Abstract
Direct T cell-to-T cell HIV-1 infection is a distinct mode of HIV-1 infection that requires physical contact between an HIV-1-infected “donor” cell and an uninfected, CD4-expressing “target” cell. In vitro studies indicate that HIV-1 cell-to-cell infection is much more efficient than infection by cell-free viral particles; however, the exact mechanisms of the enhanced efficiency of this infection pathway are still unclear. Several assays have been developed to study the mechanism of direct cell-to-cell HIV-1 transmission and to assess sensitivity to neutralizing antibodies and pharmacologic inhibitors. These assays are based on the coculture of donor and target cells. Here, we describe methods that utilize flow cytometry, which can discriminate donor and target cells and can assess different stages of entry and infection following cell-to-cell contact. HIV Gag-iGFP, a clone that makes fluorescent virus particles, can be used to measure cell-to-cell transfer of virus particles. HIV NL-GI, a clone that expresses GFP as an early gene, facilitates the measure of productive infection after cell-to-cell contact. Lastly, a variation of the β-lactamase (BlaM)-Vpr fusion assay can be used to measure the viral membrane fusion process after coculture of donor and target cells in a manner that is independent of cell-cell fusion. These assays can be performed in the presence of neutralizing antibodies/inhibitors to determine the 50 % inhibitory concentration (IC50) required to block infection specifically in the target cells.
Keywords: HIV entry, Cell-to-cell transfer, Cell-to-cell infection, Virological synapse, Neutralization assay, Fluorescent reporter virus, Gag-iGFP, β-lactamase (BlaM) fusion assay
1 Introduction
Direct cell-to-cell HIV-1 infection is a major mode of HIV-1 transmission between T cells in vitro and is likely to make a significant contribution to in vivo HIV-1 spread and pathogenesis in lymph nodes, gut-associated lymphatic tissue (GALT), and other tissues containing high numbers of HIV-1 susceptible cells. In CD4+ T cells, two major modes of HIV-1 infection have been documented—infection by cell-free virus and infection facilitated by cell-cell contact. Traditional “cell-free” infection involves the release of viral particles from a productively infected cell, followed by particle diffusion, viral attachment, and entry into an uninfected cell. The T cell-to-T cell virological synapse is distinct from the synapse formed between dendritic cells (DCs) and T cells [1, 2]. HIV-1 infection of the donor DC is not required for DC-to-T cell infection. Rather, virus particles are concentrated at the DC-T cell junction and transferred in trans to the target cell after exposure of donor DCs to virus particles [3]. T cell-to-T cell infection is mediated by a stable adhesion called a virological synapse (VS) [4], formed between a de novo HIV-1-infected donor T cell and an uninfected target T cell (also see reviews [5–7]). T cell-to-T cell infection was first observed for HTLV-1, a retrovirus that produces poorly infectious cell free virions [8]. For HIV-1, T cell-to-T cell infection has also been recognized as a much more efficient mode of HIV-1 infection compared to cell-free HIV-1 [9, 10]. HIV-1 Env and CD4 are required on the infected and uninfected T cells, respectively [4], and integrins may facilitate or reinforce the cell-cell adhesions [11–13]. Once contact is established, cell-surface Env, Gag, and CD4 polarize to the site of cell contact through actin cytoskeleton rearrangement, forming an adhesive structure that is defined as a “virological synapse,” as it resembles the immunological synapse formed during T cell activation, but with unique characteristics (reviewed in [14] and [15]).
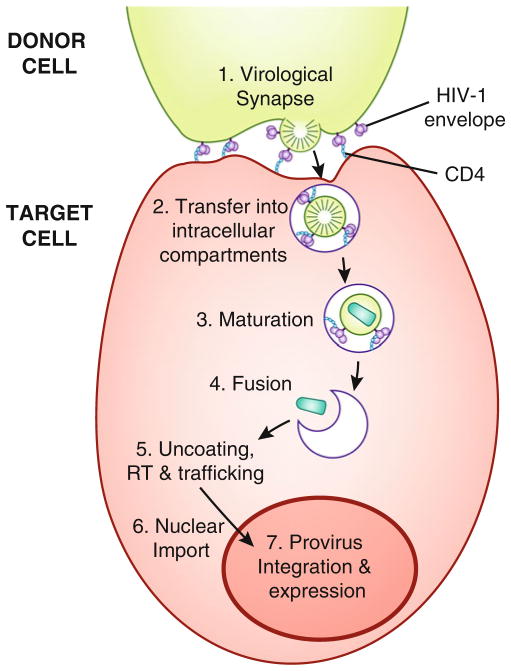
After virological synapse formation, viral particles have been described as following different pathways to viral entry. Some studies suggest that particles bud from the infected donor cell into the synaptic cleft and fuse at the plasma membrane of the uninfected target cell, similar to cell-free infection, but without extensive particle diffusion [4, 16, 17]. Alternatively, particles may be transferred directly into the target cell within intracellular compartments in a co-receptor-independent manner [18], before fusion of the viral and intracellular membranes which requires the presence of either CXCR4 or CCR5 co-receptor [19–21] (Fig. 1). Subsequent to viral fusion, the viral life cycle (uncoating, integration, and viral gene expression) is thought to be similar to cell-free infection.
Fig. 1.
