Abstract
The circulation of the South-east Asian/American (AS/AM) dengue 2 virus (DENV-2) genotype in the Americas has been associated with a high rate of severe disease. From 1993, the year DENV was reintroduced in Panama, until 2011 there were 29 dengue-associated deaths, 17 of which occurred in 2011, the most severe outbreak with a case fatality rate (CFR) of 44% (17 deaths out of 38 severe dengue cases). During this outbreak DENV-2 was reintroduced into the country, whereas over the prior five years DENV-1 and −3 were predominant. Herein, we describe the 2011 Panama outbreak and genetically characterize the Panamanian DENV-2 strains, which were associated with severe dengue disease in Panama. Our results suggest that the DENV-2 isolates from this outbreak belonged to the AS/AM genotype sub-clade 2BI and were genetically close to viruses described in the outbreaks in Nicaragua, Honduras, Guatemala and Mexico from 2006–2011. Sub-clade 2BI has previously been associated with severe disease in Nicaragua during outbreaks from 2005–2007.
Keywords: DENV-2, outbreak, phylogenetics, epidemiology, case fatality rate
Graphical abstract
During 2011, DENV-2 South-east Asian/American genotype, sub-clade 2BI was introduced in Panama, after 5 years without detection of DENV-2. This sub-clade, has been previously associated with severe disease in others Central American countries.
1. Introduction
Dengue viruses (DENV) are the most common mosquito-borne arbovirus in the world with approximately 390 million cases annually1. It is an enormous world wide economic burden1 costing the Americas 1.9–2.2 billion dollars annually2. DENV belong to the genus Flavivirus, family Flaviviridae3 and there are currently four recognized serotypes (DENV-1–4). It is transmitted by the mosquitoes Aedes aegypti and Ae. albopictus as primary and secondary vectors respectively4. Clinical disease caused by DENV is classified as dengue, dengue with warning signs and severe dengue base on the patients clinical presentation4. The underlying mechanisms for progressing to severe disease have not been fully elucidated. However these are likely influenced by both specific host and viral factors that act in concert5. Severe disease has been associated with: secondary infection with a different serotype6–8, host genetic susceptibility9, patient co-morbidities10,11, specific DENV serotypes and genotypes8,12–14, viral load15 and the immune response induced by the DENV-NS1 protein16–19.
Phylogenetic and molecular analyses have revealed extensive genetic diversity among DENVs, leading to the recognition of different genotypes within each serotype20,21. DENV-2 has 5 genotypes (Asian I, Asian II, Cosmopolitan, American and South-east Asian/American). The initial introduction of DENV-2 in Americas was caused by American genotype, then in 1975, it was replaced by the Asian/American (AS/AM) genotype in the Caribbean, with dissemination to Central and South America in the 1980–90s22. Its reintroduction in Nicaragua, Peru and Brazil since 2000’s has been associated with more severe outbreaks23–25.
Recent analysis of DENV-2 strains from Nicaragua, Honduras and Guatemala showed that the AS/AM circulating in Central America is divided into 2 clades; clade 2 is further divided in 2 sub-clades: 2A and 2B26. Sub-clade 2B has been implicated in severe cases in patients with primary exposure to DENV-1 in Nicaragua27.
Dengue was responsible for outbreaks in Panama in 190428, 191229 and 1941–194230. In 1958 Panama was declared Ae. aegypti-free after an intense vector control campaign. Panama was re-infested by Aedes aegypti in 198531, and in 1993, a limited outbreak of dengue occurred in Panama City, confirming the return of autochthonous circulation of DENV-231,32. Since then, the four DENV serotypes have circulated in Panama and continue to be a substantial public health burden33.
In Panama, during the first half of 2011, DENV-1 and DENV-3 comprised the majority of circulating DENV. However, during the second half of the year, DENV-2 was detected and became the predominant circulating serotype and a corresponding increase in dengue deaths was observed. DENV-2 previously circulated in Panama in 1993–1994 and 1999 to 2004 (www.paho.org) and each reintroduction was associated with outbreaks but none were apparently associated with an increase in mortality. A retrospective analysis of data available through the National Dengue Epidemiologic Surveillance System was performed to characterize the 2011 DENV-2 outbreak in Panama. Herein, we report the results of the outbreak investigation as well as the genetic analysis of the viral strains detected during the outbreak.
2. Materials and Methods
2.1. Ethical considerations
The permit to study arboviral outbreaks was approved by The National Committee of Bioethics in Panama, IRB number: 0277/CBI/ICGES/15. Patient information was de-identified in order to protect the confidentially of individuals involved.
2.2. Case definition
The Dengue case definition used during 2011 Dengue outbreak in Panama, was based on the 1997-World Health Organization (WHO) classification system which includes: Dengue Fever (DF), Dengue Hemorrhagic Fever (DHF) and Dengue Shock Syndrome (DSS)4. The epidemiological link criterion was defined as a case in close contact with a confirmed case within 30 days of symptoms onset.
