Nonalcoholic fatty liver disease (NAFLD) is regarded as the most frequent cause of chronic liver damage (1,2). The natural history of the disease presents a complex scenario of potential progression into severe clinical outcomes, including nonalcoholic steatohepatitis (NASH), NASH-fibrosis, cirrhosis, and hepatocellular carcinoma (1,2). In addition, NAFLD is closely associated with comorbidities of the metabolic syndrome (MetS), including type 2 diabetes, obesity, arterial hypertension, and dyslipidemia, which together aggravate the morbidity and mortality associated with the disease.
It is known that NAFLD pathogenesis is dictated by multiple factors, including the interplay between genetic predisposition and environmental exposure (1-3). Nevertheless, the molecular landscape that determines the disease progression is not sufficiently understood. This scenario suggests that the design of safe and efficient pharmacological interventions aimed at reverting liver inflammation and fibrosis—major prognostic outcomes—is an extremely complex process.
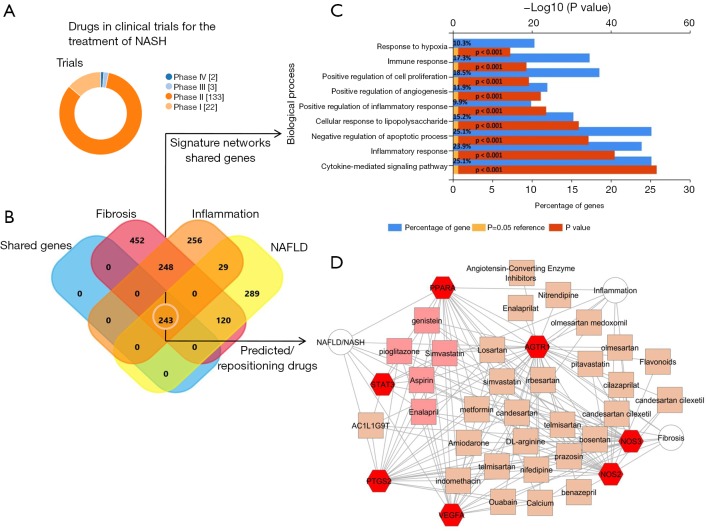
In fact, there are only a few approved drugs for the treatment of NASH that remain untested in clinical practice, although many promising molecules and novel drug candidates are currently being investigated. The large proportion of these drugs are in phase II (Figure 1A). Hence, it is expected that it will take time to establish whether any of these drugs will be useful, or identify those that might be the most effective for treating the multifaceted histological conditions associated with the disease. Most importantly, safety of any new treatment must be proven.
Figure 1.
Tackling the complexity of NASH treatment. Machine learning approaches identify candidate targets associated with NAFLD progression and suggest drug repositioning as an alternative strategy until novel drugs currently in the pipeline become available. (A) Number of drugs in clinical trials or approved for non-alcoholic fatty liver disease. Results were retrieved from The Open Targets Platform (https://www.targetvalidation.org/disease), which consists of data integration for access to and visualization of potential drug targets associated with diseases. The search was conducted using ChEMBL, a database of bioactive drug-like small molecules, and ClinicalTrials.gov (https://clinicaltrials.gov), a database of privately and publicly funded clinical studies around the world; (B) Venn diagram representation of genes/ proteins shared between NAFLD, fibrosis, and inflammation. The search of genes/proteins was performed by literature-enrichment analysis offered by the Genie web server (http://cbdm.uni-mainz.de/genie), a tool that computes associations of genes with keywords (in our case NAFLD/NASH, inflammation, and fibrosis as three distinct concepts) using biomedical literature annotations. The diagram was generated using the FunRich tool available online: http://www.funrich.org; (C) gene enrichment analysis for biological processes of the shared list of genes/proteins obtained as the intersection set from the three gene lists using the platform NetworkAnalyst (http://www.networkanalyst.ca). The analysis was conducted using the FunRich tool (http://www.funrich.org). Bonferroni, Benjamini-Hochberg, and FDR (false discovery rate) methods were used to correct for multiple testing. Pathways were ranked according to the P value (red bar) and P<0.05 was considered statistically significant. The blue bar indicates the percentage of altered genes in a whole pathway; (D) NAFLD/fibrosis/inflammation gene-drug connectivity network. The network is shown as a Cytoscape graph. The training set consisted of the lists of genes/proteins related to NAFLD/NASH, fibrosis, and inflammation, as explained above. Prediction analysis was performed by the ToppCluster resource available online: https://toppcluster.cchmc.org/. The list of predicted drug terms was manually curated to highlight examples of drug repositioning and was restricted to the P values below 1×10–10. The enrichment map shows terms corresponding to selected genes/proteins in the shared gene list (red hexagons), and predicted drugs/bioactive compounds that had significance scores (orange squares); the complete gene-drug connectivity network is shown in Figure S1. Pink squares show predicted drugs that are shared among all the selected targets. STAT3, signal transducer and activator of transcription 3; PPARA, peroxisome proliferator activated receptor alpha; PTGS2, prostaglandin-endoperoxide synthase 2; VEGFA, vascular endothelial growth factor A; AGTR1, angiotensin II receptor type 1; NOS2, nitric oxide synthase 2; NOS3, nitric oxide synthase 3; NAFLD, nonalcoholic fatty liver disease.
Drug repurposing, also known as repositioning, can circumvent the long time-to-market for drug development (4). This concept refers to repositioning already approved drugs for new indications, which may offer several advantages. Among the many opportunities, drug repositioning is economical and riskless, and avoids the need for passing lengthy clinical tests required for new drugs. Nevertheless, in our opinion, repositioning of an existing drug, rather than involving serendipity, must be biologically justified.