Schematic representation of direct T cell-to-T cell HIV-1 entry, illustrating a multistep entry model. An HIV-1-infected donor T cell (top) expressing cell-surface envelope binds to CD4 on the surface of an uninfected target T cell (bottom ), forming a virological synapse. Gag and other molecules also co-localize to the site of adhesion. Virions bud and may be transferred directly into intracellular compartments where viral maturation and co-receptor binding occur, followed by fusion of the viral and intracellular membranes, uncoating, reverse transcription (RT), trafficking to the nucleus, nuclear import, provirus integration, and HIV-1 proviral gene expression. Alternatively, virions may bud into the synaptic cleft and undergo maturation, CD4 and co-receptor binding, and fusion of the viral and cell plasma membranes, similar to cell-free infection (not shown).
Some antiviral drugs and antibodies have been described as having lower inhibitory potency when blocking cell-to-cell infection as compared to cell-free infection [22–24]. Cell-to-cell transmission may promote viral persistence when suboptimal therapy or immune responses are present. Recently, these have been tested extensively in various in vitro cell-to-cell entry/infectivity assays [9, 17, 23–30].
A challenge in measuring cell-to-cell infection is clearly distinguishing the infectious signal in the target cells from the input signal of the infected donor cells. A common feature of the assays described here is the use of inert fluorescent cell-labeling dyes to accurately distinguish between donor and target cells. On the day of the synapse-forming assay, donor cells are Ficoll purified, and target cells are differentially labeled with cell proliferation dye, cocultured for a given duration, trypsinized, fixed, and analyzed by flow cytometry. Methods that employ flow cytometry enable rapid measurements of thousands of cells per second on a highly quantitative single-cell basis. The three assays in the following sections use flow cytometry and functional fluorescent indicators to detect distinct stages of cell-to-cell infection through T cell virological synapses:
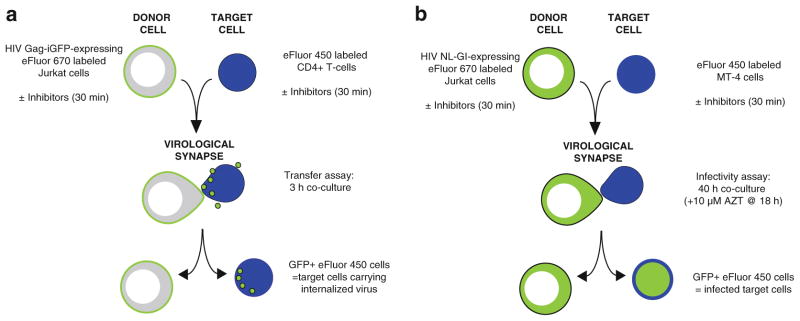
Cell-to-cell transfer assay utilizes fluorescent virus particles to measure the CD4-dependent steps that lead up to transfer of virus particles to the target cell during cell-to-cell infection (Fig. 2a).
Cell-to-cell infection assay utilizes a virus that expresses fluorescent protein as an early gene and thus measures all the steps that lead to productive viral infection, up to and including new viral gene expression (Fig. 2b).
Cell-to-cell viral membrane fusion assay measures the efficiency of viral membrane fusion with target cells by adapting a well-described BlaM-Vpr enzymatic assay for detecting viral fusion to a cell-to-cell transmission format.
Fig. 2.
Overview of cell-to-cell viral transfer and cell-to-cell infection assays. (a) In the cell-to-cell transfer assay, donor Jurkat cells are nucleofected with HIV Gag-iGFP proviral DNA 24 h prior to the assay. Donor Jurkat cells and primary CD4+ target cells are labeled with Cell Proliferation dyes eFluor 670 and eFluor 450, respectively, and may be preincubated separately for 30 min with inhibitors or antibodies before mixing at a 1:1 ratio. After 3 h coculture, GFP expression levels in target cells are detected by flow cytometry, indicating HIV-1 transfer. (b) In the cell-to-cell infectivity assay, donor Jurkat cells are nucleofected with HIV NL-GI proviral DNA 24 h prior to the assay. Donor Jurkat cells and MT-4 target cells are labeled with Cell Proliferation dyes eFluor 670 and eFluor 450, respectively, and may be preincubated separately for 30 min with inhibitors or antibodies before mixing at a 1:1 ratio. After 18 h coculture, cell culture media is replaced with media containing 10 μM AZT. 40 h after initial coculture, GFP expression levels in donor and target cells are detected by flow cytometry, indicating HIV-1 infection.
These assays can be performed with or without antibodies/inhibitors, using the 96-well plate format described here. The assays described here employ the CD4+ T cell line, Jurkat clone E6-1, as donor cells. While other T cell lines or even primary cells may be employed, we use Jurkat cells as they are a well-studied and infectable CD4+ T cell line that we have optimized for transfection by nucleofection methods. By transfecting the cells one can ensure that virus has been made de novo in donor cells, as is the case during acute infection, and is not derived from residual cell-free virus that is bound to the cell surface. The cell-to-cell transfer assay uses primary resting CD4+ T cells as target cells. The cell-to-cell infectivity and viral membrane fusion assays use MT-4 cells as target cells because of their robust infectivity. With some modification, these protocols can also be utilized with activated primary CD4+ T cells.
2 Materials
2.1 Plasmids
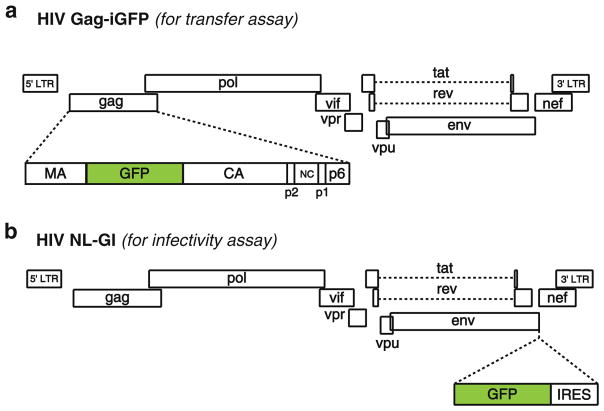
The plasmid encoding the fluorescent reporter virus HIV Gag-iGFP (Fig. 3a) is expressed in donor cells for the cell-to-cell transfer assay (Subheading 3.4). This construct expresses green fluorescent protein (GFP) as a fusion with the viral Gag protein, inserted between MA and CA, allowing the GFP to be packaged into particles at high copy number. Viral particles contain stoichiometric quantities of fluorescent protein in addition to Gag [20, 31]. Acquisition of fluorescence in the target cell is indicative of a CD4- and Env-dependent process that transfers virus to the target cell and does not require viral fusion, integration, or viral gene expression.
