA). Then, 20 ng of the bisulfite-treated DNA was used for PCR amplification with Roche HiFidelity PCR system, in a final volume of 10ul, and using 400nM of forward/reverse primers (aggaagagagTTTGTTGGGTATTTGTTTTATTTTT/cagtaatacgactcactatagggagaaggctACTAAAAACCAATATTATCTTTATATTCCC). The following PCR amplification settings were used: 1) 94°C, 15 mins; 2) 45 cycles of: 94°C, 20 secs; 62°C, 30 secs; 72°C, 1min; 3) 72°C, 3 mins; 4) hold at 4°C. A volume of 2ul of each reaction was applied to the HiSeq D1000 TapeStation assay (Agilent, Santa Clara, CA) to verify the presence of an amplicon at the expected size. The amplicons were then submitted to the UM DNA Sequencing Core for SAP, in vitro transcription/cleavage, cleanup and spotting onto a SpectroCHIP (AgenaBio, San Diego, CA) for mass spectra acquisition. The data was analyzed with the EpiTyper software (AgenaBio) and exported to an excel spreadsheet.
Results
T790M is acquired after TKIs treatment
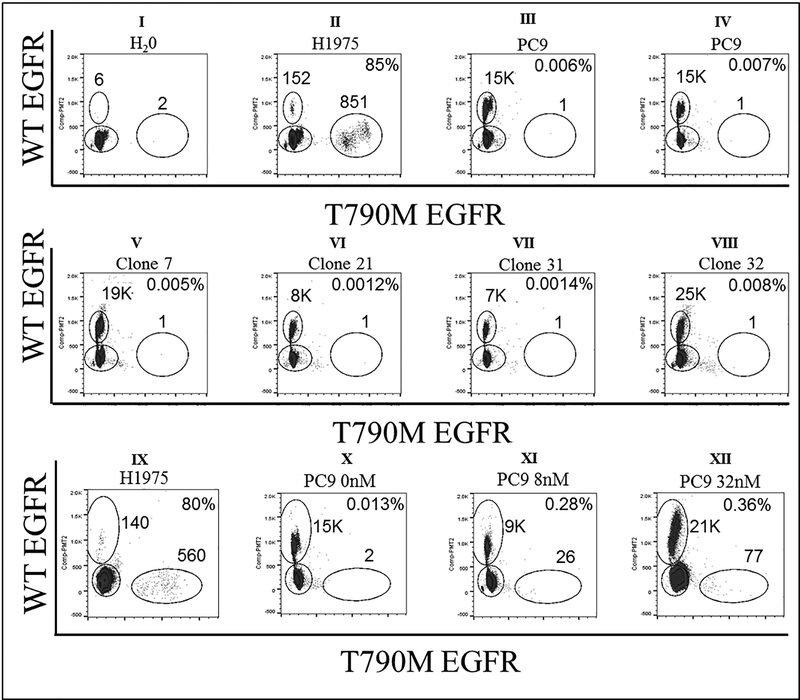
The absence of baseline EGFR T790M in contrast to its prevalence after first line EGFR TKI treatment, raises an important question about being an acquired event. In fact, previous work done by Engleman’s laboratory (20), showed that PC9, an EGFR del19 lung adenocarcinoma cell line can acquire T790M mutation from T790M-negative clone. Thus, using flow cytometry, we established PC9 single cell clones in 96 well plates. And then verified by digital droplet PCR (ddPCR) that these PC9 single cell clones had no baseline T790M mutation (Figure 1, panels I- VIII). However, upon treating these clones with an increasing dose of gefitinib, we detected the emergence of T790M mutation, confirming the acquired nature of this mutation (Figure 1, panels IX- XII).
Figure 1. EGFR mutation status by ddPCR.
Detection of WT EGFR (y-axis) and EGFR T790M mutation (x-axis). Panel I shows six WT EGFR and two T790M EGFR events in H2O (negative control) representing ddPCR assay background. Panel II shows 85% T790M mutation rate in H1975 cell line (positive control). Panels III and IV show baseline/background T790M mutations in parental PC9 cell line. Single cell PC9 clones show similar baseline/background T790M mutations as parental PC9 cell line (panels V, VI, VII, and VIII). PC9 cell line was treated with serially increasing gefitinib dose. Panel IX shows high T790M mutation rate in positive control, H1975 cell line. Panel X shows low T790M mutation rate in untreated PC9 cell line as compared to Panel XI, XII high mutation frequency after exposure to gefitinib at 8nM, 32nM, respectively.
