Abstract
Objective
Several small case series identified KCTD7 mutations in patients with a rare autosomal recessive disorder designated progressive myoclonic epilepsy (EPM3) and neuronal ceroid lipofuscinosis (CLN14). Despite the name KCTD (potassium channel tetramerization domain), KCTD protein family members lack predicted channel domains. We sought to translate insight gained from yeast studies to uncover disease mechanisms associated with deficiencies in KCTD7 of unknown function.
Methods
Novel KCTD7 variants in new and published patients were assessed for disease causality using genetic analyses, cell-based functional assays of patient fibroblasts and knockout yeast, and electron microscopy of patient samples.
Results
Patients with KCTD7 mutations can exhibit movement disorders or developmental regression before seizure onset, and are distinguished from similar disorders by an earlier age of onset. Although most published KCTD7 patient variants were excluded from a genome sequence database of normal human variations, most newly identified patient variants are present in this database, potentially challenging disease causality. However, genetic analysis and impaired biochemical interactions with cullin 3 support a causal role for patient KCTD7 variants, suggesting deleterious alleles of KCTD7 and other rare disease variants may be underestimated. Both patient-derived fibroblasts and yeast lacking Whi2 with sequence similarity to KCTD7 have impaired autophagy consistent with brain pathology.
Interpretation
Bi-allelic KCTD7 mutations define a neurodegenerative disorder with lipofuscin and lipid droplet accumulation but without defining features of neuronal ceroid lipofuscinosis or lysosomal storage disorders. KCTD7 deficiency appears to cause an underlying autophagy-lysosome defect conserved in yeast, thereby assigning a biological role for KCTD7.
Introduction
Autism, schizophrenia, dystonia, epilepsy and other disorders have been linked to several members of the gene family KCTD (potassium channel tetramerization domain).1–4 All KCTD family proteins have an N-terminal BTB domain that is most similar in sequence to voltage-gated potassium channel tetramerization (T1/BTB) domains, hence the name KCTD.5 However, KCTD proteins appear unlikely to form channels as they lack predicted transmembrane domains and any direct interaction with potassium channels remains uncertain. Although KCTDs could indirectly alter channel properties, the gene name is potentially misleading and has caused some diagnostic challenges with implied treatments for channelopathies.6 Patients with KCTD7 mutations have been diagnosed with progressive myoclonic epilepsy (EPM3),4, 7–12 neuronal ceroid lipofuscinosis 14 (CLN14),13 or opsoclonus-myoclonus syndrome (OMS).14 We sought to further define the clinical syndrome resulting from KCTD7 deficiency, to distinguish deleterious from normal variants in the general population, and to assign a function to the uncharacterized KCTD7 protein based on insights gained from our studies of the related protein in yeast, Whi2.
KCTD family proteins are relatively uncharacterized, but one common theme has begun to emerge. A subset of KCTD family proteins may be components of cullin 3 (CUL3) ubiquitin ligase complexes.15 CUL3 uses BTB-containing adaptor proteins from other protein families to recruit cargo proteins for ubiquitination. Several KCTD family proteins including KCTD7 have been reported to bind CUL3,13, 16 consistent with structure modeling for other KCTDs.15 In this capacity as potential cargo adaptors, several other KCTD family proteins have been suggested to target specific cargo proteins for degradation,1 although most await confirmation. A similar role for KCTD7 is consistent with lysosome pathway defects in several other EPM and CLN disorders,17, 18 and other neurodegenerative processes.19 However, the molecular and cellular consequences of KCTD7 deficiency are not known.
This project was prompted by our genome-wide yeast genetic screen that uncovered the KCTD-like BTB-containing protein Whi2 of Saccharomyces cerevisiae.5 Yeast Whi2 is reportedly a general stress response factor,20 and has a contested role in mitophagy.21, 22 Yeast Whi2 and autophagy regulator Atg6 (human Beclin 1) were identified in our screen for factors required to respond to low amino acid availability,5, 23 a condition known to induce catabolic processes such as autophagy in yeast and mammals.24 Because yeast Whi2 shares sequence similarity and a common domain architecture with human KCTD proteins,5 we sought to gain new insight into the disease mechanisms due to KCTD7 mutations. We found that both yeast Whi2 and human KCTD7 are required for normal autophagy in low nutrients, further supported by the accumulation of lipid bodies, defective mitochondria and abnormal autolysosomes.
Methods
Patient data collection
Clinicians were provided a form for deidentified data (Table S1, IRB exempt status).
Electron microscopy
Patient fibroblasts were fixed in 2.5% glutaraldehyde (EM grade; Electron Microscopy Sciences, Hatfield, PA) in 0.1 M sodium cacodylate buffer (pH 7.4) for 1 h at room temperature, and processed by the Yale University Electron Microscopy Core as described.25
PCR verification of KCTD7 mutations in patient fibroblasts
Genomic DNA for KCTD7 exon-2 of primary human fibroblasts was amplified from extracted DNA using 5’ Primer (TGGCACCAATCAGACCCCAGGGATTGAAGATGGAGCAGCCC) and 3’ Primer (CCCATTTATTAAATTTCATCAATATGCTATCTCCTCTTCTAGG), separated on 1% agarose gel, extracted using Qiagen Gel Extraction Kit, and sequenced at MacrogenUSA using the 5’ primer to verify patient mutations in these cell lines.
Plasmids
Wild type KCTD7 ORF was PCR amplified (Invitrogen AccuPrime Pfx) from HEK293 cDNA (Qiagen RT Kit) and cloned into a pSG5-derived vector to create N-terminal tagged proteins. Coding changes were subsequently engineered into KCTD7 and CUL3 expression vectors and verified by Sanger sequencing. prPGK-GFP-ATG8 was engineered by replacing the ATG8 5’ regulatory region with the 5’ regulatory region of PGK.
Mammalian autophagy assays
Primary patient fibroblasts (derived from 3 mm axilla skin punch) were shipped/received live and maintained in DMEM, 10–20% FBS and pen/strep. Age- and passage-matched control human fibroblasts GM05757 and 498 (Corriel Institute) were maintained in parallel and KCTD7 expression was assessed by qRT-PCR. Subconfluent cells, passage 4–8, were plated in 12-well dishes (50,000/well). The next day were washed 1x with PBS before treating. For nutrient deprivation, cells were switched to EBSS or to RPMI1640 without L-glutamine, amino acids or glucose (US Biologicals) supplemented with 5% dialyzed FBS (Thermo) with or without amino acids (R7131 Sigma) and glucose (4.5 g/L, Gibco). For lysate preparation, cells were washed with PBS, solubilized in lysis buffer (62.5 mM Tris-HCl, 2% SDS, 0.01% Bromophenol Blue, 10% glycerol, 2% β-mercaptoethanol) with protease inhibitor cocktail (Thermo Scientific) and heated to 100°C for 10 min before separation by 12% SDS-PAGE and transfer to PVDF. Immunoblots were performed using antibodies for LC3B (CST #2775 or 3868, 1:1000) and actin (MP Biologicals #691001, 1:10,000) and visualized using a Bio-Rad ChemiDoc MP system and analyzed with ImageLab 5.0 software.
Immunofluorescence microscopy
Kyoto HeLa cells (ATCC) were grown on 12 mm glass cover slips in DMEM, 10% FBS plus pen/strep, and transfected the next day with 150 ng total DNA using JetPRIME (VWR). At 16–18 h posttransfection, cells were fixed 10 min with 4% formaldehyde (Polysciences, Warrington, PA), permeabilized with 0.2% Triton X-100 in PBS, immunostained, mounted (Prolong Gold, Life Technologies) and viewed with a Zeiss AxioImager M2 (60x Olympus objective), Hamamatsu Orca R2 camera and Volocity Software, or an Applied Precision DeltaVision microscope and software (60x Olympus objective) with Hamamatsu Photonics camera.
Co-immunoprecipitations
N-terminal 3HA/3Flag-KCTD7 and 6Myc-cullin 3 (CUL3) expression vectors were transiently transfected into HEK-293 cells using BioT (Bioland Scientific) for 2 days. Cells were lysed in native lysis buffer containing AEBSF, pepstatin, and leupeptin, rotated for 60 min at 4 ºC, and remaining insoluble material was removed by centrifugation. Samples were normalized (A280 nm) and rotated at 4 ºC for 2 h with pre-equilibrated EZview Red anti-HA or anti-FLAG affinity gel (Sigma). Beads were washed 4 times in the same buffer and proteins eluted by boiling in SDS-PAGE sample buffer. Whole-cell lysates (WCL) and anti-HA precipitates (IP) were separated by SDS-PAGE, transferred to PVDF membranes and probed with antibodies against FLAG (F1804, Sigma), HA (12CA5, Roche), or α-tubulin (HRP-66031, Proteintech) and HRP-conjugated secondary antibodies. Proteins were detected with Pierce ECL Western Blotting Substrate, SuperSignal West Femto Maximum Sensitivity Substrate and autoradiography.
