Introduction
Propagation of genetic material is central to the survival and fitness of multicellular organisms. To efficiently duplicate eukaryotic genomes of sizes between ~107 bp (yeast) to ~1011 bp (plants), replication initiates from multiple start sites called “replication origins” distributed on multiple chromosomes [1]. This strategy imposes a need for cellular controls to ensure complete DNA replication while also preventing re-replication, which can cause genomic instability, leading to developmental diseases (e.g. Meier-Gorlin Syndrome) and cancer [2,3]. Once-per-cell-cycle replication requires the oscillation of cyclin-dependent-kinases (CDKs) and ubiquitin-mediated proteolysis activities [4], establishing two discrete time periods during which origin licensing (assembly of replicative helicases onto origins) and origin firing (activation of helicases) occur in a mutually exclusive manner. Recent progress in acquiring structural information on origin licensing has advanced our mechanistic understanding and regulation of this process. This review discusses current knowledge of origin licensing and compares the origin recognition machinery with other multi-subunit, ATP-driven cellular motors that are required for later stages of DNA replication.
The Function of Initiators at Replication Origins
Replicators and Initiators
In 1963, a dual-element replication initiation model was proposed to describe the Escherichia coli replicon. In this model, a cis-acting replicator acts as the genetic element (DNA sequence) that is specifically recognized by a trans-acting initiator [5]. This model was confirmed by the identification of the E. coli replicator (oriC) and initiator (DnaA) respectively [6,7]. Bacteria tend to have a single origin of replication per chromosome, but in eukaryotes, the necessity of multiple origins on each chromosome creates a more complicated mechanism for origin licensing and regulation. The eukaryotic initiator is a complex of the six-subunit Origin Recognition Complex (ORC) and the related Cdc6 ATPase [8-10]. The nature of the eukaryotic cis-acting origins, however, is variable and depends on the species studied. Initiator proteins play multiple roles: they recognize or define the start sites within the DNA/chromatin for the initiation of DNA replication, they promote the recruitment of the helicase that unwinds the double helix and they ensure that the process is highly regulated (Fig 1). In viral replication systems, the helicase itself often functions as the initiator.
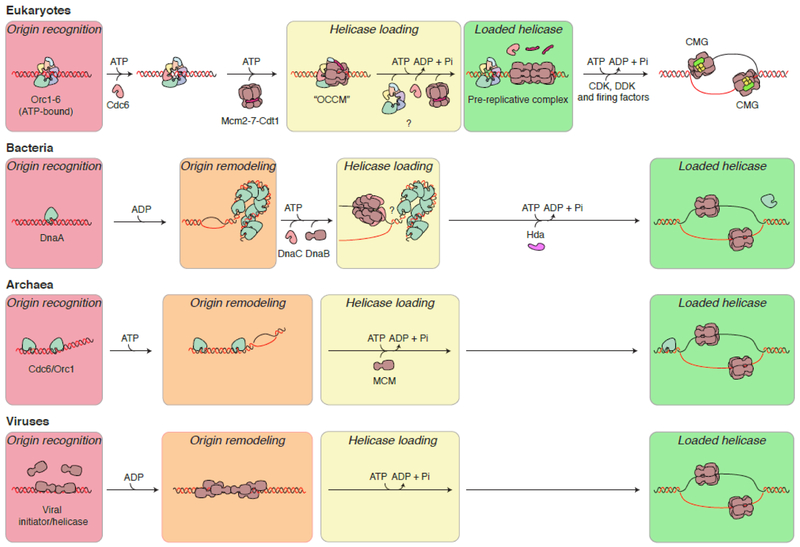
Fig. 1.
Models for origin recognition and helicase loading in the three domains of life and in viruses. In eukaryotes, bacteria and archaea, the origin is first recognized by the origin recognition machinery, with concomitant DNA remodeling occurring in bacteria and archaea. The replicative helicase is then recruited and directly interacts with the origin recognition complex. ATP binding but not hydrolysis is required for this interaction (see text). In eukaryotes, multiple origin recognition proteins may be required for loading a double hexameric form of the helicase. After helicase loading, ATP hydrolysis allows for reiterate assembly of helicases on other locations on the DNA. In viruses (human papillomavirus is shown as an example in this case), the viral helicase often serves the function of both origin recognition and self-assembly onto DNA into the mature helicase. In papillomavirus, a double-trimeric assembly of the helicase complex, linked to origin melting, is a prerequisite for the fully functional helicase complexes to be formed in subsequent steps in an ATP hydrolysis-dependent manner (for simplicity, other auxiliary factors are omitted in the diagram).
Origin Recognition in Different Domains of Life
Bacterial Origin Recognition
Genome replication in bacteria initiates from a single origin called oriC, which contains a highly AT-rich sequence required for initial melting of the DNA-unwinding element (DUE) [11]. In E. coli, the best characterized prokaryotic replication system, replication initiation involves the recognition of a 9-bp consensus sequence within the oriC by the initiator protein DnaA, a sequence-specific DNA binding protein that is also an AAA+ family ATPase. The consensus sequence [5’-TTAT(C/A)CA(C/A)A-3’] essential for initiation is called the DnaA-box [12]. Surrounded by weaker binding sites, the R1, R2 and R4 DnaA-boxes are recognized by multiple ATP-bound DnaA subunits with high-binding affinity [13]. The crystal structure of E. coli DnaA domain IV bound to the 9-bp DnaA-box showed the insertion of the highly conserved recognition helix from the helix-turn-helix (HTH) motif within DnaA into the major groove of the DnaA-box [14]. After the initial interaction of DnaA with high-affinity binding sites on oriC, multiple DnaA are then recruited to weak sites in an ATP-dependent manner. Oligomerization of multiple DnaA monomers leads to the formation of a high-order nucleoprotein assembly [13,15]. Subsequently, this ATP-bound DnaA assembly facilitates melting at the DUE region and allows for recruitment of the bacterial replicative helicase DnaB onto single-stranded DNA [16]. The ATPase activity of DnaA is activated by its interaction with a complex consisting of the Hda protein and the β-clamp of DNA polymerase that are loaded as DNA synthesis begins at oriC, limiting initiation of replication to once per cell cycle [17,18].