2.3. Dengue surveillance program and Epidemiological Data Collection
Using the WHO’s Dengue case definition4,34 the Ministry of Health of Panama (MINSA) and Gorgas Memorial Institute of Health Studies (ICGES) established the National Dengue Surveillance program in 1988 across the country. Around 10% of dengue-suspected cases from the country are sent to the ICGES laboratory for confirmation and serotype characterization, whereas most of the cases are laboratory confirmed by enzyme-linked immunosorbent assay (ELISA) of Dengue IgM/secondary IgG and NS1 (PANBIO) detection in local health care institutions. All Dengue cases seen in both public and private health care institutions are reported to National Department of Epidemiology (NDE), as regulated by the executive order N°268 from 2001. The dengue surveillance database was obtained from the National Department of Epidemiology of Ministry of Health and demographic information was available from the National Institute of Statistics and Census.
2.4. Epidemiological and Statistical methods
Epidemiological and demographic variables were analyzed using Epi-Info 7, Microsoft Excel and STATA SE software, version13.1. Dengue incidence, mortality and case fatality rate (CFR) were estimated using data from dengue surveillance database and the estimated population at July 1st of 2011 using demographic information of the General Comptroller of the Republic of Panama. To calculate the endemic channel percentiles, the epidemiological data from previous years (2004–2010) were used; this included 2 epidemic years (2005 and 2009), which increases the superior limit of the endemic channel.
2.5. Correlation of dengue cases and vector index
A systematic entomological monitoring system is in place throughout the year. Entomological data are collected from cluster surveys to establish the Ae. aegypti and Ae. albopictus infestation rates, using the survey of 100% of houses in a block. The number of containers with larvae per household was quantified. Positive houses were reported if at least one recipient has one larva. The larvae Breteau (BI) was estimated with the number of positive containers per 100 houses investigated. While the house indices (HI) index was estimated by the following equation: Using the HI index, MINSA classified the Dengue transmission risk as >4: high, 4 to 2: moderate and <2: low risk35 . Entomologic surveillance database was obtained from the Vector National Department of the Ministry of Health. This was used to analyze the relationship between vector infestation rates and dengue cases during 2011 using Spearman’s Rho correlation analysis with STATA SE software v13.1.
2.6. Laboratory Assays
2.6.1. Molecular Testing
Acute sera samples were sent to ICGES for serotype and genotype surveillance through the Dengue virus surveillance program. RNA was extracted using QIAamp RNA Viral Extraction Kit (Qiagen, Germany) and DENV viral RNA and serotype-specificity was detected using qRT-PCR specific protocol36. Positive samples were used for viral isolation.
2.6.2. Serologic Testing
Serum samples from DHF suspected cases with ≥ 5 days from onset of symptoms were tested using NS1 and DENV-IgG Indirect ELISA (PANBIO). A total of 10% of qRT-PCR dengue negative samples were also tested for antibodies against flaviviruses and alphaviruses known to circulate in Panama with the use of hemagglutination-inhibition assays37.
2.6.3. Viral Isolation
Viral isolation was attempted by inoculation of Vero cells (African green monkey-ATCC CCL-81. USA), with acute sera or 10% tissue homogenates (spleen and liver) from acute DENV or fatal cases respectively, and daily observation for cytopathic effects (CPE) was done. When CPE were evident, viral RNA was extracted using Quiagen and tested again using DENV-specific qRT-PCR36.
2.6.4. E gene and complete genome sequencing
Twenty-seven isolates of DENV-2 were obtained from acute sera. Six DENV-2 isolates that circulated in different regions of the country were randomly selected for analysis. Viral RNA was extracted and RT-PCR was performed to obtain 5 overlapping fragments for the complete viral envelope (E protein) gene using the Titan Kit (Roche) with specific primers described previously38. Amplicons were purified with the QIAquick PCR purification kit (Qiagen) and sequences were obtained using an Applied Biosystems 410 Genetic Analyzer (Foster City, CA), following the manufacturer’s protocols. Only one complete genome of the strain 250330 was obtained using overlapping primers derived from the reference strain DENV2–16881 (primers available upon request).
2.7. Phylogenetic analysis
The six Panamanian 2011 DENV-2 E sequences (1485 nt) were aligned with 1,965 sequences of DENV-239 from the Genbank library and from Virus Pathogen Database and Analysis Resource (ViPR) using SEAVIEW 4.0 software (Gouy M et al, Mol Biol Evol 2010). This alignment was used to do a Neighbor Joining (NJ) Tree using a heuristic search with Nearest Neighbor Interchange (NNI) operation using the GTR+G+I model showing that Panama 2011 DENV-2 belong to South East Asian/American (AS/AM) genotype (data not shown). The closer 63 sequences to the five Panamanian 2011 E sequences (Genbank numbers KY977455, KY977456, KY977457, KY977458, KY977459), selected from the DENV-2 AS/AM genotype, were used to construct a maximum likelihood (ML) tree under GTR+G+I model with midpoint root for clarification. The statistical significance of the tree topology was evaluated by bootstrap resampling of the sequences 1000 times. Phylogenetic and molecular evolutionary analyses were conducted using PAUP* version 4.0,10b (Swofford DL. PAUP*: Phylogenetic Analysis using Parsimony (*and Other Methods) Sinauer Assoc Sunderland, Massachusetts. 2002;1–142) and MEGA7 * Kamura, Stecher and Tamura 2015).
