Abstract
Purpose:
Osimertinib was initially approved for T790M positive NSCLC and, more recently, for first-line treatment of EGFR-mutant NSCLC. However, resistance mechanisms to osimertinib have been incompletely described.
Experimental Design:
Using cohorts from MD Anderson Lung Cancer Moonshot GEMINI and Moffitt Cancer Center Lung Cancer databases, we collected clinical data for patients treated with osimertinib. Molecular profiling analysis was performed at the time of progression in a subset of the patients.
Results:
In the 118 patients treated with osimertinib, 42 had molecular profiling at progression. T790M was preserved in 21 (50%) patients and lost in 21 (50%). EGFR C797 and L792 (26%) mutations were the most common resistance mechanism and were observed exclusively in T790M-preserved cases. MET amplification was the second most common alteration (14%). Recurrent alterations were observed in 22 genes/pathways, including PIK3CA, FGFR, and RET. Preclinical studies confirmed MET, PIK3CA, and epithelial-to-mesenchymal transition (EMT) as potential resistance drivers. Alterations of cell cycle genes were associated with shorter median PFS (4.4 vs 8.8 months, p=0.01). In 76 patients with progression, osimertinib was continued in 47 cases with a median second progression-free survival (PFS2) was of 12.6 months; 21 patients received local consolidation radiation with median PFS of 15.5 months. Continuation of osimertinib beyond progression was associated with a longer overall survival compared to discontinuation (OS 11.2 vs 6.1 months, p=0.02).
Conclusions:
Osimertinib resistance is associated with diverse, predominantly EGFR-independent genomic alterations. Continuation of osimertinib post-progression, alone or in conjunction with radiotherapy, may provide prolonged clinical benefit in selected patients.
INTRODUCTION:
Osimertinib is a third-generation tyrosine kinase inhibitor (TKI) targeting Epidermal Growth Factor Receptor (EGFR)[1], with significant efficacy against both classical EGFR sensitizing mutations (such as exon 19 deletion or L858R mutation) and EGFR resistance T790M mutation[2]. In a randomized phase III trial[3], osimertinib significantly improved both progression-free survival (PFS) and objective response rate (ORR) when compared to platinum-pemetrexed chemotherapy for T790M-positive non-small cell lung cancer (NSCLC) after progression on 1st or 2nd generation EGFR-TKIs. Moreover, when compared to erlotinib or gefitinib in the first-line setting for EGFR mutant NSCLC, osimertinib showed a significant PFS benefit in favor of osimertinib in a phase III study (FLAURA trial, NCT02296125) [4]. Despite the increasing role of osimertinib for treatment NSCLC, there is limited data regarding resistance mechanisms to this agent. Reports on individual cases or clinical series have demonstrated that EGFR exon 20 tertiary mutations, including C797S[5–7], MET amplification[8], HER2 amplification[9], small cell transformation[10], and epithelial-mesenchymal transition (EMT) [10, 11] are possible drivers of resistance. Most recently, several groups reported that a subset of cases lost T790M at the time of progression[6, 12–14]
A comprehensive understanding of resistance mechanisms to osimertinib is needed in order to develop strategies to overcome osimertinib resistance. Here, we analyzed a cohort of 118 patients with advanced EGFR-mutant NSCLC treated with osimertinib at MD Anderson Cancer Center (MDACC) and Moffitt Cancer Center and Research Institute (MCC). We evaluated clinical and molecular data and described features that are associated with differential outcome. We also identified known and novel potential resistance mechanisms upon osimertinib progression.
PATIENTS AND METHODS:
Study population
Retrospective analyses were performed at MDACC and MCC. We queried The MD Anderson Lung Cancer Moon Shot GEMINI database (MDA protocol number 13–0589), a prospective database for patients with advanced NSCLC, for patients treated with osimertinib from January 2014 - October 2017. We queried the Moffitt electronic health record, Clinical Genomic Action Committee database (an internal initiative for tumor genomic profiling across multiple tumor types) and pyrosequencing database for NSCLC patients with EGFR T790M mutation between January 2011-December 2017 and isolated patients who were treated with osimertinib. Information on patient demographics, previous lines of therapy, survival and current status were collected until February 2018 (MDACC) and January 2018 (MCC), when the dataset was locked for the outcome analysis. Written informed consents were obtained the patients, and the studies were conducted in accordance with ethical guidelines including Declaration of Helsinki and U.S. Common Rule. The study was approved by the IRBs at MDACC and University of South Florida.
Statistical analysis
Time to treatment failure (TTF) on prior EGFR-TKI was defined as time from initiation of first EGFR-TKI until discontinuation of treatment due to PD or toxicity. Progression-free survival 1 (PFS1) was defined as time from starting osimertinib until PD or death. For patients who continued treatment beyond progression, PFS2 was defined as time from starting osimertinib until second PD or death[15]. Overall survival 1 (OS1) was defined as time from starting osimertinib until death from any cause. Overall survival from diagnosis (OS Dx) was defined as time from diagnosis of recurrent or metastatic EGFR-positive NSCLC until death. Patients alive at last follow-up were censored for both OS analysis. Kaplan-Meier method was used to estimate PFS1, PFS2, OS1 and OS Dx. Exploratory subgroup analysis included cell cycle gene alterations (e.g. CDK4/6 amplification), TP53 and T790M mutational status (previously associated with worse outcomes for EGFR-TKIs[16]), and CNS metastasis at the time of starting osimertinib. Between group differences were assessed through log-rank test. Hazard ratios and 95% confidence intervals were assessed with Cox proportional-hazards model. Multivariate analysis was performed by Cox-regression method.