Archaeal Origin Recognition
Replication initiation in archaea combines features from bacteria and eukaryotes. Most archaeal genomes contain one origin, while others have several [1]. In the archaeal oriC, DNA sequences called the “origin recognition boxes” (ORBs) are the target sites recognized and bound by an initiator called the Cdc6/Orc1 protein, a AAA+ protein with a primary structure similar to the Orc1 and Cdc6 proteins of the eukaryotic initiator [19]. ORBs, which are clustered within the origin, can be categorized into two main types: long ORB motifs (22-35 bp) [20-22], and short ORB motifs (also called miniORBs) (12-13 bp) [1].
A crystal structure of Cdc6/Orc1 in complex with an ORB showed the main DNA contacts occur through the insertion of a helix from the C-terminal winged-helix domain (WHD) into the major groove, while the wing forms an unusually deep insertion into the adjacent minor groove [20,21]. The AAA+ domain makes another contact with an adjacent G-rich sequence [20,21]. These three binding events result in significant widening of both major and minor grooves, resulting in a bend of the DNA of up to 35°. This deformation may induce local unwinding of the duplex DNA, setting the stage for subsequent helicase loading onto single-stranded DNA [20,21].
Although Cdc6/Orc1 shows sequence homology to the eukaryotic ORC-Cdc6, data for Cdc6/Orc1 forming a higher order assembly are limited. However, the presence of multiple miniORBs as direct repeats suggest an interesting possibility of some degree of Cdc6/Orc1 oligomerization during origin recruitment, analogous to DnaA. Co-crystallization of an Orc1-1/Orc1-3 heterodimer bound to a miniORB supported this notion [20].
Origins and Origin Recognition in Eukaryotes
The most well-studied eukaryotic replication origins are those in the yeast Saccharomyces cerevisiae, which include a key element called the Autonomously Replicating Sequence (ARS). All ARS contain multiple sequence elements including a conserved ARS Consensus Sequence, ACS [23]. The A and B1 elements within the origin bind to ORC [8].
Although the origin sequences are not conserved, ORC, comprised of six proteins (Orc1-6) (Fig 2), and Cdc6 are found in all eukaryotes [10]. While ORC regulatory controls are species-specific [4], the general architecture and functions of ORC are similar across species. Five of the ORC subunits (Orc1-5) and Cdc6 show sequence homology to AAA+ proteins [24], while only four (Orc1, Orc4, Orc5 and Cdc6) contain the motifs (Walker-A, Walker-B and Arginine finger) necessary for ATPase activity [25,26]. Mutational analysis of ATPase motifs in ORC demonstrated that the ORC1/ORC4 ATPase site is responsible for the majority, if not all, of the ATPase activity observed in vitro [26,27]. Cdc6 is also an ATPase that controls the initiation of DNA replication [28-31]. Metazoan ORC6, the smallest subunit in ORC, shares partial structural similarity with RNA Polymerase II auxiliary factor TFIIB [32]. Apart from the AAA+ domains, ORC1-5 and CDC6 all possess C-terminal winged-helix domains (WHD) that contribute to the oligomerization of the five ORC subunits by docking onto the RecA-fold domain of a neighboring subunit [25,26]. ORC1, the largest ORC subunit, contains a bromo-adjacent homology (BAH) domain on the N-terminus implicated in gene silencing and origin recruitment via interaction with H4K20me2 [33]. Although crystal structures of the yeast and mouse ORC1 BAH-domains and part of human ORC6 were determined [32-35], structural information of the full ORC is warranted to further our understanding on how origin recognition works in eukaryotes.
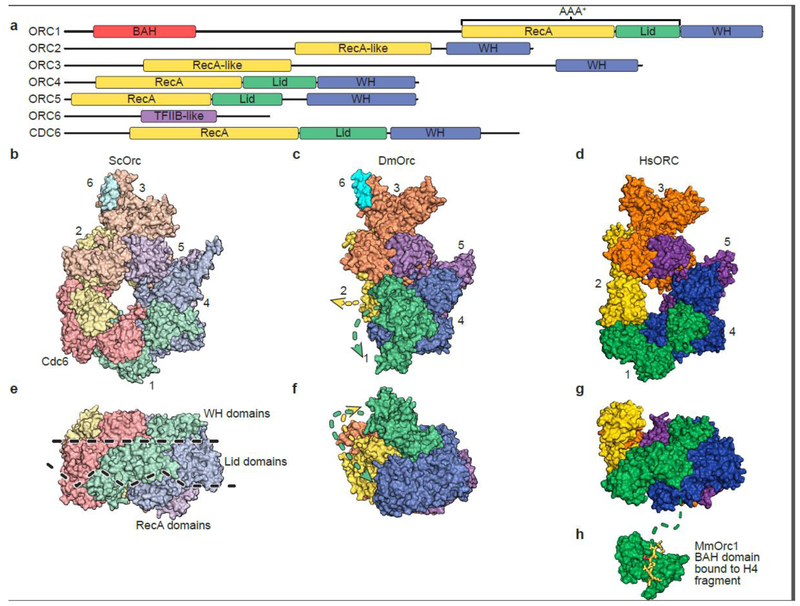
Fig. 2.
