Abstract
Antibiotics, once considered the lifeline for treating bacterial infections, are under threat due to the emergence of threatening antimicrobial resistance (AMR). These drug-resistant microbes (or superbugs) are non-responsive to most of the commonly used antibiotics leaving us with few treatment options and escalating mortality-rates and treatment costs. The problem is further aggravated by the drying-pipeline of new and potent antibiotics effective particularly against the drug-resistant strains. Multidrug efflux pumps (EPs) are established as principal determinants of AMR, extruding multiple antibiotics out of the cell, mostly in non-specific manner and have therefore emerged as potent drug-targets for combating AMR. Plants being the reservoir of bioactive compounds can serve as a source of potent EP inhibitors (EPIs). The phyto-therapeutics with noteworthy drug-resistance-reversal or re-sensitizing activities may prove significant for reviving the otherwise fading antibiotics arsenal and making this combination-therapy effective. Contemporary attempts to potentiate the antibiotics with plant extracts and pure phytomolecules have gained momentum though with relatively less success against Gram-negative bacteria. Plant-based EPIs hold promise as potent drug-leads to combat the EPI-mediated AMR. This review presents an account of major bacterial multidrug EPs, their roles in imparting AMR, effective strategies for inhibiting drug EPs with phytomolecules, and current account of research on developing novel and potent plant-based EPIs for reversing their AMR characteristics. Recent developments including emergence of in silico tools, major success stories, challenges and future prospects are also discussed.
Keywords: antimicrobial resistance, efflux pumps, efflux pump inhibitors, phyto-therapeutics, drug resistance reversal
Introduction
Antimicrobial resistance (AMR) or ineffectiveness of commonly used drugs/antibiotics against specific bacteria has emerged as one of the most threatening human health concerns and a major challenge for global drug discovery programs. AMR (also known as drug resistance) has been reported at three increasing levels, multidrug resistance (MDR), extensive drug resistance (XDR) and pan-drug resistance (PDR). By definition, MDR stands for acquisition of non-susceptibility to at least one agent in three or more antimicrobial classes, while XDR shows the non-susceptibility to at least one agent in all, except two or fewer antimicrobial classes, while PDR implies non-susceptibility to all antimicrobial agents from all available classes (Exner et al., 2017; Spengler et al., 2017). AMR is threatening millions of lives worldwide, and is rightly declared as a global risk by the World Economic Forum (World Economic Forum, 2013). Since the very first report on AMR in Enterobacteria in 1950s (Watanabe, 1963; Levy, 2001), many drug-resistant strains have been reported and their number as well as the resistance level is on the rise. Though several classes of antibiotics were discovered in the antibiotic era (Table 1), we are heading to a post-antibiotic era, where an increasing number of previously curable infections are turning into non-curable and life-threatening (Spengler et al., 2017). Though development of AMR or antibiotic resistance is a natural phenomenon, irrational use of antibiotics speed-ups the emergence of drug-resistant strains (World Health Organization, 2014). Once the AMR is gained by the bacteria, it is successively transmitted to the next progeny via vertical gene transfer or other bacteria through horizontal gene transfer process, making their treatment more difficult (Chandra et al., 2017).
Table 1.
Classes of commonly used antibiotics along with their examples and corresponding modes of action.
| Class of antibiotics | Examples | Mode of action | |||
|---|---|---|---|---|---|
| Beta Lactams | Beta lactamase inhibitors | Sulbactam, Tazobactam, Clavulanic acid, Avibactam | Cell wall synthesis inhibitors | ||
| Penicillins | Penicillinase sensible | Aminopenicillins | Ampicillin, Amoxicillin | ||
| Natural penicillins | Penicillin G, Penicillin VK | ||||
| Penicillinase resistant | Nafcillin, Dicloxacillin, Oxacillin | ||||
| Anti-pseudomonal | Carboxypenicillins | Ticarcillin, Carbenicillin | |||
| Ureidopenicillins | Piperacillin, Mezlocillin, Azlocillin | ||||
| Cephalo-sporins | 1st generation | Cephalexine, Cefadroxil, Cefazolin, Cephradrine | |||
| 2nd generation | Cefuroxime, Cefoxitin, Cefaclor, Loracarbef, Cefprozil | ||||
| 3rd generation | Cefoperazone, Cefpodoxime, Ceftriaxone, Cefotaxime, Ceftazidime | ||||
| 4th generation | Cefepime, Cefpirome | ||||
| 5th generation | Ceftaroline, Ceftolozane | ||||
| Carbapenems | Meropenem, Doripenem, Ertapenem, Imipenem | ||||
| Monobactams | Aztreonam | ||||
| No lactams | Glycopeptides | Vancomycin, Dalbavancin, Telavancin, Oritavancin | |||
| Other | Colistin, Daptomycin, Polymixin B, Isoniazid | ||||
| Amino-glycosides | Amikacin, Streptomycin, Neomycin, Gentamicin, Tobramycin | Protein synthesis inhibitors | |||
| Tetracyclins | Doxycyclin, Tetracyclin, Democlocyclin, Minocyclin, Tigecyclin | ||||
| Oxazolidonones | Linezolid, Tidezolid | ||||
| Streptogramins | Quinupristin | ||||
| Chloramphenicol | |||||
| Macrolides | Erythromycin, Clarithromycin, Azithromycin | ||||
| Lincosamides | Clindamycin, Lincomycin | ||||
| Fluorquinolones | Ciprofloxacin, Sparfloxacin, Levofloxacin, , Norfloxacin | DNA | |||
| Quinolones | Nalidixic acid | topoisomerases inhibitors | |||
| Sulfonamides | Sulfamethoxazole, Sulfasalazine, Ag sulfadiazine, Sulfisoxazole | Folic acid synthesis inhibitor | |||
| DHFR inhibitors | Trimethoprim, Pyrimethamine | ||||
| Nitroimidazoles | Metronidazole, Tinidazole | DNA Damage | |||
| Rifampin | mRNA synthesis | ||||
The drug resistance characteristics may be attributed to the abilities of such strains in fast altering their genetic make-up or inducing epigenetic changes (Davies and Davies, 2010; Motta et al., 2015; Rahman et al., 2017). Necessary adaptations are achieved by bacteria to-respond-to and to counteract the antibiotics either via procurement of foreign genetic material encoding resistance via horizontal gene transfer or mutations in drug-targets / antibiotics-degrading enzymes and alterations in permeability of the outer bacterial membrane. There is an unprecedented upsurge in bacterial strains with elevated AMR in both Gram-negative and Gram-positive phenotypes. The ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) pathogens have emerged with high degree of AMR and are major cause of life-threatening nosocomial infections (Santajit and Indrawattana, 2016). Even other strains like Escherichia coli, Shigella species, Neisseria gonorrhoeae, and Proteus mirabilis have shown significant levels of AMR (Fair and Tor, 2014; Prasch and Bucar, 2015; Cerceo et al., 2016). Development of resistance against the carbapenem, a class of highly effective antibiotics and regarded as the last line of defense against pathogenic Gram-negative bacteria hints at the alarming situation (Kumarasamy et al., 2010; Dwivedi et al., 2015).
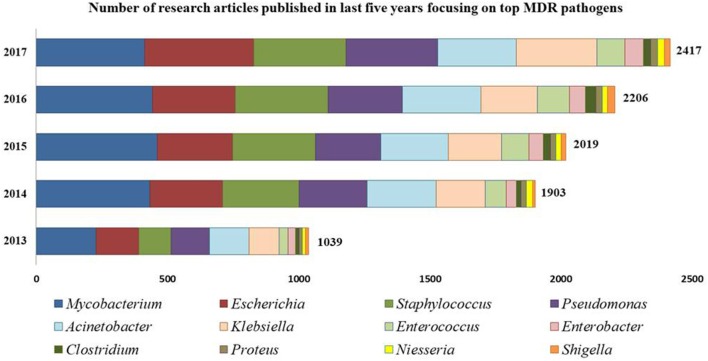
Intrinsically, AMR is more prevalent and severe in Gram-negative bacteria than their Gram-positive counterparts due mainly to the outer membranes serving as permeability barrier for drug-influx into the Gram-negative bacteria (Silhavy et al., 2010; Exner et al., 2017). To attain low sensitivity against biocidal compounds, Gram-negative bacteria reduce their outer membrane permeability by reducing the number of porins and inducing drug efflux pumps (EPs) for outward transport of drug molecules, often in a non-specific manner making the bacterial cells resistant to multiple antibiotics (Masi et al., 2017). However, despite these morphological differences, Gram-positive bacteria cannot be ignored or underestimated and noteworthy examples include methicillin resistant S. aureus (MRSA) and vancomycin resistant S. aureus (VRSA), coagulase negative Staphylococci members including S. epidermidis and S. haemolyticus, Streptococcus pneumonia, E. faecalis, E. faecium, and Clostridium difficile (Schindler and Kaatz, 2016). Figure 1 shows a gradual upsurge in the number of research articles focused on most-prevalent MDR strains.
