Abstract
Because survivors of pediatric acute lymphoblastic leukemia (ALL) are more likely to be obese than unaffected contemporaries, we compared DNA methylation profiles between normal-weight and obese survivors at adiposity-associated CpG sites previously-reported by epigenome-wide association studies (EWAS) of body mass index (BMI) in the general population. We selected 96 ALL survivors from the Childhood Cancer Survivor Study (CCSS): 48 obese (BMI ≥30.0 kg/m2) and 48 normal weight (BMI = 18.5–24.9 kg/m2). The Illumina HumanMethylation450 BeadChip was used to compare DNA methylation at 211 loci identified in EWAS of BMI in the general population. The false discovery rate (FDR) was used to account for multiple testing. In exploratory analyses, we also tested for interaction between cranial radiotherapy (CRT) status and selected CpG sites to evaluate differences by CRT exposure. Thirty-nine loci were associated (FDR <0.05) with obesity among survivors who only received chemotherapy (n = 49), including ABCG1 cg06500161. No loci were significantly associated with obesity among CRT-exposed survivors (n = 47). There was evidence (P-value <0.05) of interaction between CRT and methylation at six of the 39 methylation sites. Our results suggest that previously identified BMI-DNA methylation loci are associated with obesity in pediatric ALL survivors who were spared CRT, while no loci were significantly associated with obesity in survivors who received CRT. Given the obesogenic characteristics of ALL therapy, this study adds to the growing evidence of that the mechanisms underlying obesity in ALL survivors differ based on treatment exposures and may inform future intervention strategies among these individuals.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the most common malignancy diagnosed in those less than 15 years of age. While advances in treatment strategies have led to five-year survival rates approaching 90%,1 curative therapy for pediatric ALL is associated with an increased risk for numerous chronic health conditions, including obesity.2–8 In a previous report from the Childhood Cancer Survivor Study (CCSS), the risk of obesity among long-term survivors of pediatric ALL (diagnosed between 1970 and 1986) was increased 20% for males and 50% for females compared to population-based normative data.9 Notably, this late effect has persisted across decades of evolution in treatment strategies and is prevalent regardless of age at diagnosis, sex, or treatment exposure.10 As obesity is known to contribute to an increased risk of hypertension,11 type 2 diabetes,12 cardiovascular disease,13 cancer,14 and premature death,15 it is imperative to develop and apply interventions in this at-risk population.16 However, effective intervention strategies for ALL survivors requires a clear understanding of: 1) those who are at greatest risk of becoming obese; 2) the mechanisms of obesity in survivors; and 3) the molecular pathways that lead to adverse outcomes observed in obese survivors.
A few demographic and treatment factors, including female sex, younger age and higher body mass index (BMI) at diagnosis, Hispanic ethnicity, and exposure to cranial radiotherapy (CRT), have been linked to an increased risk of obesity among ALL survivors.9, 10, 17, 18 However, these factors do not fully explain the inter-patient variability in susceptibility to obesity following treatment for pediatric ALL. While there is mounting evidence that treatment and genetic factors jointly contribute to risk of obesity in childhood cancer survivors,19 the molecular pathways that underlie this risk are not yet understood.
Epigenome-wide analyses in the general population have revealed associations between DNA methylation at cytosine-guanine dinucleotides (CpG sites) and several metabolic traits, including BMI, waist circumference, and obesity.20–23 Collectively, studies conducted in diverse adult populations have identified obesity-associated differential methylation at >200 CpG sites;20–23 however, associations between DNA methylation and obesity phenotypes in pediatric ALL survivors have not been studied, which could shed new light on the molecular underpinnings of obesity in this population. Informed by previous studies of childhood cancer survivors,9, 16, 18, 24 our hypothesis was that obesity in survivors of ALL shares similar molecular pathogenesis to that in the general population. Therefore, we compared methylation profiles at previously-reported, adiposity-associated CpG sites between normal-weight (BMI: 18.5–25.0 kg/m2) and obese (BMI ≥30.0 kg/m2) adult survivors of pediatric ALL enrolled in the CCSS, with the objective of characterizing epigenetic profiles associated with obesity among survivors of pediatric ALL.
MATERIALS AND METHODS
Study population
Survivors were participants in the CCSS,25 a collaborative, multi-institutional study of pediatric and adolescent cancer patients who survived at least five years following a diagnosis of leukemia, bone tumor, central nervous system tumor, Hodgkin lymphoma, kidney tumor, neuroblastoma, non-Hodgkin lymphoma, or soft-tissue sarcoma. Survivors enrolled in the original CCSS cohort met the following criteria: 1) received treatment at one of 26 participating CCSS institutions; 2) cancer diagnosis between 01/01/1970 and 12/31/1986; and 3) younger than 21 years old at diagnosis. The study documents and protocols were approved by the institutional review board at each participating institution.