Overall structures of ORC from different species.
(a) Schematic alignment of ORC subunits and CDC6. The RecA (or RecA-like) and Lid domains are part of the AAA+ domain common in this clade of AAA+ ATPases. A winged helix (WH) domain is at the C-terminus in all subunits except for ORC6.
Structures of (b, e) Saccharomyces cerevisiae ORC, (c, f) Drosophila melanogaster ORC and (d, g and h) human ORC shown in perpendicular views.
Recently, three high-resolution ORC structures from different species and in different states have been reported, advancing our understanding of the helicase loading reaction. While the crystal structure of the Drosophila melanogaster ORC (DmORC) represents an interesting inactive state of ORC [25], both the HsORC [26], determined by a combination of X-ray crystallography and cryoEM, and the cryoEM structure of ScORC are in an ATP hydrolysis-competent state. In addition, ScORC is bound to Cdc6 and the MCM-Cdt1 complex, together referred to as the “OCCM” complex [36]. While the fly and human ORC structures are N-terminal truncations of ORC1 and ORC2, these regions are disordered in the ScORC that is part of the OCCM structure. In addition, fly ORC also includes a C-terminal truncation of ORC6 and the human ORC is missing this smallest subunit. However, the general arrangement of all three ORCs is very similar but some key differences distinguish them from one another.
All three structures revealed ORC as having a two-tiered configuration with the AAA+ domains forming one tier and the WHDs a second tier arranged in a right-handed helical architecture. The WHDs of each subunit interact with the AAA+-fold of the neighboring subunit. Nucleotides are nestled at the subunit interfaces between the RecA domains of ORC1 and 4, ORC4 and 5 and ORC5 and 3 [26,36] as would be expected for AAA+ ATPases. A central channel for DNA binding in ORC is clearly visible in the human and yeast structures (Figure 2b,d). In the case of DmORC – ORC1 is rotated away from the interface with ORC4, disrupting that important ATP-binding site, rendering DmORC inactive for ATP hydrolysis (Figure 2c,f). ORC1 as well as ORC2 would have to rearrange considerably to reform the ATP-binding site and open the central channel for DNA binding. This state of ORC may represent an inactive conformation to prevent futile ATPase activity or premature DNA replication initiation prior to Drosophila egg fertilization [25]. The presence of ATP might promote the transition from this inactive to an ATP-hydrolysis competent state [37].
The HsORC structure appears to be in an active conformation poised for ATP hydrolysis [26]. A homology model of HsCDC6 was built based on the HsORC1 structure, leading to a structural model of ORC1-5-CDC6 to explain the enhanced ORC DNA-binding activity upon associating with CDC6 [26,30,38]. Importantly, this model was consistent with the ScORC-Cdc6 structure [36]. The interface between ORC1 and CDC6 indeed constitutes an additional ATP-binding site, with residues poised for ATP hydrolysis [36]. The Cdc6-Orc2 interface on the other side of Cdc6 appears to be somewhat open compared to other subunit interfaces [36]. The absence of the lid domain in Orc2 may contribute to this more open interface. Notably, the disordered N-terminus of Orc2 also emanates from that region. Since the Cdc6-Orc2 interface of the ORC tier nearly lines up with the Mcm2-5 interface on the MCM tier, it raises an interesting possibility that this more open interface plays a role in MCM docking onto DNA and Mcm2-5 gate opening during MCM loading (see below) [39].
The initiator-specific motif (ISM) and the WHD of ScCdc6 (and Orc2) were observed to contact DNA, thus contributing to DNA-binding directly apart from forming a topological linkage with ORC [36]. In all structures, ORC1, ORC3, ORC4, ORC5, and CDC6 contain hairpin loops in their WHDs that face the core cavity [25], and in the case of ScORC, they appear to contact DNA. In ScOrc4, a helix is positioned in the major groove of the DNA and may have an important role in DNA recognition and specificity (Fig 3). This helix corresponds to an insertion in the sequence of ScOrc4 compared to the metazoan complexes. Coincidentally, Orc4 in Schizosaccharomyces pombe is also the determinant factor in origin recognition. In this case, the helix is missing and instead, nine AT-hook motifs in Orc4 interact with the AT-rich sequence at origins [40]. Together with Orc1, Orc4 is part of the major active ATPase interface in ORC [26,27]. Since addition of a replication origin (ARS1) was shown to inhibit ORC ATPase activity in vitro [27], it is possible that ORC4 acts as a switch to monitor DNA-binding status of ORC by regulating its ATPase activity. Finally, the relative positions of Orc3-Orc6 and the Cdt1 moieties in the “OCCM” structure are almost aligned, suggesting the possibility that Orc6 might act as a scaffold to bridge Orc3 and Cdt1 during the MCM loading process and stabilize this reaction intermediate. This notion is consistent with the absolute requirement of Orc6-Cdt1 interaction for MCM loading [41].