Yeast autophagy assays
Yeast strains (BY4741) transformed with autophagy reporter plasmids were grown overnight in synthetic SCCSH medium, refed for 1 h in fresh SCCSH medium at 1 OD600/ml, washed once and switched to low amino-acid medium SCME as described.5 Cells corresponding to 2 OD600 units were collected, lysed for immunoblot analysis.5
Results
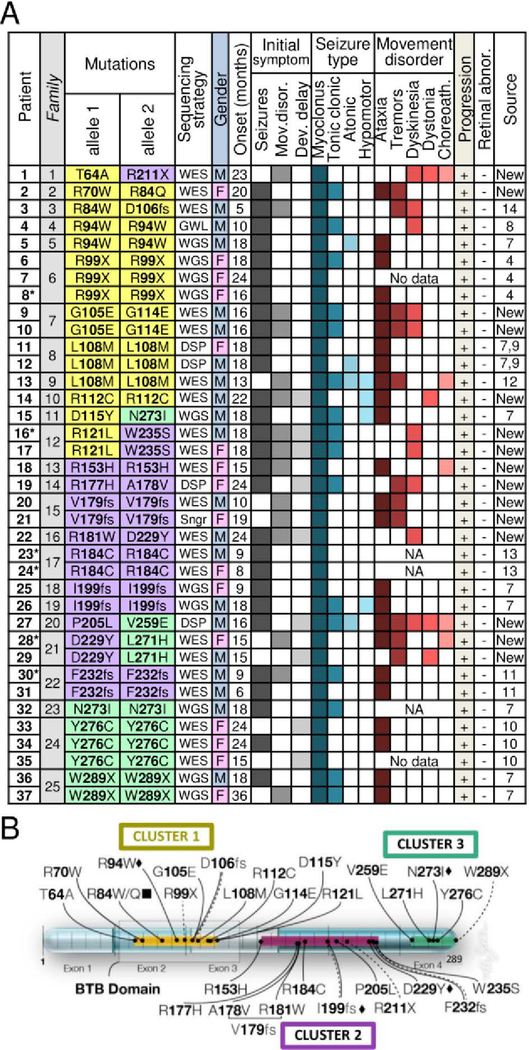
To better define the disorder caused by KCTD7 mutations, we identified 18 novel mutations in 15 patients (11 families) from several sequencing centers and institutions (Fig 1A). A total of 30 novel KCTD7 variants from 37 new and published patients can be grouped into three protein regions, the N-terminal BTB domain, a C-terminal cluster and a less defined middle region (Fig 1B). All 37 patients have homozygous or compound heterozygous variants in KCTD7 (23 missense, 3 stopgain and 4 frameshifts). However, their association with disease does not constitute proof of pathogenicity for each patient variant. To address this point, we first characterized the clinical syndrome.
Fig 1. Genetics and clinical features for all KCTD7 patients.
(A) KCTD7 protein changes and clinical features for 37 new and published patients numbered by KCTD7 variant position and color-coded by protein region as in panel B. Initial DNA sequencing strategies: whole genome (WGS), whole exome (WES), clinical diagnostic sequencing panel (DSP), genome-wide linkage/autozygosity mapping (GWL), Sanger sequencing (Sngr), and the heat map of clinical features for all new patients are derived from deidentified clinical data (Table S1) and from published cases as cited by reference number.4, 7–11, 13, 14 Note, L108M patient 13 was confirmed by the authors to be distinct from patients-11 and −12. Estimated age of onset (months) for males vs. females is not significant, p=0.055 (two-tailed t-test). *Deceased. (B) Map of human KCTD7 isoform-1 (289 amino acids) encoded on 4 color-shaded exons (scale units = 10 residues); 30 patient variants are grouped in color-coded clusters as in panel A. BTB domain (transparent box); missense mutations (solid line), nonsense mutations (dashed), frameshift (double line), mutations occurring in >1 family (diamond), different amino acid mutation at the same position in >1 family (square).
KCTD7 mutations are associated with a progressive neurodegenerative disorder
Overt seizures mark the recorded age of onset for 76% of KCTD7 patients, many with accompanying developmental delays and movement disorders, predominantly ataxia, tremors and dyskinesia (Fig 1A). The remaining 24% first develop movement disorders or developmental delays prior to seizure onset. Genetic testing for KCTD7 variants at earlier ages could potentially identify more patients with movement disorders before seizure onset. Other prominent clinical features include the loss of normal developmental milestones achieved in early childhood, difficulty walking, loss of speech and fine motor skills, and severe cognitive decline. While EEG findings are often positive, brain MRI is typically normal at onset but may detect diffuse or focal brain atrophy after disease progression, for example as observed for new patient-1 (T64A/R211X) but not for new patient-2 (Table S1). Patient-2 underwent a complete corpus callosotomy at age 6.9 years, and was seizure-free for at least 18 months, with some improvement in motor control. All patients progressed to develop myoclonic epilepsy, and all with available data developed movement disorders. Most become wheelchair-bound and non-verbal, and 6 of 37 died at ages 3–18 years (Fig 1A, asterisks). The few ambulatory patients now in their 20’s (patients-9, −10 and −19) have significant motor and cognitive deficits and exhibit autism, obsessive compulsive disorder or schizophrenia, in addition to epilepsy (Table S1).
Early onset without retinal degeneration distinguishes KCTD7 patients
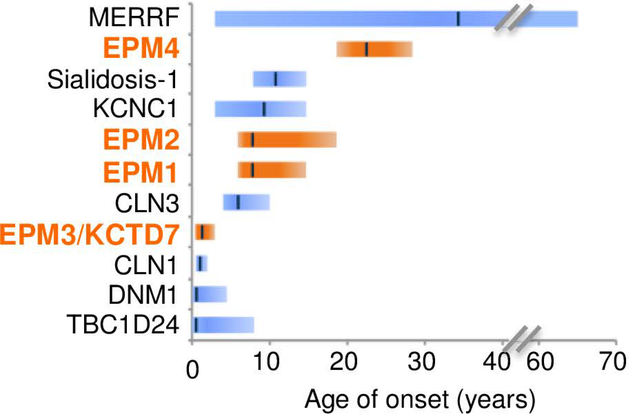
Distinguishing KCTD7/EPM3 patients from related disorders is the early age of onset, consistent with published case reports (Fig 2). Average onset age for all 37 patients is ~17 months, range 5–24 months except one of a sibling pair lacking C-terminal residue W289 with disease onset reported at 36 months (patient-37) (Fig 1A). No gender bias is present for incidence (49% males) although males tended to be diagnosed at a younger age (mean onset 15.1 mo males, 19.1 mo females). This early onset age for KCTD7/EPM3 patients does not overlap with other early onset myoclonic epilepsies (EPM1A, EPM2, EPM4), with the exception of infantile CLN1 (onset 6–24 months) caused by mutations in lysosomal enzyme PPT1 (Fig 2).26, 27 However, KCTD7 patients uniformly lacked the characteristic CLN1-associated retinal abnormalities at onset and associated blindness (Fig 1A).18, 26 These findings shift the age downward for considering the diagnosis of EPM3, which is typically diagnosed in childhood or adolescence.
Fig 2. Earlier disease onset distinguishes KCTD7/EPM3 from other myoclonic epilepsies.
Range (bars) and median (line) age of disease onset for early-onset disorders with myoclonic seizures. TBC1D24 calculated from Supplementary data in Balestrini et al.,29 DNM1,30 CLN1/CLN3,17 KCNC1,28 MERRF,50 EPM1A/EPM2/EPM4,27 sialidosis/mucolipidosis-I https://emedicine.medscape.com/article/948704-overview, and for KCTD7/EPM3 from Fig 1A.