Figure 1.
Number of research articles reported from 2013 to 2017 focusing on MDR bacterial strains. [Source: PubMed; Key words used: Multidrug resistant “Genus name”].
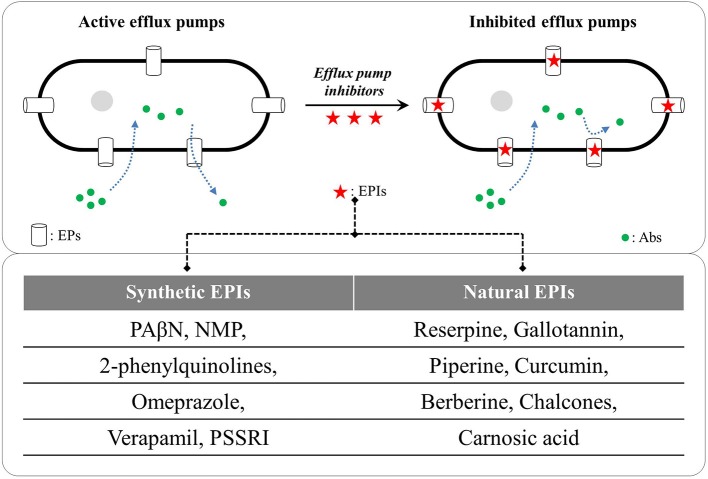
In recent years, EPs have emerged as key drivers for AMR in Gram-negative and Gram-positive bacteria, and therefore, are looked upon as potent and universal targets for containing the drug-resistant phenotypes. EPs are vital in other physiological processes also including stress-adaptations, virulence, pathogenicity and transportation of essential nutrients (Piddock, 2006; Fernandez and Hancock, 2012; Costa et al., 2013; Kourtesi et al., 2013; Sun et al., 2014). Identifying novel and potent EP inhibitors (EPIs) to revert the AMR is therefore gaining momentum. EPIs are the compounds with capability to reduce resistance or a complete reversal of AMR against otherwise ineffective antibiotics via inhibiting the EPs (Sun et al., 2014; Gill et al., 2015; Wright, 2016; Spengler et al., 2017). The first EPI against RND-type EPs was reported by Lomovskaya et al. (2001) a phenylalanine-arginine β-naphthylamide (PAβN) effective against Mex pumps in P. aeruginosa and AcrAB-TolC pump in E. coli. Since then, various synthetic and natural compounds have been screened for their EPI capabilities (reviewed by Prasch and Bucar, 2015; Spengler et al., 2017; Shin et al., 2018; Yang et al., 2018). Figure 2 illustrates the inhibition of microbial drug efflux via synthetic and natural EPIs.
Figure 2.
Examples of synthetic and natural efflux pump inhibitors (EPIs); PAβN: phenyl-arginine-β-naphthylamide, NMP: 1-(1- naphthylmethyl)-piperazine, PSSRIs: phenylpiperidine selective serotonin re-uptake inhibitors (German et al., 2008; Hannula and Hänninen, 2008; Li et al., 2015; Ni et al., 2016; Willers et al., 2016; Sabatini et al., 2017). Abs, antibiotics; EPs, efflux pumps.
Medicinal plants with antimicrobial properties have the potential to serve as the reservoir of novel and effective EPIs (Newman and Cragg, 2012). Though there are several reports on assessing medicinal plants for their antimicrobial properties (crude extracts and occasionally pure molecules), few investigations were aimed against MDR/XDR strains and fewer on deciphering the underlying resistance mechanisms targeted by the plant products (Kumar et al., 2013). But newer studies are coming up for identifying novel phytomolecules capable of reversing EP-mediated AMR. Some striking phytochemicals which have recently been identified for their EPI potentials include catechol, pinene, gingerol, capsaicin, resveratrol and the number is increasing (Prasch and Bucar, 2015).
In this review, we are presenting an account of major EPs, their roles in imparting bacterial AMR, strategies for identifying plant-based EPIs emphasizing on the potent phyto-EPIs active at relatively lower concentrations, reported during the last 8 years. High throughput screening and in silico approaches for predicting the EPIs and their binding targets/sites are also discussed.
Physiological Roles Played by Bacterial Efflux Pumps
Bacterial genome comprises of EP genes, expressed under tight regulation of global/local transcription factors (e.g., BmrR: transcriptional regulator of efflux pump Bmr in B. subtilis; QacR: transcriptional repressor of QacA transporter in S. aureus; AcrR: transcription repressor of acrB efflux pump in E. coli) proposing the important physiological roles the EPs play during cell development, stress adaptations and bacterial pathogenesis (Sun et al., 2014). The knowledge about these regulatory mechanisms may advance the understandings of physiologically originated AMR, frequently observed in nature (Sun et al., 2014). As discussed above, bacterial EPs have a tremendous capacity to extrude the variety of toxic compounds, needed for the cell survival in a given physiological niche and are vital for maintaining pathogenicity. This is further supported by the studies showing reduced pathogenicity in the bacterial strains lacking EPs. Buckley et al. (2006) showed that S. typhimurium acrB or tolC deficient mutant poorly colonized in the avian gut, highlighting the requirement of complete AcrAB-TolC system for virulence. The S. typhimurium strain lacking all the drug efflux assemblies became avirulent, when tested in a mouse model (Nishino et al., 2006). To confirm the role of EPs in bacterial pathogenesis, Hirakata et al. (2002) assessed the ability of EP (MexAB-OprM, MexCD-OprJ, MexEF-OprM, and MexXY-OprM) mutants of P. aeruginosa to invade Madin-Darby canine kidney cells. The findings revealed that except mexCD-OprJ, all other systems evidenced decreased bacterial invasion abilities.
EPs are also known to effect the bacterial cell communication during the stress-responses, especially in the quorum-sensing (González and Keshavan, 2006). As transportation of auto-inducers (chemical signals generated during quorum sensing) is a key-event during cell-cell interactions via quorum-sensing, drug EPs assist their transport across the membrane (Liang et al., 2016). Moore et al. (2014) confirmed a vital role played by MexAB-OprM efflux system in the secretion of a major auto-inducer N-acylated L-homoserine lactone by P. aeruginosa cells. The study also postulated this auto-inducer as a substrate for MexAB-OprM system (Moore et al., 2014). Further, Martinez et al. (2009) advocated the EP-mediated ceasing of quorum-sensing via augmented efflux of auto-inducers, facilitating quick bacterial response to stress signals. One more physiological role attributed to EPs is in the biofilm formation. Recent studies confirm the involvement of many well-characterized efflux systems, AcrAB-TolC of E. coli, AcrD of S. enterica, AdeFGH of A. baumannii and MexAB-OpeM of P. aeruginosa in biofilm formation (Alav et al., 2018). Kvist et al. (2008) reported an up-regulation of 20 genes encoding EP-transporters in E. coli during the growth of biofilm. Similarly, the EP-mediated export of colanic acid for capsule-matrix formation was observed along with up-regulated TetA(C) in E. coli, facilitating the biofilm formation (May et al., 2009). Collectively, the physiological roles of EPs are vital for pathogenic stability and virulence maintenance in bacteria.
The synthetic EPIs namely carbonylcyanide m-chlorophenylhydrazone (CCCP), chlorpromazine and PAβN were reported to prevent biofilm formation in E. coli, P. aeruginosa and S. aureus (Baugh et al., 2014). However, investigations on evaluating phytochemicals for their anti-biofilm potencies via inhibiting EPs are few. Fiamegos et al. (2011) isolated 4,5-O-dicaffeoylquinic acid from Artemisia absinthium which was proved to be a potent inhibitor of MFS pumps in E. coli and E. faecalis and as an anti-biofilm agent (Fiamegos et al., 2011). Recent reports advocate that nanomaterials in combination with phyto-EPIs can also be an effective therapy for containing drug-resistant infections (Gupta et al., 2017).
Bacterial Efflux Pumps: The Warheads in AMR Strains
Though bacterial AMR has several origins and many adaptive mechanisms are employed by drug-resistant strains against the antibiotics, the intrinsic EPs hold the key. Recent clinical and laboratory data establish that bacterial EPs are not only critical for drug-extrusion but also contribute to their virulence and adaptive responses (Du et al., 2018). Often, antimicrobial drug exposure induces intricate bacterial reactions including altered expressions of several genes encoding the transporters, as revealed by the phenotypic profiling of E. coli (Nichols et al., 2011).
Bacterial EPs are acknowledged either as primary active transporters using ATPs as an energy source, or as secondary active transporters acquired due to electrochemical potential difference created by pumping out Na+ and H+ outside the membrane (Dwivedi et al., 2017a).