Amino acid sequences of complete E gene of the 6 Panamanian 2011 DENV-2 and the complete ORF of KY977454 isolate were analyzed and compared to the closest isolates from 2B and 2A DENV-2 subclades using MEGA7.
3. Results
3.1. Outbreak Description
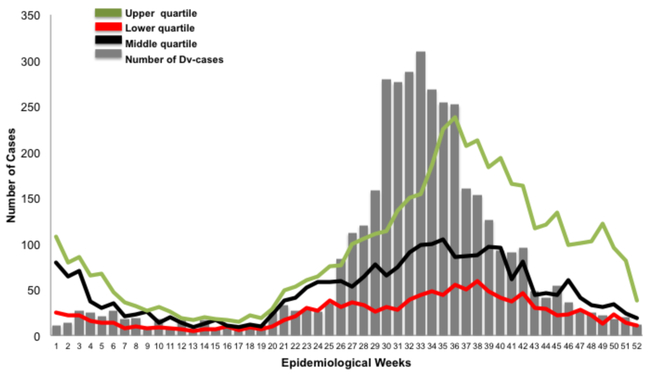
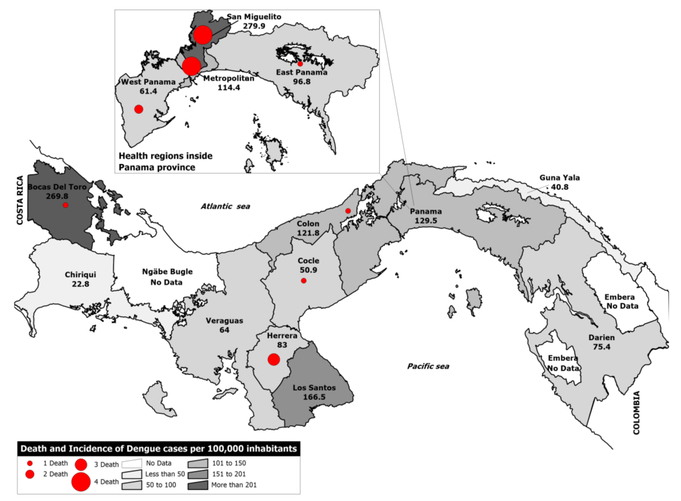
A total of 3,850 dengue cases were reported in 2011. An increase of dengue cases that surpassed the upper quartile of the endemic channel was observed during epidemiological week 26 (June) through week 36 (September), when 62.2% of the cases occurred (Figure 1). The total incidence rate of dengue in the country was 103.4 per 100,000 inhabitants. The majority of cases occurred in Bocas del Toro, San Miguelito and Panama City areas with an incidence rate of 269.8 per 100,000, 279.9 per 100,000 and 114.4 per 100,000 inhabitants respectively (Table 1, Figure 2). Persons 0–14 years old had an incidence of dengue of 69.9 per 100,000 inhabitants, 15–29 was 114.6 per 100,000 inhabitants, 30–44 was 121.8 per 100,000 inhabitants, 45–59 was 102.4 per 100,000 and over 60 was 109.2 per 100,000 (Table 2). An univariate analysis comparing dengue cases by sex and age range revealed no statistical difference.
Figure 1. Endemic channel of Dengue cases during 2011 outbreak.
In grey bars, number of dengue cases per epidemiological week: lines show the endemic channel with green representing the upper quartile, red the lower quartile, black the middle quartile. Since epidemiological week 26, an increase of cases is shown; which is maintained for 11 weeks, with a sustained increase during epidemiological weeks 30 to 33. The endemic channel was constructed using 7 years of data, which included two Dengue epidemic years 2005 and 2009.
Table 1.
Dengue confirmed cases by laboratory and epidemiological link, and total incidence rate by province during 2011.
| Province | Percentage of Dengue cases confirmed by epidemiological link %(n) | Percentage of Dengue confirmed cases by Laboratory tests %(n) | Total of Dengue cases | Incidence rate* (x105 inhabitants, July 1st, 2011) |
|---|---|---|---|---|
| Total | 18.7 (721) | 81.3 (3129) | 3850 | 103.4 |
| Bocas del Toro | 45.3 (170) | 54.7 (205) | 375 | 269.8 |
| Code | 27.8 (35) | 72.2 (91) | 126 | 50.9 |
| Colon | 0.6 (2) | 99.4 (314) | 316 | 121.8 |
| Chiriqui | 1 (1) | 99 (99) | 100 | 22.8 |
| Darien | 35.9 (14) | 64.1 (25) | 39 | 75.4 |
| Herrera | 0 (0) | 100 (97) | 97 | 83.0 |
| Los Santos | 12.1 (19) | 87.9 (138) | 157 | 166.5 |
| Panama city | 24.5 (286) | 75.5 (879) | 1165 | 114.4 |
| West Panama | 45.3 (139) | 54.7 (168) | 307 | 61.4 |
| East Panama | 2 (1) | 98% (48) | 49 | 96.8 |
| San Miguelito | 5.3 (50) | 94.7 (899) | 949 | 279.9 |
| Verag lias | 0 (0) | 100% (154) | 154 | 64.0 |
| Guna Yala | 25 (4) | 75 (12) | 16 | 40.8 |
| Embera | 0 (0) | 0 (0) | 0 | 0 |
| Ngäbe-Bugle | 0 (0) | 0% (0) | 0 | 0 |
Figure 2. Map of dengue cases in Panama during 2011.