Genomic profiling
Genomic profiling data was collected through test reports or medical charts. At MDACC, Molecular Diagnostics Labs-MDACC (MDL, 50 gene-panel, Supple. Methods) was used for tissue samples and digital-droplet PCR (ddPCR) for blood samples. Biodesix GeneStrat, Pyrosequencing of EGFR gene, and Moffitt Illumina TruSight Tumor 26 (TST26) sequencing (Supple. Methods) were used at MCC. Commercially available next-generation sequencing platforms such as FoundationOne (Foundation Medicine) and Guardant360 (Guardant Health) were used at both sites.
Cell lines, drug treatment and western blotting
HCC827, HCC4006 and H1975 cells were maintained in RPMI with 10% FBS. Western blot, drug treatment, and cell proliferation measurements were previously described (Supple. Methods).
RESULTS
Patient characteristics and efficacy of osimertinib
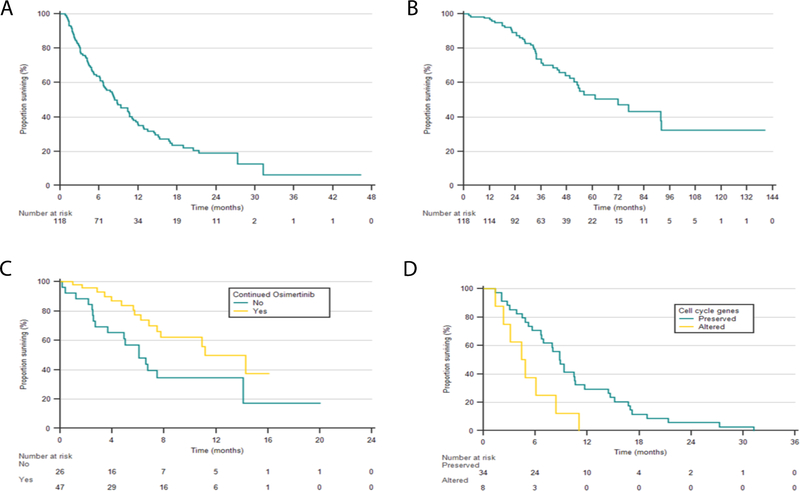
A total of 118 patients met the study criteria (Consort diagram shown in Supple. Fig 1). Median age was 63 years, 72% were female, 68% were never smokers, and 95% received previous EGFR-TKI (erlotinib: 78%). Median TTF on prior EGFR-TKI was 14 months (Table. 1). Median follow-up on osimertinib was 13 months, with 63% alive at the time of analysis. Median PFS1 was 8.4 months (95%CI, 6.7 to 10.7, Fig. 1A) and median OS1 was 25.2 months (95% CI, 17.5 to 29.2, Supple. Fig. 2). Three patients developed treatment-related pneumonitis and two patients developed treatment-related cardiotoxicity with decreased ejection.
Table 1.
Patient demographics and clinical characteristics
| Patients | 118 | |
|---|---|---|
| Age - years | ||
| Median | 63 | |
| Range | 36–88 | |
| Gender – N (%) | ||
| Male | 33 (28) | |
| Female | 85 (72) | |
| Smoking status – N (%) | ||
| Never | 80 (68) | |
| Former | 38 (32) | |
| Current | 0 (0) | |
| Histology – N (%) | ||
| Adenocarcinoma | 116 (98) | |
| Squamous cell carcinoma | 1 (1) | |
| Mixed | 1 (1) | |
| Disease stage – N (%) | ||
| Recurrent | 21 (18) | |
| Metastatic | 97 (82) | |
| Performance status – N (%) | ||
| 0–1 | 78 (66) | |
| 2 | 5 (4) | |
| 3–4 | 2 (2) | |
| NA | 33 (28) | |
| Central nervous system disease – N (%) | ||
| Yes | 31 (26) | |
| No | 85 (72) | |
| NA | 2 (2) | |
| Previous lines of therapy – N (%) | ||
| 0 | 6 (5) | |
| 1 | 45 (38) | |
| 2 | 36 (30) | |
| 3 | 15 (13) | |
| ≥ 4 | 16 (14) | |
| Previous EGFR TKI treatment – N (%) | ||
| Erlotinib | 92 (78) | |
| Other | 20 (17) | |
| None | 6 (5) | |
| TTF on previous TKI - months | ||
| Median | 14 | |
| Range | 3 – 88 | |
| Previous cytotoxic chemotherapy – N (%) | ||
| Yes | 54 (46) | |
| No | 64 (54) | |
| Previous immunotherapy – N (%) | ||
| Yes | 16 (14) | |
| No | 102 (86) | |
| Overall response rate – N (%) | ||
| Deep response | 1 (1) | |
| Overall response | 59 (50) | |
| Stable disease | 31 (26) | |
| Progressive disease | 24 (20) | |
| NA | 3 (3) | |
| Cause for PFS1 – N (%) | ||
| Disease progression | 76 (64.4) | |
| Death | 11 (9.3) | |
| No event | 31 (26.3) | |
| Site of progression – N (%) | ||
| Systemic | 66 (87) | |
| CNS | 7 (9) | |
| Both | 3 (4) | |
| Status at analysis – N (%) | ||
| Alive | 74 (63) | |
| Dead | 44 (37) | |
Figure 1.