This may be considered for classifying EPs into two broad super-families namely; ATP-binding cassette (ABC) multidrug transporters and secondary transporters using proton motive force (PMF) as an energy source (Putman et al., 2000). The second super-family can again be categorized in four subclasses, the major facilitator superfamily (MFS), resistance-nodulation-cell division (RND), multidrug and toxic compound extrusion (MATE) and small-MDR (SMR) family (Fernandez and Hancock, 2012; Sun et al., 2014). RND and MFS pumps are the most common in bacteria. With relatively narrow spectrum of specificity, MFS pumps are found in both Gram-negative and Gram-positive bacteria; while poly-selective RND pumps are exclusive to Gram-negative bacteria (Ward et al., 2001; Molnár et al., 2010). The SMR transporters show specificity for broad-spectrum polyaromatic cations convening resistance for compounds sharing similar chemical description. The MATE transporters are similar in size to MFS transporters but they do not share any sequence similarity with them (Jack et al., 2001). Apart from the classified super-families and sub-families, the MATE, SMR, and RND classes are distributed uniquely to prokaryotes whereas MFS and ABC transporters are dispersed in both prokaryotes and eukaryotes.
The large MFS is one of the most functionally diverse transporter families comprising multiple transportation types for drugs as well as sugars. These transporters comprise ~400 amino acids arranged as membrane-spanning helices (Saier et al., 1998). Based on the helical structure, MFS transporters can be classified as either 12-[e.g., TatA(B): class B tetracyclin transporter from E. coli] or 14-helix transporters [e.g., TatA(K): class K tetracyclin transporter from S. aureus], and TetA(B) is one of the most extensively studied members of the family (Lynch, 2006).
The SMR pumps represent smallest multidrug transporters, possessing only four trans-membrane helices without any extra membrane domain. But single minimal functional SMR unit represents eight helices as these are functionally active in dimeric form (Higgins, 2007). Well-illustrated example from this family is the electromagnetic antiporter EmrE from E. coli, responsible for resistance to a range of cationic-hydrophobic entities including antibiotics.
The latest structurally characterized class of EPs is MATE, involved in various vital biological functions (He et al., 2010). These transporters are equivalent to MFS transporters with a typical composition of ~450 amino acids putatively arranged in 12 helices; but with no sequence similarity with MFS counterparts (Jack et al., 2001). Some characterized MATE members include NorM from N. gonorrhoeae and N. meningitides and YdhE from E. coli. However, limited structural and functional knowledge is available about this family (Dwivedi et al., 2017a).
Though EPs from other families contribute to the AMR against certain antibiotics, RND pumps are the most potent drug efflux systems conferring resistance against clinically important antibiotics and biocides. Members of this family are known for their roles against a wide range of molecules with dissimilar structures including antibiotics, biocides, organic solvents, antimicrobial peptides, detergents, dyes, and bile salts (Poole, 2004). The tripartite complex of pumps from this family comprises inner membrane protein (IMP), outer membrane protein (OMP) along with a periplasmic membrane fusion protein (MFP) as a connector (Venter et al., 2015). The best understood tripartite complexes include AcrA-AcrB-TolC from E. coli and MexA-MexB-OprM from P. aeruginosa (Du et al., 2013, 2014). Greater structural and functional similarities between IMPs of these two systems are described by Welch et al. (2010). Exploration on biochemical and structural aspects of AcrB has revealed that these IMPs contain proximal and distal binding pockets, divided by G-loop (with 614–621 residues). Conformational flexibility of G-loop is crucial for movement of substrate along the binding site (Eicher et al., 2012; Cha et al., 2014). Table 2 lists various EPs belonging to the major families from prevalent pathogenic bacteria.
Table 2.
Examples of various efflux pumps belonging to major efflux pump families from prevalent pathogenic bacterial strains.
| Pathogen | EP Family | Example | Substrate | References |
|---|---|---|---|---|
| Acinetobacter baumannii | ABC | MacAB-TolC | ML | Okada et al., 2017 |
| MATE | AbeM | ACR, AG, DAU, DOR, FQ | Su et al., 2005 | |
| MFS | CraA | CHL | Roca et al., 2009 | |
| RND | AdeABC, AdeFGH, AdeIJK | AG, BL, FQ, ML, TET, BIO | Coyne et al., 2010, Rajamohan et al., 2010 |
|
| Campylobacter jejuni | RND | CmeABC, CmeDEF | AG, BL, CHL, FQ, ML, RIF, TET, EB | Lin et al., 2002; Akiba et al., 2006 |
| Enterococcus faecalis | ABC | EfrAB | ACR, CIP, DAU, DOR, DOX, NOR, TPP | Lee et al., 2003 |
| Escherichia coli | ABC | MacAB-TolC | ML | Kobayashi et al., 2001 |
| MFS | MdfA | CHL, DOR, NOR, TET | Nishino et al., 2006 | |
| QepA/QepA2 | FQ | Cattoir et al., 2008 | ||
| RND | AcrAB-TolC | BL, CHL, FQ, ML, NOV, RIF, TET, TGC, R6G | Swick et al., 2011, Lennen et al., 2013 |
|
| OqxAB | CHL, FQ | Hansen et al., 2004 | ||
| SMR | EmrE | ACR, EB, QAC | Schuldiner, 2009, Beketskaia et al., 2014 |
|
| Klebsiella pneumoniae | MATE | KetM | DAPI | Ogawa et al., 2015 |
| MFS | KpnGH | CAZ, CEF, STR, TET | Srinivasan et al., 2014 | |
| RND | OqxAB | CHL, FQ | Hansen et al., 2004 | |
| SMR | KnpEF | BAC, CEF, CHX, ERY, STR, TET, TRI | Srinivasan and Rajamohan, 2013 | |
| Mycobacterium tuberculosis | ABC | Rv1218c | BAP, BPD, PRI, PYR | Balganesh et al., 2010, Balganesh et al., 2012 |
| MFS | Tap | PAS, SPE, TET | Ramón-García et al., 2012 | |
| SMR | Mmr | CAB, CLA, TPP | Balganesh et al., 2012, Rodrigues et al., 2013 |
|
| Pseudomonas aeruginosa | RND | MexAB-OprM, MexXY-OprM/A, MexCD-OprJ, MexEF-OprN | AG, BL, CHL, FQ, ML, SUL, TET, TGC, TMP, BIO, EB | Poole et al., 1996, Köhler et al., 1997 |
| Staphylococcus aureus | ABC | Isa(E) | LIN, PLE, STA | Wendlandt et al., 2015 |
| Msr(A) | ML, TEL | Vimberg et al., 2015 | ||
| MATE | MepA | BIO, EB, FQ, TIG | Kaatz et al., 2005 | |
| MFS | NorA | FQ | Yoshida et al., 1990 | |
| QacA | ACR, CHX, EB, QAC | Littlejohn et al., 1992 | ||
| Streptococcus pneumoniae | ABC | PatAB | FQ | Marrer et al., 2006 |
| MFS | MefE | ML | Tait-Kamradt et al., 1997 | |
| Vibrio spp. | ABC | VcaM | CIP, DAU, DOR, NOR, TET | Huda et al., 2003 |
| MATE | NorM | AG, EB, FQ | Morita et al., 1998 | |
| MFS | EmrD-3 | EB, LNZ, ERY, CHL | Bruns et al., 2017 |
ACR, acriflavine; AG, aminoglycosides; BAC, benzalkonium chloride; BAP, biaryl-piperazines; BL, β-lactams; BIO, biocides; BPD, bisanilino-pyridines; CAB, cetyl-trimethyl-ammoniumbromide; CAP, cationic antibacterial peptides; CAZ, ceftazidime; CEF:cefepime; CHL, chloramphenicol; CHX, chlorhexidine; CIP, ciprofloxacin; CLA, clofazimine; DAU, daunomycin; DAPI, 4',6-diamidino-2-phenyl indole; DOR, doxorubicin; DOX, doxycycline; EB, ethidium bromide; ERY, erythromycin; FQ, fluoroquinolones; FUA, fusidic acid; LIN:lincosamide; LNZ, linezolid; ML, macrolides; NOR, norfloxacin; NOV, novobiocin; PAS, p-aminosalicylate; PLE, pleuromutilin; PRI, pridones; PYR, pyrroles; QAC, quaternary ammonium compounds; SPE, spectinomycin; STA, streptogramin A; STR, streptomycin; SUL, sulfonamides; TEL, telithromycin; TET, tetracycline; TGC, tigecycline; TMP, trimethoprim; TPP, tetraphenylphosphonium, TRI, triclosan.