Survivors enrolled in the original cohort completed baseline (1992–2002) and follow-up (2000–2002, 2002–2005, 2007–2009) questionnaires, capturing detailed demographic, employment, educational, health, and medical information. Participants were asked to report their height and weight without shoes, which was used to calculate BMI (kg/m2). National Heart, Lung, and Blood Institute criteria were applied to define the following BMI categories: underweight (BMI <18.5 kg/m2), normal weight (BMI: 18.5–24.9 kg/m2), overweight (BMI: 25.0–29.9 kg/m2), and obese (BMI ≥30.0 kg/m2).26 For the current report, 96 survivors were selected from the pool of ALL survivors with an available DNA sample using “extreme phenotype” sampling: 48 obese (BMI ≥ 30.0 kg/m2) and 48 normal weight (18.5 ≤ BMI ≤ 24.9 kg/m2) based on the CCSS 2007 Follow-up survey. Specifically, eligible participants included adult (age ≥20 years at 2007 survey) survivors of ALL (with no history of bone marrow transplant, recurrence, or subsequent malignant neoplasms), genotyped as part of the CCSS and National Cancer Institute collaborative genome-wide association study and genetically-defined as being predominantly of European ancestry (i.e., ≥80%) with details on therapeutic exposures.27 To form the obese group, we randomly selected 24 obese survivors who had CRT doses of ≥18 Gy and 24 obese survivors who did not receive CRT (i.e., chemotherapy only), in order to be able to evaluate the methylation-association differences by CRT status, one of the strongest risk factors for obesity among adult survivors of pediatric ALL.18 An equal number of normal-weight survivors was selected and frequency matched on: sex (male, female); age at diagnosis (<10 years, ≥10 years); age at follow up (20–30 years, >30 years); and CRT dose (≥18 Gy, none). Although certain treatment-associated endocrine abnormalities may contribute to obesity risk in specific survivor populations,28 no participants were excluded on the basis of reported endocrinopathies because information on the age of onset and whether the condition was controlled by medication was limited for many of the conditions.
Treatment information
Medical records were used to abstract dose information for chemotherapy agents. Radiation records were centrally reviewed and dosimetry estimated by the Radiation Physics Center at MD Anderson Cancer Center (Houston, Texas). Study questionnaires and medical record abstraction forms are available at: https://ccss.stjude.org/.
Sample collection and DNA methylation profiling
Buccal cell samples were collected from study participants using a 45 mL mouthwash kit at baseline. As noted, the outcome was defined using the CCSS 2007 Follow-up survey to evaluate DNA methylation profiles prior to the endpoint (late-onset obesity). Kits were mailed to participants along with instructions for collecting the specimen and returning the container to the CCSS Biorepository at Cincinnati Children’s Hospital (Cincinnati, OH), where samples were processed and stored. DNA was extracted from buccal cell samples and sent to Baylor College of Medicine (BCM) for bisulfite treatment using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA). DNA methylation levels were interrogated using the Infinium HumanMethylation450 BeadChip Array (Illumia, Inc., San Diego, CA) in the Laboratory of Translational Genomics at BCM. Raw DNA methylation data were processed in R using the ChAMP Bioconductor package (http://bioconductor.org/biocLite.R). Quality control and processing of the 485,512 CpG sites included on the array removed sites with a detection P-value >0.01 (n = 2,310 probes removed), a bead-count <3 in ≥5% of the samples (n = 373 probes removed), cross-reactive with non-CpG sites (n = 3,078 probes removed), associated with a single nucleotide polymorphism (n = 50,162 probes removed), or aligned to multiple positions (n = 7,141 probes removed). Non-autosomal CpG sites were removed to facilitate comparisons across sexes (n = 10,138 probes removed). Beta-mixture quantile (BMIQ) normalization and Combat batch effect correction was performed on the remaining 412,310 CpG sites to obtain batch-corrected, normalized Beta-values, which provides an estimate of the proportion of cells methylated at a particular site (range: 0–1). Overall, 184 of the 211 BMI-associated CpG sites identified in recent epigenome-wide analyses passed quality control and were retained for statistical analyses.20–23
Statistical analysis
Descriptive statistics provided for the normal-weight and obese survivor groups included: 1) means and standard deviations for continuous variables; and 2) counts and proportions for categorical variables. Transformed DNA methylation Beta-values (i.e., M-values) at each of the 184 quality-controlled, BMI-associated CpG sites were compared between normal-weight and obese survivors using linear regression, with the M-value serving as the dependent variable: adjustment for demographic and treatment variables did not notably change the effect estimates obtained from unadjusted models and, therefore, only unadjusted estimates are presented. Secondary analyses stratifying on CRT status were performed to evaluate potential differences in effect due to this treatment exposure. Additionally, product terms between CRT and obesity status were included in the linear regression models to assess differences in the methylation-obesity association between those who received CRT and those who received chemotherapy only. Finally, because of previous reports indicating differences in genetic risk of obesity by sex among CRT-exposed survivors,24 an exploratory analysis was performed stratifying on sex among the CRT-exposed group.
False discovery rates (FDR) were calculated to account for multiple comparisons. Quantile-quantile (Q-Q) plots were generated to compare the distribution of observed P-values to the expected distribution under the null hypothesis of no association between CpG methylation and obesity.