Kaplan-Meier estimates of survivals for osimertinib-treated patients and subgroups (A) Progression-free survival 1 (PFS1) 8.8 months (CI 95% 6.7–10.7); (B) overall survival since diagnosis (OS DX) was 71.9 months (95% CI, 51.4–92.1) (C) OS1 continued vs. discontinued osimertinib 11.2 months vs. 6.1 months, (HR 0.45, 95% CI 0.2 – 0.9), log-rank p=0.02); (D) PFS1 in patients with preserved and altered cell cycle gene alterations, 8.8 months vs 4.4 months (HR 2.8, 95% CI 1.2 – 6.4, log-rank p=0.01).
On exploratory subgroup analysis, CNS metastasis prior to osimertinib was associated with inferior outcome (PFS1 10.4 vs 4.6 months; HR 1.9, 95% CI, 1.2 – 3.0, log-rank p=0.01, Supple. Fig. 3). Seven patients with asymptomatic CNS diseases deferred brain radiation, among those, osimertinib CNS disease control rate was 86% (6/7), with one patient developing leptomeningeal disease. Cell cycle gene alterations at the time of PD were associated with worse outcome when compared to wild-type (PFS1 8.8 vs. 4.4 months; HR 2.8, 95% CI 1.2 – 6.4, log-rank p=0.01, Fig. 1D, Supple. Methods), whereas TP53 mutation and T790M loss were not (Supple. Fig. 4)
Subsequent treatment post-progression on osimertinib
Osimertinib was continued beyond PD in 62% of patients (47/76). Median PFS2 was 12.6 months (95% CI, 8.3 to 15.5, Supple. Fig. 5), with 10 patients (21%) without an event at data cutoff (Table. 2). Among the osimertinib-continued patients, 21 (45%) received palliative radiation for oligometastatic progressing lesions (Table. 2), which was the most common practice in this cohort. There was a trend for improved outcomes in the radiated population compared to non-radiated patients (PFS2 15.5 vs 8.2 months; HR 0.5; 95% CI 0.3 to 1.0, log-rank p=0.05, Supple Fig. 6) with PD occurred in 62% (13/21;) of radiated and 77% (20/26) of non-radiated patients. Of the 21 patients who received radiation therapy, the most commonly radiated sites were lung, mediastinal lymph nodes, bone and brain. Two patients received radiation only to the brain (one with stereotactic radiosurgery, also known as GammaKnife, the other with whole brain radiation therapy (WBRT)). Eight had radiation to bone metastases, 8 to the lung, and 5 to lymph nodes, with three cases having radiation to more than one organ. One case received radiation to a hepatic lesion. Majority of the cases (13/21, 62%) had radiation only to one site (Supple. Table 1).
Table 2.
Treatment after progression on osimertinib
| Continued osimertinib beyond progression – total 76 - N (%) | ||
| Yes | 47 (62) | |
| No | 26 (34) | |
| Not evaluable | 3 (4) | |
| Additional treatment with osimertinib continuation – total 47 - N (%) | ||
| Radiation | 21 (45) | |
| Cryoablation | 1 (2) | |
| Osimertinib 160mg/day | 1 (2) | |
| Bevacizumab | 1 (2) | |
| Erlotinib | 1 (2) | |
| Gefitinib | 1 (2) | |
| Nivolumab | 1 (2) | |
| None | 20 (43) | |
| Cause for PFS2 – total 47 - N (%) | ||
| Disease progression | 33 (70) | |
| Death | 4 (9) | |
| No event | 10 (21) | |
| Additional treatment without osimertinib – total 26 - N (%) | ||
| Yes | 24 (92) | |
| No | 2 (8) | |
| Types of additional treatment without osimertinib – total 24 - N (%) | ||
| PD-1/PD-L1 checkpoint inhibitors | 10 (41.7) | |
| Chemotherapy | 8 (33.3) | |
| Phase 1/2 clinical trials | 3 (12.5) | |
| TKI | 2 (8.3) | |
| Radiation | 1 (4.2) | |
For one patient (2%), bevacizumab was added to the treatment regimen and best response was stable disease (Pt. 38, Supple. Table 2) for 8 months. Two patients developed C797S mutation and were treated with osimertinib and first-generation inhibitors. The combination was well-tolerated. One patient had disease progression after 4 months (gefitinib) and the other died from influenza pneumonia without response assessment (erlotinib). Twenty-six patients discontinued osimertinib at the time of progression and 24 (92%) received additional lines of therapy. The most commonly used agents were PD-1/PD-L1 checkpoint inhibitors (10/24; 41.7%) and chemotherapy (8/24; 33.3%) (Table 2). When compared to osimertinib-continued patients, the discontinued group had shorter survival (OS 11.2 vs 6.1 months; HR 0.45, 95% CI 0.2–0.9, log-rank p=0.02, Fig. 1C). This difference is likely a result of multiple contributing factors, including difference in tumor biology, extent of disease progression, and physician/patient preference.
With median follow-up of 39 months, OS Dx for the entire cohort was 71.9 months (95% CI 51.4 – 92.1, Fig. 1B), a strikingly long survival compared to median OS of 19–28 months with first- or second-generation EGFR TKIs[17–19].