AICDA is induced upon EGFR TKIs exposure
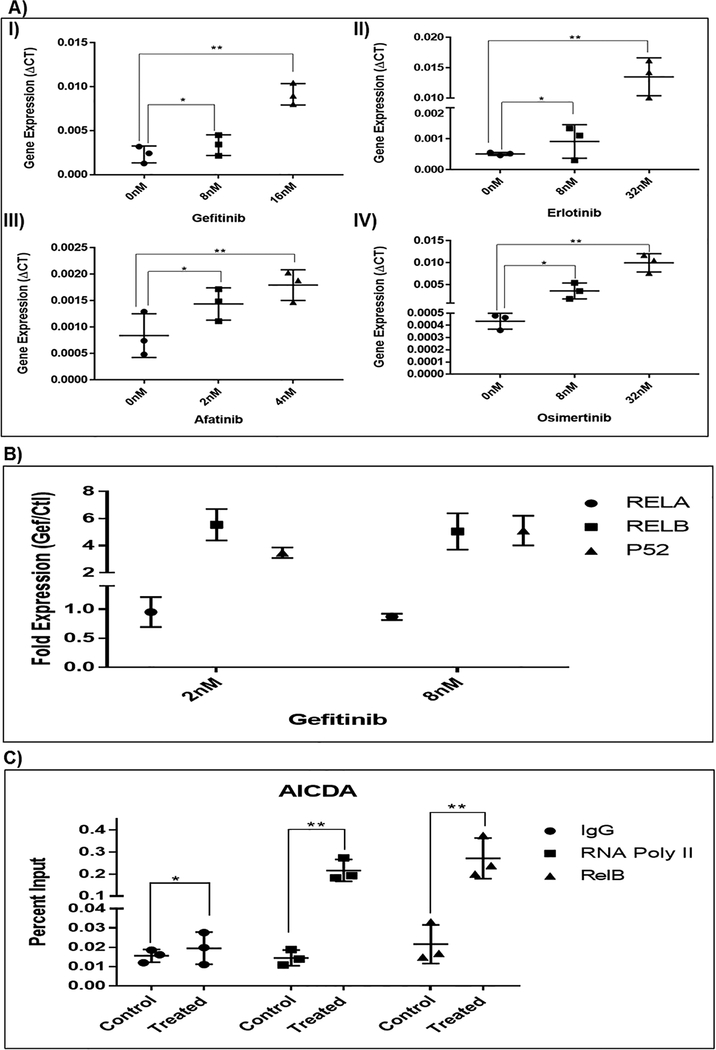
AICDA cytosine deamination capability has been implicated in resistance to imatinib in CML where its overexpression can induce blast crisis as well the C>T gate keeper resistant mutation, T315I (23). We thus explored the possibility that exposure to EGFR TKI induced a cytosine deamination process, promoting the cytosine to thymine mutation. We screened for multiple known cytosine deaminases mRNA expression after exposure to gefitinib, a first line EGFR TKI. We found that gefitinib mainly increased AICDA expression (Supplementary figure 3). To determine whether this is a unique response to gefitinib or a class effect, we used other known EGFR TKIs, first generation, erlotinib, second generation, afatinib and third generation osimertinib. As shown in figure 2A, there is significant increase in the mRNA expression of AICDA upon treatment with different TKIs in PC9 cell line. This is also seen in other EGFR mutated lung adenocarcinoma cell lines (Supplementary figure 3).
Figure 2. Changes in AICDA and NFkB pathway expression.