RESULTS
The demographic and clinical characteristics of the study population are presented in Table 1. Obesity status was ascertained at a mean age of 33.8 years and an average of 12.3 years after the collection of the DNA sample.
TABLE 1.
Demographic characteristics of the study participants
| Normal (n = 48) |
Obese (n = 48) |
P-value* | |
|---|---|---|---|
| Sex, n (%) | |||
| Male | 24 (50) | 24 (50) | 1.00 |
| Female | 24 (50) | 24 (50) | |
| Age at diagnosis, mean (SD) | 6.1 (4.3) | 6.0 (4.4) | 0.93 |
| Age at sample, mean (SD) | 21.4 (6.5) | 21.7 (6.9) | 0.81 |
| Age at last visit, mean (SD) | 33.5 (6.3) | 34.1 (6.7) | 0.62 |
| CRT status, n(%) | |||
| Chemotherapy only | 25 (52.1) | 24 (50) | 0.84 |
| CRT+Chemotherapy | 23 (47.9) | 24 (50) | |
P-Values calculated using Pearson’s Chi-squared test or T-Test
For each of the 184 CpG sites included in the analysis, the chromosome location, proximal gene, difference in methylation Beta-values between obese and normal-weight survivors (Δβ), and statistical significance of the observed differences are presented in Supporting Information Table 1. In the overall cohort (n = 96), there was statistically significant differential DNA methylation at cg06500161 (Δβ = 0.0335, uncorrected P-value = 6.24×10−5; FDR P-value = 0.012), a well-established methylation locus located on chromosome 21 near the ABCG1 gene (Table 2). Methylation at this site was significantly associated with obesity in the chemotherapy only group (Δβ = 0.0450, uncorrected P-value = 4.25×10−5; FDR P-value = 0.003) but not statistically significantly associated with obesity in the CRT-exposed group (Δβ = 0.0281, uncorrected P-value = 0.010; FDR P-value = 0.149). In models stratified on CRT status, 39 sites demonstrated statistically significant (FDR P-value <0.05) differential methylation among the chemotherapy only group, while no methylation loci reached statistical significance in the CRT-exposed group after accounting for multiple comparisons (results for these 39 sites are presented in Table 2 overall and by CRT status). This was true of both male and females treated with CRT (data not shown). The strongest evidence of differential methylation in those who received chemotherapy only was for cg00431050 (Δβ = 0.0308, uncorrected P-value = 4.29×10−6; FDR P-value = 0.001), which is located on chromosome 10 near the ELOVL3 gene.
TABLE 2.
Significant obesity-DNA methylation associations overall and by CRT status
| Loci characteristics | All subjects | Chemotherapy only | CRT+Chemotherapy | Obesity-CRT interaction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG Site | Chr | Gene | Δβ | P-value | FDR | Δβ | P-value | FDR | Δβ | P-value | FDR | P-value |
| cg00431050 | 10 | ELOVL3 | 0.0169 | 9.37E-03 | 0.103 | 0.0308 | 4.29E-06 | 0.001 | 0.0017 | 8.61E-01 | 0.943 | 0.040 |
| cg03523676 | 14 | CPNE6 | 0.0257 | 1.20E-02 | 0.103 | 0.0502 | 1.09E-05 | 0.002 | −0.0013 | 9.28E-01 | 0.971 | 0.023 |
| cg06500161 | 21 | ABCG1 | 0.0335 | 6.24E-05 | 0.012 | 0.0450 | 4.25E-05 | 0.003 | 0.0281 | 1.00E-02 | 0.149 | 0.379 |
| cg07728579 | 15 | FSD2 | 0.0101 | 2.19E-02 | 0.133 | 0.0181 | 1.14E-04 | 0.006 | 0.0008 | 9.07E-01 | 0.959 | 0.072 |
| cg08972190 | 7 | MAD1L1 | 0.0059 | 6.68E-02 | 0.236 | 0.0161 | 6.12E-04 | 0.014 | −0.0031 | 4.17E-01 | 0.691 | 0.019 |
| cg01101459 | 1 | NA | 0.0181 | 7.80E-02 | 0.261 | 0.0456 | 6.60E-04 | 0.014 | −0.0047 | 7.37E-01 | 0.875 | 0.034 |