Map shows, using a grey color scale, the incidence of dengue cases with red dots corresponding to the number of fatal cases per province or indigenous reservation (Guna Yala, Embera, Ngäbe Bugle). No cases were reported from other indigenous reservations besides the Guna Yala.
Table 2.
Incidence, Mortality and Case Fatality Rate of Dengue during 2011.
| Age group (yr) | *Incidence of Dengue **(cases/population) | Mortality rate of Dengue **(deaths / total population) | (%) CFR of DHF (deaths/DHF cases) |
|---|---|---|---|
| 0–14 | 69.9 (746/1,071,235) | 0.19 (2/1,071,235) | 66.7 (2/3) |
| 15–29 | 114.6 (1072/935,364) | 0.43 (4/935,364) | 30.8 (4/13) |
| 30–44 | 121.8 (984/807,909) | 0.37 (3/807,909) | 33.3 (3/9) |
| 45–59 | 102.4 (552/539,286) | 0.93 (5/539,286) | 62.5 (5/8) |
| >60 | 109.2 (404/370,027) | 0.81 (3/370,027) | 60 (3/5) |
| Total | 103.4 (3850/3,723,821) | 0.46 (17/3,723,821) | 44.7 (17/38) |
Incidence calculated per 100 000 inhabitants
Total population estimated to July 1st of 2011.
From the 3,850 dengue cases there were 38 cases of DHF (0.99%). Of these, there were 17 fatal cases across the country (Supplementary Table 1 and Figure 2). Two fatal cases occurred in the 0–14 years of range, 4 in those 15–29, 3 in those 30–44, 5 in those 45–59, and 3 in those over 60 (Table 2). The mortality of dengue cases during this year was 0.46% (17 death/3,723,821 total inhabitants) per 100,000 inhabitants, whereas CFR of deaths to DHF were 44.7% (17 death/38 DHF cases). The CFR of DHF was high (>60%) in children (0–14 years old), and those more than 45 years old (Table 2). Fatal cases affected more males than females, with 64.7% males and 35.3% females respectively.
Of total dengue cases 3,129 (81.3%) were confirmed by laboratory tests: 2,843 (90.9)% by ELISA or rapid antibody test detecting IgM, NS1, IgG-indirect or tissue culture at local health institutions or ICGES, 286 (9.1%) by qRT-PCR at ICGES; and 721 (18.7%) cases were confirmed by epidemiological link. In 2011, as part of the national dengue surveillance program, ICGES received 1,197 acute sera and 18 homogenized tissue samples (liver and spleen), of which only 6 had paired acute sera. Of the acute sera samples 23.9% (286/1197) were positive for DENV by qRT-PCR and/or viral isolation. Of these 286 confirmed cases, 197 were identified by qRT-PCR as DENV-2, 59 as DENV-1 and 30 as DENV-3. Of the 18 tissue samples received from suspected dengue cases, one was positive for Varicella Zoster virus by RT-PCR and viral isolation from spleen and liver and was negative for dengue. From the subsequent 17 fatal cases due to dengue, only one was diagnosed by epidemiological link, 10 were diagnosed by serological methods (IgM/IgG Dengue ELISA or NS1 ELISA), and 6 were diagnosed by qRT-PCR (Supplementary Table 1). Of these 6 cases, 5 were identified as DENV-2 and one as DENV-3. Viral isolation was attempted for all fatal cases; however, it was unsuccessful due to multiple breaks in the cold chain. For this study, from the seventeen fatal cases, only seven had corresponding clinical data which was available for analysis. This clinical data was similar to that previously described in the literature40–42 (Supplementary Table 1).
Around 10% of dengue negative acute samples (97/911 negative samples), were tested for other arboviruses, which have circulated in Panama. Alphaviruses (VEEV and EEEV) and flaviviruses (Yellow fever virus: YFV, Ilheus virus: ILHV, West Nile virus:WNV and Saint Louis virus:SLEV) was tested using hemaglutination inhibition test (HI). HI activity was detected in 53 samples. Of these samples, 14 were positive for antibodies against DENV (10 for DENV-2, two for DENV-1 and two for DENV-3) without cross-reactivity with any of the other tested arboviruses. Twelve samples presented an HI pattern for secondary response against Dengue, five samples showed titles for other flaviviruses, 2 for flavivirus and alphavirus and 20 sera showed cross-reactivity between different flaviviruses (Supplementary Table 2).