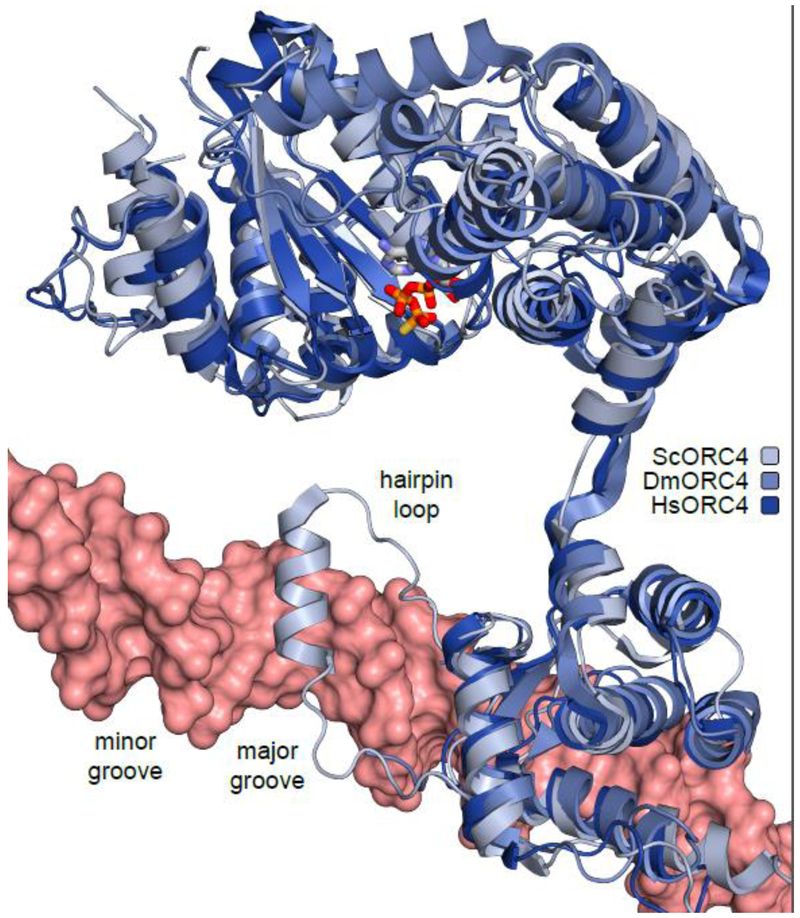
Fig. 3.
Comparison of the ORC4 subunit from S. cerevisiae, D. melanogaster, and H. sapiens. ScOrc4 contains an α-helix insertion located in the major groove of DNA. This α-helix is missing in human and Drosophila ORC4.
In metazoans, replication origins are more challenging to identify because the cis-acting origins show very loose sequence requirements [42]. Metazoan ORC was shown to bind DNA without sequence specificity [43,44]. As mentioned, neither the α-helix insertion in budding yeast Orc4 nor the AT-hook motifs in fission yeast Orc4 exist in metazoan ORC4 (Fig 3). Parts of the WHDs of ORC1, ORC4 and ORC5 oriented towards the central cavity of the ORC1-5 complex, implicating these WHDs in non-specific DNA interactions during origin recognition [26]. Genome-wide identification of initiation sites suggests other potential determinants for ORC-DNA binding. For example, negative supercoiling of DNA might be a feature that ORC recognizes [43]. Another feature involves the BAH domain at the N-terminus of ORC1, which interacts with modified histones [33]. Metazoan ORCs were also shown to be enriched in nucleosome-free regions (NFRs) in the genome [45], implicating CpG islands and G-rich sequences in origin recognition [10]. Further investigations are warranted to verify these predictions.
Helicase loading
General strategies of helicase assembly in various species appear to be conserved. The main approach is to employ the origin-recruited helicase loader to pry open the helicase ring, hold it in a conformation compatible with being assembled onto either single-stranded or double-stranded DNA, and release upon completing the reaction.
In bacteria, the helicase loading reaction begins with initial melting of the duplex DNA by DnaA. The initial melting at the origin by DnaA oligomerization results in the formation of a bubble containing two ssDNA strands onto which two DnaB hexameric complexes are loaded respectively. This molecular event is dependent on a direct interaction between DnaA and DnaB [46]. Structural studies revealed details of how DnaB loading is accomplished by the DnaA and DnaC loaders. The crystal structure of DnaC AAA+-ATPase domain showed that it is a close paralog of DnaA and can form a helical assembly in a similar manner to DnaA [47]. The EM structure of the DnaBC complex showed that the N-terminal region of hexameric DnaC associates with hexameric DnaB’s RecA domains, implicating DnaC as a ring-breaker in the helicase loading reaction [48]. Critically, this DnaBC complex is in an open-ring conformation with a right-handed spiral, allowing ssDNA entry into the central channel of DnaB [48]. Association of DnaC with DnaB also induces structural changes at the N-terminal tier of DnaB, which might be important to keep DnaB in the open-ring conformation during the helicase loading process.
Recently, an archaeal MCM in vitro assembly system demonstrated that Orc1/Cdc6 loads MCM onto DNA by directly interacting with both the DNA and the MCM in an ATP-dependent manner [49]. Interestingly, the loaded MCM was found to be in open-ring conformation, raising the possibility that other factors are required to turn the open-ring structure into the closed ring observed in structural studies. The archaeal MCM complexes exist in a double-hexameric architecture, reminiscent of the Simian Virus 40 T antigen and papillomavirus E1 proteins that function as both origin recognition proteins and DNA helicases [50-52], and the eukaryotic Mcm2-7 double hexamer complexes after loading [53-56].