Collectively, the complex EP assemblies are critical for bacterial pathogenesis, virulence, biofilm formation, and adaptive-responses ultimately conferring and defining bacterial AMR (Piddock, 2006; Martinez et al., 2009; Baugh et al., 2012, 2014; Du et al., 2018). EPs are critical for bacterial AMR as they exclude most of the unwanted entities until the cell gets required time for acquiring resistance (Piddock, 2014; Venter et al., 2015).
Bacterial Efflux Pumps: The Urgent Threats Requiring Immediate Remedy
It is well-established that EPs comprise one of the most vital systems in bacteria responsible for both innate and acquired AMR (Blair et al., 2015). There are reports of EPs from different superfamilies and the occurrence of various types of EPs from the same superfamily in a single bacterial species (Piddock, 2006). For instance, whole genome sequencing of the colistin resistant Enterobacter cloacae showed presence of multiple EPs (Norgan et al., 2016). Differential substrate profiles of EPs are also a characteristic feature which may diverge between or within the superfamily (Poole, 2005, 2007). Although the core motive of EPs related studies is focused on AMR, several reports however confirmed other but significant functions of bacterial EPs including quorum-sensing, biofilm formation, virulence, pathogenicity and bacterial behavior (Piddock, 2006; Yang et al., 2006; Fahmy et al., 2016).
Up-regulation of gene expression levels are one of the main drivers for chromosomally acquired AMR. This can be triggered due to the gene induction, activated transcription, or due to regulatory mutations (Grkovic et al., 2002). The coding region for an EP is usually found contiguous to the regulatory proteins controlling the expression levels of pump gene in response to substrates. For example, AdeL, an LTTR (LysR-type transcriptional regulator) family protein exists opposite to the adeFGH operon that regulates the expression of genes encoding RND efflux system in A. baumannii (Liu et al., 2018). The expression levels of EP-associated proteins along with porins is mutually controlled by several global regulatory elements, modifying the transcription patterns of EP-family transcripts either directly or through a cascade of regulatory events (Warner and Levy, 2010; Sun et al., 2014). Further, the expression of MexAB-OprM efflux system is governed by repressor protein mexR, encoded by a gene located upstream of the mexAB-oprM operon in P. aeruginosa (Suresh et al., 2018). Similarly, the acrAB operon system is regulated by regulator acrR in E. coli, located 140 bp up-stream of the acrAB operon (Ma et al., 1996).
Another striking bacterial character adding to the AMR nature is heteroresistance, the occurrence of differential responses to antibiotics by the bacterial cells from the same population, a phenomenon first reported in S. aureus (Kayser et al., 1970). Interestingly, drug resistant and sensitive bacterial cells may co-exist in a single culture (Morand and Mühlemann, 2007). The mechanism underlying heteroresistance acquirements are yet to be fully understood, however, the active EPs are strongly linked to heteroresistance (Chen et al., 2017). Designing a treatment course against such strains is difficult as there are high chances of increase in the frequency of resistant-bacterial-population and stimulation of cross-resistance to antimicrobial lysozymes of the host system (Napier et al., 2014; Telke et al., 2017). Up-regulation of OpxAB gene in Salmonella typhimurium (Chen et al., 2017) and AdeABC gene in A. calcoaceticus-A. baumannii (Ruzin et al., 2007) are attributed for mediating the tigecyclin heteroresistance. Similarly, colistin associated heteroresistance is also reported in E. asburiae LH74 and E. cloacae NH52 (Telke et al., 2017), showing its association with overexpression of acrAB-tolC EPs under the regulation of soxRS genes.
AMR phenotypes may result from concurrent acquisition of several AMR mechanisms simultaneously. It may include a combination of phenomena like chromosomally acquired resistance, multiple chromosomal changes with time, and/or a single mutational event activating the AMR mechanisms including the EPs (Lister et al., 2009). The over-expressions of EPs and their corresponding genes have been reported to contribute to MDR in P. aeruginosa (Shigemura et al., 2015). Recent studies have confirmed the role of EPs in fluoroquinolone resistant E. coli (Amabile-Cuevas et al., 2010; Swick et al., 2011; Yasufuku et al., 2011). Similarly, two fluoroquinolone resistant clinical isolates of Shigella showed overexpression of the TolC channels, part of AcrAB-TolC tripartite responsive to ciprofloxacin (Kim et al., 2008). These findings confirm that the up-regulation of EP genes contribute significantly to diminish intracellular antibiotics level, with a selectivity of the efflux transporter.
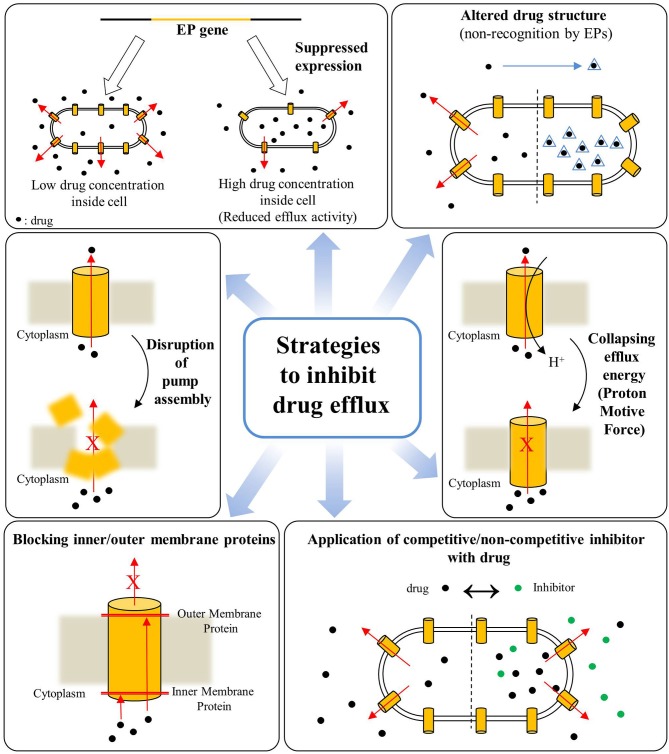
Overall, the poly-specificity of EPs, their overexpression in response to drugs along with the phenomenon of heteroresistance seem key factors responsible for drug-resistance in a wide-range of bacterial species, especially in Gram-negative bacteria making them difficult to treat with conventional drug arsenal. The drug-efflux mediated bacterial AMR is a mounting threat to global healthcare, therefore EPs are gaining unprecedented attention not only from the perspectives of basic understandings that how they work and impart drug-resistance but also as emerging targets for development of novel and potent adjunct-therapies for combating AMR in community and nosocomial infections. As a result, inhibition of drug efflux from bacterial cells via inhibiting or disrupting the EPs is an emerging approach for combating the threatening AMR. Various approaches have been developed in recent past and a schematic for these strategies for inhibition or disruption of bacterial drug efflux is illustrated in Figure 3.
Figure 3.
Various strategies for inhibition of drug efflux from bacterial cells for combating antimicrobial resistance (Based on reviews by Pagès and Amaral, 2009; Venter et al., 2015).
Phytotherapeutics–The Potent Efflux Pump Inhibitors
Phytochemicals are critical for human health-care since ancient times. Medicinal plants are hailed as a reservoir for phytochemicals capable of providing new and potent drug leads to contain the AMR via targeting the principal determinants of drug-resistance including EPs (Newman and Cragg, 2012; Prasch and Bucar, 2015). This review focuses mainly on EPIs of plant origin (phyto-EPIs) reported in the running decade. We are discussing some important and successful case studies on phyto-EPIs effective against AMR phenotypes. Table 3 summarizes the list of phytochemicals, their source and effective concentrations used for inhibiting the efflux pumps of AMR bacterial strains.
Table 3.