KCTD7 patients are also distinguished from other related disorders. They have more severe cognitive decline and earlier onset than patients with a BTB domain variant in Kv3.1/ KCNC1 (onset age 3–15yr),28 but a later average onset than infantile spasms due to autosomal dominant mutations in DNM1 (onset typically 4–7 months) or autosomal recessive mutations in TBC1D24 (GTPase-activating ARF6-binding protein) that cause pleiotropic neurologic disorders with myoclonic seizures (median onset 2–3 months, often at birth) (Fig 2).29, 30
Prevalence of patient KCTD7 mutations in the general population
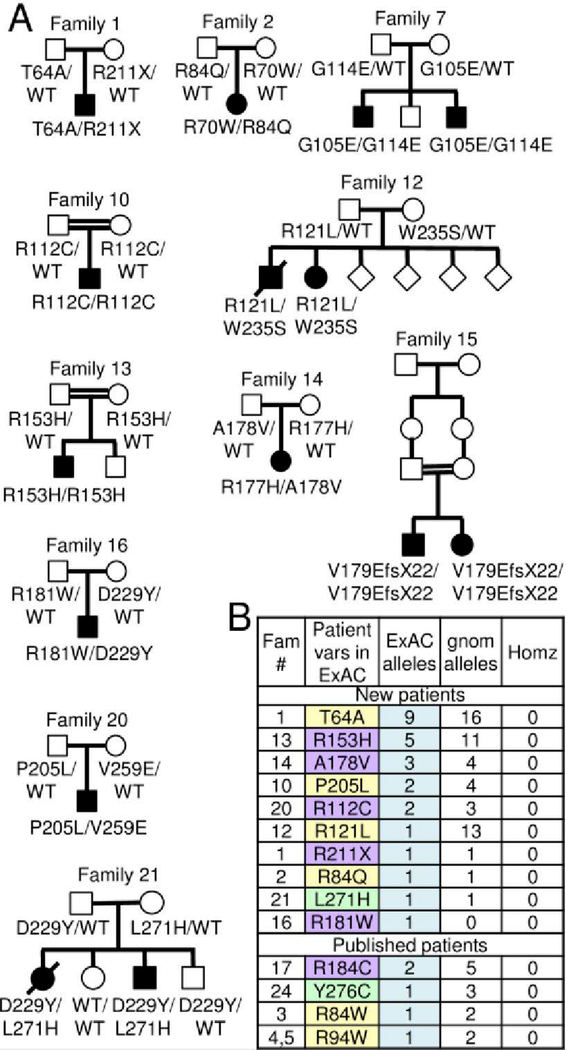
To address disease causality of KCTD7 variants found in new patients, family pedigrees were constructed for the 15 new patients, revealing all variants were inherited (Fig 3A). For all patients (except published patient-15), 57 unaffected family members with available data were either wild type (2 of 10 sequenced siblings) or heterozygous for patient mutations (41 parents, 8 siblings, and 1 grandparent) based on sequence data, and 5 parents based on kinship records of consanguinity (Table S2). The gene damage index GDI-Phred value for KCTD7 (1.235, medium damage prediction) and the selective pressure assessed by the McDonald-Kreitman neutrality index (0.004, moderate purifying)31 are also consistent with a monogenic autosomal recessive disease with complete penetrance. However, cautious causality assignments may still be warranted for specific KCTD7 variants given ~500–20,000 protein-altering variants per individual.32
Fig 3. KCTD7 family pedigrees and mutation frequency.
(A) Pedigrees of 11 new families from Fig 1A with 18 novel KCTD7 variants in 15 bi-allelic patients; birth order unknown for family 12. (B) All patient variants listed in the ExAC32 and gnomAD sequence databases http://gnomad.broadinstitute.org/ for the correct KCTD7 transcript ENST00000275532 (Table S2).
If all 30 KCTD7 variants are disease-causing, each is expected to be rare in the general population. In an effort to catalog normal genetic variation in healthy individuals, the exome aggregation consortium ExAC database of ~60,000 unrelated healthy individuals excluded cancer genomes and cohorts with severe pediatric diseases.32 Thus, most of the KCTD7 patient variants listed at ExAC (10 of 14) are from previously unpublished patients (Table S2). Among all KCTD7 variant alleles identified, T64A (patient-1, T64A/R211X) is the most frequent in ExAC, where it is reported in 9 individuals (0.010% to 0.019% allele frequency in European and African populations, respectively), and in 16 heterozygous individuals in the aggregate gnomAD database of ~130,000 individuals (Fig 3B). However, this frequency is still rare and to date no homozygotes for any amino acid change in KCTD7 has been reported in healthy individuals.32 Some disease variants reported to cause other CLN disorders (e.g. autosomal recessive CLN1 and CLN6) were subsequently challenged because of their prevalence in the ExAC database; for example CLN6 variant R252H.33 However, this CLN6 variant remains a disease candidate as none of the 21 normal individuals with this variant is homozygous.33
KCTD7 patient mutations cause altered protein behavior
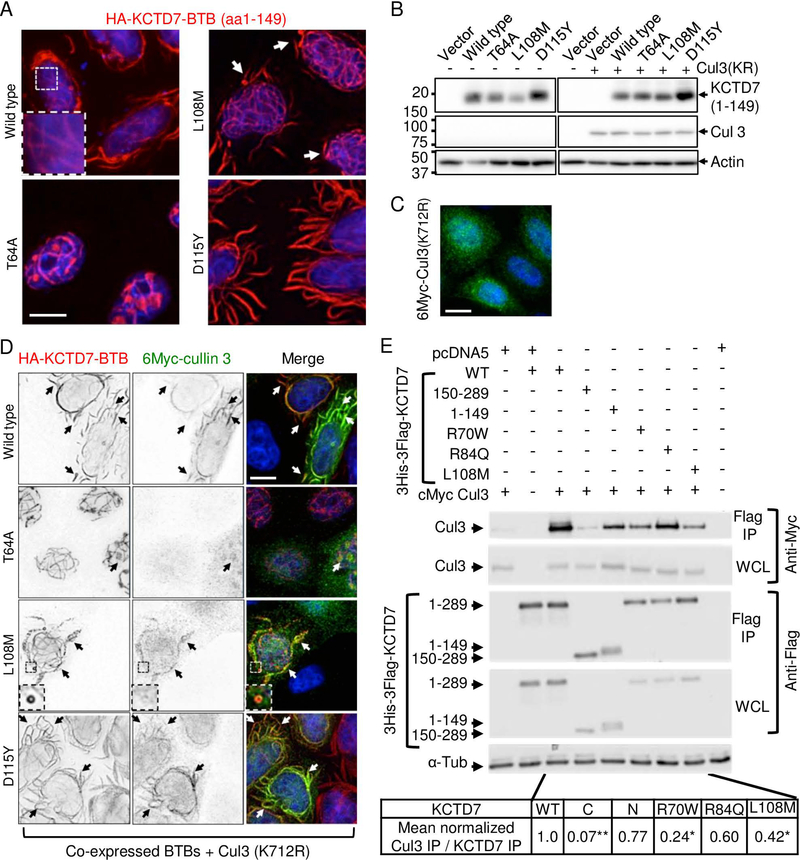
To acquire further evidence that the most prevalent KCTD7 variant T64A (patient-1) is pathogenic, we sought a functional assay. As KCTD7 has no established biochemical activities, we tested for altered subcellular localization as an alternative strategy. The N-terminal amino acids 1–149 of wild type KCTD7 containing the BTB domain, when expressed in HeLa (or other) cells, forms unusual flowing filament-like structures of unknown relevance in the cytoplasm and forms similar smaller structures in the nucleus (Fig 4A). Taking advantage of these elaborate structures to distinguish the effects of patient mutations, we found that expression of T64A(1–149) abolishes these cytoplasmic structures and instead localizes predominantly in a nuclear ball-and-stick pattern (Fig 4A). Two other BTB domain mutations from published patients cause other distinguishable morphologies. L108M (patients 11–13) increases the occurrence of mini-circles at filament termini (Fig 4A, arrows), and D115Y (patient-15, lacking family genetics) causes massive filament-like structures (Fig 4A), which is likely a cause or consequence of protein stabilization (Fig 4B). When co-expressed with CUL3, an E3 ubiquitin ligase component and reported binding partner of KCTD7,13, 16 both wild type and D115Y, and to a lesser extent L108M, were capable of recruiting CUL3 from its more diffuse localization to KCTD7 structures, except the T64A mutant that only rarely co-localizes with CUL3 in fuzzy nuclear spots (Fig 4C and D). Thus T64A, as well as L108M and D115Y are likely to alter KCTD7 function, consistent with functional-effect prediction algorithms PolyPhen2, SIFT and PROVEAN, and L108M and D115Y are predicted damaging by two of these algorithms (Table S2).