| cg27184903 | 15 | APBA2 | 0.0176 | 1.24E-02 | 0.103 | 0.0275 | 5.62E-04 | 0.014 | 0.0082 | 4.28E-01 | 0.698 | 0.230 |
| cg14017402 | 2 | NA | 0.0167 | 2.06E-02 | 0.133 | 0.0296 | 5.09E-04 | 0.014 | 0.0087 | 3.87E-01 | 0.684 | 0.315 |
| cg13708645 | 12 | KDM2B | 0.0347 | 6.47E-03 | 0.103 | 0.0484 | 5.28E-04 | 0.014 | 0.0235 | 2.07E-01 | 0.47 | 0.383 |
| cg10927968 | 11 | NA | 0.0082 | 2.76E-01 | 0.494 | 0.0318 | 1.11E-03 | 0.017 | −0.0122 | 2.48E-01 | 0.537 | 0.024 |
| cg27243685 | 21 | ABCG1 | 0.0138 | 4.09E-02 | 0.193 | 0.0256 | 1.17E-03 | 0.017 | 0.0023 | 8.15E-01 | 0.915 | 0.117 |
| cg02711608 | 19 | SLC1A5 | −0.0182 | 9.49E-03 | 0.103 | −0.0287 | 8.98E-04 | 0.017 | −0.0143 | 1.37E-01 | 0.442 | 0.571 |
| cg04577162 | 7 | RFC2 | 0.0266 | 1.47E-03 | 0.057 | 0.0368 | 9.86E-04 | 0.017 | 0.0222 | 5.73E-02 | 0.33 | 0.705 |
| cg13922488 | 19 | PKN1 | 0.0141 | 5.27E-02 | 0.222 | 0.0300 | 1.91E-03 | 0.024 | −0.0006 | 9.46E-01 | 0.979 | 0.062 |
| cg27547344 | 1 | TIE1 | 0.0200 | 1.54E-03 | 0.057 | 0.0267 | 1.95E-03 | 0.024 | 0.0115 | 1.57E-01 | 0.442 | 0.435 |
| cg11024682 | 17 | SREBF1 | 0.0222 | 1.43E-01 | 0.354 | 0.0533 | 2.39E-03 | 0.026 | 0.0005 | 9.80E-01 | 0.986 | 0.138 |
| cg01243823 | 16 | NOD2 | −0.0337 | 6.59E-04 | 0.057 | −0.0333 | 2.35E-03 | 0.026 | −0.0361 | 1.05E-02 | 0.149 | 0.867 |
| cg18307303 | 5 | IL12B | 0.0150 | 1.58E-02 | 0.112 | 0.0237 | 2.63E-03 | 0.027 | 0.0075 | 3.81E-01 | 0.681 | 0.315 |
| cg26403843 | 5 | RNF145 | 0.0458 | 2.62E-03 | 0.069 | 0.0530 | 2.84E-03 | 0.028 | 0.0491 | 2.87E-02 | 0.22 | 0.812 |
| cg11832534 | 1 | WDR8 | 0.0105 | 1.24E-01 | 0.344 | 0.0243 | 3.56E-03 | 0.029 | 0.0014 | 8.83E-01 | 0.947 | 0.169 |
| cg07037944 | 15 | DAPK2 | −0.0162 | 1.15E-02 | 0.103 | −0.0260 | 3.12E-03 | 0.029 | −0.0113 | 1.56E-01 | 0.442 | 0.303 |
| cg10438589 | 4 | NA | 0.0168 | 1.69E-01 | 0.366 | 0.0430 | 3.53E-03 | 0.029 | −0.0024 | 8.85E-01 | 0.947 | 0.353 |
| cg05720226 | 7 | ST7 | 0.0086 | 1.46E-01 | 0.354 | 0.0192 | 3.25E-03 | 0.029 | 0.0053 | 5.06E-01 | 0.752 | 0.467 |
| cg25217710 | 1 | NA | 0.0136 | 2.06E-01 | 0.426 | 0.0354 | 3.98E-03 | 0.03 | −0.0057 | 7.20E-01 | 0.866 | 0.120 |
| cg26952928 | 8 | SLC45A4 | 0.0110 | 9.51E-02 | 0.292 | 0.0207 | 3.97E-03 | 0.03 | 0.0045 | 6.38E-01 | 0.811 | 0.229 |
| cg23232188 | 3 | EAF2 | 0.0143 | 3.61E-01 | 0.573 | 0.0530 | 5.08E-03 | 0.033 | −0.0131 | 5.42E-01 | 0.762 | 0.070 |
| cg08443038 | 16 | CBFA2T3 | −0.0031 | 4.52E-01 | 0.655 | −0.0130 | 5.10E-03 | 0.033 | 0.0051 | 3.90E-01 | 0.684 | 0.071 |
| cg07682160 | 19 | UPF1 | 0.0189 | 2.14E-02 | 0.133 | 0.0262 | 5.37E-03 | 0.033 | 0.0171 | 1.53E-01 | 0.442 | 0.784 |
| cg10549088 | 3 | NA | 0.0345 | 1.24E-03 | 0.057 | 0.0415 | 4.68E-03 | 0.033 | 0.0347 | 1.01E-02 | 0.149 | 0.968 |
| cg04927537 | 17 | LGALS3BP | 0.0447 | 3.58E-03 | 0.075 | 0.0519 | 5.14E-03 | 0.033 | 0.0451 | 2.76E-02 | 0.22 | 0.998 |
| cg06192883 | 15 | MYO5C | 0.0195 | 2.24E-02 | 0.133 | 0.0331 | 6.02E-03 | 0.035 | 0.0075 | 4.68E-01 | 0.731 | 0.279 |
| cg22012981 | 3 | ACOX2 | 0.0305 | 1.84E-03 | 0.057 | 0.0307 | 5.80E-03 | 0.035 | 0.0306 | 2.99E-02 | 0.22 | 0.812 |
| cg03050965 | 1 | S1PR1 | 0.0025 | 2.81E-01 | 0.495 | 0.0081 | 7.04E-03 | 0.039 | −0.0035 | 2.96E-01 | 0.593 | 0.032 |
| cg19373099 | 2 | NA | 0.0239 | 2.31E-01 | 0.443 | 0.0631 | 7.05E-03 | 0.039 | −0.0192 | 4.89E-01 | 0.738 | 0.072 |
| cg26687842 | 13 | LOC646982 | 0.0111 | 1.67E-01 | 0.366 | 0.0253 | 7.47E-03 | 0.039 | −0.0017 | 8.82E-01 | 0.947 | 0.149 |