Most of our knowledge of eukaryotic MCM loading comes from S. cerevisiae. Structural and biochemical studies indicated that MCM complexes are assembled as inactive double hexamers around dsDNA at origins [57-59]. Loaded MCM complexes are closed rings, while soluble MCMs exist in an equilibrium between closed and open rings. This phenomenon is attributed to the presence of a “gate” at the Mcm2/Mcm5 interface [60]. Both ATP- and Cdt1-binding contribute to modulate these two MCM conformations, particularly at the loading step [61-63].
In vitro MCM loading requires only three other factors: ORC, Cdc6 and Cdt1 [57,59]. However, the helicase loading reaction is very complex, involving a multitude of interactions, attachments and release factors at different reaction stages. The process begins with initial recruitment of an ORC-Cdc6 complex to the origin in an ATP-dependent manner [10]. Then, the MCM-Cdt1 complex is recruited to the ORC-Cdc6 complex via direct interaction, forming the OCCM intermediate complex [36]. The recent cryoEM structure of the OCCM complex revealed important molecular details of the MCM loading reaction. ORC-Cdc6, which showed a near-planar arrangement encircling dsDNA, was engaged with four C-terminal WHDs from MCM: Mcm3, Mcm4, Mcm6 and Mcm7, forming a right-handed spiral. The structure also explains the absolute requirement of the Mcm3 C-terminus during MCM loading [64]. Interestingly, this arrangement is similar to that observed in the DnaBC complex (see above) and clamp-loading reaction (see below) [65]. Another feature of the OCCM is the partial opening of the NTD-tier of the MCM ring. Whereas the CTD-tier (in contact with ORC-Cdc6) is closed, the NTD tier was partially open at the Mcm2/5 gate interface [36]. This arrangement is consistent with the notion that dsDNA would pass through this gate during the loading reaction. This OCCM intermediate was in complex with the slowly-hydrolyzable ATPγS, implying that ATP hydrolysis is not required for the partial engagement of MCM with dsDNA. This notion correlates with previous observations that ATP hydrolysis by ORC-Cdc6 is not required for loading of MCM, but rather for release of loading factors upon reaction completion, as in the clamp-loading situation (see below) [31,64,66].
Replicative Helicase Loaders and Clamp Loaders
Both ORC-Cdc6 and the DNA polymerase clamp loaders are AAA+-ATPases complexes responsible for assembling ring-shaped molecules onto DNA in the cells. Whether the helicase loader and the clamp loader employ similar ring-loading mechanisms has been controversial. Here, we describe a number of similarities in mechanics shared by both of these machineries, suggesting a conservation of mechanisms in these pathways.
The subunits of both loaders are structurally very similar and when combined, they are arranged in a helical fashion, facilitating the loading of ring-shaped proteins onto DNA (Fig 4) [26,67,68]. This is especially apparent when considering the motor module of ORC comprised of the AAA+ ORC1, ORC4 and ORC5 subunits. Interestingly, the helical parameters for the various clamp loaders whose structures are available more closely match those of duplex DNA, while ORC appears somewhat shallower [26,68]. The substrates to be loaded are either homooligomers (e.g. PCNA is a homotrimer; DnaB and archaeal MCM are homohexamers) or heterooligomers (eukaryotic Mcm2-7 is a hetero-hexamer), with their final state on DNA that of a closed ring. The presence of a gap (e.g. in RFC: between RFC1 and RFC5 and in ORC: between ORC1 and ORC2) allows DNA to enter the ring [26,67]. However, the gap in ORC is essentially closed by Cdc6 during the loading reaction [36], whereas the gap in the clamp loader remains open throughout the reaction.
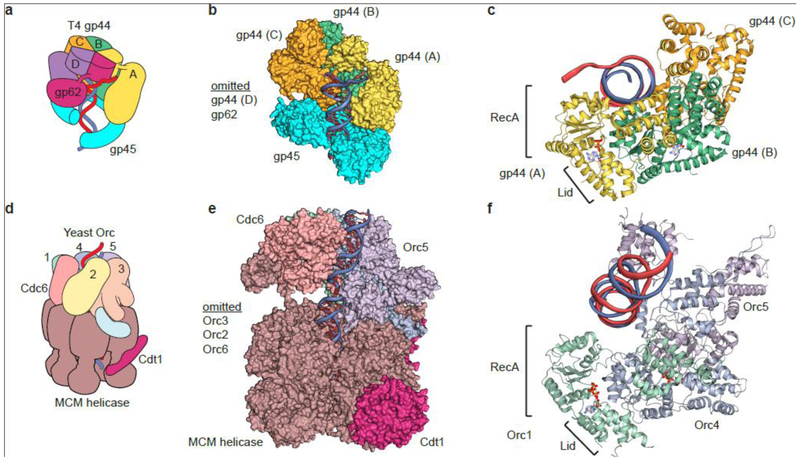
Fig. 4.
Comparison of loaders of ring-shaped proteins from bacteriophage T4 and S. cerevisiae.
(a) Schematic of the T4 DNA polymerase clamp loader (gp44 and gp62) bound to the gp45 clamp [an analogue of the eukaryotic PCNA]. (b) Surface representation of the structure of the T4 clamp loader bound to the clamp. (c) Cartoon representation of gp44 subunits A,B,C bound to primer-template DNA (red and blue strands) (d) Schematic of the S. cerevisiae helicase loader (ScORC+Cdc6) and pre-helicase complex (MCM+Cdt1). (e) Surface representation of the structure of ScOrc-Cdc6 bound to the MCM complex and Cdt1 on double strand origin DNA (f) Cartoon representation of ScORC subunits 1,4,5 bound to double-stranded DNA [red and blue strands]. (c) and (f) illustrate the corkscrew orientation of the subunits in the clamp loader and in ORC, both of which surround dsDNA.