A summarized list of phytochemicals, their source and effective concentrations for inhibiting efflux pumps from antimicrobial resistant bacteria.
| Active Compound | Verified effective concentration/s | Plant | Antimicrobial resistant microbes | Targeted efflux pumps | References |
|---|---|---|---|---|---|
| 1′-S-1′-acetoxyeugenol acetate | 3.12–25 mg L−1 | Alpinia galanga | Mycobacterium smegmatis mc2 155 | EtBr EP | Roy et al., 2012 |
| 4-hydroxy-α-tetralone | 125 μg mL−1 | Ammannia spp. | MDR E. coli | YojI | Dwivedi et al., 2014b |
| Baicalein | 16 μg mL−1 | Scutellaria baicalensis | S. aureus SA-1199B | NorA | Chan et al., 2011 |
| Berberine and Palmatine |
250–1,000 μg mL−1
500–100 μg mL−1 |
Berberis vulgaris | MDR P. aeruginosa (clinical isolates) | MexAB-OprM | Aghayan et al., 2017 |
| Capsaicin | 25 μg mL−1 | Capsicum spp. | S. aureus SA-1199B | NorA | Kalia et al., 2012 |
| Catechol | 5 and 10 mg mL−1 | Acer saccharum |
P. aerugenosa ATCC 15692 and UCBPPPA14, E. coli ATCC 700928, P. mirabilis HI4320 |
EtBr EP | Maisuria et al., 2015 |
| Catharanthine | 25 mg L−1 | Catharanthus roseus | P. aeruginosa | EtBr EP | Dwivedi et al., 2017b |
| Conessine | 20 mg L−1 | Holarrhena antidysenterica | P. aeruginosa | MexAB-OprM | Siriyong et al., 2017 |
| Cumin-methanol extract | 5 mg mL−1 | Cuminum cyminum | S. aureus MRSA OM505 | LmrS | Kakarla et al., 2016 |
| Essential oil | 5 μl mL−1 | Salvia fruticosa | S. epidermidis (clinical isolates) | Tet(K) EP | Chovanova et al., 2015 |
| Essential oil | 170.6 μl mL−1 |
Chenopodium Ambrosioides |
Staphylococcus aureus IS-58 | Tet(K) EP | Limaverde et al., 2017 |
| Gallotannin | 12.1–97.5 μg mL−1 | Terminalia chebula | MDR uropathogenic E. coli | EtBr EP | Bag and Chattopadhyay, 2014 |
| Indirubin | 1.25 and 2.5 mg L−1 | Wrightia tinctoria | S. aureus SA-1199B | NorA | Ponnusamy et al., 2010 |
| Kaempferolrhamnoside | 1.56 mg L−1 | Persea lingue | S. aureus SA-1199B | NorA | Holler et al., 2012 |
| Lysergol | 10 μg mL−1 | Ipomoea muricata | E. coli MTCC1652 and KG4 | YojI | Maurya et al., 2013 |
| Olympicin A | 50 μM | Hypericum olympicum | S. aureus 1199B | NorA | Shiu et al., 2013 |
| Sarothrin | 100 μM | Alkanna orientalis | Staphylococcus aureus NCTC 8325-4 | NorA | Bame et al., 2013 |
| Ursolic acid and derivatives | 25 and 50 μg mL−1 | Eucalyptus tereticornis | MDR E. coli (KG4) | AcrA/B, MacB, TolC and YojI | Dwivedi et al., 2014a |
One of the potent EPIs, the anti-hypersensitive alkaloid reserpine was isolated from Rauwolfia vomitoria (Stavri et al., 2007). Similarly, EP inhibitory activity of gallotannin (1,2,6-tri-O-galloyl-b-D-glucopyranose) isolated from hydro-alcoholic extracts of Terminalia chebula fruits was demonstrated by Bag and Chattopadhyay (2014) against MDR uropathogenic E. coli. Gallotannin induced a 2- to 4-fold reduction in minimal inhibitory concentration (MIC) of test antibiotics via inhibiting the ethidium bromide (EtBr) pump (Table 3). As EtBr is a known EP-substrate, inhibition of EtBr efflux backs the postulated EP-inhibitory activity of gallotannin (Bag and Chattopadhyay, 2014). Methanolic sap extracts of Acer saccharum was evaluated for its drug efflux inhibitory potentials against P. aeruginosa (ATCC 15692 and UCBPPPA14), E. coli (ATCC 700928) and P. mirabilis (HI4320) confirmed via monitoring the EtBr efflux (Maisuria et al., 2015).
A clavine alkaloid lysergol from Ipomoea muricata was evaluated against AMR E. coli strains (MTCC1652 and KG4) to test its EP inhibitory potentials, and strong activities (higher than the standard reserpine) were exhibited by lysergol and its derivative, 17-O-3″,4″,5″-trimethoxybenzoyllysergol (Maurya et al., 2013). Authors also reported the inhibitory activities of this compound against ABC pump YojI in E. coli (Maurya et al., 2013). Similarly, falcarindiol, isolated from Levisticum officinale exhibited EPI activities against the Gram-negative strains (Garvey et al., 2011).
On the similar lines, Dwivedi et al. (2017b) reported the antibiotic-potentiating activities of catharanthine against superbug P. aeruginosa. The investigation involved in silico docking followed by the in vitro evaluation revealed that catharanthine potentiates the activity of tetracyclin and streptomycin, as confirmed by a reduced MIC, and acts as a potent EPI (Dwivedi et al., 2017b). A pentacyclic triterpenoid ursolic acid from leaves of Eucalyptus tereticornis described as a precursor of putative EPI was evaluated against MDR E. coli (KG4), two promising semi-synthetic, esterified derivatives of ursolic acid, 3-O-acetyl-urs-12-en-28-isopropyl ester and 3-O-acetyl-urs-12-en-28-n-butyl ester and the parent compound exhibited better EP inhibitory potencies than the standard reserpine (Dwivedi et al., 2014a). The molecular docking confirmed the targets of these compounds as AcrA/B, MacB, TolC, and YojI (Dwivedi et al., 2014a). Similarly, two alkaloids isolated from roots and rhizomes of Berberis vulgaris, the barberine and palmatine showed potent EP inhibitory efficacies against P. aeruginosa isolated from burn infections (Aghayan et al., 2017).
Phenylpropanoids from the n-hexane and chloroform fractions of Alpinia galanga exhibited EP inhibitory activities against Mycobacterium smegmatis mc2 155ATCC 700084 (Roy et al., 2012). A dose-dependent EP inhibition was observed with 1′-S-1′-acetoxyeugenol acetate (Roy et al., 2012). Mukanganyama et al. (2012) examined another mycobacterial member Mycobacterium aurum A+ against a naphthoquinone diospyrine isolated from Diospyros montana along with its derivatives. Two derivatives proved highly potent EPI and allowed bacterial cells to accumulate high concentrations of ciprofloxacin (Mukanganyama et al., 2012).
The acylphloroglucinol isolated from n-hexane fractions from Hypericum olympicum, olympicin-A showed promising activities against S. aureus (Shiu et al., 2013). The radiometric accumulation assay of the strain overexpressing NorA pump indicated the enhanced accumulation of (14)C-enoxacin, thus confirming efflux inhibition (Shiu et al., 2013). Two coumarins [5,7-dihydroxy-6-(2-methylbutanoyl)-8-(3-methylbut-2-enyl)-4-phenyl-2H-chromen-2-one and 5,7-dihydroxy-8-(2-methylbutanoyl)-6-(3-methylbut-2-enyl)-4-phenyl-2H-chromen-2-one] obtained from floral buds of Mesua ferrea were assessed against NorA-overexpressing S. aureus 1199B and the clinical isolate MRSA 831 (Roy et al., 2013). Linoleic acid isolated from ethanolic extracts of Portulaca oleracea showed efflux inhibitory potential at 64 mg L−1 concentration, equivalent to reserpine when quantified against MRSA (RN4220/pUL5054: erythromycin resistant, over-expressing MsrA ABC EP, Chan et al., 2015). In search of the drug-resistance reversal agents, dos Santos et al. (2018) assessed caffeic acid and gallic acid against four strains of S. aureus; 1199 as a wild type strain, 1199B as NorA harboring fluroquinolone resistant, IS-58 possessing TetK pump and RN4220 possessing MrsA pump. The study confirmed caffeic acid as a potent AMR-reversal agent, as it effectively inhibited MrsA and NorA EPs from S. aureus strains RN4220 and 1190B, respectively (dos Santos et al., 2018). In another interesting study, Kakarla et al. (2016) reported LmrS inhibitory activities of Cuminum cyminum. The study revealed that the cumin inhibits the LmrS mediated transport of drugs resulting in growth inhibition of MRSA clinical isolate in a dose-dependent manner (Kakarla et al., 2016).
Traditionally, most of the investigations were aimed at identifying EPIs for Gram-positive strains for reversing their AMR characters with very few reports against Gram-negative members. This can be because Gram-negative bacteria are more difficult targets then their positive counterparts due mainly to the presence of powerful EPs and other effective membrane barriers (lipophilic layer) averting them from external impacts (Stavri et al., 2007; Prasch and Bucar, 2015). Though, some approaches have emerged in recent years for improving antibiotic-penetration across the permeability membranes of Gram-negative bacteria such as the inhibition of new accessible target, identification of uptake pathways and the “Trojan Horse” approach (achieving fast or facilitated antibiotics uptake), establishing the rules of permeation (for predicting whether elevated uptake or reduced efflux would be the most efficient way for increasing the potency of specific antimicrobial class) and identifying potent EPIs, last one being probably the most potent (Zgurskaya et al., 2015).
Recently, Bruns et al. (2017) successfully inhibited EmrD-3 pump-mediated drug efflux from a Gram-negative bacterium Vibrio cholerae by garlic extract and its bioactive compound, allyl sulfide. At relatively low concentrations, the extract seems to target the EmrD-3 pump, but at higher garlic extract concentrations, the respiratory chain was affected. This example confirms targeting the energization of the efflux system by plant compounds as a potential strategy for drug efflux inhibition (Bruns et al., 2017).