Fig 4. Patient mutations in the BTB domain affect cullin 3 (CUL3) interactions.
(A) Immunofluorescence microscopy of N-terminal HA-tagged KCTD7-BTB (amino acids 1–149) in Kyoto HeLa cells transfected 18 h and stained with 1:1000 anti-HA (Santa Cruz HA Y-11) and 1:2000 anti-Myc (Calbiochem Ab-1 OP10L) (similar results without tag and in other cell types tested). Representative of >3 independent experiments. (B) Immunoblots (12% SDS-PAGE, PVDF) of samples described for panels A, C and D probed for anti-HA (1:1000; Santa Cruz Y11), anti-Myc (1:2000; Calbiochem), and anti-actin as loading control (1:10,000; MP Biomedicals 691001), representative of two independent experiments each with both WT and inactive K712R mutant CUL3 yielding similar results. (C) Immunofluorescence microscopy of Kyoto HeLa cells transfected with N-terminal 6Myc-tagged CUL3 (K712R) alone and detected with anti-Myc (similar results for wild type 6Myc-CUL3). (D) Parallel samples to panels B and C co-transfected with HA-KCTD7(1–149) and N-terminal 6Myc-cullin3(K712R) and dual-stained with anti-Myc and anti-HA. Individual gray scale and color merged immunofluorescence microscopy images shown. Representative of >3 independent experiments per condition. Scale bar = 10 μm in all panels. (E) Co-immunoprecipitation of WT 6Myc-Cul3 from HEK293 whole cell lysates (WCL) after transient co-transfection with 3His-3Flag-KCTD7 using anti-Flag M2 affinity matrix for immunoprecipitation (IP). The strength of the interaction between CUL3 and KCTD7 variants was quantified as a ratio of the IP cMyc signal to the IP Flag signal and normalized to the WT KCTD7/CUL3 ratio in the total IP. Representative of 3 independent experiments is shown. *P<0.000001, *P<0.03
Providing further evidence that patient mutations can alter interactions with CUL3, co-immunoprecipitation assays revealed that the N-terminal BTB-containing region but not the C-terminus of KCTD7 is required for CUL3 interaction, consistent with a role for KCTD7 as a CUL3 adaptor. Furthermore, the patient BTB domain mutations tested in full-length KCTD7 (R70W, L108M and likely R84Q) impair binding to CUL3 (Fig 4E).
The only patients predicted to have functionally benign variants by PolyPhen2, SIFT and PROVEAN also have less debilitating disease (sibling patients-9/−10, Table S1). However, their nucleotide change corresponding to G105E (c.314G>A) is located at the exon2-intron junction and is predicted to affect normal splicing (SpliceSiteFinder-like, MaxEntScan, NNSPLICE, GeneSplicer and Human Splicing Finder). Other KCTD7 patient variants have discordant functional predictions between the different algorithms (Table S2), reinforcing that pathogenicity predictions are inherently limited without 3D structures and functional biochemical assays.
KCTD7 heterozygosity in other neurological disorders
We also identified 16 novel heterozygous KCTD7 variants in 18 additional unrelated individuals with phenotypes related to bi-allelic KCTD7EPM3 patients. These heterozygous patients with predominantly unsolved disease etiologies (mean onset age 8.6 years) have neurological phenotypes including developmental delays, seizures, disease progression, movement disorders and/or intellectual disabilities (Table S3). Only one of these 18 variants occurs more frequently than T64A in the population (Y86H occurs in 53 normal heterozygotes).32 Two of the 18 are also found in EPM3 patients with bi-allelic KCTD7 mutations, R121L and R153H. However, any role for heterozygous KCTD7 variants as genetic modifiers is not known.
Conversely, we cannot rule out the possibility that non-KCTD7 variants act as genetic modifiers of more complex traits affecting onset age or other variations between bi-allelic KCTD7 patients. Patient-9 (G105E/G114E) also has a heterozygous pathogenic variant in GALC that is reported in patients with autosomal recessive, late onset neurodegenerative Krabbe disease, and a heterozygous variant of unknown significance (VUS) in ARID1A, a conserved gene responsible for autosomal dominant intellectual disability (Coffin-Siris syndrome). Patient-14 (R112C/R112C) has mutations in three other genes linked to epilepsy, including a homozygous predicted damaging VUS in the glutamate receptor GRIN2A. These and other noted variants in bi-allelic KCTD7 deficient patients (Table S1) are currently not predicted to be disease-related, but only on the basis that all were inherited from one heterozygous parent and therefore insufficient to cause disease.
Evidence for lysosome-pathway defects without characteristic neuronal ceroid lipofuscinosis
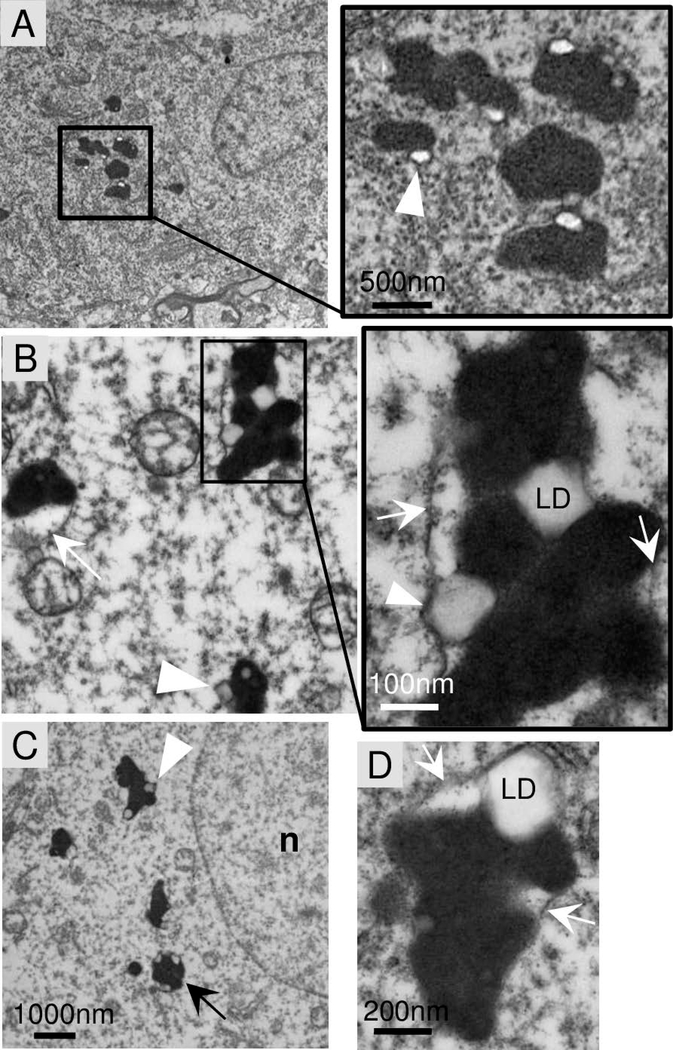
Electron microscopy analysis of a frontal lobe brain biopsy from new patient-1 (T64A/R211X) at age 8 years revealed neuronal pathology (Fig 5A) that was absent in the brain of a neurologically normal 6.7 yr child (not shown). The prominent brain pathology in patient-1 was the electron-dense lipofuscin (lysosomes) (Fig 5A). Similar structures are commonly observed in normal aging brain but not in children. A potential feature of KCTD7 brain lipofuscin is the late autophagosome-like structures (Fig 5B-D, arrows) engorged with both electron-dense lysosomes and electron-lucent structures presumed to be lipid droplets (Fig 5A-D, arrowheads). Soft lipid droplets are compressed between lysosomes or bulging against a delimiting membrane (Fig 5C and D, arrowheads). Others have observed similar structures in autophagy-deficient cells, and in cells overfed with oleic acid.34 The persistence of lipid droplets in brain tissue could potentially reflect an underlying defect related to lipophagy, a form of autophagy that is required for utilization of lipid stores as an energy source and that requires components of the autophagy pathway.35
Fig 5. Abnormal phagolysosomes with lipid droplets in patient brain biopsy.
A-D. Electron micrographs of frontal lobe brain biopsy from patient-1 (T64A/R211X) at age 8 years. Electron-dense lipofuscin (lysosome) structures typically contain electron-lucent lipid droplets (arrowheads) that may be encased within the delimiting membrane (arrow) of phagosome-like structures also containing lysosomes, but no curvilinear, fingerprint inclusions, GROD or other characteristics of neuronal ceroid lipofuscinosis (NCL). Images presented are from two independent preparations.