Although both loaders are AAA+ family proteins, not all subunits can hydrolyze ATP. In fact, among the four potential ATP sites in RFC, only three are needed for PCNA loading [69]. In ORC, the main ATP site required for MCM loading was proposed to be between ORC1 and ORC4 [26,27]. The ScORC in the “OCCM” structure was shown to be in the ATP-bound state. Normally, upon completion of helicase recruitment, ATP hydrolysis by the ORC-Cdc6 complex is believed to trigger release of the loaded MCM complex [31,36,64]. For instance, when the loading reaction could not be completed, e.g. when ORC is phosphorylated by CDK, this event also helps to ensure that the loading machinery does not get stuck on chromosomes [64]. Importantly, ATP hydrolysis by ORC-Cdc6 was suggested to trigger a conformational change on both the MCM N-terminal tier and Cdt1, thus allowing the second MCM complex to be recruited and form a double hexamer with the first loaded MCM complex [31,36,70]. In both cases, the conversion of ATP to ADP is expected to trigger conformational changes, allowing the reaction to proceed to the next stage. Similarly, the clamp loader is in an ATP-bound state when bound to the opened clamp. ATP hydrolysis by the clamp loader results in the ejection of the loaded clamp [66]. This is a critical step in the loading pathway because both the clamp loader and DNA polymerase bind at the same surface of the clamp [65]. Thus, ATP hydrolysis is mainly required during the final step rather than the beginning step of both reactions. Finally, the stoichiometry of these reactions is also different. For the clamp-loading reaction, one clamp loader assembles one clamp in each reaction. As for MCM loading, one hypothesis is that two ORC-Cdc6 complexes are required to load the two Mcm2-7 hexamers [71].
RFC interacts with the DNA backbone rather than a specific nucleotide sequence [65]. This is of particular importance because the clamp loader assembles clamps at the primer-template junctions of replication forks. Bacterial, archaeal and yeast ORC display sequence specificity for DNA binding (see above). As for ORC in metazoans, little is known about how it recognizes origins. The involvement of direct binding to histone modifications [33] suggests possible mechanisms for origin definition. However, the general theme of DNA binding is the same: manipulating a central channel to encircle DNA. In this respect, key differences exist between the two loaders. The clamp loader associates with DNA after it binds to and pries open the clamp, thus it is the reaction intermediate that associates with the DNA [68]. By contrast, ORC-Cdc6 associates with the DNA before binding to MCM. Although MCM was reported to exist in an equilibrium of open (lockwasher) and closed rings in the cell [60], it is only after being bound by ORC-Cdc6 that the MCM ring would be stabilized in the open conformation to be loaded onto the DNA [36,57,59]. Interestingly, the T4 bacterial clamp frequently exists in an open conformation in solution (and PCNA to a lesser extent) [72], which is similar to the MCM in its lockwasher state.
Concluding Remarks
It is clear that the mechanisms of origin recognition and recruitment of the helicase core subunits is substantially different between bacteria and eukaryotes. But even within eukaryotes, origin recognition has rapidly evolved and there is still much research needed to understand the cis-determinates for origin specification in mammalian and plant genomes and how the initiators license these origins. The structural conservation of ORC-Cdc6 and Mcm2-7 suggests that although origin recognition varies between species, helicase loading and its regulation are remarkably similar.
Note
A paper appeared after acceptance of this review describing the structure of S. cerevisiae ORC bound to origin DNA [73]. The paper confirms previous findings about the evolution of origin specificity and describes new details about how ORC binds specific nucleotide. It also reports on additional DNA interactions with the Orc1 and Orc6 subunits that are absent in the ORC present in the OCCM complex [36].
Acknowledgements
The authors apologize to authors whose work was not included due to space limitations. B.S. is supported by the National Institutes of Health GM45463 and CA13106. L.J. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
**of outstanding interest
*of special interest
- 1.Leonard AC, Mechali M: DNA replication origins. Cold Spring Harb Perspect Biol 2013, 5:a010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbas T, Keaton MA, Dutta A: Genomic instability in cancer. Cold Spring Harb Perspect Biol 2013, 5:a012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klingseisen A, Jackson AP: Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 2011, 25:2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddiqui K, On KF, Diffley JF: Regulating DNA replication in eukarya. Cold Spring Harb Perspect Biol 2013, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob F, Brenner S: [On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon]. C R Hebd Seances Acad Sci 1963, 256:298–300. [PubMed] [Google Scholar]
- 6.Fuller RS, Kornberg A: Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc Natl Acad Sci U S A 1983, 80:5817–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuda S, Hirota Y: Cloning and mapping of the replication origin of Escherichia coli. Proc Natl Acad Sci U S A 1977, 74:5458–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **8.Bell SP, Stillman B: ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 1992, 357:128–134. [DOI] [PubMed] [Google Scholar]; Landmark paper of the discovery, isolation and biochemical characterisation of the eukaryotic origin recognition complex in Saccharomyces cerevisiae. It demonstrates that yeast ORC interacts with yeast replication origins in a sequence-specific manner.