A second reporter virus HIV NL-GI (Fig. 3b) is expressed in donor cells for the cell-to-cell infectivity assay (Subheading 3.5). This reporter construct expresses GFP in place of the viral nef gene, providing an indicator of early viral gene expression, which requires viral integration [32]. GFP is not tagged to a viral protein, so these viral particles are nonfluorescent. To restore Nef expression in this clone, an internal ribosome entry site (IRES) is inserted upstream of the Nef open reading frame.
Full-length HIV-1 molecular clone pNL4-3 [33] for cell-to-cell viral membrane fusion assay (Subheading 3.6).
pMM310 encodes the BlaM-Vpr fusion protein [34, 35] that packages the β-lactamase Vpr fusion protein into virus particles when cotransfected with an HIV proviral plasmid, e.g., pNL4- 3. The cell-to-cell viral membrane fusion assay described in Subheading 3.6 is a variation of the FRET-based virion fusion assay described by Cavrois et al. [34, 36]. A green to blue shift in the BlaM fluorescent substrate CCF2-AM indicates fusion of cell-associated virus with target cells.
Fig. 3.
Fluorescent HIV-1 reporter virus clones. (a) HIV Gag-iGFP proviral DNA contains the fluorescent protein GFP in the gag gene between matrix (MA) and capsid (CA), flanked by viral protease cleavage sites, as described in [20 ]. Viral particles contain GFP in addition to MA, CA, and the other viral structural proteins. (b) HIV NL-GI proviral DNA contains GFP and an internal ribosomal entry site (IRES) directly upstream of the nonstructural gene nef [32 ]. GFP is expressed in infected cells in place of nef, during early viral gene expression. Nef expression is driven from the IRES.
2.2 Cell Culture and Nucleofection Reagents
Nucleofection media: RPMI-1640 medium supplemented with 10 % heat inactivated fetal bovine serum (FBS).
RPMI complete: RPMI-1640 medium supplemented with 10 % FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 200 μM L-glutamine.
Phosphate-buffered saline (PBS).
Trypsin-EDTA (0.05 % trypsin, 1 mM ethylenediaminetet-raacetic acid).
2 % paraformaldehyde (PFA) solution in PBS.
Recombinant Human IL-2 (IL-2).
Cell Proliferation dye eFluor 450, 10 mM stock in DMSO.
Cell Proliferation dye eFluor 670, 5 mM stock in DMSO.
Amaxa Cell Line Nucleofector Kit V containing Supplement V; stored at 4 °C, warmed to room temperature before use.
Ficoll-Paque PLUS.
Jurkat Clone E6-1 cells.
MT-4 cells (for cell-to-cell infectivity assay).
0.4 % trypan blue solution.
Unactivated (resting) primary human CD4+ T cells (for cell-to-cell transfer assay).
6-well flat-bottom tissue culture plates.
96-well round-bottom tissue culture plates.
Zidovudine (AZT).
2.3 Additional Reagents for Cell-to-Cell Viral Membrane Fusion Assay
CO2-independent media.
CCF2-AM substrate and β-lactamase loading solutions.
Probenecid stock solution: 250 mM probenecid in 250 mM NaOH.
Solution B: 100 mg/mL of Pluronic-F127 and 0.1 % acetic acid.
Loading solution (1 mL): 1 μL of 1 mM CCF2-AM stock+9 μL Solution B + 1 mL CO2-independent media.
Development media (1 mL): 10 μL probenecid stock solution+100 μL FBS + 1 mL CO2-independent media.
2.4 Additional Reagents for Intracellular p24 Staining of Donor Cells for Viral Membrane Fusion Assay (Optional)
1 % PFA solution in PBS.
FIX & PERM Cell Permeabilization Kit (Invitrogen).
Wash Buffer: 1 % FBS in PBS; at 4 °C.
Anti-p24-PE or anti-p24-FITC monoclonal antibody.
96-well V-bottom tissue culture plates.
2.5 Specialized Equipment
Nucleofector Device (single cuvette-based model) (Amaxa, Lonza).
BD LSR II Flow Cytometer (BD Biosciences).
3 Methods
Subheadings 3.1–3.3 describe the shared protocol for the preparation of donor Jurkat cells and target cells used in the subsequent assays (Subheadings 3.4–3.6). The plasmid DNA used for nucleo-fection in Subheading 3.1 will depend on the assay being performed (see above and see Note 1). The cell-to-cell assays in Subheadings 3.4–3.6 are performed in a 96-well round-bottom tissue culture plate and can be adapted to include neutralizing antibody/inhibitor titrations, as described in Subheading 3.4.