Potential mechanisms of resistance to osimertinib
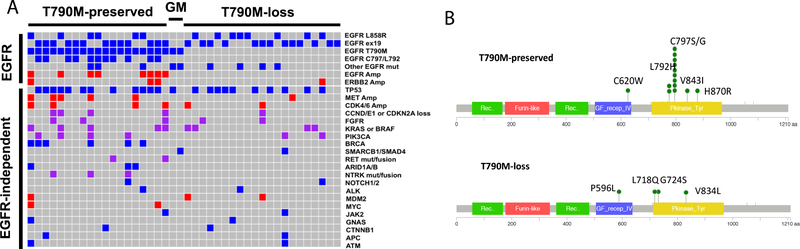
In our cohort, six patients received osimertinib as their first-line treatment, including four cases with inferred germline T790M due to near 50% allelic frequency and two with baseline T790M (Supple. Table 3). Genomic profiling at the time of progression was available for 42 patients (Supple. Table 2). Majority of the genetic tests (32/42) were obtained at the time of first progression (PFS1) or at both first and second progression (3/42) (Supple. Table 4). Of the acquired T790M cases, 47% (19/40) preserved and 53% (21/40) lost T790M at progression (Fig. 2A). Both T790M-preserved and -loss groups had similar frequency with next-generation sequencing based paneled tests (14/19 vs. 15/21, Chi-Square p=0.87). In one patient, two genetic profiling results (4 weeks apart) showed discrepancy between T790M status (Pt. 4, Suppl. Table 2).
Figure 2.
Recurrent genomic alterations associated with osimertinib resistance (A) co-occurring mutations for 42 cases having genetic profiling at progression to osimertinib. Blue box: mutations; red box: amplification; purple box: mutation and/or amplification. GM: germline T790M. (B) EGFR tertiary mutations in T790M-preserved cases (C) EGFR tertiary mutations in T790M-loss cases.
In the T790M-preserved group, previously defined osimertinib-resistant EGFR mutations (11/19; C797S: 8; C797G: 1; L792H: 2 [20] [21]) were the most common resistance mechanisms. These previously defined osimertinib-resistant EGFR mutations were exclusively observed in T790M-preserved cases; this association was statistically significant (11/19 vs. 0/21, p<0.001, Fisher exact test Fig. 2B vs. 2C). Other than C797S/L792 mutations, we observed other acquired EGFR mutations (Fig. 2A-C and Supple. Table 5). Among those, 2 co-occurred with germline T790M (Pt. 20, 21, Supple. Table 2), 6 were acquired after osimertinib. We performed in silico predictions (Combined Annotation Dependent Depletion score (CADD)) to determine their potential oncogenic function. All variants had CADD≥20, which indicated high likelihood of biological function (among the 1% most deleterious mutations for the EGFR gene, Supple. Table 5). The four cases where acquired EGFR mutations emerge when T790M was lost were particularly interesting, as some of the resulted changes might be overcome by first- and/or second-generation EGFR inhibitors. L718 is located within the p-loop and directly interacts with bound inhibitors. Substitution of leucine for glutamine was shown to result in steric hindrance reducing binding ability of third-generation EGFR TKI, such as WZ4002 [20] and lead to resistance. However, in vitro work has also demonstrated that L718Q may still respond to first- and second-generation of EGFR inhibitors, especially when T790M is lost, as is the case for the patient No. 29[20]. G724S also resides in the p-loop, and previous studies have shown that this mutation results in resistance to osimertinib in the presence of T790M[22]. Other mutations shown to prevent inhibitor stabilization include L844V where the shortening of amino acid prevents hydrophobic interactions with WZ4002 [20]. Similarly, we predict that V834L which results in a larger hydrophobic amino acid at V834 may result in steric hindrance of the drug near the anisole (methoxybenzene) group.
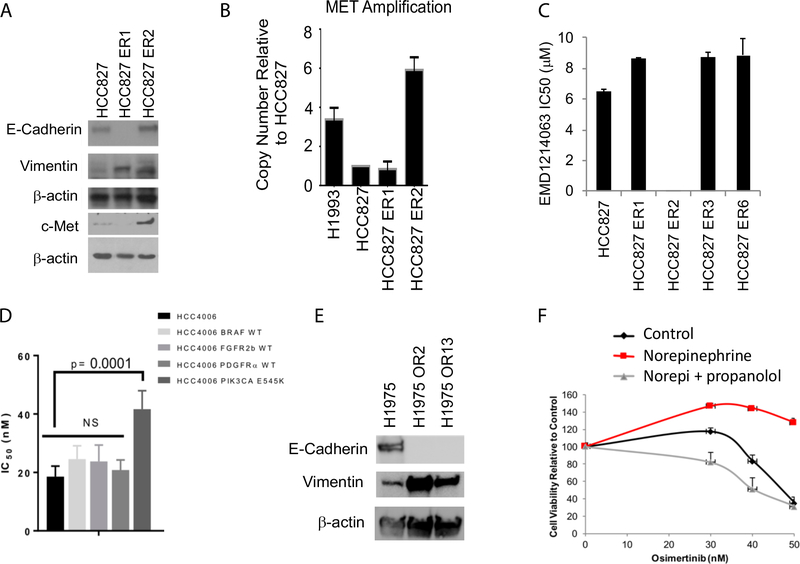
MET amplification has been reported as a mechanism of resistance to gefitinib[8] and osimertinib[23]. In our cohort, it was identified in T790M-preserved (5/19) and T790M-loss (1/21) cases, and the second most common resistance mechanism after EGFR tertiary mutations (5/42, 14%). We cultured HCC827 cells (EGFR Del745_760) in erlotinib to develop EGFR TKI resistance and identified one clone (ER2) having increased c-Met protein expression and MET copy-number gain (Fig. 3A-3B). This clone was resistant to osimertinib (Supple. Fig. 7A) without resistant mutations in the EGFR exon 20 (Supple. Fig 7C), but exquisitely sensitive to c-met inhibitors (tepotinib, EMD Serono, Fig. 3C) and crizotinib (Supple. Table. 5), suggesting that c-met inhibition can overcome MET amplification mediated osimertinib resistance. Among MET-amplified patients, CDK6 (4/5) and BRAF amplification (3/5) were common events (Fig. 2A and Pt. 1, 4, 5, 9, 15, Suppl. Table 2). Because of the relative proximity of the three genes in chromosome 7q (CDK6 at Chr7q21.2, MET at Chr7q31.2 BRAF at Chr7q34), it is possible that these amplifications arose from a single genomic event.