| cg22488164 | 12 | PLBD1 | 0.0227 | 8.14E-02 | 0.263 | 0.0414 | 7.29E-03 | 0.039 | 0.0171 | 3.72E-01 | 0.675 | 0.417 |
| cg05648472 | 11 | PRDM11 | 0.0143 | 2.33E-02 | 0.134 | 0.0216 | 8.13E-03 | 0.041 | 0.0065 | 4.08E-01 | 0.689 | 0.310 |
| cg25649826 | 17 | USP22 | 0.0110 | 1.41E-01 | 0.354 | 0.0223 | 8.36E-03 | 0.041 | 0.0058 | 6.18E-01 | 0.81 | 0.594 |
| cg00673344 | 3 | NA | −0.0114 | 6.68E-02 | 0.236 | −0.0196 | 8.96E-03 | 0.043 | −0.0041 | 5.82E-01 | 0.792 | 0.182 |
In our exploratory analyses, six of the 39 sites (15%) that reached statistical significance in the population who received chemotherapy only also had statistical evidence of interaction (P-value <0.05) between obesity and CRT, indicating that the association between obesity and methylation at these loci differed significantly between survivors who received chemotherapy only and those who were also treated with CRT (Table 2). Specifically, there were differences in effect for ELOVL3 cg00431050 (P-value = 0.04); CPNE6 cg03523676 (P-value = 0.02); MAD1L1 cg08972190 (P-value = 0.02); locus cg01101459 on chromosome 1 (P-value = 0.03); locus cg10927968 on chromosome 11 (P-value = 0.02); and S1PR1 cg03050965 (P-value = 0.03). For each of these six loci, the difference in methylation levels between obese and normal-weight survivors was greater in those who received chemotherapy only compared to the CRT-exposed. In fact, for five of the six, the effect was in the opposite direction.
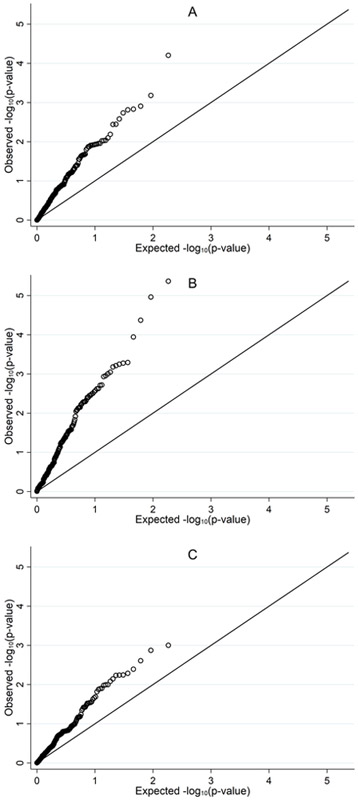
Q-Q plots depict the observed distribution of the group-specific P-values for each of the 184 BMI-associated CpG sites (Figure 1A-C). Overall, there was a departure from expectation (Figure 1A), indicating that previously identified BMI-associated CpG sites were enriched among obese survivors. However, the departure between the observed versus expected was attenuated in the CRT-exposed survivors (Figure 1C) compared to survivors who received chemotherapy only (Figure 1B).
FIGURE 1.
A, Q-Q plot showing distribution of observed P-values for the 184 BMI-DNA methylation loci compared to the expected distribution among all subjects. B, Q-Q plot showing distribution of observed P-values for the 184 BMI-DNA methylation loci compared to the expected distribution among subjects who only received chemotherapy. C, Q-Q plot showing distribution of observed P-values for the 184 BMI-DNA methylation loci compared to the expected distribution among subjects who received CRT and chemotherapy.
DISCUSSION
While several studies have demonstrated an association between DNA methylation and obesity in the general population,20–23 to our knowledge this is the first attempt to replicate these previously identified BMI-DNA methylation loci among adult survivors of pediatric ALL, a population at greater risk of obesity compared to their unaffected contemporaries.2–9 Our assessment sought to determine: 1) if BMI-DNA methylation loci identified in the general population were similarly associated with obesity among ALL survivors, and 2) if the association between DNA methylation and obesity varied by treatment exposure (i.e., CRT). Notably, methylation at the CpG site for which there is the strongest evidence in the general population of a causal link with BMI (NFATC2IP cg26663590)23 was not implicated in our assessment, suggesting that if ALL therapy induces epigenetic changes that lead to obesity, it is through other loci.