Further, the MFS conserved sequence motifs, present across the entire superfamily, provide vital information regarding alignments of MFS transporter sequences (at least motif containing region), which may help in understanding the structural templates and actual binding events achieved via these transporters. Molecular dynamic simulation (MDS) studies of VMAT2 multidrug transporter (MFS family) revealed the presence of two domains of six trans-membrane helices (Yaffe et al., 2013). The trans-membrane residues at anchoring sites are identified as hinge points, at which straightening and flexing movement of helices occur, required for transport. These anchor point residues are highly conserved throughout the MFS family (Yaffe et al., 2013) and are emerging targets for drug efflux inhibition. Recent advances in scientific and technological arena have added significant in-depth understandings of the structural and biochemical basis of drug efflux, substrate profiles, molecular regulation and inhibition of major EPs.
Active EPs play a critical role in intrinsic and elevated drug resistance acquired via overproduction or over-activation of pumps in Gram-negative bacteria, and the development of clinically useful EPIs or new antibiotics to bypass pump-effects continues to be a challenge in combating Gram-negative bacterial infections (Li et al., 2015). As practically all the antibiotics are susceptible to active drug efflux, the potent EPIs can target these pumps antagonistically and can make old antibiotics effective again (the phenomenon known as re-sensitization). Besides, considering the fact that several antimicrobial agents like lipophilic penicilines, many glycopeptides, oxazolidinones, macrolides and lipopeptide daptomycin are effective in treating only Gram-positive bacterial infections and their poor potencies against Gram-negative pathogens is at least partially due to their active drug efflux, novel and potent EPIs are needed to significantly broaden the range of these antimicrobial agents. All this clearly indicates that EPIs have tremendous potential in adjunctive therapies along with the known but otherwise ineffective antibiotics ultimately reducing the emergence of AMR and virulence (Opperman and Nguyen, 2015). But developing novel and potent EPI is difficult and needs to overcome several hurdles such as choice of antibiotics for potentiation and matching the pharmacological properties of EPI-antibiotics pair (Opperman and Nguyen, 2015; Zgurskaya et al., 2015).
Considering the serious threats posed by the Gram-negative bacteria and their drug-resistance nature, more investigations aiming to target them with the novel, alternative and effective approaches including exploration of natural products are coming up. Though there are limited success stories, but they may lay the foundation for developing potent EPIs to avert the AMR phenotypes with the help of natural sources.
In silico Molecular Dynamics Simulations (MDS) to Screen and Develop EPIs
The MDS is a commanding approach for computational validation and to support the hypothesized mechanisms of EPs and EPIs (Nikaido and Takatsuka, 2009). It has made possible to simulate the membrane protein complex structures with micro-second time-scales. The MDS approach along with molecular docking and other in silico tools are successfully utilized for screening and prediction of molecular interactions between potential EPs and their corresponding inhibitors (Jamshidi et al., 2016). This has led to the unraveling of the mechanism how drug efflux systems recognize and transfer specific molecules; thus helping researchers in challenging the efflux-mediated resistance and finding appropriate EPIs for improvement in antibiotics efficacy during AMR (MDR/XDR) infections (Collu et al., 2012; Nakashima et al., 2013). There are several recent reports describing the successful applications of these in silico approaches for screening and identifying potent EPIs of plant origin (Bhaskar et al., 2016; Jamshidi et al., 2016; Mangiaterra et al., 2017; Verma et al., 2017).
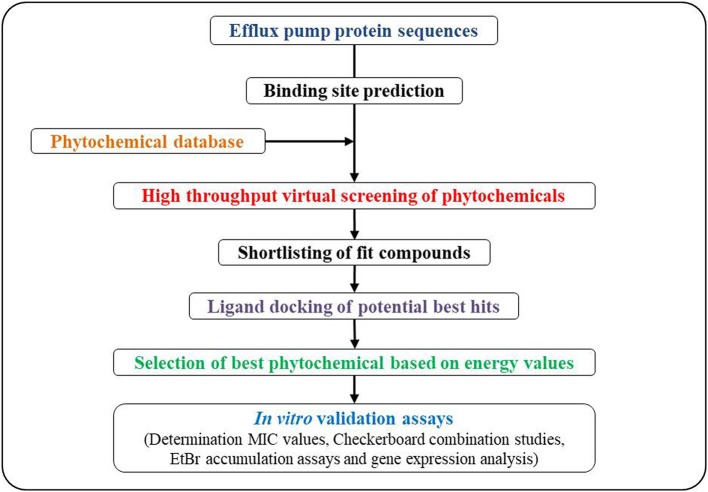
A general scheme for MDS-based approaches is depicted in Figure 4. Briefly, it starts with the identification of three-dimensional structures of potential EP-binding sites (pockets). The next step is the prediction of trans-membrane segments from protein sequences. The predicted structure can then be checked for its stereochemical properties by analyzing the overall and residue-by-residue geometry. The modeled protein structure is then reduced with the solvent implied by chimera programs (http://www.cgl.ucsf.edu/chimera/, Pettersen et al., 2004) and the projected protein structure can be used for its interaction with potential EPI molecules. The three-dimensional structure of the EPI is then explored to attain perfect and stable EPI-EP complex. Potent tools for docking studies include AutoDock (http://autodock.scripps.edu/, Morris et al., 2009), and SwissDock (http://www.swissdock.ch/, Grosdidier et al., 2011). Such automated docking tools can predict the exact binding position of the candidate drug-molecule to the receptors, and provides vital information about exact amino residues taking part in bond-formation with potential drug(s), their bond lengths and type and other interactions adding to the stability of the docking complexes.
Figure 4.
A general scheme for in silico molecular dynamics simulations approach for screening and developing plant based efflux pump inhibitors.
Recently, Kesherwani et al. (2017) used high throughput virtual screening of natural compounds against NorM, a MATE transporter from N. gonorrhea followed by flexible docking. Authors performed molecular simulation in a membrane environment for investigating the stability and binding energy of top lead compounds, and identified a phytomolecule from Terminalia chebula with higher binding free energy than the substrates (rhodamine 6 g, ethidium). The compound successfully blocked the disruption of Na+-coordination along with an equilibrium state bias toward occlude state of NorM transporter, ultimately blocking the extrusion of antimicrobial drugs via inhibiting the NorM transporter in drug-resistant N. gonorrhea.
Similarly, Suriyanarayanan and Sarojini (2015) analyzed EPI potentials of plant-derived flavonoid quercetin in bringing down the drug efflux via inhibiting the EmrE, a transporter belonging to SMR family from E. coli. Authors used in silico approaches and molecule docking approaches. The docking analysis of quercetin with EP-protein showed the importance of residues for function and stability, and notably quercetin showed best interactions as compared to the compounds like verapamil, reserpine, chlorpromazine, and carbonyl cyanide m-chloro phenylhydrazone. MDS confirmed the stability of quercetin-Mmr complex, which insights the potential of quercetin as a non-antibiotics adjuvant for treatment of bacterial infection via reducing the drug efflux from bacteria.
Mangiaterra et al. (2017) identified two phyto-EPIs using in silico high-throughput virtual screening. Molecular docking revealed these two compounds morelloflavone and pregnan-20-one derivative as inhibitors of MexAB-OprM EPs of P. aeruginosa and supportive in vitro assays confirmed their synergism with ciprofloxacin (Mangiaterra et al., 2017).
Molecular docking plays a crucial role and help in defining drug-protein interactions which determines whether compounds act as substrates for EP proteins. Therefore, inhibitors or modulators of EPs are well-recognized along with their comparative binding efficiencies via detailed docking analyses (Collu et al., 2012). Putative EPI activities of quercetin, plumbagin, nordihydroguaretic acid, shikonion and mangiferin were confirmed (Ohene-Agyei et al., 2014). Similarly, docking of reserpine, salvin, totarol, ferruginol along with known antibiotics to NorA revealed that all the tested compounds showed binding at large hydrophobic cleft, suggesting the substantial interactions with key-residues (Bhaskar et al., 2016). Notably, all these investigations were backed by the bioassays confirming the validity of information generated via in silico screening.
Owing to the importance of an instantaneous requirement of curing the XDR/MDR strains with utmost specificity, a greater understanding of exact drug-identification and its transport by MDR-EPs is important. The in silico MDS approach along with virtual docking and wet laboratory validation therefore can be considered as an imperative path in identifying potent phyto-EPIs.