Figure 3.
EGFR-independent potential mechanisms of resistance (A) HCC827 parental cells and cells with acquired resistance to EGFR inhibitors were tested for protein levels of E-cadherin, vimentin and c-Met (B) HCC827 parental cells and cells with acquired resistance to EGFR inhibitors were evaluated for MET copy number changes. H1993 cells were used as positive control (C) MET small molecule inhibitor (EMD 1214063) IC50 in HCC827 parental cells and cells with acquired resistance to EGFR inhibitors (D) Osimertinib IC50 to HCC4006 cells with overexpression of BRAF, FGFR1b, PDGFRa and PIK3CA E545K (E) H1975 parental cells and cells with acquired resistance to osimertinib (OR2 and OR13) were tested for protein levels of E-cadherin and vimentin (F) Norepinephrine (1uM) increases HCC827 cells proliferation in the presence of osimertinib, and propranolol (1uM) overcomes such resistance.
The 21 cases with T790M-loss had more diverse genetic alterations. One tumor with pre-existing RB1/TP53/PIK3CA alterations underwent neuroendocrine trans-differentiation (Pt. 27, Supple Table 2). Two cases acquired PIK3CA E418K/E542K and E542K/E545K mutations (Pt. 35, 40, Supple Table 2), two cases acquired KRAS alterations, one with CDK4/KRAS/MDM2 amplification and the other with KRAS Q61R mutation (Pt. 22, 39, Supple Table 2), one had FGFR3/FGFR19 amplification (Pt. 32, Supple Table 2), one had MET amplification (Pt. 36; FISH ratio: 7.4, Supple Table 2) and one had ERBB2 amplification (Pt. 40, Supple Table 2). The remaining cases had unknown resistance mechanisms (Fig. 2A, Supple Table 2).
We overexpressed PIK3CA E545K, BRAF and FGFR2b in HCC4006 cells (EGFR Del747_749, A750P), and evaluated response to osimertinib. We found that PIK3CA E545K overexpression confers moderate resistance to osimertinib (IC50 increased from 25nM to 67nM, p<0.01), whereas BRAF, FGFR2b, PDGFR overexpression did not significantly altered osimertinib sensitivity (Fig. 3D). This data suggests that acquired PIK3CA hotspot mutations may drive contribute to osimertinib resistance, and that BRAF, FGFR, or PDGFR amplifications may be “passenger” events instead of “drivers”, or may not be able to promote resistance alone without additional concurrent alterations.
There were 14 cases without potential resistance genetic drivers, 12 in T790M-preserved group, which highlights our limited understanding of T790M-negative resistance mechanisms. One case with pre-existing TP53 mutation and RB1 loss displayed neuroendocrine transformation (Pt. 27; Supple Table 2). Epithelial‐mesenchymal transition (EMT) has also been indicated as one mechanism of resistance to EGFR-TKIs[10]. We observed an AXL mutation in one case with acquired T790M/C797S (Pt. 12 Supple Table 2)[24], suggesting association with EMT. In cell lines, we observed E-cadherin loss and vimentin gain in HCC827 EGFR-TKI resistant cells (HCC827-ER1, Fig 3A) and in H1975 osimertinib-resistant cells (H1975-OR3 and OR13, Fig. 3E, Supple Fig. 7B), indicating that EMT could be a potential resistance mechanism to osimertinib. Stress hormone induced beta adrenergic pathway activation has recently been identified as a driver of resistance to first-generation TKIs and can be overcome by beta-adrenergic blockade[25] (Supple. Fig. 7D) therefore, we hypothesize that stress hormones may induce resistance in osimertinib-treated lung cancers. In HCC827 cells, norepinephrine (10uM) significantly increased proliferation in the presence of osimertinib (from 10–50nM), suggesting resistance (red line, Fig. 3F). Propranolol (1uM), a non-selective beta-adrenergic blocker, restored osimertinib sensitivity in HCC827 cells (gray line, Fig. 3F), supporting that stress hormones induce proliferation in the presence of osimertinib. Furthermore, such resistance can be overcome through beta-adrenergic blockade in EGFR-mutant T790M-negative lung cancer cells. Taken together, these preclinical data provide evidence that EGFR-independent mechanisms of resistance observed for first- and second-generation EGFR TKIs (MET amplification, PIK3CA mutation, EMT, beta adrenergic activation) may promote osimertinib resistance as well.