3.1 Nucleofection of Jurkat Donor Cells
Virus-producing Jurkat donor cells are generated by nucleofection of proviral DNA the day before the cell coculture. This method yields high transfection efficiencies and ensures that virus examined in the assay was produced in the donor cell. Nucleofection should be performed using Amaxa Cell Line Nucleofector Solution V containing Supplement V. Allow the solution to warm to room temperature before use. Cell survival after nucleofection will vary depending on the amount of DNA transfected. In general, approximately 50 % cell death is expected. Perform additional nucleofection reactions as needed, to prepare sufficient numbers of live cells for subsequent assays:
For each reaction, place 8 mL of nucleofection media per well in a 6-well tissue culture plate (see Note 2). Incubate at 37 °C for 10 min to warm media.
Count Jurkat cells. For each nucleofection reaction, spin 7 × 106 cells at 400 × g for 5 min to pellet cells. Carefully remove as much supernatant as possible and discard (see Note 3).
Resuspend cell pellet in 140 μL of room temperature Amaxa Cell Line Nucleofector Solution V containing Supplement V (see Note 4).
Immediately add DNA to cell suspension and pipette several times to mix (see Notes 1 and 5).
Immediately transfer suspension to Amaxa cuvette and cover with cap (see Note 6).
Select the program “S-18” on the Nucleofector Device, insert cuvette, and press the start button to transfect cells.
After successful transfection (i.e., “OK” message on the Nucleofector Device), remove cuvette. Immediately remove the cells gently with a single-use pipette provided with the Nucleofector Kit. Slowly add the cell suspension dropwise to the pre-warmed nucleofection media.
Use the single-use pipette to carefully rinse the cuvette with pre-warmed media to remove residual cells. Add cells to the 6-well plate.
Incubate overnight at 37 °C (see Note 7).
3.2 Density Gradient Purification of Nucleofected Jurkat Donor Cells Using Ficoll
The day after nucleofection, donor cells are spun through a Ficoll density gradient to remove dead cells:
For each 8 mL nucleofection reaction, place 4 mL of Ficoll in a sterile 15 mL centrifuge tube. Carefully layer nucleofected cells onto Ficoll using a 10 mL pipette.
Use 1 mL of RPMI complete to rinse the well and remove residual cells. Add cells to the 15 mL tube.
Spin at 400 × g for 20 min. Set centrifuge brake to “low” or off to prevent disruption of layers when centrifuge brakes at the end of the spin.
Carefully remove tubes from centrifuge. A layer of live cells should be visible at the interface of Ficoll and media.
Remove and discard a few mL of media from the top of the tube. Carefully remove the layer of live cells from the Ficoll- media interface using a pipette, and transfer to a clean 15 mL centrifuge tube. Avoid removing excess Ficoll.
Fill the tube containing live cells with RPMI complete, cap, and invert several times to mix. Spin at 400 × g for 5 min to wash cells (see Note 8). Discard supernatant.
Resuspend cell pellet in 1 or 2 mL of RPMI complete. Remove a sample of cell suspension and determine cell count (see Notes 9 and 10).
3.3 Cell Labeling
Cell labeling should be optimized for the cells and dye combination that one chooses to use. We often use Jurkat donor cells labeled with 4 μM Cell Proliferation dye eFluor 670 and MT-4 or CD4+ target cells labeled with 6 μM Cell Proliferation dye eFluor 450, except in the viral membrane fusion assay where donor cells are unlabeled and target cells are labeled with eFluor 670. Labeling cells with eFluor 670 and eFluor 450 allows detection of distinct donor and target populations by flow cytometry using APC and Pacific Blue filter sets, respectively, with minimal spectral compensation required. Depending upon the flow cytometer you use, you may also choose to use other inert fluorescent dyes from other manufacturers that are compatible with formaldehyde-based fixatives. As a flow cytometry control in the assays, it is useful to also label uninfected Jurkat cells:
Spin Ficoll-purified Jurkat donor cells and the desired amount of target cells at 400 × g for 5 min to pellet (see Note 11). Use a 15 or 50 mL sterile centrifuge tube for target cells, as appropriate. Discard supernatant and resuspend cell pellet in PBS.
Fill tube with PBS and spin at 400 × g for 5 min to pellet cells. Discard supernatant.
Prepare a working solution of 6 μM Cell Proliferation dye eFluor 450 in PBS for labeling target cells. Use 1 mL for a maximum of 10 × 106 cells.
Prepare a working solution of 4 μM Cell Proliferation dye eFluor 670 in PBS for labeling donor cells. Use 1 mL for a maximum of 10 × 106 cells.
Resuspend target cells in the appropriate volume of 6 μM Cell Proliferation dye eFluor 450. Resuspend donor cells in the appropriate volume of 4 μM Cell Proliferation dye eFluor 670.
Incubate cells at 37 °C for 10 min in the dark.
Add 4–5 volumes of RPMI complete to stop the labeling reaction. Spin at 400 × g for 5 min.
Remove supernatant and resuspend cells in 4–5 volumes of RPMI complete. Spin at 400 × g for 5 min to wash cells (wash #1).
Remove supernatant and resuspend cell pellet in 1 or 2 mL of RPMI complete. Remove a sample of cell suspension and determine live cell count using trypan blue (see Note 12).
Add to 4–5 volumes of RPMI complete and spin at 400 × g for 5 min to pellet cells (wash #2).
Resuspend each cell type at 2.5 × 106 cells/mL in RPMI complete.
3.4 Cell-to-Cell Transfer and Neutralization Assay
Steps 1–3 describe the preparation of eight dilutions of antibody/inhibitors for neutralization of cell-to-cell transfer from infected Jurkat donor cells to uninfected resting CD4+ human T cells. A positive control antibody such as Leu3a (an anti-CD4, HIV-blocking antibody) should be included to ensure the assay is performed correctly. Leu3a should neutralize both cell-to-cell transfer and cell-to-cell infection at 0.5 μg/mL. A negative control of RPMI complete (no antibody) should also be included in the assay:
Prepare 150 μL of each antibody/inhibitor at twice the highest desired concentration in RPMI complete. For example, prepare 150 μL of Leu3a at 1 μg/mL in RPMI complete. Mix and add to well A1 of a 96-well round-bottom tissue culture plate. Repeat for additional inhibitors, using wells B1 to H1, as needed (see Note 13).