Bi-allelic KCTD7 mutations that define a diagnosis of EPM3, also define the diagnosis of neuronal ceroid lipofuscinosis 14 (CLN14) based on a study of patient-24 (R184C/R184C).13 That study reported characteristic ultrastructural features of fingerprint-like profiles and granular osmiophilic deposits (GROD) in fibroblasts and neurons from a skin biopsy and in lymphocytes.13, 36 However, we did not observe these or other features considered to be characteristic of neuronal ceroid lipofuscinosis, such as cytosomes, rectilinear profiles (RLP) or curvilinear morphologies described for other CLN subtypes.36 Clinical records indicate that new patient-1 also lacked such ultrastructural features in lymphocytes, skin and rectal biopsy histology, recommended sites for detection of neuronal ceroid lipofuscinosis.36 Although detection can be challenging,36 electron microscopy and/or light microscopy of skin and muscle biopsies from published patients-4, 5, 11, 15, 26, and 30/31,7, 8, 11 as well as new patients-1, 20, 22 and 28 also lack these specific features (Table S1). In addition, no storage material resembling other lysosomal storage disorders was detected in neurons, vascular endothelial or smooth muscle cells of patient-1. Broader diagnostic criteria may be needed to include KCTD7 as a CLN disorder.
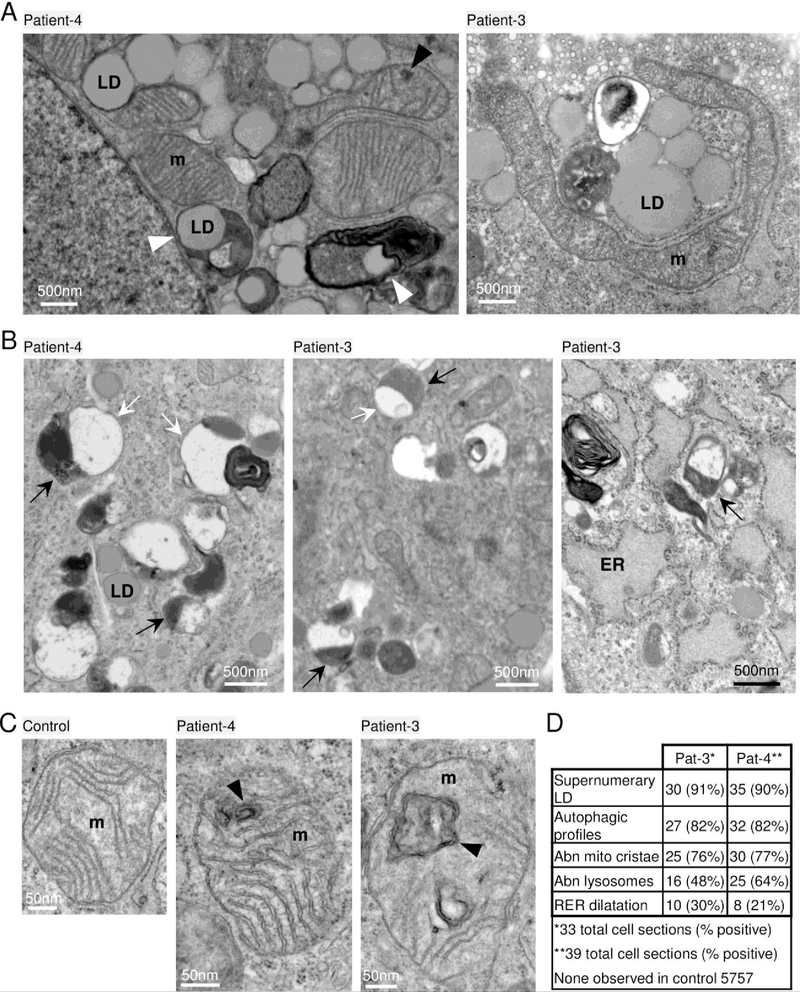
Cultured skin fibroblasts naturally lack some of the lysosomal substrates enriched in the brain such as specific glycosphingolipids, and are therefore not expected to reveal some brain pathologies. However, fibroblast cultures are useful for evaluating other autophagosome-lysosome functions and ultrastructure. In contrast to an age- and passage-matched control, prominent features of early-passage skin-derived fibroblast cultures from patient-3 and patient-4 with BTB domain mutations include the supernumerary lipid droplets often in close proximity to mitochondria and ER (Fig 6A). Some lipid droplets appeared to be engulfed in membrane-bound structures (Fig 6A, arrowheads), reminiscent of the lipid droplets in patient brain, possibly suggesting a stalled catabolic process.
Fig 6. Lipid droplets and abnormal phagolysosomes and mitochondria in patient fibroblasts.
Electron microscopy of low (~6) passage cultured skin fibroblasts taken from patients-3 and −4 at ages 4 years and 6 years, respectively. (A) Supernumerary lipid droplets (LD) near mitochondria (m) common to both patients unlike age/passage-matched controls not depicted. LDs engulfed in membrane-bound structures (white arrowheads); normal caveolae pits and caveolae vesicles typical of normal healthy fibroblasts (patient-3, top). (B) Abnormal membrane-bound lysosome structures (black arrows) associated with larger single or partial double membrane phagosome-like compartments sparsely filled with lightly-stained undegraded material (white arrows) and occasional electron-dense lipid whirls (example in left and right panels); lipid droplets (LD) detected in all 3 panels; swollen endoplasmic reticulum (ER, right panel). (C) Age- and passage-matched control fibroblasts 5757 (left panel) lack abnormal mitochondria (m) containing double membrane structures (black arrowheads) common to both patient fibroblasts. (D) Quantification of abnormal morphologies presented as the percent of randomly selected cells (n=33, patient-3) and (n=39, patient-4), and ~100 fields of control 5757. Cells with supernumerary LDs typically contain over 100 LDs often close to mitochondria.
Other prominent features that distinguish both patient fibroblasts from age- and passage-matched control fibroblasts include hybrid structures resembling late single-membrane or partially double-membrane autolysosomes.37 These hybrid structures contain local electron-densities typical of lysosomes (Fig 6B, black arrows) that are associated with larger autophagosome-like structures containing residual undegraded material (Fig 6B, white arrows) and occasional electron dense lipid whirls (Fig 6B). Similar hybrid structures are also characteristics of diverse neurodegenerative disorders including mucolipidosis type IV, Alzheimer’s disease, CLN types and autophagy-deficient cell lines.37–39
Although mitochondrial organelles were not sufficiently preserved in patient biopsy preparations, most cultured fibroblasts from both patients contained a fraction of mitochondria with internal, closed double membrane structures presumed to reflect cristae malformations (Fig 6C). These malformations are somewhat reminiscent of mitochondria in mitofilin-deficient cells with concentric cristae.40 Ultrastructural abnormalities were prevalent in patient cells but absent in control cells (Fig 6D). The apparent accumulation of several abnormal organelles in patient-derived fibroblasts and in a brain biopsy is consistent with a defect in the phagolysosome pathway.