- 9.Liang C, Weinreich M, Stillman B: ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 1995, 81:667–676. [DOI] [PubMed] [Google Scholar]
- 10.Parker MW, Botchan MR, Berger JM: Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol 2017, 52:107–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalski D, Eddy MJ: The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J 1989, 8:4335–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zakrzewska-Czerwinska J, Jakimowicz D, Zawilak-Pawlik A, Messer W: Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev 2007, 31:378–387. [DOI] [PubMed] [Google Scholar]
- 13.Rozgaja TA, Grimwade JE, Iqbal M, Czerwonka C, Vora M, Leonard AC: Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol Microbiol 2011, 82:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **14.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S: Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res 2003, 31:2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]; A Crystal structure of DnaA-bound to DNA was presented, showing how one helix and another loop in a helix-turn-helix insert into the major groove of the DNA, facilitating origin sequence recognition by directly binding to the strong Dna-boxes at oriC.
- 15.Erzberger JP, Mott ML, Berger JM: Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 2006, 13:676–683. [DOI] [PubMed] [Google Scholar]
- 16.Kaguni JM: Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol 2011, 15:606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JS, Nanfara MT, Chodavarapu S, Jin KS, Babu VMP, Ghazy MA, Chung S, Kaguni JM, Sutton MD, Cho Y: Dynamic assembly of Hda and the sliding clamp in the regulation of replication licensing. Nucleic Acids Res 2017, 45:3888–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima T, Nishida S, Kurokawa K, Katayama T, Miki T, Sekimizu K: Negative control of DNA replication by hydrolysis of ATP bound to DnaA protein, the initiator of chromosomal DNA replication in Escherichia coli. EMBO J 1997, 16:3724–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wigley DB: ORC proteins: marking the start. Curr Opin Struct Biol 2009, 19:72–78. [DOI] [PubMed] [Google Scholar]
- *20.Dueber EL, Corn JE, Bell SD, Berger JM: Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 2007, 317:1210–1213. [DOI] [PubMed] [Google Scholar]
- *21.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB: Structural basis of DNA replication origin recognition by an ORC protein. Science 2007, 317:1213–1216. [DOI] [PubMed] [Google Scholar]; Crystal structures of archaeal Orc1 (21) and the Orc1-1-Orc1-3 (20) were reported. The modes of DNA-binding involve a non-canonical type of insertion by the WHDs into both the major and the minor grooves of origin DNA. Unexpectedly, AAA+-domains also contribute to DNA-binding by directing binding to origin DNA, resulting in extensive bending of the DNA which might promote origin melting.
- 22.Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, Atanassova N, Wigley DB: Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J Mol Biol 2006, 363:355–369. [DOI] [PubMed] [Google Scholar]
- 23.Marahrens Y, Stillman B: A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 1992, 255:817–823. [DOI] [PubMed] [Google Scholar]
- 24.Erzberger JP, Berger JM: Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct 2006, 35:93–114. [DOI] [PubMed] [Google Scholar]
- **25.Bleichert F, Botchan MR, Berger JM: Crystal structure of the eukaryotic origin recognition complex. Nature 2015, 519:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first crystal structure of a metazoan ORC, from Drosophila melanogaster, was reported at high resolution. This ORC configuration represents an inactive state (incompatible with ATP hydrolysis) that might be unique to the egg stored form of the complex.
- **26.Tocilj A, On KF, Yuan Z, Sun J, Elkayam E, Li H, Stillman B, Joshua-Tor L: Structure of the active form of human origin recognition complex and its ATPase motor module. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]; The structure of human ORC determined by a combination of crystallography and cryoEM. This paper described eukaryotic ORC competent for ATP hydrolysis, potentially represent a state prior to MCM engagement for origin licensing reaction.
- 27.Klemm RD, Austin RJ, Bell SP: Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell 1997, 88:493–502. [DOI] [PubMed] [Google Scholar]
- 28.Chang F, Riera A, Evrin C, Sun J, Li H, Speck C, Weinreich M: Cdc6 ATPase activity disengages Cdc6 from the pre-replicative complex to promoteDNA replication. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randell JC, Bowers JL, Rodriguez HK, Bell SP: Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell 2006, 21:29–39. [DOI] [PubMed] [Google Scholar]
- 30.Speck C, Stillman B: Cdc6 ATPase activity regulates ORC x Cdc6 stability and the selection of specific DNA sequences as origins of DNA replication. J Biol Chem 2007, 282:11705–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP: Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 2015, 161:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Balasov M, Wang H, Wu L, Chesnokov IN, Liu Y: Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc Natl Acad Sci U S A 2011, 108:7373–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O: The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 2012, 484:115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu HC, Stillman B, Xu RM: Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc Natl Acad Sci U S A 2005, 102:8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong D, DePamphilis ML: Site-specific ORC binding, pre-replication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. EMBO J 2002, 21:5567–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al. : Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol 2017, 24:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]; A high-resolution cryoEM structure of the budding yeast ORC, trapped in an MCM-loading reaction intermediate state, was reported. With both the DNA and other protein components (Cdc6, MCM and Cdt1), this paper revealed important details about subunit interactions during MCM loading.