Add 120 μL of RPMI complete to columns 2–8 for each row being used.
Make fivefold titrations by adding 30 μL of antibody/inhibitor from column 1 to column 2. Mix and repeat using a multi-channel pipette, making serial fivefold dilutions until column 8. Discard 30 μL from column 8.
Mix donor cells gently to resuspend cells. Add 50 μL of donor cells (i.e., 0.125 × 106 cells) to each well of a clean 96-well round-bottom tissue culture plate (plate 1). Reserve remaining cells.
Repeat step 4 for target cells, using a clean 96-well round- bottom tissue culture plate (plate 2) (see Note 14).
Add 50 μL of antibody/inhibitor to the corresponding wells in plate 1 containing donor cells. Pipette several times to mix (see Note 15).
Repeat step 6 for plate 2.
Incubate plate 1 and plate 2 for 30 min at 37 °C.
During incubation, fix samples of donor and target cells to determine infectivity levels at the time of coculture. Add 50 or 100 μL of donor or target cells to separate wells in a clean 96-well round-bottom tissue culture plate (plate 3). Samples can be prepared in duplicate. Proceed to steps 12–17.
After 30 min, pipette donor cells (plate 1) to resuspend cells, and transfer the entire suspension to corresponding wells containing target cells (plate 2).
Incubate for 3 h at 37 °C.
Spin plate at 500 × g for 5 min to pellet cells. Discard supernatant by quickly flicking the plate contents once into a waste container or by carefully using a multichannel pipette without disrupting the cell pellet.
Add 50 μL 0.05 % trypsin-EDTA to each well and mix to resuspend cells. Incubate for 4 min at 37 °C (see Note 16).
Add 150 μL of RPMI complete to each well to stop reaction.
Spin plate at 500 × g for 5 min. Discard supernatant.
Add 200 μL of PBS to each well and mix to resuspend cells. Spin plate at 500 × g for 5 min to wash cells. Discard supernatant.
Add 200 μL of 2 % PFA to each well to fix cells. Mix to resuspend cells.
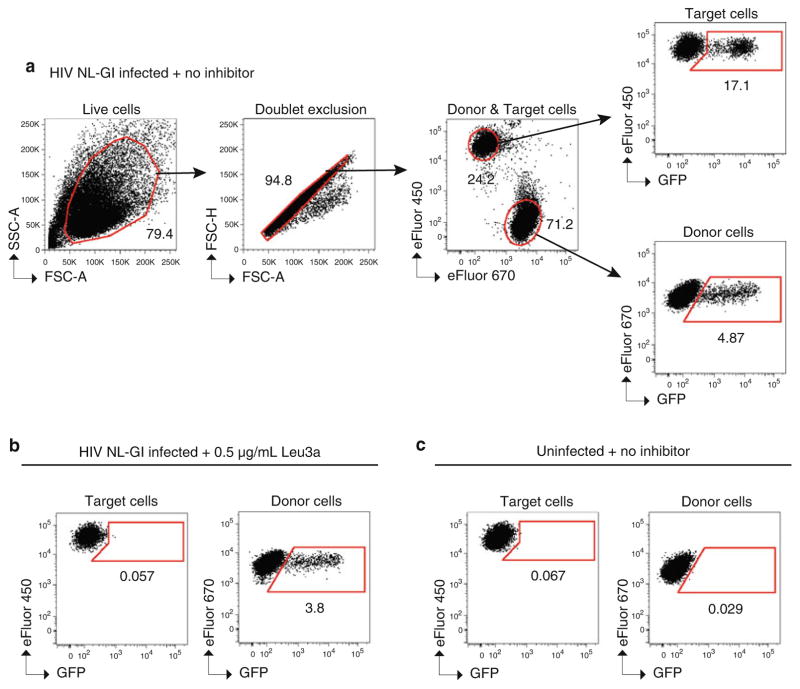
Seal plate and store at 4 °C in the dark until samples are analyzed by flow cytometry for up to 48 h. Figure 4 shows example data and a suggested gating strategy for data analysis.
-
If using neutralizing antibodies/inhibitors, calculate % inhibition:
100−[(% infected target cells with inhibitor/% infected target cells without inhibitor) × 100].
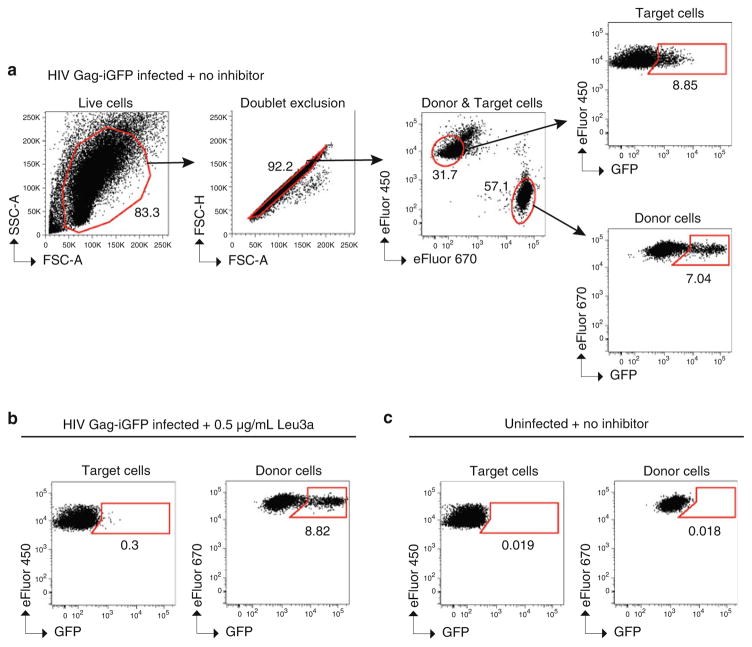
Fig. 4.