Molecular Interactions Underlying Inhibition of Efflux Pumps by Phyto-Therapeutics
The inhibition of active drug efflux by EPIs results into the elevated intracellular antimicrobial concentrations, and lowered or complete reversal of efflux-mediated bacterial drug resistance, prevention of microbial invasiveness by inhibiting the export of virulence-factors and shortened adaptation-time required for bacteria, prohibiting the emergence of mutant strains with high AMR (Bhardwaj and Mohanty, 2012; Sun et al., 2014). Major strategies developed for drug efflux inactivation are, first, alterations in regulatory mechanisms for activation/repression of EP gene expressions (Purssell and Poole, 2013), second, deprivation of motive forces required for working of pumps by diminishing the proton gradient (Viveiros et al., 2005; Martins et al., 2008), third structural modifications in existing antimicrobials to bypass the chemophore recognition by the EPs (Chollet et al., 2004; Rice et al., 2005), fourth disrupting the pump-functionality by averting assembly of pumps by targeting protein interfaces (Tikhonova et al., 2011); interaction between protein motifs (Hobbs et al., 2012); obstructing the exit duct (Zeng et al., 2010), and fifth, the trapping of EPs in the inactivated form by competitive binding of EPIs and cytoplasmic membrane proteins (Nakashima et al., 2013; Opperman et al., 2014; Nguyen et al., 2015; Opperman and Nguyen, 2015; Figure 3). In addition, targeting the molecular hinge structures by the conserved sequence motifs is also an emerging strategy for EP inhibition (Abdali et al., 2017). The conserved sequence motifs (7–13 residues) are characteristics of MFS family and these motifs on c-terminal end of trans-membrane helix are rich in glycine and proline, vital for promotion of hinge formation. These conserved residues are considered as major contributors in binding and transportation of respective substrates (Luo and Parsons, 2010), and therefore targeting them holds significance for drug efflux inhibition.
To identify the phyto-EPIs, some authors tried to decipher the physiological and molecular interactions involved in EP dysfunction. Sharma et al. (2010) described piperine as an inhibitor of Rv1258c, an efflux protein transporter present on cytoplasmic membrane [encoding for tetracyclin/aminoglycoside resistance (TAP-2)-like EP] in M. tuberculosis H37Rv. After structural prediction of the protein, further investigation revealed the binding pocket of Rv1258c. Authors showed H-bond interaction (2.06 Å) with Arg141 residue and piperine provided stable protein-ligand interaction. The findings confirmed the role of piperine in augmenting rifampicin sensitivity in M. tuberculosis (Sharma et al., 2010). Capsaicin also proved a potent EPI, inhibiting NorA pump of S. aureus (Kalia et al., 2012). The study showed the involvement of Arg98 and Ile23 residues from active binding site in the key binding interactions. The stable interaction between capsaicin and active site at the proposed orientation allows an aliphatic chain of capsaicin, extending in a hydrophobic cleft (containing residues Pro24, Phe140, Ile244, and Phe303) permitting strong hydrophobic interactions due to a lesser distance between ligand and molecule (1.7–3.2 Å). A weak H-bond formation between OH-group (from aryl moiety of capsaicin) and Arg98 was attributed for providing extra-stability to capsaicin/NorA complex (Kalia et al., 2012). Another study by Zhang et al. (2014) described the interactions between ginsenoside 20(S)-Rh2 and NorA from S. aureus. The stable H-bond formation between ginsenoside 20(S)-Rh2 and Gln51/Asn340/Ser226 residues at active binding site in the central cavity of protein was attributed for the inhibition of NorA pump, thus promoting accumulation of ciprofloxacin inside the bacterial cell (Zhang et al., 2014). In the similar vein, Ohene-Agyei et al. (2014) assessed five phytochemicals (plumbagin, shikonin, quercetin, mangiferin and nordihydroguaretic acid) for their EPI potentials against AcrB protein from AcrAB-TolC drug transporter. The stable H-bond formation between T monomer of AcrB with minocycline attached to binding pocket and phytochemicals was responsible for efflux inhibition by the phytochemicals (Ohene-Agyei et al., 2014). Further, the authors also postulated that the considered natural compound act as a substrate and compete with the antibiotics for drug-resistance reversal (similar to PAβN). Therefore, these natural products act as high-affinity substrate inhibitors rather than substances for trapping the EPs in an inactive state.
Concluding Remarks
Increasing AMR in community and nosocomial settings is a big threat to human healthcare and accounts for a large number of mortalities and morbidities globally. Bacterial EPs make up a major warhead of the drug-resistant pathogens and increase and maintain the AMR via extruding or reducing the intracellular concentrations of applied antibiotics, often in a non-specific manner. The drug EPs are also emerging as chemical tools to understand molecular mechanisms underlying drug extrusion from the bacterial cells. EPs play several important physiological and molecular roles in bacterial cell survival and stress-responses. The necessity to overcome AMR has encouraged investigators to characterize resistance-inhibiting or modulating EPIs to block the drug extrusion, restoring antibacterial susceptibility and returning existing antibiotics into the clinic. The severity of the AMR is higher in Gram-negative bacteria, owing to their superior capabilities in maintaining high drug efflux levels coupled with lower intracellular levels of toxic drugs including antibiotics. MDR/XDR strains maintain their intrinsic and acquired resistance via overproduction of pumps. The development of clinically useful EPIs to bypass pump effects continues to be a challenge. Though there are some noteworthy developments in recent past aimed at reversing the AMR phenotypes including facilitation of better drug-penetration across the outer membranes of Gram-negative bacteria, and establishing new rules of permeation, identifying new and powerful EPIs seems best approach that can be explored as drug leads or in adjunctive therapies.
Several recent developments in in-silico MDS approaches have enabled the researchers to computationally validate and support the hypothesized mechanisms of EPs and EPIs (Suriyanarayanan and Sarojini, 2015; Ramaswamy et al., 2017; Vargiu et al., 2018). It is now possible to simulate membrane protein complex structures with micro-second time-scales. The MDS approach along with molecular docking and other in silico tools are successfully utilized for screening and prediction of the molecular interactions between potential EPs and their corresponding EPIs, ultimately helping in identification of the potent EPIs particularly from plant origin. However, this field is yet to be explored fully.
Considering the fact that practically all the antibiotics are susceptible to active drug-efflux, use of the potent EPIs to target and block these pumps can help in potentiating the old antibiotics effective again against a range of drug-resistant bacteria. EPIs are being looked as promising adjunctive therapies with the known antibiotics to improve their antibacterial potency at low concentrations, reduce the emergence of AMR and virulence. But developing novel and potent EPI is not easy and needs to overcome several hurdles such as choice of antibiotics for potentiation and matching the pharmacological properties of EPI-antibiotic(s) pair. More comprehensive and deeper investigations are therefore needed that involve the exploring the high-throughput screening assisted by in silico tools for identifying the potent EPI phytomolecules and their corresponding targets. Newer studies are being undertaken for identifying phytomolecules effective in inhibiting bacterial efflux pumps via potentiation of antibiotics against pathogenic bacteria including Gram-negative pathogens. This may pave the way for identification of phyto-EPIs that can head toward clinical phases and ultimately clinical practices with an aim to contain the AMR.
Author Contributions
VK conceived the idea. VS, TK, RB, RS, and VK wrote the manuscript. All the authors made substantial contribution to the work and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The support under Star College Scheme of Department of Biotechnology, Government of India implemented at Modern College, Ganeshkhind, Pune is gratefully acknowledged. The use of facilities created under FIST program of Department of Science and Technology (DST), Government of India, implemented at Prof. Ramkrishna More College, Akurdi, Pune and Modern College, Ganeshkhind, Pune are gratefully acknowledged.