Conserved autophagy defects as a potential mechanism of disease pathogenesis
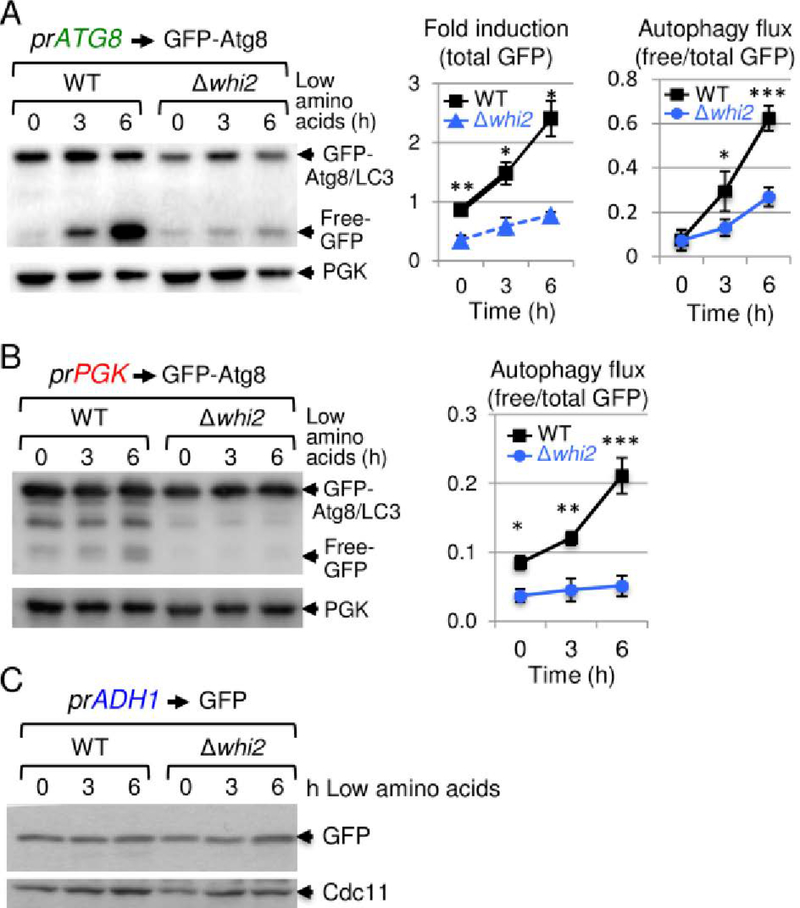
We initiated this KCTD7 project based on insights gained from studying the poorly characterized yeast protein Whi2, which shares sequence similarity with KCTD7 and harbors a homologous BTB structural domain.5 Yeast lacking WHI2 are sensitive to multiple cell stresses and fail to halt the cell cycle in response to low amino acid levels in the media, conditions known to induce autophagy.5, 20, 23 Taken together with our ultrastructural findings in patient samples, we asked if whi2-deficient yeast have a general autophagy defect using an established reporter for yeast Atg8,41 homolog of mammalian autophagy marker LC3. In low amino acid medium, the autophagy-responsive ATG8 promoter is induced to express the reporter, and during autophagy flux undergoes lysosomal/vacuolar processing that cleaves the protease-sensitive Atg8 moiety from the more stable GFP protein in wild type cells.41 In striking contrast to wild type, whi2-deletion strains are profoundly defective for autophagy induction and flux (Fig 7A). Similar but less dramatic results were obtained with an autophagy flux-specific reporter expressed by the constitutive PGK promoter, indicating that Whi2 is required for normal autophagy induction and flux after switching to low amino acids (Fig 7B). This autophagy defect was not due to a general defect in reporter expression or global protein translation, as free GFP expressed via an autophagy-independent promoter (ADH1) was expressed indistinguishably in wild type and whi2-deficient yeast (Fig 7C).
Fig 7. Defective autophagy in whi2-deficient yeast.
(A) Immunoblots of wild type and whi2 knockout strains of Saccharomyces cerevisiae (BY4741) expressing autophagy reporter fusion protein GFP-Atg8 expressed by autophagy-responsive yeast ATG8/LC3 promoter (pr), before and after switching from high to low amino acid medium as described,5, 23 probed with 1:1000 anti-GFP (Santa Cruz sc-9996), loading control anti-Pgk (Abcam 113687), and HRP-conjugated secondary antibodies (GE Healthcare, 1:20,000). Induction of the ATG8 promoter (total GFP in upper+lower bands relative to PGK loading control) and autophagy flux (protease-resistant free GFP liberated by vacuolar protease-dependent digestion of the Atg8 moiety relative to total GFP) are presented in separate graphs as mean +/−SD for 3 independent experiments with/without switching to low amino acids. (B) Experiment and analysis as described for panel A except using the constitutive PGK promoter (pr) to express the GFP-Atg8 autophagy reporter. For panels A and B, results are presented as mean+/−SD for 3 independent experiments. Two-tailed t-test: WT vs. Δwhi2, *p<0.05, **p<0.005, ***p<0.001). (C) As described in panel A except using the transcription/translation control GFP reporter (no Atg8) expressed by the constitutive ADH1 promoter, probed with anti-GFP and loading control anti-Cdc11 (Novus Biologicals NB100–81019).
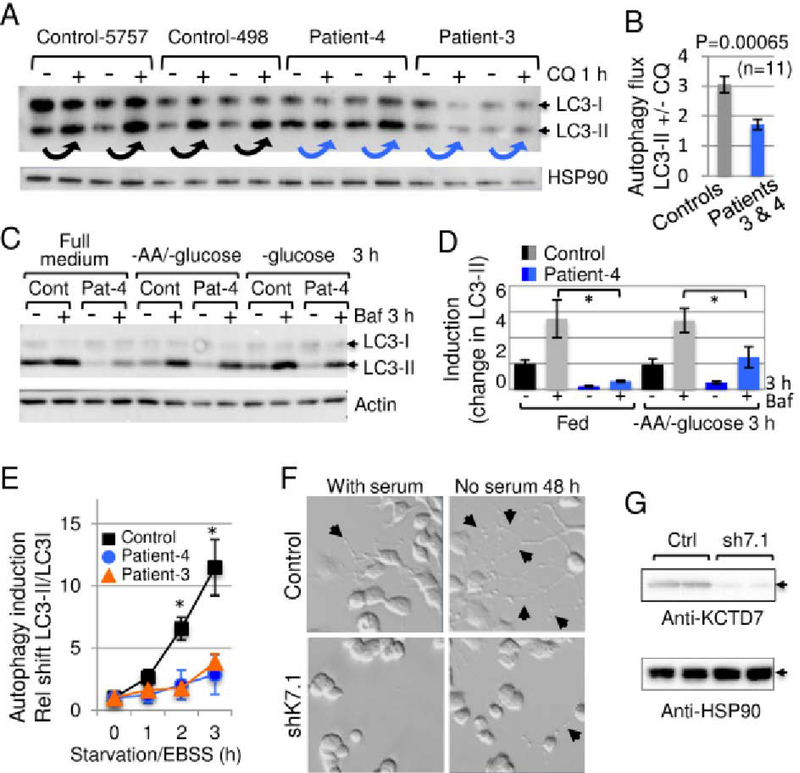
To extend these findings from yeast, the skin-derived fibroblast lines from patients-3 and −4 with BTB domain mutations were analyzed for autophagy defects. The accumulation of endogenous lipidated/mature LC3-II following treatment with chloroquine to inhibit lysosome function that otherwise degrades LC3-II.42 Consistently, LC3-II accumulated less efficiently in low-passage patient fibroblasts versus age- and passage-matched controls following chloroquine treatment to assess basal autophagy flux (Fig 8A and B), and when autophagy was induced by withdrawing amino acids and glucose (Fig 8B and C). Autophagy induced by more severe starvation was also significantly lower in patient fibroblasts assessed by conversion of LC3I to LC3II (Fig 8E). Thus, both WHI2-deficient yeast and patient cells have a defect in autophagy based on Atg8/LC3 assays following nutrient depletion. To confirm the origins of our fibroblast cell lines, genomic DNA isolated from patient and control cells used in these studies was sequenced, revealing the expected variants for patients-3 and −4.
Fig 8. Conservation of autophagy defect in KCTD7 patient fibroblasts.
(A) Immunoblot of control age-/passage-matched human fibroblasts and fibroblasts derived from patient-3 and −4 (passage 6–8) for endogenous autophagy protein LC3 (1:1000 anti-LC3, Cell Signaling 2775) and HSP90 loading control (BD Biosciences 610419) detected with HRP-conjugated secondary antibodies (GE Healthcare, 1:20,000), developed using ECL-Prime (GE Healthcare). Compare LC3-II levels before versus after 1 h with 15 μM chloroquine (Sigma C6628) or carrier control to assess basal autophagy (arrows). (B) Quantified data for panel A, pooling 5757 and 498 control cells versus patient cells from 3 independent experiments, each performed in duplicate or triplicate for each of the 4 cell lines (n=11 per condition) and presented as the ratio of LC3-II levels +/−15 μM chloroquine after correcting for loading; mean+/−SE, 2-tailed t-test, p = 0.00065. (C) Autophagy assay as in panel A except cells were treated 3 h +/−glucose, +/−amino acids, +/−100 nM Bafilomycin A1 (Enzo). (D) Quantification for panel C presented as mean+/−SE of LC3-II levels adjusted to relative loading controls and calculated as the fold change over control 5757 in full medium for 6–8 replicates in 4 independent experiments. Two-tailed t-test, *p<0.05. (E) Autophagy assay as in panel A except cells were incubated in amino acid-free/serum-free Earle’s balanced salt solution (Thermo Fisher). The change in LC3-II/LC3-I ratios relative to time 0 is presented as mean+/−SE for 3 independent experiments; 2-tailed t-test for control-5757 vs. patient-3 at 2 h and 3 h (p=0.002 and p=0.018), and patient-4 (p=0.023 and p=0.020, respectively). (F) Light microscopy (Nikon TE200) of control and mouse Kctd7-specific shK7.1 shRNA knockdown (TRCN0000069304 transfected with FuGENE or Lipofectamine2000) in N2a mouse neuroblastoma cells refed +/−serum 48 h to monitor neurite outgrowths (arrows). (G) Immunoblot confirmation of endogenous Kctd7 knockdown by shK7.1 with 1:1000 anti-KCTD7 (Abcam ab83237) and anti-HSP90 (BD Biosciences 610419) loading control. Lysates and immunoblots were prepared as described for panel A, except visualized using Amersham Hyperfilm ECL (GE Healthcare).