Detection of cell-to-cell transfer and inhibition by flow cytometry. Example flow cytometry plots of a typical cell-to-cell transfer assay using HIV Gag-iGFP. Gating strategy to determine live cells, doublet exclusion, eFluor 670 (APC)-positive Jurkat donor cells vs. eFluor 450 (Pacific Blue)-positive primary CD4+ target cells, GFP-positive (i.e., HIV+) CD4+ target cells, and GFP-positive (i.e., HIV+) Jurkat donor cells. CD4+ target cells are cocultured for 3 h with (a ) Jurkat donor cells expressing HIV Gag-iGFP in the absence of neutralizing antibody, (b ) donor cells expressing HIV Gag-iGFP in the presence of the antibody Leu3a at 0.5 μg/mL, and (c) uninfected donor cells.
3.5 Cell-to-Cell Infectivity and Neutralization Assay
Label Jurkat donor cells with eFluor 670 and MT-4 target cells with eFluor 450 as described in Subheading 3.3.
Prepare antibodies/inhibitors, infected Jurkat donor cells, and uninfected MT-4 target cells as described in Subheading 3.4, steps 1–10.
Incubate for 18 h at 37 °C.
18 h after coculture, spin plate at 500 × g for 5 min to pellet cells. Discard supernatant.
Resuspend cell pellet in 200 μL per well of RPMI complete containing 10 μM AZT. Mix well. Incubate for an additional 22 h at 37 °C (i.e., for a total of 40 h after coculture).
Spin plate at 500 × g for 5 min to pellet cells. Discard supernatant.
Treat cells with trypsin-EDTA and fix with 2 % PFA as described in Subheading 3.4, steps 13–19. Figure 5 shows example data and a suggested gating strategy for data analysis.
Fig. 5.
Detection of cell-to-cell infection and inhibition by flow cytometry. Example flow cytometry plots of a typical cell-to-cell infectivity assay using NL-GI. Gating strategy to determine live cells, doublet exclusion, eFluor 670 (APC)-positive Jurkat donor cells vs. eFluor 450 (Pacific Blue)-positive MT-4 target cells, GFP- positive (i.e., HIV+) MT-4 target cells, and GFP-positive (i.e., HIV+) Jurkat donor cells. MT-4 target cells are cocultured for 40 h with (a) Jurkat donor cells expressing HIV NL-GI in the absence of neutralizing antibody, (b) donor cells expressing HIV NL-GI in the presence of the antibody Leu3a at 0.5 μg/mL, and (c) uninfected donor cells.
3.6 Cell-to-Cell Viral Membrane Fusion Assay
Label MT-4 target cells with 4 μM Cell Proliferation dye eFluor 670 as described in Subheading 3.3.
Prepare antibodies/inhibitors, infected Jurkat donor cells, and uninfected MT-4 target cells as described in Subheading 3.4, steps 1–10 (see Note 17).
Incubate for 5 h at 37 °C.
Prepare appropriate amount of loading solution and development media.
5 h after coculture, spin plate at 800 × g for 5 min to pellet cells. Discard supernatant.
Resuspend cells in 200 μL per well of CO2-independent media to wash cells. Spin plate at 800 × g for 5 min to pellet cells. Discard supernatant.
Resuspend cells in 100 μL per well of loading solution, and incubate for 1–1.5 h in the dark at room temperature.
Spin plate at 800 × g for 5 min to pellet cells. Discard supernatant.
Resuspend cells in 200 μL per well of Development media to wash cells.
Spin plate at 800 × g for 5 min to pellet cells. Discard supernatant.
Resuspend cell pellet in 200 μL per well of Development media and incubate at room temperature in the dark for 16–18 h.
Spin plate at 800 × g for 5 min to pellet cells. Discard supernatant.
Treat cells with trypsin-EDTA, stop trypsinization, and wash cells with PBS as described in Subheading 3.4, steps 13–16.
Add 200 μL of 1 % PFA to each well to fix cells. Mix to resuspend pellet.
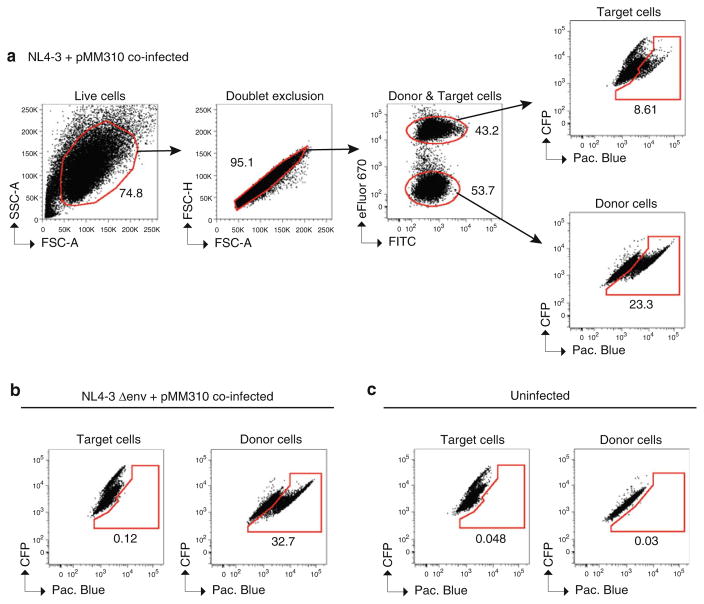
Seal plate and store at 4 °C in the dark until samples are analyzed by flow cytometry for up to 48 h. Figure 6 shows example data and a suggested gating strategy for data analysis.