3.2. Aedes vector surveillance
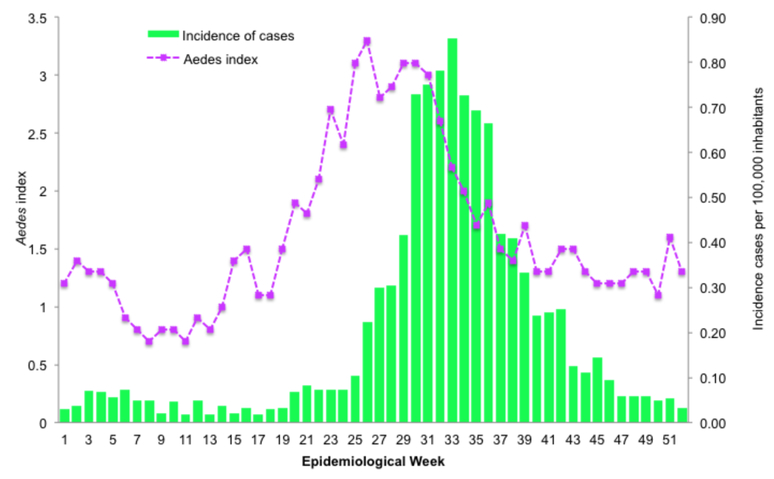
A moderate increase of the Aedes infestation index (Ae. aegypti and Ae. albopictus) occurred during epidemiological week 22, four weeks before Dengue clinical cases started to rise (Figure 3). This increase in Aedes circulation reached its highest point at week 30, and slowly decreased to a low risk index of <2 during week 35. The decrease in Aedes infestation rate is followed by a decrease in dengue reported cases, 5 weeks later. A correlation analysis of rho= 0.69 (p<0.001) between incidence rate and Aedes index infestation by week showed a moderate relationship between Aedes infestation and the incidence of dengue during the 2011 outbreak.
Figure 3. Incidence of dengue cases and Ae. infestation index during 2011.
In green the incidence of dengue cases per 100,000 inhabitants and in purple the Aedes infestation index per epidemiological week. The Aedes infestation index are classified as <2=low risk; 2–4=medium risk; >4=high risk.
3.3. Phylogenetic analysis
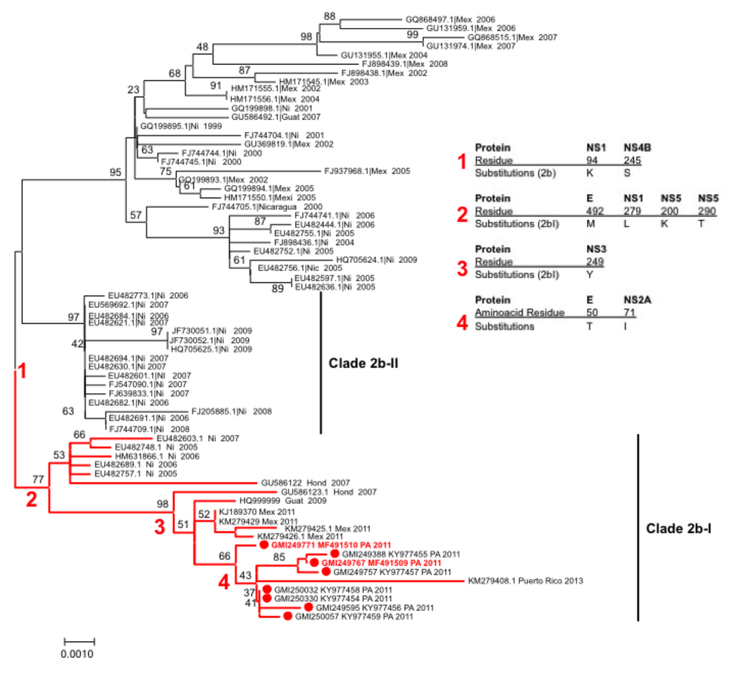
Six complete E gene sequences and one complete genome of the Panama DENV-2 isolates from the 2011 outbreak were aligned with representative sequences of all DENV-2 genotypes. Phylogenetic analysis of DENV-2 E gene alignment using NJ and ML methods created phylogenetic trees with identical topologies showing that Panama DENV-2 isolates clustered together and originated from the AS/AM lineage similar to previously reported DENV-2 isolates found in the Americas since 197922 (Figure 4). Using the same classification as Añez G et al26, the Panamanian isolates cluster in the AS/AM genotype clade 2 sub-clade 2b.I. This is confirmed by the full genome analysis with Panamanian strain 250330, KY977454 (Figure 4).
Figure 4. Panamanian Dengue virus type 2 Phylogeny.
Maximum likelihood tree (MLT) of DENV-2 based on complete E gene nucleotide sequences of South-east Asian American genotype from GenBank (n=60) and Panamanian strains (n=8). MLT was constructed using MEGA 7. Bootstrap values (1000 replications) above 70 were showed along the branch. The tree is mid-point rooted. Red branches represent clade 2B-I. Red spots represents Panamanian DENV-2 strains. The two Panamanian DENV-2 strains in red correspond to the isolates from the fatal cases.
The alignment between the six Panamanian DENV-2 2011 E gene sequences with the fourteen closest sequences from DENV-2 (five from Mexico, 2011; two from Guatemala, 2007 and 2009; one from Honduras 2007; five from Nicaragua, one from 2005, three from 2006, one from 2007; and one from Puerto Rico 2013) was done to look for specific mutations. Among the six Panamanian sequences from DENV-2 2011, there were only 8 nucleotide changes and all of them were synonymous. Among the Panamanian and the other analyzed sequences, 11 nucleotide differences. Only one of them was non-synonymous and resulted in an A50T amino acid substitution in PA-2011 and PR-2013 strains (Figure 4).