Only ABCG1 cg06500161 was significantly associated with obesity in the overall survivor population after adjusting for multiple comparisons. However, 39 of the 184 previously identified BMI-DNA methylation loci (21%) were significantly associated with obesity in survivors who received chemotherapy only. The differences observed in the CRT-exposed survivors and those who received chemotherapy only are not likely attributable to sample size, as a similar number of individuals were included in each group. In fact, this finding could point to differences in the mechanisms of obesity between these groups. For instance, one mechanism proposed to explain the association between CRT and obesity in survivors of pediatric ALL is radiation-induced damage to the hypothalamic-pituitary axis, resulting in leptin insensitivity.18, 19, 24 This is consistent with one report indicating the association between genetic variation in the leptin receptor (LEPR) gene and obesity was specific to those who received CRT.24 Another potential underlying pathology in those exposed to CRT is growth hormone deficiency,29 which could result in differences in DNA methylation profiles in these individuals. Although a detailed evaluation of endocrine abnormalities was not feasible in this study of 96 ALL survivors, it is possible that CRT-related endocrinopathies are partly responsible for the elevated risk of obesity observed among radiated survivors as well as differences in epigenetic profiles. Our results are consistent with the previous literature indicating radiation-induced obesity may involve different molecular mechanisms compared to obesity in survivors who receive chemotherapy only.
Notably, among the survivors who received chemotherapy only, the direction of associations for the 39 replicated BMI-DNA methylation loci were consistent with previous assessments.20–23 The strongest association was observed for the ELOVL3 locus. ELOVL3 (ELOVL Fatty Acid Elongase 3) is located on the long arm of chromosome 10 and is a protein coding gene.30 The ELOVL3 protein is involved in the metabolism of lipids and lipoproteins.30–32 The ELOVL3 BMI-DNA methylation locus cg00431050 has been reported in the epigenome-wide association study (EWAS) of BMI conducted by Wahl et al.23 While the function of this locus remains unknown, it has also been implicated in chronic obstructive pulmonary disease among African-Americans33 and serum levels of C-reactive protein,34 which suggests this locus may be involved in systemic and chronic inflammation, a potential driver of cardiovascular disease.35
As noted, the only association that remained statistically significant in the entire cohort was at the ABCG1 cg06500161 locus. This BMI-DNA methylation locus has been identified and replicated in several EWAS across multiple tissues.21, 23, 36 ABCG1 is involved in macrophage cholesterol and phospholipids transport, as well as cellular regulation of lipid homeostasis.37 Notably, ABCG1 promoter hypermethylation is strongly associated with coronary artery disease,38 and ABCG1 cg06500161 has been identified in an epigenome-wide assessment of insulin-related traits.39 As ABCG1 is involved in cholesterol transport37 and regulates insulin secretion,39 these observations may provide insight into the pathways that may link obesity to other adverse outcomes including type 2 diabetes, cardiovascular disease, and even subsequent cancers.23 Understanding the biology of BMI-DNA methylation loci may elucidate the molecular mechanisms underlying adverse outcomes that are a consequence of obesity in survivors of ALL.
Our study must be considered in the light of certain limitations. First, emerging evidence suggests that differential DNA methylation at a majority of the BMI-associated CpG sites identified thus far appear to be secondary to increased adiposity rather than a “cause”.23 However, given the central role many of the identified loci play in important biologic processes, such as lipid metabolism and inflammation, the results of this study provide a potential molecular link between obesity and subsequent adverse outcomes even if the associations identified reflect epigenetic consequences of obesity. Therefore, these findings could eventually inform intervention strategies for obesity-related chronic health conditions among childhood cancer survivors. Second, while our population was well characterized, our sample was limited in size (n = 96). Specifically, because of the sample size and the absence of an independent population for replication, we were restricted to evaluating previously identified BMI-DNA methylation loci rather than conducting an epigenome-wide assessment in this at-risk population. Nonetheless, we were able to replicate several loci associated with obesity in the general population among those ALL survivors who received chemotherapy only, using the targeted selection in the extreme phenotype design. Additionally, we were limited in our ability to explore differences in estimates of adiposity other than BMI or evaluate differences by demographic and clinical characteristics beyond CRT status. Patients included in this study were likely exposed to numerous treatment protocols (diagnosed: 1970–1986), though 98% were treated with glucocorticoids (results not shown). While no differences were observed by chemotherapy exposures or demographic factors, including sex, in exploratory analyses, future studies should also evaluate treatment- and sex-specific effects in contemporary populations.24 Moreover, BMI may not accurately estimate total or central adiposity in all cancer survivors.40, 41 Future studies should consider alternative, perhaps more sensitive measures of adiposity. Third, a limitation for several studies evaluating the role of DNA methylation on adverse outcomes is the use of biologically relevant tissues.42 In our case, DNA methylation profiles from buccal cells may not be the most relevant for studies of obesity. However, the BMI-DNA methylation loci evaluated in this study have been replicated across several tissues including blood and adipose.21, 23 Strengths of this study include well-annotated treatment information on ALL survivors included in the CCSS, which allowed the assessment of BMI-DNA methylation loci by CRT dose, a strong predictor of obesity in this population.9, 18 Because of the larger sample size of the CCSS, we were able to select a balanced population in terms of key demographic and treatment variables, which limited confounding by these factors.