Platform-matched samples in pre- and post-osimertinib treatment are especially valuable to understand acquired genetic events that lead to clinical resistance. Among the 42 cases, we identified 18 cases having platform-matched profiling before and at the time of progression to osimertinib treatment (Supple Table 7 and Supple Fig. 8), 11 by Guardant360, 1 by FoundationOne and 6 by MD Anderson Molecular Diagnostic Laboratory test (MDL, Supple. Methods). Although small, this platform-matched paired sample cohort is most informative for understanding acquired resistance mechanisms. Of the 18 cases, 9 (50%) preserved T790M with 6 acquired EGFR C797S/L792H mutations and one with concurrent EGFR amplification (Pt. 10). One patient acquired NCOA4-RET fusion as well as EGFR and CDK4 amplification (Pt.19), which might represent different resistant clones. Only two cases with preserved T790M did not acquire new resistant genetic alterations. Nine cases lost the T790M resistant clones at the time of progression. Four did not acquire any new genetic alterations. One case lost T790M but acquired EGFR V834L (Pt. 34). One case acquired co-amplification of FGF3/FGF19/EMSY at chromosome 11q13.3–13.5 locus (Pt. 32). One acquired PIK3CA E418K/E542K mutations (Pt. 35); one acquired TSC1 E1044fs, and one with ERBB2 R340Q. The 18 platform-matched cases represent the common resistance mechanisms seen in our 42 cases cohort, confirming that the T790M-preserved cases frequently use EGFR reactivation or bypass pathway for resistance; whereas T790M-loss cases engage in EGFR-independent mechanisms for resistance.
We also investigated cases in which there was a discrepancy in the NGS results from blood ctDNA and tumor biopsies, to determine whether these may represent somatic mutations from hematopoietic stem cells instead of NSCLC tumor cells, otherwise known as clonal hematopoiesis[26]. There were four cases having both Guardant360 tests for more than one time point, and a tissue test (Pt. 28, 35, 39, and 42). In three of these cases, mutations potential resulting from clonal hematopoiesis were observed: TP53 C135W in Pt. 28, JAK2 V617F in Pt. 35, and MYC N353S in Pt. 42 (Supple Table 7). Both JAK2 V617F and TP53 mutations are known to be common mutations in clonal hematopoiesis [27]. This data is consistent with the recent report from Circulating Cell-Free Genome Atlas (CCGA) study showing that up to 54% of the somatic mutations detected in the blood samples from lung cancer patients were from hematopoietic cells’ somatic mutations, but not from tumors[28]. This real-world experience highlighted that thoracic oncologists need to be aware of the phenomenon of clonal hematopoiesis when interpreting blood biopsy reports for lung cancer patients.
Discussion
In this study, we reviewed a cohort of patients with advanced NSCLC treated with osimertinib at MDACC and MCC. Patients who continued osimertinib beyond progression had improved clinical outcomes compared with those who discontinued it. Due to the nature of retrospective analysis, this result may be impacted by selection bias, where physicians had to choose discontinuation of osimertinib in cases with obvious progression and clinical deterioration. Nevertheless, the practice of continuation of TKI beyond progression is supported by previous reports of patients with indolent and small volume progression benefiting from continuation of TKIs[15, 29]. The biological mechanism underlying this observation is hypothesized to be that in the case of localized disease progression, the drug may continue to suppress the majority of tumor cells; furthermore, compensatory pathways may be upregulated during treatment and drug withdrawal may therefore result in a “tumor flare”. In our cohort, about half of cases that continued osimertinib also received local consolidation radiation to the progressing sites. This practice is supported by randomized phase II trials showing that aggressive local consolidation therapy (LCT) increases time to progression compared to maintenance or observation in patients with oligometastatic NSCLC[30]. The rationale for this approach is that local consolidation therapies decrease the resistant sub-clone tumor burden, while continuation of osimertinib suppresses sensitive cells. We are currently evaluating the role of LCT in EGFR-mutant lung cancers treated with osimertinib in a randomized phase II study (NCT03410043).
We then analyzed genomic profiling data at the time of osimertinib progression. We found that cell cycle gene alterations (CDK4/6 or CCND/E1 amplifications, or CDKN2A loss) were associated with worse outcome[31], highlighting the need for novel therapeutic options for these patients. In our cohort, we identified both known and novel resistance mechanisms to osimertinib that, based on prior studies, may be divisible into three groups: 1) reactivation of EGFR pathway through tertiary mutations; 2) activation of known bypass signaling pathways, such as MET or ERBB2 amplifications, or PIK3CA mutation; 3) Alterations that likely promote “rewiring” and diminished dependence on EGFR or bypass signaling (e.g. EMT, SCLC, etc). In our cohort, tertiary EGFR mutations were the most common resistance mechanism (11/42, 26%). As for bypass signaling, MET amplification was observed in both T790M-preserved and T790M-loss, and the second most common resistance mechanism (6/42, 14%). Other than MET, we also observed cases with amplification or mutations in other possible driver oncogenes, including PIK3CA, BRAF, FGFR, and PDGFR. In our preclinical models, PIK3CA hotspot mutation, but not BRAF, FGFR, and PDGFR overexpression alone, was found to contribute to osimertinib resistance. Whether PIK3CA mutations determine treatment response to EGFR TKIs remains controversial [32] [16] [33] [34] [35]. The role of PI3K pathway activation in osimertinib-treated resistance EGFR-mutant lung cancer warrants further evaluation. Other than oncogene driver-mediated resistance, we showed that neuroendocrine and mesenchymal cellular rewiring and cytokine changes in the tumor microenvironment can also impact tumor cell’s sensitivity to therapy. We found that epinephrine reduces the sensitivity of EGFR mutant cells to osimertinib in vitro. Beta-adrenergic blockade with propranolol reversed this resistance phenotype. These data highlighted the importance of understanding T790M-negative resistance mechanisms, with a focus on cytokine mediated changes.