The amino acid sequence of the complete polyprotein of Panama DENV-2 2011 strain was analyzed. The Panama DENV-2 sequence had the two expected mutations specific of the clade 2b (compared to clade 2a): serine S in residue 245 from NS4B and lysine K in residue 94 from NS126 (Figure 4). It was confirmed that it had some of the mutations specific of 2b.I sub-clade like the M492 from E, L279 from NS1, K200 from NS5 and T290 from NS5. There is one mutation that substituted H249Y in NS3 in the PA-2011 as well as MX-2011, which is not found in the subclade 2b.I. Finally, there was one amino change, V71I from NS2A, specific to PA-2011 compared to the other available complete genome of Dengue 2b.I.
4. Discussion
During 2011, Panama experienced a dengue outbreak associated with the re-introduction of DENV-2. This study describes the characteristics of this outbreak and analyzes for the first time the genetic variation of DENV-2 strains that circulated in the country in 2011. Although the circulation of DENV-2 was detected in April, the majority of cases occurred from July to September. Similar to Vargas et al43, the Aedes infestation index (HI) was predictive of an increase in dengue cases with a preceding interval of six weeks. Our analysis showed a moderate correlation between the Aedes infestation index and when dengue cases surpassed the third quartile of the endemic channel. This suggests that the Aedes infestation index (HI) could be predictive of human dengue transmission. Other studies showed Aedes adult index and pupae index are better predictors of human dengue transmission44 however for this specific outbreak larvae index were enough.
Before 2011, the major dengue outbreak reported in Panama occurred in 2009 with a total of 7,459 Dengue cases. During this year the dengue mortality rate was 0.19 per 100,000 inhabitants. The CFR of DHF was 15%, (7 deaths / 46 DHF cases). During this outbreak DENV-1 and DENV-3 were the circulating serotypes. In 2010, the Americas experienced a large dengue outbreak with some countries having >200 cases per 100,000 inhabitants23. During this time DENV-2 was present throughout endemic countries in the Americas except in the USA, Panama and several Caribbean islands. Panama reported a decrease of cases in 2010, with a total of 2,002 cases, 3 DHF and no fatalities. At that time DENV-1 and DENV-3 were still co-circulating.
The Panama 2011 outbreak had a dengue mortality rate of 0.46 per 100,000 inhabitants and a case fatality rate of 44.7%, both the highest since the 1993 reemergence of dengue. When compared to other countries with a predominance of DENV-2 during the 2010 outbreak Panama had the highest CFR. In Colombia, the CFR was 2.3% (217 deaths/9,482 DHF) and in Honduras it was 2.54% (83 deaths/3,266 severe)23. However the number of DHF/total dengue cases is 1% in Panama, much lower than the 6% and 4.9% in Colombia and Honduras respectively. During 2011, the mortality rate in Panama was eighth in the Americas, just below Bolivia (0.47%) and above Brazil (0.24%). This could be due to improved healthcare infrastructure, less virulent DENV strain, errors in epidemiologic data collection or an underreporting of DHF cases. In addition, differential detection of severe dengue cases due to the recent change in the clinical dengue classification systems could have artificially decreased DHF cases and thus increased the Panama CFR.
Severe dengue and death is associated with host health status, genetic and previous immunity as well as viral features24. The presence of co-infections also plays a role in the development of severe dengue infections. Martins VDCA, et al41 did not find any difference between dengue serotype and severity of the disease during an outbreak with all four serotypes; however the severity of the disease was higher in patients infected with multiple serotypes when compared to those mono-infected. We analyzed 1,197 acute dengue cases, and no co-infection was detected by viral isolation or qRTPCR. The role of dengue serotype and genotype in the development of severity is controversial, however recent studies have shown that some specific clinical manifestations could be overrepresented by a specific serotype even in primary infections8,14. Our study reports that the Panamanian DENV-2 strain, circulating during 2011 outbreak, belongs to Asian/American genotype (AS/AM). The introduction of this genotype has been linked with outbreaks with high mortality rates41,45,46 in Peru47, Nicaragua27 and Brasil48. Our phylogenetic analysis clustered Panama 2011 DENV-2 strains in clade 2 sub-clade 2b.I using previous classification of DENV-2 AS/AM circulating in Central America26, closely related with strains from Mexico 2011 and Guatemala 2009. OhAinle et al27 reported the dominance of sub clade 2b after 2005/06 epidemic seasons in Nicaragua, explained by its higher median relative fitness than clade 1 and the higher viremia compared with clades 1 and 2A. It was observed in this study that DENV-3 immunity was associated with more severe disease among clade DENV-2 2b infections, while the DENV-2 clade I was more virulent in children immune to DENV-127. The high CFR during Panama 2011 outbreak could be explained by an introduction of this DENV2 sub-clade 2b, when DENV-1 and DENV-3 were co-circulating, considering this DENV-2 clade characteristics and its association with outbreaks in several countries of the region. The observed mutations in the 2011 Panamanian strains not previously described (A50T from E protein, V71I from NS2A, H249Y from NS3) need to be studied to determine if they are implicated in viral fitness or immune responses. Indeed A50 is one of the residues from the C chain of the E protein that interacts with a potent DENV antibody49, thus the A50T substitution could play a role in antibody recognition or other immune responses.