Fig. 6.
Detection of cell-to-cell viral membrane fusion by flow cytometry. Example flow cytometry plots of a typical cell-to-cell viral membrane fusion assay using donor Jurkat cells co-expressing NL4-3 and pMM310 and MT-4 target cells. Gating strategy to determine live cells, doublet exclusion, eFluor 670 (APC)-positive MT-4 target cells and eFluor 670 (APC)-negative Jurkat donor cells, and Pacific Blue-positive (i.e., cleaved CCF2-AM) MT-4 target cells and Pacific Blue-positive (i.e., cleaved CCF2-AM) Jurkat donor cells. MT-4 target cells are cocultured for 5 h with (a) Jurkat donor cells co-expressing NL4-3 and the plasmid pMM310, which encodes BlaM-Vpr, (b) donor cells co-expressing NL4-3 Δenv and pMM310, and (c) uninfected donor cells.
Acknowledgments
We thank members of the B. K. Chen Lab for helpful comments and the Flow Cytometry Shared Resource Facility, Icahn School of Medicine at Mount Sinai, for assistance. This work was supported by NIH/NIDA DA028866 and NIH/NIAID A1074420.
Footnotes
When preparing donor cells for the cell-to-cell transfer assay (Subheading 3.4), use HIV Gag-iGFP proviral DNA. If donor cells will be used in the cell-to-cell infectivity assay (Subheading 3.5), use HIV NL-GI proviral DNA. Other versions of these proviral constructs that contain another fluores-cent protein, e.g., mCherry, instead of GFP, can also be used. If donor cells will be used in the cell-to-cell viral membrane fusion assay (Subheading 3.6), use a 3:1 ratio of pNL4-3 pro-viral DNA to pMM310 DNA. Other pNL4-3 genetic variants can also be tested, for example, pNL4-3 Δenv, by co-transfection with pMM310 (see Fig. 6b).
Up to two (i.e., duplicate) reactions can be placed in the same well containing 8 mL of media.
If cells are ≥1×106/mL at the time of counting, wash cells once with PBS before step 3.
Steps 3–8 should be performed separately for each nucleofection reaction, to avoid keeping cells in Nucleofector Solution for longer than necessary.
Use endotoxin-free purified HIV-1 proviral DNA, prepared at a concentration of approximately 1 μg/μL. The amount of DNA used will vary depending on the desired transfection efficiency. Typically 5 μg of fluorescent reporter proviral DNA will result in approximately 10–20 % positive cells when detected by flow cytometry the day after nucleofection.
Avoid air bubbles when adding the cell suspension to the cuvette to prevent an “error” message during nucleofection. This is easiest if the exact volume of the suspension (approximately 160 μL) is transferred to the cuvette with a 1 mL pipette. Gently tap the bottom of the cuvette to dispense air bubbles and ensure the suspension covers the bottom of the cuvette.
For nucleofection of proviral DNA encoding a fluorescent reporter virus, fluorescent virus-expressing cells should be readily visible with a fluorescent microscope the day after transfection.
If excess Ficoll was removed in step 5 and is visible after washing in step 6, remove and discard the top layer of RPMI complete, resuspend remaining volume up to 15 mL, invert tube several times to mix, and repeat spin.
The cell count here is used to determine the volume required for cell labeling.
An aliquot of cells can be fixed in a 96-well round-bottom tissue culture plate (see Subheading 3.4, steps 12–17) and analyzed by flow cytometry to determine the percentage of HIV-1-positive donor cells. If several HIV-1 genetic variants are being compared, it may be desirable to normalize samples to equal transfection efficiency if the efficiency of nucleofection varies between samples. Measure the transfection efficiency by flow cytometry and add uninfected cells to the nucleofected donor cells to achieve uniform transfection efficiency before proceeding to Subheading 3.3.
Label approximately 1.5× or 2× more target cells than needed, as cells are frequently lost during labeling wash steps.
Cells can be incubated at 37 °C in sterile centrifuge tubes for up to 2 h before proceeding to step 10.
You may also use wells B2-B11 through G2-G11 and fill outside wells with PBS during coculture to avoid sample evaporation from outer wells of the plate.
If no antibody/inhibitor is being tested, target cells can be added directly to donor cells. Add 100 μL of RPMI complete, pipette several times to mix, and proceed to step 11.
Mixing the antibody/inhibitor with an equal volume of cells dilutes it in half, to the desired final concentration, e.g., from 1 to 0.5 μg/mL for Leu3a.
Treating samples with 0.05 % trypsin-EDTA reduces the number of cell doublets before flow cytometry. In the cell-to-cell transfer assay, it also removes cell-surface bound fluorescent virus, ensuring that any fluorescent signal is from internalized virus only [9].
Optional: to determine the transfection efficiency of donor Jurkat cells, stain for intracellular p24 using anti-p24-PE or anti-p24-FITC monoclonal antibodies.