Our real-world study has several limitations due to its retrospective nature. In the clinical outcome analysis, the disease progression was based on physician assessment and not specific formal criteria. Therefore, the benefit of continuation of osimertinib beyond progression must be validated in future prospective studies, several of which are ongoing. In the genetic profiling analysis, in some cases different platforms were used for the pre-treatment and post-progression profiling; however, analysis of the platform-matched cases shows similar results as the overall analysis. In addition, the majority of the patients in our cohort received osimertinib as second- or later-lines of therapy (Table 1), with only six patients with baseline EGFR T790M or germline T790M receiving osimertinib as the first-line treatment (Supple. Table 3). As osimertinib is increasingly being used as first-line therapy, additional studies will be needed to determine whether similar alterations are associated with osimertinib resistance in the first-line, TKI-naïve setting.
Despite these limitations, our cohort is one of the largest analyzing both real-world clinical outcomes of osimertinib treated EGFR-mutant NSCLC, as well as genetic profiles associated with resistance. We showed that many patients with EGFR-mutant NSCLC might have prolonged benefit from osimertinib beyond their initial progression event, especially when radiation was used at progression, suggesting a potential role for local consolidative therapy in this setting. Furthermore, we showed that T790M loss is common in osimertinib-resistant cases, and that these cases demonstrated a different pattern of resistance compared with the T790M-preserved cases. Specifically, in most of the T790M-preserved cases, resistance is associated with continued EGFR activation through known resistance tertiary mutations (e.g. C797S) or activation of bypass signaling pathways, whereas resistance in T790M-loss cases occurs through diverse, and predominantly EGFR-independent mechanisms. Given this pattern, and the effectiveness of osimertinib at suppressing T790M positive subclones, it is conceivable that resistance to osimertinib in the first line setting will also be associated with a greater frequency of EGFR-independent mechanisms. These issues will require further study; nevertheless, given the increasing role for osimertinib in the treatment of EGFR-mutant advanced NSCLC, these data represent an important step for understanding resistance to this agent and developing subsequent treatment strategies.
Supplementary Material
Translational relevance.
The mechanisms underlying resistance to osimertinib, a third-generation EGFR inhibitor, has been investigated by several other groups. Here, we characterized molecular landscape of osimertinib resistance in NSCLC and clinical practices that might prolong osimertinib benefit. We found that in contrast to first-generation EGFR inhibitors, resistance mechanisms to osimertinib are predominantly EGFR-independent, including MET amplification, PI3K pathway activation and epithelial-mesenchymal transition. EGFR-dependent resistance occurs more frequently in T790M preserved cases through C797/L792 mutations, whereas EGFR mutations can also occur in EGFR-T790M clonal depletion settings. Therefore, treatment strategies for osimertinib-resistant patients will require tailoring for these diverse subgroups. At the time of progression, continuation of osimertinib alone or in combination with local consolidative therapy, was associated with clinical benefit in selected patients. Our analysis of molecular profiles and clinical outcomes of osimertinib-treated lung cancers underscored the importance of EGFR-independent resistance mechanisms to osimertinib and provided potential treatment strategies for prolonging clinical benefit in selected patients.
Acknowledgements:
This work is supported by the generous philanthropic contributions to the University of Texas MD Anderson Lung Moon Shot Program and the MD Anderson Cancer Center Support Grant P30 CA01667. NIH R01 CA190628. UT Lung SPORE P50 CA70907, the Rexanna Foundation for Fighting Lung Cancer, Bruton Endowed Chair in Tumor Biology, Standing Fund for EGFR inhibitor resistance, the Hallman fund, and the Fox Lung EGFR Inhibitor Fund. Dr. Gray receives research funding from Boehringer Ingelheim, AstraZeneca and Genentech.
Footnotes
Conflict of interest: Dr. Le received a consultant fee from Eli Lilly AstraZeneca and Boehringer Ingelheim. Dr. Papadimitrakopoulou receives a consultant fee from AstraZeneca. Dr. Gray is an advisor for AstraZeneca, Boehringer Ingelheim and Genentech. Dr. Heymach is an advisor for AstraZeneca, Boehringer Ingelheim, EMD Serono, Genentech, Eli Lily, Merck, Roche, Spectrum, Guardant, Janssen, Novartis, Foundation Medicine. Other authors do not report relevant conflict of interest.