A) AICDA expression is significantly increased after treatment with four different TKIs in PC9 cell line: I) first generation TKI, gefitinb; II) first generation TKI, erlotinib; III) second generation TKI, afatinib; IV) third generation TKI, osimertinib. B) Expression of NFĸB pathway components after TKI treatment. No change in RelA expression (canonical pathway). The expression of RelB & P52 (non-canonical pathway) increased by multiple folds. C) NFkB ChIP assay. RELB and RNA polymerase II are recruited to the AICDA promoter region after 6hrs exposure to 8nM gefitinib. AICDA qPCR results are reported as a percentage of the total chromatin input. (* p>0.05; ** p<0.05)
AICDA is induced through non-canonical NFĸB
It is intriguing that after EGFR TKI exposure and before T790M resistance development, a tolerant cell population persists. Blakely et al have shown that initial cell survival occurs due to activation of NFĸB pathway (29) which is a known AICDA transcription factor. Thus, we tested whether NFĸB pathway was induced after EGFR treatment and whether its components bind to AICDA promoter. As shown in figure 2B, the expression of RelB and p52 (non-canonical) were increased after 24hrs exposure to TKI. This indicates that the NFĸB non-canonical pathway is activated by TKI inhibition of EGFR. Using Bay11–7802, a known inhibitor of NFĸB pathway, the expression of AICDA was abrogated after TKI treatment (Supplementary figure 4). We then used a ChIP assay to evaluate whether RelB interacts directly with AICDA in PC9 cells after TKI exposure. RelB and RNA polymerase II antibodies were used for immunoprecipitation with IgG as control. Then qPCR was performed with primers covering the AICDA promoter region. As seen in figure 2C, there is a significant increase in binding of RelB to the AICDA promoter after exposure to TKI. This indicates that the non-canonical NFĸB pathway induces AICDA expression after EGFR inhibition. RNA polymerase II is also recruited to AICDA promoter, further supporting the increase in AICDA transcription after EGFR inhibition.
AICDA knock down decreases T790M frequency
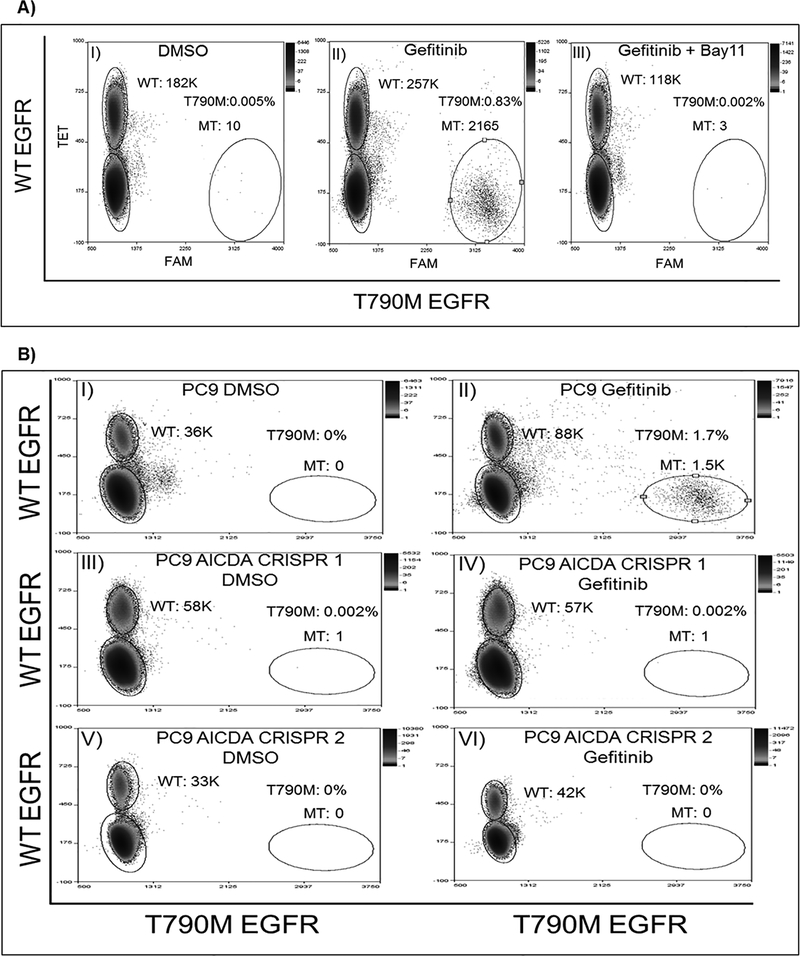
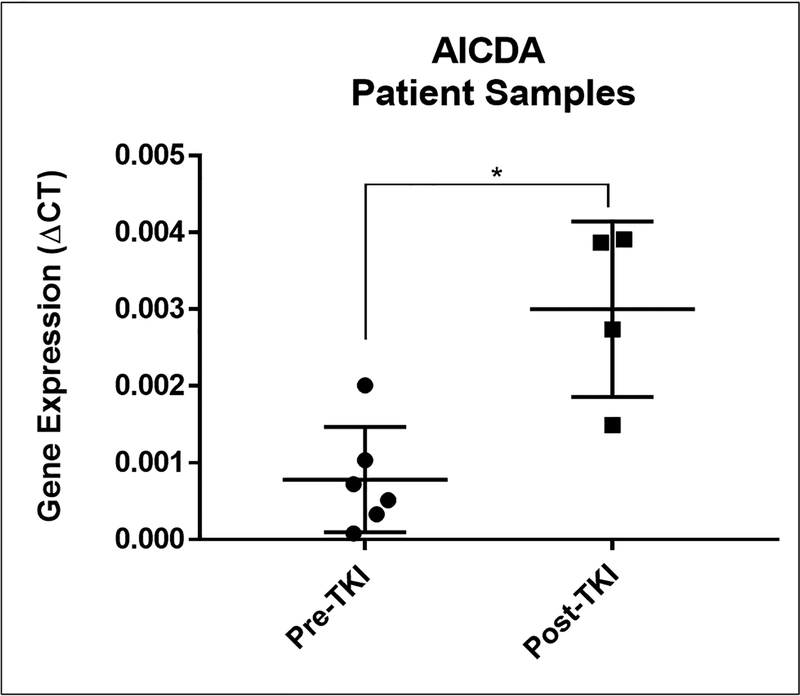