References
- 1.McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300:1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 2.Cameron PU, Freudenthal PS, Barker JM, Gezelter S, Inaba K, Steinman RM. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 3.Arrighi JF, Pion M, Garcia E, Escola JM, van Kooyk Y, Geijtenbeek TB, Piguet V. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J Exp Med. 2004;200:1279–1288. doi: 10.1084/jem.20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 2004;199:283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piguet V, Sattentau Q. Dangerous liaisons at the virological synapse. J Clin Invest. 2004;114:605–610. doi: 10.1172/jci22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dale BM, Alvarez RA, Chen BK. Mechanisms of enhanced HIV spread through T-cell virological synapses. Immunol Rev. 2013;251:113–124. doi: 10.1111/imr.12022. [DOI] [PubMed] [Google Scholar]
- 7.Puigdomenech I, Massanella M, Cabrera C, Clotet B, Blanco J. On the steps of cell-to-cell HIV transmission between CD4 T cells. Retrovirology. 2009;6:89. doi: 10.1186/1742-4690-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Igakura T, Stinchcombe JC, Goon PK, Taylor GP, Weber JN, Griffiths GM, Tanaka Y, Osame M, Bangham CR. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, Hubner W, Spinelli MA, Chen BK. Predominant mode of human immu-nodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol. 2007;81:12582–12595. doi: 10.1128/jvi.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol. 2007;81:1000–1012. doi: 10.1128/jvi.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 12.Rudnicka D, Feldmann J, Porrot F, Wietgrefe S, Guadagnini S, Prevost MC, Estaquier J, Haase AT, Sol-Foulon N, Schwartz O. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J Virol. 2009;83:6234–6246. doi: 10.1128/jvi.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jolly C, Mitar I, Sattentau QJ. Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. J Virol. 2007;81:13916–13921. doi: 10.1128/jvi.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller C, Fackler OT. HIV-1 at the immunological and T-lymphocytic virological synapse. Biol Chem. 2008;389:1253–1260. doi: 10.1515/bc.2008.143. [DOI] [PubMed] [Google Scholar]
- 15.Vasiliver-Shamis G, Dustin ML, Hioe CE. HIV-1 virological synapse is not simply a copycat of the immunological synapse. Viruses. 2010;2(5):1239–1260. doi: 10.3390/v2051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sattentau Q. Cell-to-cell spread of ret-roviruses. Viruses. 2010;2:1306–1321. doi: 10.3390/v2061306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. Virological synapse- mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol. 2010;84:3516–3527. doi: 10.1128/jvi.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta P, Balachandran R, Ho M, Enrico A, Rinaldo C. Cell-to-cell transmission of human immunodeficiency virus type 1 in the presence of azidothymidine and neutralizing antibody. J Virol. 1989;63:2361–2365. doi: 10.1128/jvi.63.5.2361-2365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dale BM, McNerney GP, Thompson DL, Hubner W, de Los RK, Chuang FY, Huser T, Chen BK. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe. 2011;10:551–562. doi: 10.1016/j.chom.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sloan RD, Kuhl BD, Mesplede T, Munch J, Donahue DA, Wainberg MA. Productive entry of HIV-1 during cell-to-cell transmission via dynamin-dependent endocy-tosis. J Virol. 2013;87:8110–8123. doi: 10.1128/jvi.00815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature. 2011;477:95–98. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 23.Abela IA, Berlinger L, Schanz M, Reynell L, Gunthard HF, Rusert P, Trkola A. Cell- cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog. 2012;8:e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durham ND, Yewdall AW, Chen P, Lee R, Zony C, Robinson JE, Chen BK. Neutralization resistance of virological synapse-mediated HIV-1 Infection is regulated by the gp41 cytoplasmic tail. J Virol. 2012;86:7484–7495. doi: 10.1128/jvi.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swartz TH, Esposito AM, Durham ND, Hartmann B, Chen BK. P2X-selective purinergic antagonists are strong inhibitors of HIV-1 fusion during both cell-to-cell and cell- free infection. J Virol. 2014;88:11504. doi: 10.1128/jvi.01158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massanella M, Puigdomenech I, Cabrera C, Fernandez-Figueras MT, Aucher A, Gaibelet G, Hudrisier D, Garcia E, Bofill M, Clotet B, Blanco J. Antigp41 antibodies fail to block early events of virological synapses but inhibit HIV spread between T cells. AIDS. 2009;23:183–188. doi: 10.1097/QAD.0b013e32831ef1a3. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Palomino S, Massanella M, Carrillo J, Garcia A, Garcia F, Gonzalez N, Merino A, Alcami J, Bofill M, Yuste E, Gatell JM, Clotet B, Blanco J. A cell-to-cell HIV transfer assay identifies humoral responses with broad neutralization activity. Vaccine. 2011;29:5250–5259. doi: 10.1016/j.vaccine.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, Scheid JF, Eden C, Mouquet H, Nussenzweig MC, Schwartz O. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J Exp Med. 2013;210:2813–2821. doi: 10.1084/jem.20131244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agosto LM, Zhong P, Munro J, Mothes W. Highly active antiretroviral therapies are effective against HIV-1 cell-to-cell transmission. PLoS Pathog. 2014;10:e1003982. doi: 10.1371/journal.ppat.1003982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titanji BK, Aasa-Chapman M, Pillay D, Jolly C. Protease inhibitors effectively block cell-to-cell spread of HIV-1 between T cells. Retrovirology. 2013;10:161. doi: 10.1186/1742-4690-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol. 2007;81:12596–12607. doi: 10.1128/jvi.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 33.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 35.Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J Virol. 2003;77:10645–10650. doi: 10.1128/JVI.77.19.10645-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavrois M, Neidleman J, Bigos M, Greene WC. Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol Biol. 2004;263:333–344. doi: 10.1385/1-59259-773-4:333. [DOI] [PubMed] [Google Scholar]