Recent discoveries resulting from OMICS-evolving technologies have challenged the classical single-level conception of “disease”, replacing it with a more multidimensional concept of “pathophenotypes”. It is then plausible to presume that the underlying mechanisms associated with chronic liver damage, specifically inflammation and fibrogenesis, share common perturbed pathways as well as similar genetic and epigenetic modifiers (5-7). Commonality of risk and genetic factors across NAFLD and MetS has also been suggested (8).
This novel approximation implies the concept of pathobiological processes interacting in an intricate network. As a proof-of-concept, we have explored to what extent the concept of repurposing of established medicines or active substances can be used for the treatment of NASH until new drugs become available (Figure 1). We then combined machine learning approaches and system biology resources to identify candidate targets associated not only with NAFLD, but also with inflammation and fibrosis as systemic pathobiological processes, rather than “liver-specific” processes. Further analysis was prioritized to the list of genes/proteins common to NAFLD, inflammation, and fibrosis (see footnote of Figure 1 and http://hbsn.amegroups.com/public/system/hbsn/supp-hbsn.2018.09.06-1.pdf). Not surprisingly, shared molecular targets were predicted to be involved in major distinctive pathways, including hypoxia, immune response, cytokine signaling, and angiogenesis (Figure 1C). Using molecular targets shared by NAFLD, inflammation, and fibrosis (http://hbsn.amegroups.com/public/system/hbsn/supp-hbsn.2018.09.06-1.pdf, 247 genes), we found a very complex gene-drug connectivity network (Figure S1), which we manually edited to show interesting examples of potential repositioning-pharmacological candidates (Figure 1D). For example, five drugs/compounds (pioglitazone, simvastatin, enalapril, aspirin, and genistein) were predicted to share the same gene and protein network associated with the NAFLD, inflammation, and fibrosis (Figure 1D). This analysis reinforces the concept that treatment of the NAFLD-associated conditions, including arterial hypertension, type 2 diabetes and dyslipidemia, with recommended and approved/existing drugs is not only relevant for reducing cardiovascular mortality, but represents a rational option for partially ameliorating the liver histological outcomes associated with poor prognosis. This approach could be systematically adopted to overcome the issue of long testing and approval process associated with the new drugs that are on the horizon. For example, could low-dose aspirin be recommended to patients with NASH not only for preventing ischemic heart disease but also for preventing liver inflammation? Even diet supplements, such as calcium, genistein (a natural isoflavone) or L-arginine, or a variety of sartans (AT1R antagonists), as well other inhibitors of the renin-angiotensin system with pleiotropic effects, may be used. A more systematic search can be done by looking at other examples in the complex-gene-drug network depicted in Figure S1.
In conclusion, it seems that NASH pharmacological treatment will not be successfully achieved by monotherapy. On the contrary, a dedicated multicomponent therapeutic approach might have a better chance of achieving NASH resolution and/or reversion of severe histological outcomes, and of reducing not only liver-related but also MetS-associated morbimortality. Nevertheless, any multimodal therapeutic strategy must find synergy with the biological processes that underlie the disease pathogenesis. While the search for the ideal pharmacological agent continues, given the unmet medical need for the treatment of NASH, we should optimize all available options. Identifying repurposing opportunities should be encouraged and all options should be explored, including mining of existing scientific databases, in silico approaches, in vivo experiments, and post-hoc analysis of trials on drugs that are already approved.
Figure S1.
NAFLD/fibrosis/inflammation gene-drug connectivity network. The network is shown as a Cytoscape graph. Details of this network are provided in the legend of Figure 1. NAFLD, nonalcoholic fatty liver disease.
Acknowledgements
Funding: Agencia Nacional de Promoción Científica y Tecnológica, Fondo para la Investigación Científica y Tecnológica (FonCyT) (PICT 2014-0432 and PICT 2015-0551 to SS and PICT 2014-1816 and 2016-0135 to CJ Pirola).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Brunt EM, Wong VW, Nobili V, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers 2015;1:15080. 10.1038/nrdp.2015.80 [DOI] [PubMed] [Google Scholar]
- 2.Friedman SL, Neuschwander-Tetri BA, Rinella M, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018;24:908-22. 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sookoian S, Pirola CJ. Genetic predisposition in nonalcoholic fatty liver disease. Clin Mol Hepatol 2017;23:1-12. 10.3350/cmh.2016.0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 2004;3:673-83. 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- 5.Greuter T, Malhi H, Gores GJ, et al. Therapeutic opportunities for alcoholic steatohepatitis and nonalcoholic steatohepatitis: exploiting similarities and differences in pathogenesis. JCI Insight 2017. [Epub ahead of print]. 10.1172/jci.insight.95354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sookoian S, Pirola CJ. Systems biology elucidates common pathogenic mechanisms between nonalcoholic and alcoholic-fatty liver disease. PLoS One 2013;8:e58895. 10.1371/journal.pone.0058895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sookoian S, Arrese M, Pirola CJ. Genetics meets Therapy? Exome-wide association study reveals a loss-of-function variant in HSD17B13 (17-beta-hydroxysteroid dehydrogenase 13) that protects patients from liver damage and NAFLD-progression. Hepatology 2018. [Epub ahead of print]. 10.1002/hep.30209 [DOI] [PubMed] [Google Scholar]
- 8.Sookoian S, Pirola CJ. Nonalcoholic fatty liver disease and metabolic syndrome: Shared genetic basis of pathogenesis. Hepatology 2016;64:1417-20. 10.1002/hep.28746 [DOI] [PubMed] [Google Scholar]