Since AICDA is induced after EGFR TKI treatment, we evaluated its ability to deaminate cytosine in EGFR gene. To this end, we purified AICDA protein and tested cytosine deamination of an EGFR oligomer designed to contain the cytosine of interest, c.2369, twice. We found that AICDA does recognize EGFR sequence and induces deamination of cytosine leading to hydrolysis of the oligomer at these nucleotides (Supplementary figure 5). We next asked if inhibiting AICDA expression would have an impact on the development of T790M mutation. We used two approaches to interrogate AICDA role. First, we inhibited NFĸB pathway induction by using Bay11–7082 and found that combination treatment of gefitinib and Bay11–7082, abrogated the development of T790M mutation after long term exposure to EGFR TKI (Figure 3A). Second, we generated CRISPR/Cas9 AICDA knock out clones verified by sanger sequencing (Supplementary figure 6) and treated with an increasing dose of gefitinib. As seen in figure 3B, using ddPCR we didn’t detect T790M mutations in the AICDA knock out clones (panels IV and VI) at a gefitinib dose of 64nM in comparison to PC9 control (panel II). Thus, AICDA is necessary to deaminate cytosine at position 2369 in PC9 cells in response to TKIs. We further evaluated AICDA expression in primary lung cancer tissues from patients before and after the development of resistance EGFR TKI treatment. As shown in figure 4, AICDA expression significantly increased upon progression on first line TKI treatment and detection of the T790M resistant mutation, supporting the induction of AICDA after TKI exposure.
Figure 3. T790M mutation frequency after AICDA activity manipulation.
A) T790M mutation rate detected after gefitinib exposure (Panel II), is reduced to baseline (Panel I), after combination treatment of gefitinib and Bay11–7802 (Panel III). B) T790M mutation is abrogated in PC9 AICDA CRISPR knock out cell lines (Panels IV, VI) compared to parental PC9 cell line (Panel II) after exposure to 64nM gefitinib. The corresponding DMSO treated controls Panels I, III and V.
Figure 4. Patients’ AICDA expression pre and post TKI exposure.
AICDA is significantly increased from baseline (n=6) in patients who received TKI and progressed with evidence of T790M mutation (n=4).