- 37.Bleichert F, Leitner A, Aebersold R, Botchan MR, Berger JM: Conformational control and DNA-binding mechanism of the metazoan origin recognition complex. Proc Natl Acad Sci U S A 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speck C, Chen Z, Li H, Stillman B: ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol 2005, 12:965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riera A, Speck C: Opening the gate to DNA replication. Cell Cycle 2015, 14:6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang RY, Kelly TJ: The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci U S A 1999, 96:2656–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, de Vries MA, Bell SP: Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2-7 loading. Genes Dev 2007, 21:2897–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mechali M, Kearsey S: Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 1984, 38:55–64. [DOI] [PubMed] [Google Scholar]
- 43.Remus D, Beall EL, Botchan MR: DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J 2004, 23:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC: Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 2003, 17:1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM: Chromatin signatures of the Drosophila replication program. Genome Res 2011, 21:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bleichert F, Botchan MR, Berger JM: Mechanisms for initiating cellular DNA replication. Science 2017, 355. [DOI] [PubMed] [Google Scholar]
- 47.Mott ML, Erzberger JP, Coons MM, Berger JM: Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell 2008, 135:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arias-Palomo E, O'Shea VL, Hood IV, Berger JM: The bacterial DnaC helicase loader is a DnaB ring breaker. Cell 2013, 153:438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samson RY, Abeyrathne PD, Bell SD: Mechanism of Archaeal MCM Helicase Recruitment to DNA Replication Origins. Mol Cell 2016, 61:287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enemark EJ, Joshua-Tor L: Mechanism of DNA translocation in a replicative hexameric helicase. Nature 2006, 442:270–275. [DOI] [PubMed] [Google Scholar]
- 51.Enemark EJ, Joshua-Tor L: On helicases and other motor proteins. Curr Opin Struct Biol 2008, 18:243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gai D, Wang D, Li SX, Chen XS: The structure of SV40 large T hexameric helicase in complex with AT-rich origin DNA. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Abid Ali F, Renault L, Gannon J, Gahlon HL, Kotecha A, Zhou JC, Rueda D, Costa A: Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun 2016, 7:10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell SD, Botchan MR: The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol 2013, 5:a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li N, Zhai Y, Zhang Y, Li W, Yang M, Lei J, Tye BK, Gao N: Structure of the eukaryotic MCM complex at 3.8 A. Nature 2015, 524:186–191. [DOI] [PubMed] [Google Scholar]
- 56.Noguchi Y, Yuan Z, Bai L, Schneider S, Zhao G, Stillman B, Speck C, Li H: Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc Natl Acad Sci U S A 2017, 114:E9529–E9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *57.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C: A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A 2009, 106:20240–20245. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper, along with Remus et al. reported the reconstitution of the pre-replicative complex in vitro with purified proteins and was a milestone in reconstitution of the initiation of DNA replication in eukaryotes.
- 58.Gambus A, Khoudoli GA, Jones RC, Blow JJ: MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem 2011, 286:11855–11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *59.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF: Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 2009, 139:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper, along with Evrin et al. reported the reconstitution of the pre-replicative complex in vitro with purified proteins and was a milestone in reconsititution of the initiation of DNA replication in eukaryotes.
- 60.Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM: The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol 2011, 18:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coster G, Frigola J, Beuron F, Morris EP, Diffley JF: Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell 2014, 55:666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frigola J, He J, Kinkelin K, Pye VE, Renault L, Douglas ME, Remus D, Cherepanov P, Costa A, Diffley JFX: Cdt1 stabilizes an open MCM ring for helicase loading. Nat Commun 2017, 8:15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai Y, Li N, Jiang H, Huang X, Gao N, Tye BK: Unique Roles of the Non-identical MCM Subunits in DNA Replication Licensing. Mol Cell 2017, 67:168–179. [DOI] [PubMed] [Google Scholar]
- 64.Frigola J, Remus D, Mehanna A, Diffley JF: ATPase-dependent quality control of DNA replication origin licensing. Nature 2013, 495:339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao NY, O'Donnell M: The RFC clamp loader: structure and function. Subcell Biochem 2012, 62:259–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bertram JG, Bloom LB, Hingorani MM, Beechem JM, O'Donnell M, Goodman MF: Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when gamma complex loads beta on DNA. J Biol Chem 2000, 275:28413–28420. [DOI] [PubMed] [Google Scholar]
- *67.Bowman GD, O'Donnell M, Kuriyan J: Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 2004, 429:724–730. [DOI] [PubMed] [Google Scholar]; A crystal structure of a clamp loader-clamp-DNA complex, in an ATP-bound state, was reported. This structure revealed a spiral conformation that the clamp loader and the clamp, both in an open conformation, closely matches the helical symmetry of DNA. A model of how ATP-hydrolysis is coupled to clamp release was proposed.
- 68.Kelch BA, Makino DL, O'Donnell M, Kuriyan J: How a DNA polymerase clamp loader opens a sliding clamp. Science 2011, 334:1675–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gomes XV, Burgers PM: ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem 2001, 276:34768–34775. [DOI] [PubMed] [Google Scholar]
- 70.Zhai Y, Tye BK: Structure of the MCM2-7 Double Hexamer and Its Implications for the Mechanistic Functions of the Mcm2-7 Complex. Adv Exp Med Biol 2017, 1042:189–205. [DOI] [PubMed] [Google Scholar]
- 71.Coster G, Diffley JFX: Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science 2017, 357:314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kelch BA: Review: The lord of the rings: Structure and mechanism of the sliding clamp loader. Biopolymers 2016, 105:532–546. [DOI] [PubMed] [Google Scholar]
- **73.Li N, Lam WH, Zhai Y, Cheng J, Cheng E, Zhao Y, Gao N, Tye BK: Structure of the origin recognition complex bound to DNA replication origin. Nature 2018, 559:217–222. [DOI] [PubMed] [Google Scholar]; A high-resolution cryoEM structure of the budding yeast ORC bound to a yeast replication origin was reported. It described ORC-DNA interactions critical for origin recognition by ORC, which agrees with previous experimental findings of ORC-DNA binding and uncovers novel DNA-binding sites on ORC absent in the OCCM structure.