The functional consequences of autophagy deficiency were assessed by determining the effects of KCTD7 on neurite outgrowth triggered by serum withdrawal in mouse neuroblastoma N2a cells. The maturation of N2a cells to produce extensive neuron-like processes requires autophagy, as knockdown of conserved Beclin 1/Atg6 blocks neurite extension.43 We found that partial knockdown of endogenous mouse Kctd7 severely reduces neurite outgrowth triggered by serum withdrawal, indicating a critical role for KCTD7 in neurite maturation (Fig 8F and G).
Discussion
Manifestations of KCTD7 mutations
Guided by our findings in yeast, we investigated the clinical and cellular consequences of KCTD7 mutations. Bi-allelic KCTD7 mutations cause an early onset (16.8±6 months) progressive myoclonic epilepsy previously ascribed only to older children.17, 18 These patients are characterized by movement disorders and developmental delays that may precede onset of intractable myoclonic seizures more often than appreciated based on recorded parental comments. The few ambulatory patients exhibit severe cognitive and psychosocial impairments. All patients exhibit disease progression indicative of an underlying degenerative process despite apparently normal initial developmental milestones. Ultrastructural analysis of a brain biopsy or skin fibroblasts from a total of three different patients revealed shared features, most notably the accumulation of lipid droplets and abnormal phagolysosomes containing undegraded material. Misshapen mitochondrial cristae membranes were also prominent features of skin fibroblasts where sample preparation is more amenable to organelle preservation (Figs 5 and 6). The constituents of lipofuscin granules in patient brain appear to be residual bodies derived from lysosomes and could potentially be produced by a partial degradation of unsaturated lipids. Brain biopsies for future patients may provide more valuable information. The ultrastructural features in patient samples together with defective autophagy responses in patient fibroblasts and the corresponding yeast deletion strain are consistent with the possibility that an autophagy-lysosome pathway defect underlies the disease caused by bi-allelic KCTD7 mutations. Although lysosome pathway defects are implicated in a growing number of neurological disorders, each can manifest differently, presumably owing to the molecular details not yet known. New therapies will be needed that enhance autophagosome-lysosome function without worsening defective bottlenecks downstream in the pathway.
Two reported phenotypes for KCTD7/EPM3 patients were not found in the new patient cohort reported here. Clonic eye movements reported for patient-3 diagnosed with opsoclonus-myoclonus syndrome (OMS)14 were not detected though not formally tested. We also did not detect specific neuronal ceroid lipofuscinosis pathology reported for patient-24, the founder case for CLN14 designation, although this patient potentially had more severe disease (onset 8 mo, died 17 yr).13
Causality of KCTD7 variants
The low frequency in the general population of each of the 30 unique KCTD7 patient variants, including T64A, and the lack of homozygous patient variants in healthy individuals,32 indicate that these 30 variants are causal for EPM3/CLN14. This disorder occurs worldwide and the patients in this study have diverse ancestry (e.g. Moroccan, Syrian Sephardi, European, Native American, French Canadian). Genome sequence data from the general population suggest at least 0.05% of healthy unrelated individuals may carry a heterozygous pathogenic KCTD7 variant.32 The relatively higher than average arginine content of the KCTD7 protein (7.3% vs. 4–6%) may contribute to the mutation frequency (12 of 30 variants change an Arg, Table S1) given that arginine codons have the highest proportion of CpG sequences and CpG mutations (affecting DNA methylation) are by far the most prevalent in the population.32
Additional rare heterozygous KCTD7 variants were identified in 17 additional patients with related yet clinically distinct disorders (Table S3), although any contribution to disease is unknown. However, when considering the genetic complexities of neurobehavioral disorders such as autism, other genetic modifiers with discernable clinical phenotypes may exist. KCTD7 sibling patients-9/10 were diagnosed with autism. The KCTD7-related KCTD13 gene at 16p11.2 is thought to have a role in a small subset of autism cases.1 Any manifestations of heterozygous KCTD7 mutations in late onset disorders analogous to heterozygous loss of progranulin in older adults with frontotemporal dementia/FTD versus bi-allelic mutations that cause lipofuscinosis CLN1144 are unexplored. Based on yeast studies that first identified the KCTD-like yeast Whi2,5 KCTD7 variants could potentially compensate for more deleterious mutations. Spontaneous mutations in the analogous gene WHI2 can compensate in part for the more detrimental lack of mitochondrial fission factor Fis1 or several other genes.5, 23
Biological and biochemical roles for KCTD7
The biochemical function of KCTD7 is not known and little is understood about the other 24 human family members (KCTD1–21, TNFAIP1, KCNRG, SHKBP1 and BTBD10). Inspired by our finding that yeast Whi2 is required to suppress cell growth in low amino acid conditions,5, 23 it was reasonable to consider that whi2-deficient yeast were also defective for autophagy induction. Therefore, we tested for an evolutionarily conserved function between Whi2 and KCTD7 in autophagy. We found that both yeast Whi2 and KCTD7 are required for normal basal autophagy and low nutrient-induced autophagy (Figs 7 and 8). Both yeast Whi2 and the KCTD7 homolog, KCTD11, were recently shown to suppress TORC1/mTORC1, a known inhibitor of autophagy.45 The same study failed to detect an effect of KCTD7 on TORC1/mTORC1 activity in yeast and primate COS7 cells. However, a role for KCTD7 in autophagy-lysosome function is consistent with our ultrastructural studies revealing abnormal autophagosome-lysosome structures, mitochondrial cristae and supernumerary lipid droplets potentially reflecting impaired lipophagy (Fig 5 and 6). Related pathologies are observed with aging-related lysosomal dysfunction and progressive decline in chaperone-mediated autophagy rates in late-onset Alzheimer disease,37 perpetuated by oxidation of partially degraded macromolecules derived from mitochondria, glycosphingolipids and other components in autolysosomes resulting in reactive oxygen species that interact with lysosomal iron.46
Consistent with our finding of abnormal mitochondrial cristae in patient-derived cells, one study investigated the role of yeast Whi2 in mitophagy (a subtype of autophagy). They reported that the spontaneous WHI2 mutation in FIS1 knockout strains, rather than loss of the FIS1 mitochondrial fission gene, causes a defect in the degradation of yeast mitochondrial organelles.21 However, a subsequent study challenged this conclusion, reporting that mitochondrial fission mediated by FIS1 rather than WHI2 is required for normal mitophagy.22 Thus, the question remains open.
A possible role for KCTD7 in protein turnover is consistent with having an N-terminal BTB domain, where almost half of the patient mutations identified thus far occur (Fig 1B), as BTB proteins can serve as adaptor proteins that retrieve substrates for the CUL3 ubiquitin ligase complex. 47 Interesting, several other KCTD family members were identified in screens for cullin-ARIH1 complex components,48 consistent with serving as an E3 ubiquitin ligase adaptor. CUL3 has many critical roles in cells, and a role for CUL3 in autophagy has gained recent attention. CUL3 and its BTB-Kelch adaptor protein KLHL20 were found to prevent overzealous autophagy by direct ubiquitination and degradation of ULK1, a key upstream positive regulator of autophagy induction.49 Although KCTD family proteins have not been demonstrated to have a molecular role in autophagy, defective autophagy-lysosome pathways are consistent with their causal roles in neurodegenerative disorders.