EGFR c.2369 cytosine is methylated
EGFR T790M resistant mutation is a single nucleotide transition of cytosine to thymine, C>T. Since deamination of 5-methylcytosine leads to C to T transition (Supplementary figure 1), we proposed that cytosine at position c.2369 is methylated. Using methylation specific primers and mass spectrometry, we found that c.2369 cytosine is methylated in number of lung cancer cell lines (Supplementary table 2) including PC9. Our findings suggest that deamination of the 5-methylcytosine to thymidine at position c.2369 causes the threonine/methionine amino acid change that alters TKI binding affinity and causes resistance.
Discussion
The impact of targeted therapy in oncology is undeniable since the 2001 FDA approval of imatinib for CML. This opened the door to the discovery of multiple drugs directed at known specific driver mutations in various cancers. Drugs against EGFR mutations in NSCLC were among the first to be FDA approved after imatinib. Interestingly, the resistant mutations to EGFR TKIs were predicted based on imatinib CML resistance where the discovered EGFR T790M mutation has the same “gate keeper” role of T315I mutation in BCR-ABL. Moreover, it is very intriguing that a similar mutational profile is also seen in GIST and hypereosinophilic syndrome (HES) tumors after treatment with imatinib. In fact, a single nucleotide C>T mutation is detected in BCR-ABL (CAC/CAT), c-KIT (CAC/CAT) and FIP1L1–PDGFRA (CAC/CAT) in imatinib resistant CML (30), GIST (31) and HES (32) respectively.
In all these tumors, the resistant mutation is highly prevalent, is not detected at baseline, it occurs after several months of treatment and is always C to T point mutation. Although a random mutation event followed by clonal selection can’t be completely ruled out, a specific mechanism responsible for this event is worth interrogating.
AICDA is expressed in germinal center B-lymphocytes and induced upon antigen exposure, causing cytosine deamination and point mutations in immunoglobulin genes. These acquired mutations process, known as somatic hypermutation, provide the needed diversified antibody repertoire (33). AICDA has been implicated in cytosine deamination in non-immunoglobulin genes as well (34) and in the development of resistance to imatinib (23). In CML, overexpression of AICDA was associated with increased frequency of C>T mutations and imatinib resistance (23).
In this report, we investigated whether AICDA plays a role in the development of T790M mutation after EGFR TKI treatment. Using ddPCR, one of the most sensitive mutation assays, we first verified that the T790M mutation is not detected at baseline in the tested single cell PC9 clones; however, T790M mutation is identified upon exposure to TKI, suggesting that it is an acquired event. We also show that AICDA is induced after treatment with first, second and third generation EGFR TKIs. Interestingly, verifying that cytosine at position c.2369 is methylated, provides strong explanation to the deamination mechanism of C>T mutation. Finally, knocking down AICDA pharmacologically through inhibiting NFĸB pathway or genetically through CRISPR precluded T790M development after TKI exposure.
This manuscript explicitly links EGFR TKIs to the development of a specific DNA resistant mutation through AICDA activity. Thus, targeting AICDA directly by developing specific inhibitors or indirectly by inhibiting the NFĸB pathway, provide an opportunity to delay/prevent T790M mutation. Furthermore, AICDA was shown to increase genome instability in B-cells after Phosphatidylinositol 3-kinase δ blockade (35). We performed whole exome sequencing (WES) at an average depth of 100X comparing mutation profile before and after treatment with gefitinib. We found an increase in frequency of variants and specifically of SNPs after gefitinib exposure (Supplementary table 2). This finding is similar to previously reported results by Jia et. al., where an increase in mutation frequency was detected in cell lines after EGFR TKI treatment (36). In addition, the direct effect of EGFR TKIs on base excision repair through the degradation of HSP70 was recently described (37). The consequences of deamination might even go beyond facilitating the generation of resistance mutations to perhaps contributing to tumorigenesis. Consistent with this idea, the development of small cell lung cancer transformation after TKI exposure is associated with APOBEC mutation profile in lung EGFR driven tumors (38). Our findings suggest that specific DNA mutations are the sequelae of a dynamic response to an environmental stress. The activation of the NFĸB pathway ensures cancer cells’ tolerance and induces AICDA driven mutagenesis leading to resistant mutation acquisition and eventually the selection of the best-fit clone, reminiscent of antibody affinity maturation. Targeting AICDA activity directly (developing specific inhibitors) or indirectly with NFĸB pathway inhibitors as well as using DNA repair inhibitors might lead to the delay or prevention of T790M resistance.