This study has several limitations. Epidemiological data analyzed was obtained from the dengue surveillance program, which does not contain variables such as symptoms, comorbidities, immunologic disorders, laboratory findings or travel history which all of which may have affected outcomes. In addition, acute serum was not always available and the cold chain for tissue samples was insufficient. This may have resulted in the inability to culture and/or detect dengue serotypes in all cases. We were not able to determine if severe or fatal DENV-2 infection were secondary; a cohort study in Panama would be necessary to characterize the dynamic interaction of circulating serotypes and the effect on dengue clinical manifestations.
This descriptive study of the 2011 dengue outbreak in Panama suggests that the introduction of DENV-2 AS/AM genotype subclade 2b.I contributed to the increase in cases and the high case fatality rate of deaths over DHF cases. The recent introduction of dengue vaccines make understanding severe dengue more important as these vaccines may promote severe disease in certain subpopulations50. An improved understanding of dengue and its dynamic circulation are required for successful public health policies and vaccination programs.
Supplementary Material
Highlights.
During the outbreak, a moderate correlation between the Aedes infestation index and dengue cases was observed when cases surpassed the third quartile of the endemic cannel.
During 2011, the introduction of DENV-2 AS/AM genotype subclade 2b.I after five years without DENV-2 circulation might have contributed to the increase in cases and the high case fatality rate of deaths over DHF cases.
Two mutations in the E gene and NS2A 2011 Panamanian strains have not been described previously.
Acknowledgments
We would like to thanks Jose Antonio Valenzuela for laboratory support. Mrs. Dennys Rodriguez De Gracia for epidemiological information. Claudia Gonzalez and Leyda Abrego for sequencing technical support and Alberto Cumbrera for Panama map figure. Sandra López Vergès and Blas Armien are part of the Sistema Nacional de Investigación (SNI) de SENACYT from Panama.
Financial support
This work was supported by grant 9044.020 of the Ministry of Finance of Panama (YD) and partially supported by NIH contract HHSN272201000040I/HHSN27200004/D04 (RBT, HG, ATdR, NV), NIH grant R24AI120942 (SCW), SENACYT FID-09–2013 (JPC) and SLV had UNESCO-L’OREAL international fellowship for young women in science 2014.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors of this research have no conflicts of interest.
References
- 1.Bhatt S et al. The global distribution and burden of dengue. Nature 496, 504–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shepard DS, Coudeville L, Halasa Y. a., Zambrano B & Dayan GH Economic impact of dengue illness in the Americas. Am. J. Trop. Med. Hyg 84, 200–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers TJ, Hahn CS, Galler R & Rice CM Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol 44, 649–88 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Who. Dengue: guidelines for diagnosis, treatment, prevention, and control. Spec. Program. Res. Train. Trop. Dis 147 (2009). doi:WHO/HTM/NTD/DEN/2009.1 [PubMed] [Google Scholar]
- 5.Yacoub S & Wills B Predicting outcome from dengue. BMC Med. 12, 147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mongkolsapaya J et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med 9, 921–927 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Halstead SB Pathogenesis of dengue: challenges to molecular biology. Science 239, 476–81 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Balmaseda A et al. Serotype-specific differences in clinical manifestations of dengue. Am. J. Trop. Med. Hyg 74, 449–456 (2006). [PubMed] [Google Scholar]
- 9.Stephens HAF HLA and other gene associations with dengue disease severity. Curr. Top. Microbiol. Immunol 338, 99–114 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Saqib MA, Rafique I, Bashir S & Salam A A retrospective analysis of dengue fever case management and frequency of co-morbidities associated with deaths. BMC Res. Notes 7, 205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toledo J et al. Relevance of Non-communicable Comorbidities for the Development of the Severe Forms of Dengue: A Systematic Literature Review. PLoS Negl. Trop. Dis 10, e0004284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fried JR et al. Serotype-specific differences in the risk of dengue hemorrhagic fever: An analysis of data collected in Bangkok, Thailand from 1994 to 2006. PLoS Negl. Trop. Dis 4, 1–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yung C-F et al. Dengue Serotype-Specific Differences in Clinical Manifestation, Laboratory Parameters and Risk of Severe Disease in Adults, Singapore. Am. J. Trop. Med. Hyg 92, 999–1005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halsey ES et al. Correlation of serotype-specific dengue virus infection with clinical manifestations. PLoS Negl. Trop. Dis 6, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longdon B et al. The causes and consequences of changes in virulence following pathogen host shifts. PLoS pathogens 11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halstead SB & Cohen SN Dengue Hemorrhagic Fever at 60 Years: Early Evolution of Concepts of Causation and Treatment. Microbiol. Mol. Biol. Rev 79, 281–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libraty DH et al. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis 186, 1165–8 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Chen H-R et al. Dengue Virus Nonstructural Protein 1 Induces Vascular Leakage through Macrophage Migration Inhibitory Factor and Autophagy. PLoS Negl. Trop. Dis 10, e0004828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modhiran N et al. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med 7, 304ra142–304ra142 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Rico-Hesse R Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174, 479–93 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Weaver SC & Vasilakis N Molecular evolution of dengue viruses: Contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol 9, 523–540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allicock OM et al. Phylogeography and population dynamics of dengue viruses in the Americas. Mol. Biol. Evol 29, 1533–1543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dick OB et al. Review: The history of dengue outbreaks in the Americas. Am. J. Trop. Med. Hyg 87, 584–593 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.OhAinle M et al. Dynamics of Dengue Disease Severity Determined by the Interplay Between Viral Genetics and Serotype-Specific Immunity. Sci. Transl. Med 3, 114ra128–114ra128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nogueira RM et al. Dengue epidemic in the stage of Rio de Janeiro, Brazil, 1990–1: co-circulation of dengue 1 and dengue 2 serotypes. Epidemiol. Infect 111, 163–70 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Añez G, Morales-Betoulle ME & Rios M Circulation of different lineages of dengue virus type 2 in Central America, their evolutionary time-scale and selection pressure analysis. PLoS One 6, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.OhAinle M et al. Dynamics of Dengue Disease Severity Determined by the Interplay Between Viral Genetics and Serotype-Specific Immunity. Sci. Transl. Med 3, 114ra128–114ra128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpenter Dudley and Sutton R Dengue in the Isthmian Canal Zone. J. Am. Med. Assoc 1, 214–216 (1903). [Google Scholar]
- 29.Rosen L Observations on the epidemiology of Dengue in Panama. Am. J. Trop. Med. Hyg 68, 45–58 (1958). [DOI] [PubMed] [Google Scholar]
- 30.Fairchild Laurence M.. Dengue-like fever on the isthmus of Panama. From Med. Serv. Gorgas Hosp. Ancon, Canal Zo 397–401 (1945). [DOI] [PubMed] [Google Scholar]
- 31.Quiroz E et al. Dengue en Panamá, 1993. Rev. Cubana Med. Trop 49, 86–93 (1997). [PubMed] [Google Scholar]
- 32.Ministerio de Salud de Panama. Boletin Epidemilogico. (1994).
- 33.Armien B et al. Clinical characteristics and national economic cost of the 2005 dengue epidemic in Panama. Am. J. Trop. Med. Hyg 79, 364–371 (2008). [PubMed] [Google Scholar]
- 34.WHO. Dengue haemorrhagic fever: Diagnosis, treatment, prevention and control. 84 (1997). doi:10.1002/9781444340051 [Google Scholar]
- 35.Panama M de S. de. Guías para el abordaje integral del dengue en panamá, 2014. (2014).
- 36.Leparc-Goffart I et al. Development and validation of real-time one-step reverse transcription-PCR for the detection and typing of dengue viruses. J. Clin. Virol 45, 61–66 (2009). [DOI] [PubMed] [Google Scholar]
- 37.CLARKE DH & CASALS J Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg 7, 561–573 (1958). [DOI] [PubMed] [Google Scholar]
- 38.Carrillo-Valenzo E et al. Evolution of dengue virus in Mexico is characterized by frequent lineage replacement. Arch. Virol 155, 1401–1412 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Chen R & Vasilakis N Dengue-Quo tu et quo vadis? Viruses 3, 1562–1608 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braga JCD et al. Clinical, molecular, and epidemiological analysis of dengue cases during a major outbreak in the midwest region of minas Gerais, Brazil. J. Trop. Med 2014, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins VDCA et al. Clinical and virological descriptive study in the 2011 outbreak of dengue in the Amazonas, Brazil. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Low JGH et al. The early clinical features of dengue in adults: Challenges for early clinical diagnosis. PLoS Negl. Trop. Dis 5, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vargas WP et al. Association among house infestation index, dengue incidence, and sociodemographic indicators: surveillance using geographic information system. BMC Public Health 15, 746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cromwell EA et al. The relationship between entomological indicators of Aedes aegypti abundance and dengue virus infection. PLoS Negl. Trop. Dis 11, e0005429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mamani E et al. Circulación de un linaje diferente del virus dengue 2 genotipo America / Asia en la región Amazónica de Perú, 2010. Rev. Peru. Med. Exp. Salud Publica 28, 72–77 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Williams M et al. Lineage II of southeast Asian/American DENV-2 is associated with a severe dengue outbreak in the Peruvian Amazon. Am. J. Trop. Med. Hyg 91, 611–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mamani E et al. Circulacion de un linaje diferente del virus dengue 2 genotipo America / Asia en la region Amazonica de Peru, 2010. Rev. Peru. Med. Exp. Salud Publica 28, 72–77 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Romano CM et al. Characterization of dengue virus type 2: New insights on the 2010 brazilian epidemic. PLoS One 5, 5–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty S Computational analysis of perturbations in the post-fusion Dengue virus envelope protein highlights known epitopes and conserved residues in the Zika virus. F1000Research 5, 1150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Godói IP et al. CYD-TDV dengue vaccine: systematic review and meta-analysis of efficacy, immunogenicity and safety. J. Comp. Eff. Res 6, 165–180 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.