Supplementary Material
Supporting clinical data
KCTD7/EPM3 genetic data
KCTD7heterozygous variant data
Acknowledgements
We thank Drs. Rachel Kneen and Lorraine Potocki for clinical information, Drs. Constantin D’Ydewalle and Charlotte Sumner for assistance with qRT-PCR on patient fibroblasts, medical illustrator Heidi Hildebrandt for the mutation map (Fig 1B), and Drs. Jodi Nunnari and Joseph Heitman for kindly providing the autophagy plasmids prATG8-GFP-ATG8 and prADH1-GFP, respectively. This work was supported by NIH R01 NS083373 (JMH), NIH R01 NS037402 (JMH), CURE Epilepsy Foundation (JMH), NIH K08 NS070931 (ALH), NIH R01 GM089778 (JAW), National Natural Science Foundation of China 31401197 (XT), Natural Science Foundation of Jiangsu Province BK20140318 (XT), and Jiangsu Key Laboratory of Neuropsychiatric Diseases BM2013003 (XT). This work was performed while ALH was a full-time employee of Johns Hopkins University.
Footnotes
Potential Conflicts of Interest: The authors have declared that no conflict of interest exists.
References
- 1.Escamilla CO, Filonova I, Walker AK, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551:227–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mencacci NE, Rubio-Agusti I, Zdebik A, et al. A missense mutation in KCTD17 causes autosomal dominant myoclonus-dystonia. Am J Hum Genet. 2015;96:938–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Bogaert P, Azizieh R, Desir J, et al. Mutation of a potassium channel-related gene in progressive myoclonic epilepsy. Ann Neurol. 2007;61:579–86 [DOI] [PubMed] [Google Scholar]
- 5.Teng X, Dayhoff-Brannigan M, Cheng WC, et al. Genome-wide consequences of deleting any single gene. Mol Cell. 2013;52:485–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyrer J, Maljevic S, Scheffer IE, Berkovic SF, Petrou S, Reid CA. Ion Channels in Genetic Epilepsy: From Genes and Mechanisms to Disease-Targeted Therapies. Pharmacol Rev. 2018;70:142–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kousi M, Anttila V, Schulz A, et al. Novel mutations consolidate KCTD7 as a progressive myoclonus epilepsy gene. J Med Genet. 2012;49:391–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krabichler B, Rostasy K, Baumann M, et al. Novel Mutation in Potassium Channel related Gene KCTD7 and Progressive Myoclonic Epilepsy. Ann Hum Genet. 2012;76:326–31 [DOI] [PubMed] [Google Scholar]
- 9.Lemke JR, Riesch E, Scheurenbrand T, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. 2012;53:1387–98 [DOI] [PubMed] [Google Scholar]
- 10.Farhan SM, Murphy LM, Robinson JF, et al. Linkage analysis and exome sequencing identify a novel mutation in KCTD7 in patients with progressive myoclonus epilepsy with ataxia. Epilepsia. 2014;55:e106–11 [DOI] [PubMed] [Google Scholar]
- 11.Moen MN, Fjaer R, Hamdani EH, et al. Pathogenic variants in KCTD7 perturb neuronal K+ fluxes and glutamine transport. Brain. 2016;139:3109–20 [DOI] [PubMed] [Google Scholar]
- 12.Seaby EG, Gilbert RD, Pengelly RJ, Andreoletti G, Clarke A, Ennis S. Progressive myoclonic epilepsy with Fanconi syndrome. JRSM Open. 2016;7:2054270415623145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staropoli JF, Karaa A, Lim ET, et al. A Homozygous Mutation in KCTD7 Links Neuronal Ceroid Lipofuscinosis to the Ubiquitin-Proteasome System. Am J Hum Genet. 2012;91:202–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blumkin L, Kivity S, Lev D, et al. A compound heterozygous missense mutation and a large deletion in the KCTD7 gene presenting as an opsoclonus-myoclonus ataxia-like syndrome. J Neurol. 2012;259:2590–8 [DOI] [PubMed] [Google Scholar]
- 15.Pinkas DM, Sanvitale CE, Bufton JC, et al. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem J. 2017;474:3747–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azizieh R, Orduz D, Van Bogaert P, et al. Progressive myoclonic epilepsy-associated gene KCTD7 is a regulator of potassium conductance in neurons. Mol Neurobiol. 2011;44:111–21 [DOI] [PubMed] [Google Scholar]
- 17.Mole SE, Cotman SL. Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim Biophys Acta. 2015;1852:2237–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carcel-Trullols J, Kovacs AD, Pearce DA. Cell biology of the NCL proteins: What they do and don’t do. Biochim Biophys Acta. 2015;1852:2242–55 [DOI] [PubMed] [Google Scholar]
- 19.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–97 [DOI] [PubMed] [Google Scholar]
- 20.Radcliffe P, Trevethick J, Tyers M, Sudbery P. Deregulation of CLN1 and CLN2 in the Saccharomyces cerevisiae whi2 mutant. Yeast. 1997;13:707–15 [DOI] [PubMed] [Google Scholar]
- 21.Mendl N, Occhipinti A, Muller M, Wild P, Dikic I, Reichert AS. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci. 2011;124:1339–50 [DOI] [PubMed] [Google Scholar]
- 22.Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, Hardwick JM. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ. 2008;15:1838–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017;36:397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coppens I, Joiner KA. Host but not parasite cholesterol controls Toxoplasma cell entry by modulating organelle discharge. Mol Biol Cell. 2003;14:3804–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RE, Aberg L, Autti T, Goebel HH, Kohlschutter A, Lonnqvist T. Diagnosis of the neuronal ceroid lipofuscinoses: an update. Biochim Biophys Acta. 2006;1762:865–72 [DOI] [PubMed] [Google Scholar]
- 27.Franceschetti S, Michelucci R, Canafoglia L, et al. Progressive myoclonic epilepsies: definitive and still undetermined causes. Neurology. 2014;82:405–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver KL, Franceschetti S, Milligan CJ, et al. Myoclonus epilepsy and ataxia due to KCNC1 mutation: Analysis of 20 cases and K+ channel properties. Ann Neurol. 2017;81:677–89 [DOI] [PubMed] [Google Scholar]
- 29.Balestrini S, Milh M, Castiglioni C, et al. TBC1D24 genotype-phenotype correlation: Epilepsies and other neurologic features. Neurology. 2016;87:77–85 (reanalysis of supplemental table) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Spiczak S, Helbig KL, Shinde DN, et al. DNM1 encephalopathy: A new disease of vesicle fission. Neurology. 2017;89:385–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itan Y, Shang L, Boisson B, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112:13615–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleat DE, Gedvilaite E, Zhang Y, Lobel P, Xing J. Analysis of large-scale whole exome sequencing data to determine the prevalence of genetically-distinct forms of neuronal ceroid lipofuscinosis. Gene. 2016;593:284–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JM, Wagner M, Xiao R, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo AY, Lau PW, Feliciano D, et al. AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife. 2017;6:e21690 DOI: 10.7554/eLife. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson GW, Goebel HH, Simonati A. Human pathology in NCL. Biochim Biophys Acta. 2013;1832:1807–26 [DOI] [PubMed] [Google Scholar]
- 37.Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–22 [DOI] [PubMed] [Google Scholar]
- 38.Vergarajauregui S, Connelly PS, Daniels MP, Puertollano R. Autophagic dysfunction in mucolipidosis type IV patients. Hum Mol Genet. 2008;17:2723–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23:896–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.John GB, Shang Y, Li L, et al. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 2011;30:2101–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5 [DOI] [PubMed] [Google Scholar]
- 43.Zeng M, Zhou JN. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell Signal. 2008;20:659–65 [DOI] [PubMed] [Google Scholar]
- 44.Ward ME, Chen R, Huang HY, et al. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med. 2017;9:pii: eaah5642. doi: 10.1126/scitranslmed.aah5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen X, Wang G, Zhang Y, et al. Whi2 is a conserved negative regulator of TORC1 in response to low amino acids. PLOS Genetics. 2018;in press:available online at http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurz T, Eaton JW, Brunk UT. Redox activity within the lysosomal compartment: implications for aging and apoptosis. Antioxid Redox Signal. 2010;13:511–23 [DOI] [PubMed] [Google Scholar]
- 47.Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott DC, Rhee DY, Duda DM, et al. Two Distinct Types of E3 Ligases Work in Unison to Regulate Substrate Ubiquitylation. Cell. 2016;166:1198–214 e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu CC, Lin YC, Chen YH, et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol Cell. 2016;61:84–97 [DOI] [PubMed] [Google Scholar]
- 50.Mancuso M, Orsucci D, Angelini C, et al. Phenotypic heterogeneity of the 8344A>G mtDNA “MERRF” mutation. Neurology. 2013;80:2049–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting clinical data
KCTD7/EPM3 genetic data
KCTD7heterozygous variant data