Supplementary Material
Statement of Significance.
Findings identify the mechanism behind acquisition of a common resistance mutation to TKI treatment in lung cancer.
Acknowledgments
This work was supported in part by NCI 5K08 CA158425–03, ALA Lung Cancer Discovery Award, UM Thoracic Oncology Program Elizabeth A. Crary Fund, and HOPE Foundation to KHassan. Research reported in this manuscript was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA046592 by the use of the Rogel Cancer Center Flow cytometry, vector, DNA sequencing, Bioinformatics, and Epigenomics core facilities.
Footnotes
Gregory P. Kalemkerian has received research funding from Merck, Takeda, Abbvie, GlaxoSmithKline and Pfizer. The remaining authors declare no potential conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Translational lung cancer research. 2015;4:36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature reviews. Molecular cell biology. 2001;2:127–37. [DOI] [PubMed] [Google Scholar]
- 5.Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, Bell DW, Huberman MS, Halmos B, Rabin MS, Haber DA, Lynch TJ, Meyerson M, Johnson BE, Janne PA. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–14. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sanchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M, Spanish Lung Cancer G. Screening for epidermal growth factor receptor mutations in lung cancer. The New England journal of medicine. 2009;361:958–67. [DOI] [PubMed] [Google Scholar]
- 7.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. [DOI] [PubMed] [Google Scholar]
- 8.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–39. [DOI] [PubMed] [Google Scholar]
- 9.Cortot AB, Janne PA. Molecular mechanisms of resistance in epidermal growth factor receptor-mutant lung adenocarcinomas. European respiratory review : an official journal of the European Respiratory Society. 2014;23:356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 11.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, Aoe K, Toyooka S, Kiura K, Fernandez-Cuesta L, Fidias P, Yang JC, Miller VA, Riely GJ, Kris MG, Engelman JA, Vnencak-Jones CL, Dias-Santagata D, Ladanyi M, Pao W. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E, Ohashi K, Janjigian YY, Spitzler PJ, Melnick MA, Riely GJ, Kris MG, Miller VA, Ladanyi M, Politi K, Pao W. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer discovery. 2012;2:922–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2005;352:786–92. [DOI] [PubMed] [Google Scholar]
- 15.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M, Riely GJ. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riely GJ, Pao W. Combining EGFR targeted therapy with chemotherapy in pancreatic cancer: is timing important? Cancer biology & therapy. 2005;4:1096–7. [DOI] [PubMed] [Google Scholar]
- 17.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, Ho BC, Chang GC, Shih JY, Yu SL, Yang PC. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:433–40. [DOI] [PubMed] [Google Scholar]
- 19.Ye X, Zhu ZZ, Zhong L, Lu Y, Sun Y, Yin X, Yang Z, Zhu G, Ji Q. High T790M detection rate in TKI-naive NSCLC with EGFR sensitive mutation: truth or artifact? Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2013;8:1118–20. [DOI] [PubMed] [Google Scholar]
- 20.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HE, Krishnamurthy Radhakrishna V, Siravegna G, Hu H, Raoof S, Lockerman E, Kalsy A, Lee D, Keating CL, Ruddy DA, Damon LJ, Crystal AS, Costa C, Piotrowska Z, Bardelli A, Iafrate AJ, Sadreyev RI, Stegmeier F, Getz G, Sequist LV, Faber AC, Engelman JA. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesan S, Myles PR, McCann G, Kousoulis AA, Hashmi M, Belatri R, Boyle E, Barcroft A, van Staa TP, Kirkham JJ, Nguyen Van Tam JS, Williams TJ, Semple MG. Development of processes allowing near real-time refinement and validation of triage tools during the early stage of an outbreak in readiness for surge: the FLU-CATs Study. Health technology assessment. 2015;19:1–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandra V, Bortnick A, Murre C. AID targeting: old mysteries and new challenges. Trends in immunology. 2015;36:527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, Henke N, Li Z, Hoffmann TK, Kim YM, Hofmann WK, Jumaa H, Groffen J, Heisterkamp N, Martinelli G, Lieber MR, Casellas R, Muschen M. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kou T, Marusawa H, Kinoshita K, Endo Y, Okazaki IM, Ueda Y, Kodama Y, Haga H, Ikai I, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. International journal of cancer. 2007;120:469–76. [DOI] [PubMed] [Google Scholar]