Reference:
- 1.Cross DA, et al. , AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov, 2014. 4(9): p. 1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janne PA, et al. , AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med, 2015. 372(18): p. 1689–99. [DOI] [PubMed] [Google Scholar]
- 3.Mok TS, et al. , Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med, 2017. 376(7): p. 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soria JC, et al. , Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med, 2017. [DOI] [PubMed] [Google Scholar]
- 5.Thress KS, et al. , Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med, 2015. 21(6): p. 560–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, et al. , Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res, 2018. [DOI] [PubMed] [Google Scholar]
- 7.Oxnard GR, et al. , Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, et al. , MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science, 2007. 316(5827): p. 1039–43. [DOI] [PubMed] [Google Scholar]
- 9.Takezawa K, et al. , HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov, 2012. 2(10): p. 922–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sequist LV, et al. , Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med, 2011. 3(75): p. 75–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byers LA, et al. , An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res, 2013. 19(1): p. 279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oxnard GR, Osimertinib Resistance Mediated by Loss of EGFR T790M Is Associated with Early Resistance and Competing Resistance Mechanisms World Conference on Lung Cancer, 2017.
- 13.Sonam Puri JKH, Knepper TC, Smith M, Boyle TA, Jhanelle E Gray, Genomic Profiling of EGFR T790M Mutated Non-Small Cell Lung Cancer to Evaluate the Mechanisms of Resistance to Osimertinib. World Conference on Lung Cancer, 2017. [Google Scholar]
- 14.Lin CC, et al. , Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med, 2018. 6(2): p. 107–116. [DOI] [PubMed] [Google Scholar]
- 15.Park K, et al. , First-Line Erlotinib Therapy Until and Beyond Response Evaluation Criteria in Solid Tumors Progression in Asian Patients With Epidermal Growth Factor Receptor Mutation-Positive Non-Small-Cell Lung Cancer: The ASPIRATION Study. JAMA Oncol, 2016. 2(3): p. 305–12. [DOI] [PubMed] [Google Scholar]
- 16.VanderLaan PA, et al. , Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer, 2017. 106: p. 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou C, et al. , Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol, 2015. 26(9): p. 1877–83. [DOI] [PubMed] [Google Scholar]
- 18.Sequist LV, et al. , Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol, 2013. 31(27): p. 3327–34. [DOI] [PubMed] [Google Scholar]
- 19.Rosell R, et al. , Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol, 2012. 13(3): p. 239–46. [DOI] [PubMed] [Google Scholar]
- 20.Ercan D, et al. , EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin Cancer Res, 2015. 21(17): p. 3913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K, et al. , Novel Mutations on EGFR Leu792 Potentially Correlate to Acquired Resistance to Osimertinib in Advanced NSCLC. J Thorac Oncol, 2017. 12(6): p. e65–e68. [DOI] [PubMed] [Google Scholar]
- 22.Oztan A, et al. , Emergence of EGFR G724S mutation in EGFR-mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer, 2017. 111: p. 84–87. [DOI] [PubMed] [Google Scholar]
- 23.Ou SI, Agarwal N, and Ali SM, High MET amplification level as a resistance mechanism to osimertinib (AZD9291) in a patient that symptomatically responded to crizotinib treatment post-osimertinib progression. Lung Cancer, 2016. 98: p. 59–61. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, et al. , Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet, 2012. 44(8): p. 852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson MB, et al. , Stress hormones promote EGFR inhibitor resistance in NSCLC: Implications for combinations with beta-blockers. Sci Transl Med, 2017. 9(415). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaiswal S, et al. , Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med, 2014. 371(26): p. 2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Y, et al. , False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin Cancer Res, 2018. [DOI] [PubMed] [Google Scholar]
- 28.Charles Swanton OV, Alex Aravanis, Earl Hubbell, Tara Maddala, Beausang John F., Filippova Darya, Gross Samuel, Jamshidi Arash, Shen Ling, Valouev Anton, Zhang Nan, Bolton Kelly L, Yeatman Timothy Joseph, Seiden Michael, Oxnard Geoffrey R., Liu Minetta C., Williams Richard Thomas, Hartman Anne-Renee, Baselga Jose. Prevalence of clonal hematopoiesis of indeterminate potential (CHIP) measured by an ultra-sensitive sequencing assay: Exploratory analysis of the Circulating Cancer Genome Atlas (CCGA) study. in ASCO Annual Meeting 2018. [Google Scholar]
- 29.Yap TA, Macklin-Doherty A, and Popat S, Continuing EGFR inhibition beyond progression in advanced non-small cell lung cancer. Eur J Cancer, 2017. 70: p. 12–21. [DOI] [PubMed] [Google Scholar]
- 30.Gomez DR, et al. , Local consolidative therapy versus maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer without progression after first-line systemic therapy: a multicentre, randomised, controlled, phase 2 study. Lancet Oncol, 2016. 17(12): p. 1672–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blakely CM, et al. , Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet, 2017. 49(12): p. 1693–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludovini V, et al. , Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol, 2011. 6(4): p. 707–15. [DOI] [PubMed] [Google Scholar]
- 33.Yu HA, et al. , Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res, 2018. 24(13): p. 3108–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eng J, et al. , Impact of Concurrent PIK3CA Mutations on Response to EGFR Tyrosine Kinase Inhibition in EGFR-Mutant Lung Cancers and on Prognosis in Oncogene-Driven Lung Adenocarcinomas. J Thorac Oncol, 2015. 10(12): p. 1713–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu SG, et al. , The Role of PIK3CA Mutations among Lung Adenocarcinoma Patients with Primary and Acquired Resistance to EGFR Tyrosine Kinase Inhibition. Sci Rep, 2016. 6: p. 35249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.