Overall, our study demonstrated: 1) previously identified BMI-DNA methylation loci appeared to be more strongly associated with obesity among ALL survivors who received chemotherapy only compared to CRT-exposed ALL survivors, which contributes to a growing body of evidence that the molecular mechanisms and consequences of obesity are distinct in these groups;16, 24 and 2) BMI-DNA methylation loci associated with obesity in survivors who received chemotherapy only are those suspected to be a consequence of BMI, which may prove useful in studies evaluating the molecular mechanisms underlying adverse outcomes associated with obesity in this population.23 We recommend a future EWAS of BMI among survivors of ALL that includes those exposed to CRT. This may elucidate the role of novel epigenetic mechanisms on obesity. Additionally, the integration of data on inherited genetic variation may help establish BMI-DNA methylation loci that are a consequence of obesity versus those that may have a causal influence. Ultimately, better understanding of the mechanisms of obesity in survivors of ALL, as well as the adverse outcomes that are a consequence of obesity, will aid in precision prevention and treatment efforts in this at-risk population.
Supplementary Material
ACKNOWLEDGEMENTS:
We thank the participants of the Childhood Cancer Survivor Study (CCSS).
Funding Information: This work was supported by National Cancer Institute Grant No. CA55727 (G.T.A.), National Cancer Institute Grant No. CA218362 (A.L.B.), the Childhood Cancer Survivor Study (CCSS) Career Development Award (P.J.L), and the Cancer Prevention & Research Institute of Texas RP140258 (P.J.L.).
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest.
REFERENCES
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med 2015;373(16):1541–1552. [DOI] [PubMed] [Google Scholar]
- 2.Birkebaek NH, Clausen N. Height and weight pattern up to 20 years after treatment for acute lymphoblastic leukaemia. Arch Dis Child. 1998;79(2):161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craig F, Leiper AD, Stanhope R, et al. Sexually dimorphic and radiation dose dependent effect of cranial irradiation on body mass index. Arch Dis Child. 1999;81(6):500–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Didi M, Didcock E, Davies HA, et al. High incidence of obesity in young adults after treatment of acute lymphoblastic leukemia in childhood. J Pediatr 1995;127(1):63–67. [DOI] [PubMed] [Google Scholar]
- 5.Mayer EI, Reuter M, Dopfer RE, et al. Energy expenditure, energy intake and prevalence of obesity after therapy for acute lymphoblastic leukemia during childhood. Horm Res 2000;53(4):193–199. [DOI] [PubMed] [Google Scholar]
- 6.Odame I, Reilly JJ, Gibson BE, et al. Patterns of obesity in boys and girls after treatment for acute lymphoblastic leukaemia. Arch Dis Child. 1994;71(2):147–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oeffinger KC, Buchanan GR, Eshelman DA, et al. Cardiovascular risk factors in young adult survivors of childhood acute lymphoblastic leukemia. J Pediatr Hematol Oncol 2001;23(7):424–430. [DOI] [PubMed] [Google Scholar]
- 8.Sklar CA, Mertens AC, Walter A, et al. Changes in body mass index and prevalence of overweight in survivors of childhood acute lymphoblastic leukemia: role of cranial irradiation. Med Pediatr Oncol 2000;35(2):91–95. [DOI] [PubMed] [Google Scholar]
- 9.Meacham LR, Gurney JG, Mertens AC, et al. Body mass index in long-term adult survivors of childhood cancer: a report of the Childhood Cancer Survivor Study. Cancer. 2005;103(8):1730–1739. [DOI] [PubMed] [Google Scholar]
- 10.Zhang FF, Kelly MJ, Saltzman E, et al. Obesity in pediatric ALL survivors: a meta-analysis. Pediatrics. 2014;133(3):e704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauss RM, Winston M, Fletcher RN, et al. Obesity: impact of cardiovascular disease. Circulation. 1998;98(14):1472–1476. [PubMed] [Google Scholar]
- 12.Sinaiko AR, Donahue RP, Jacobs DR Jr., et al. Relation of weight and rate of increase in weight during childhood and adolescence to body size, blood pressure, fasting insulin, and lipids in young adults. The Minneapolis Children’s Blood Pressure Study. Circulation. 1999;99(11):1471–1476. [DOI] [PubMed] [Google Scholar]
- 13.Rosengren A, Stegmayr B, Johansson I, et al. Coronary risk factors, diet and vitamins as possible explanatory factors of the Swedish north-south gradient in coronary disease: a comparison between two MONICA centres. J Intern Med 1999;246(6):577–586. [DOI] [PubMed] [Google Scholar]
- 14.Ligibel JA, Alfano CM, Courneya KS, et al. American Society of Clinical Oncology position statement on obesity and cancer. J Clin Oncol 2014;32(31):3568–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masters RK, Reither EN, Powers DA, et al. The impact of obesity on US mortality levels: the importance of age and cohort factors in population estimates. Am J Public Health. 2013;103(10):1895–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tonorezos ES, Oeffinger KC. Obesity Following Childhood Cancer: Mechanisms and Consequences In: Mittelman SD, Berger NA, editors. Energy Balance and Hematologic Malignancies. New York City: Springer; 2012. p. 141–158. [Google Scholar]