- 25.Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470–6. [DOI] [PubMed] [Google Scholar]
- 26.Endo Y, Marusawa H, Kou T, Nakase H, Fujii S, Fujimori T, Kinoshita K, Honjo T, Chiba T. Activation-induced cytidine deaminase links between inflammation and the development of colitis-associated colorectal cancers. Gastroenterology. 2008;135:889–98, 98 e1–3. [DOI] [PubMed] [Google Scholar]
- 27.Shinmura K, Igarashi H, Goto M, Tao H, Yamada H, Matsuura S, Tajima M, Matsuda T, Yamane A, Funai K, Tanahashi M, Niwa H, Ogawa H, Sugimura H. Aberrant expression and mutation-inducing activity of AID in human lung cancer. Annals of surgical oncology. 2011;18:2084–92. [DOI] [PubMed] [Google Scholar]
- 28.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic acids research. 2014;42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I, Hrustanovic G, Chan E, Lin L, Neel DS, Newton W, Bobb KL, Fouts TR, Meshulam J, Gubens MA, Jablons DM, Johnson JR, Bandyopadhyay S, Krogan NJ, Bivona TG. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell reports. 2015;11:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. [DOI] [PubMed] [Google Scholar]
- 31.Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, Leversha MA, Jeffrey PD, Desantis D, Singer S, Brennan MF, Maki RG, DeMatteo RP. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res. 2005;11:4182–90. [DOI] [PubMed] [Google Scholar]
- 32.Cools J, Stover EH, Boulton CL, Gotlib J, Legare RD, Amaral SM, Curley DP, Duclos N, Rowan R, Kutok JL, Lee BH, Williams IR, Coutre SE, Stone RM, DeAngelo DJ, Marynen P, Manley PW, Meyer T, Fabbro D, Neuberg D, Weisberg E, Griffin JD, Gilliland DG. PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFRalpha-induced myeloproliferative disease. Cancer Cell. 2003;3:459–69. [DOI] [PubMed] [Google Scholar]
- 33.Meffre E, Catalan N, Seltz F, Fischer A, Nussenzweig MC, Durandy A. Somatic hypermutation shapes the antibody repertoire of memory B cells in humans. The Journal of experimental medicine. 2001;194:375–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–6. [DOI] [PubMed] [Google Scholar]
- 35.Compagno M, Wang Q, Pighi C, Cheong TC, Meng FL, Poggio T, Yeap LS, Karaca E, Blasco RB, Langellotto F, Ambrogio C, Voena C, Wiestner A, Kasar SN, Brown JR, Sun J, Wu CJ, Gostissa M, Alt FW, Chiarle R. Phosphatidylinositol 3-kinase delta blockade increases genomic instability in B cells. Nature. 2017;542:489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia P, Jin H, Meador CB, Xia J, Ohashi K, Liu L, Pirazzoli V, Dahlman KB, Politi K, Michor F, Zhao Z, Pao W. Next-generation sequencing of paired tyrosine kinase inhibitor-sensitive and -resistant EGFR mutant lung cancer cell lines identifies spectrum of DNA changes associated with drug resistance. Genome Res. 2013;23:1434–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao X, Zhou Y, Sun H, Xu M, Bi X, Zhao Z, Shen B, Wan F, Hong Z, Lan L, Luo L, Guo Z, Yin Z. EGFR-TKI-induced HSP70 degradation and BER suppression facilitate the occurrence of the EGFR T790M resistant mutation in lung cancer cells. Cancer Lett. 2018;424:84–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JK, Lee J, Kim S, Kim S, Youk J, Park S, An Y, Keam B, Kim DW, Heo DS, Kim YT, Kim JS, Kim SH, Lee JS, Lee SH, Park K, Ku JL, Jeon YK, Chung DH, Park PJ, Kim J, Kim TM, Ju YS. Clonal History and Genetic Predictors of Transformation Into Small-Cell Carcinomas From Lung Adenocarcinomas. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2017:JCO2016719096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.