- 17.Brown AL, Lupo PJ, Danysh HE, et al. Prevalence and predictors of overweight and obesity among a multiethnic population of pediatric acute lymphoblastic leukemia survivors: A cross-sectional assessment. J Pediatr Hematol Oncol 2016;38(6):429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oeffinger KC, Mertens AC, Sklar CA, et al. Obesity in adult survivors of childhood acute lymphoblastic leukemia: a report from the Childhood Cancer Survivor Study. J Clin Oncol 2003;21(7):1359–1365. [DOI] [PubMed] [Google Scholar]
- 19.Wilson CL, Liu W, Yang JJ, et al. Genetic and clinical factors associated with obesity among adult survivors of childhood cancer: A report from the St. Jude Lifetime Cohort. Cancer. 2015;121(13):2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aslibekyan S, Demerath EW, Mendelson M, et al. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring). 2015;23(7):1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demerath EW, Guan W, Grove ML, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet 2015;24(15):4464–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–1998. [DOI] [PubMed] [Google Scholar]
- 23.Wahl S, Drong A, Lehne B, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541(7635):81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross JA, Oeffinger KC, Davies SM, et al. Genetic variation in the leptin receptor gene and obesity in survivors of childhood acute lymphoblastic leukemia: a report from the Childhood Cancer Survivor Study. J Clin Oncol 2004;22(17):3558–3562. [DOI] [PubMed] [Google Scholar]
- 25.Robison LL, Mertens AC, Boice JD, et al. Study design and cohort characteristics of the Childhood Cancer Survivor Study: a multi-institutional collaborative project. Med Pediatr Oncol 2002;38(4):229–239. [DOI] [PubMed] [Google Scholar]
- 26.National Heart L, and Blood Institute,. Why Obesity is a Health Problem. Available at: http://www.nhlbi.nih.gov/health/public/heart/obesity/wecan/healthy-weight-basics/obesity.htm. Accessed February 6, 2018.
- 27.Morton LM, Sampson JN, Armstrong GT, et al. Genome-wide association study to identify susceptibility loci that modify radiation-related risk for breast cancer after childhood cancer. J Natl Cancer Inst 2017;109(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mostoufi-Moab S, Seidel K, Leisenring WM, et al. Endocrine Abnormalities in Aging Survivors of Childhood Cancer: A Report From the Childhood Cancer Survivor Study. J Clin Oncol 2016;34(27):3240–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibson TM, Ehrhardt MJ, Ness KK. Obesity and metabolic syndrome among adult survivors of childhood leukemia. Curr Treat Options Oncol 2016;17(4):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerberg R, Tvrdik P, Unden AB, et al. Role for ELOVL3 and fatty acid chain length in development of hair and skin function. J Biol Chem 2004;279(7):5621–5629. [DOI] [PubMed] [Google Scholar]
- 31.Westerberg R, Mansson JE, Golozoubova V, et al. ELOVL3 is an important component for early onset of lipid recruitment in brown adipose tissue. J Biol Chem 2006;281(8):4958–4968. [DOI] [PubMed] [Google Scholar]
- 32.Zadravec D, Brolinson A, Fisher RM, et al. Ablation of the very-long-chain fatty acid elongase ELOVL3 in mice leads to constrained lipid storage and resistance to diet-induced obesity. FASEB J. 2010;24(11):4366–4377. [DOI] [PubMed] [Google Scholar]
- 33.Busch R, Qiu W, Lasky-Su J, et al. Differential DNA methylation marks and gene comethylation of COPD in African-Americans with COPD exacerbations. Respir Res 2016;17(1):143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun YV, Lazarus A, Smith JA, et al. Gene-specific DNA methylation association with serum levels of C-reactive protein in African Americans. PLoS One. 2013;8(8):e73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 2005;352(16):1685–1695. [DOI] [PubMed] [Google Scholar]
- 36.Al Muftah WA, Al-Shafai M, Zaghlool SB, et al. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenetics. 2016;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol 2010;30(2):139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng P, Wang L, Yang X, et al. A preliminary study of the relationship between promoter methylation of the ABCG1, GALNT2 and HMGCR genes and coronary heart disease. PLoS One. 2014;9(8):e102265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hidalgo B, Irvin MR, Sha J, et al. Epigenome-wide association study of fasting measures of glucose, insulin, and HOMA-IR in the Genetics of Lipid Lowering Drugs and Diet Network study. Diabetes. 2014;63(2):801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlage RE, Wilson CL, Zhang N, et al. Validity of anthropometric measurements for characterizing obesity among adult survivors of childhood cancer: A report from the St. Jude Lifetime Cohort Study. Cancer. 2015;121(12):2036–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller TL, Lipsitz SR, Lopez-Mitnik G, et al. Characteristics and determinants of adiposity in pediatric cancer survivors. Cancer Epidemiol Biomarkers Prev 2010;19(8):2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.