References
- Abdali N., Parks J. M., Haynes K. M., Chaney J. L., Green A. T., Wolloscheck D., et al. (2017). Reviving antibiotics: efflux pump inhibitors that interact with acra, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS Infect Dis. 3, 89–98. 10.1021/acsinfecdis.6b00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghayan S. S., Mogadam H. K., Fazli M., Darban-Sarokhalil D., Khoramrooz S. S., Jabalameli F., et al. (2017). The effects of berberine and palmatine on efflux pumps inhibition with different gene patterns in Pseudomonas aeruginosa isolated from burn infections. Avicenna J. Med. Biotechnol. 9:2. [PMC free article] [PubMed] [Google Scholar]
- Akiba M., Lin J., Barton Y. W., Zhang Q. (2006). Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57, 52–60. 10.1093/jac/dki419 [DOI] [PubMed] [Google Scholar]
- Alav I., Sutton J. M., Rahman K. M. (2018). Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 73, 2003–2020. 10.1093/jac/dky042 [DOI] [PubMed] [Google Scholar]
- Amabile-Cuevas C. F., Arredondo-Garcia J. L., Cruz A., Rosas I. (2010). Fluoroquinolone resistance in clinical and environmental isolates of Escherichia coli in Mexico City. J. Appl. Microbiol. 108, 158–162. 10.1111/j.1365-2672.2009.04401.x [DOI] [PubMed] [Google Scholar]
- Bag A., Chattopadhyay R. R. (2014). Efflux-pump inhibitory activity of a gallotannin from Terminalia chebula fruit against multidrug-resistant uropathogenic Escherichia coli. Nat. Prod. Res. 28, 1280–1283. 10.1080/14786419.2014.895729 [DOI] [PubMed] [Google Scholar]
- Balganesh M., Dinesh N., Sharma S., Kuruppath S., Nair A. V., Sharma U. (2012). Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob. Agents Chemother. 56, 2643–2651. 10.1128/AAC.06003-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balganesh M., Kuruppath S., Marcel N., Sharma S., Nair A, Sharma U. (2010). Rv1218c, an ABC transporter of Mycobacterium tuberculosis with implications in drug discovery. Antimicrob. Agents Chemother. 54, 5167–5172. 10.1128/AAC.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bame J. R., Graf T., Junio H., Bussey R., Jarmusch S., El-Elimat T., et al. (2013). Sarothrin from Alkanna orientalis is an antimicrobial agent and efflux pump inhibitor. Planta Med. 79, 327–329. 10.1055/s-0032-1328259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh S., Ekanayaka A. S., Piddock L. J., Webber M. A. (2012). Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 67, 2409–2417. 10.1093/jac/dks228 [DOI] [PubMed] [Google Scholar]
- Baugh S., Phillips C. R., Ekanayaka A. S., Piddock L. J., Webber M. A. (2014). Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob.Chemother. 69, 673–681. 10.1093/jac/dkt420 [DOI] [PubMed] [Google Scholar]
- Beketskaia M. S., Bay D. C., Turner R. J. (2014). Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux. J. Bacteriol. 196, 1483–1414. 10.1128/JB.01483-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A. K., Mohanty P. (2012). Bacterial efflux pumps involved in multidrug resistance and their inhibitors: rejuvenating the antimicrobial chemotherapy. Recent Pat. Antiinfect. Drug Discov. 7, 73–89. 10.2174/157489112799829710 [DOI] [PubMed] [Google Scholar]
- Bhaskar B. V., Babu T. M. C., Reddy N. V., Rajendra W. (2016). Homology modeling, molecular dynamics, and virtual screening of NorA efflux pump inhibitors of Staphylococcus aureus. Drug Des. Devel. Ther. 10, 3237. 10.2147/DDDT.S113556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. 10.1038/nrmicro3380 [DOI] [PubMed] [Google Scholar]
- Bruns M. M., Kakarla P., Floyd J. T., Mukherjee M. M., Ponce R. C., Garcia J. A., et al. (2017). Modulation of the multidrug efflux pump EmrD-3 from Vibrio cholerae by Allium sativum extract and the bioactive agent allyl sulfide plus synergistic enhancement of antimicrobial susceptibility by A. sativum extract. Arch. Microbiol. 199, 1103–1112. 10.1007/s00203-017-1378-x [DOI] [PubMed] [Google Scholar]
- Buckley A. M., Webber M. A., Cooles S., Randall L. P., La Ragione R. M., Woodward M. J., et al. (2006). The AcrAB–TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8, 847–856. 10.1111/j.1462-5822.2005.00671.x [DOI] [PubMed] [Google Scholar]
- Cattoir V., Poirel L., Nordmann P. (2008). Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob. Agents Chemother. 52, 3801–3804. 10.1128/AAC.00638-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerceo E., Deitelzweig S. B., Sherman B. M., Amin A. N. (2016). Multidrug-resistant gram-negative bacterial infections in the hospital setting: overview, implications for clinical practice, and emerging treatment options. Microb. Drug Resist. 22, 412–431. 10.1089/mdr.2015.0220 [DOI] [PubMed] [Google Scholar]
- Cha H. J., Muller R. T., Pos K. M. (2014). Switch-loop flexibility affects transport of large drugs by the promiscuous AcrB multidrug efflux transporter. Antimicrob. Agents Chemother. 58, 4767–4772. 10.1128/AAC.02733-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B. C., Han X., Lui S., Wong C., Wang T., Cheung D., et al. (2015). Combating against methicillin-resistant Staphylococcus aureus—two fatty acids from purslane (Portulaca oleracea L.) exhibit synergistic effects with erythromycin. J. Pharm. Pharmacol. 67, 107–116. 10.1111/jphp.12315 [DOI] [PubMed] [Google Scholar]
- Chan B. C., Ip M., Lau C. B., Lui S. L., Jolivalt C., Ganem-Elbaz C., et al. (2011). Synergistic effects of baicalein with ciprofloxacin against NorA over-expressed methicillin-resistant Staphylococcus aureus (MRSA) and inhibition of MRSA pyruvate kinase. J. Ethnopharmacol. 137, 767–773. 10.1016/j.jep.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Chandra H., Bishnoi P., Yadav A., Patni B., Mishra A. P., Nautiyal A. R. (2017). Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials—a review. Plants 6:16. 10.3390/plants6020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Hu D., Zhang Q., Liao X-P., Liu Y-H., Sun J. (2017). Efflux Pump Overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell. Infect. Microbiol. 7:37. 10.3389/fcimb.2017.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R., Chevalier J., Bryskier A., Pagès J. M. (2004). The AcrAB-TolC pump is involved in macrolide resistance but not in telithromycin efflux in Enterobacter aerogenes and Escherichia coli. Antimicrob. Agents Chemother. 48, 3621–3624. 10.1128/AAC.48.9.3621-3624.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovanova R., Mezovska J., Vaverkova S., Mikulasova M. (2015). The inhibition the Tet(K) efflux pump of tetracycline resistant Staphylococcus epidermidis by essential oils from three Salvia species. Lett. Appl. Microbiol. 61, 58–62. 10.1111/lam.12424 [DOI] [PubMed] [Google Scholar]
- Collu F., Vargiu A. V., Dreier J., Cascella M., Ruggerone P. (2012). Recognition of imipenem and meropenem by the RND transporter MexB studied by computer simulations. J. Am. Chem. Soc. 134, 19146–19158. 10.1021/ja307803m [DOI] [PubMed] [Google Scholar]
- Costa S. S., Viveiros M., Amaral L., Couto I. (2013). Multidrug efflux pumps in Staphylococcus aureus: an update. Open Microbiol. J. 7, 59–71. 10.2174/1874285801307010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne S., Rosenfeld N., Lambert T., Courvalin P., Perichon B. (2010). Overexpression of resistance- nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4389–4393. 10.1128/AAC.00155-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos J. F., Tintino S. R., de Freitas T. S., Campina F. F., Irwin R. D. A., Siqueira-Júnior J. P., et al. (2018). In vitro e in silico evaluation of the inhibition of Staphylococcus aureus efflux pumps by caffeic and gallic acid. Comp. Immunol. Microbiol. Infect. Dis. 57, 22–28. 10.1016/j.cimid.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Du D., Venter H., Pos K. M., Luisi B. F. (2013). The machinery and mechanism of multidrug efflux in Gram-negative bacteria, in Microbial Efflux Pumps: Current Research, eds Yu E. W., Zhang Q., Brown M. H. (Ames, IA: Caister Academic Press; ), 35–48. [Google Scholar]
- Du D., Wang Z., James N. R., Voss J. E., Klimont E., Ohene-Agyei T., et al. (2014). Structure of the AcrAB-TolC multidrug efflux pump. Nature 509, 512–515. 10.1038/nature13205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D., Wang-Kan X., Neuberger A., van Veen H. W., Pos K. M., Piddock L. J., et al. (2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. 10.1038/s41579-018-0048-6 [DOI] [PubMed] [Google Scholar]
- Dwivedi G., Maurya A., Yadav D., Khan F., Darokar M., Srivastava S. (2014a). Drug resistance reversal potential of ursolic acid derivatives against nalidixic acid- and multidrug-resistant Escherichia coli. Chem. Biol. Drug Des. 86, 272–283. 10.1111/cbdd.12491 [DOI] [PubMed] [Google Scholar]
- Dwivedi G. R., Gupta S., Maurya A., Tripathi S., Sharma A., Srivastava S. K., et al. (2015). Synergy potential of indole alkaloids and its derivative against drug resistant Escherichia coli. Chem. Biol. Drug Des. 86, 1471–1481. 10.1111/cbdd.12613 [DOI] [PubMed] [Google Scholar]
- Dwivedi G. R., Singh D. P., Sharma S. A., Darokar M. P. (2017a). Efflux Pumps: Warheads of Gram-Negative Bacteria and Efflux Pump Inhibitors in New Approaches in Biological Research, eds Sinha R. P., Richa (New York, NY: Nova Science Publishers; ), 35–72. [Google Scholar]
- Dwivedi G. R., Tyagi R., Sanchita Tripathi S., Pati S., Srivastava S. K., et al. (2017b). Antibiotics potentiating potential of catharanthine against superbug Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. [Epub ahead of print]. 1–15. 10.1080/07391102.2017.1413424. [DOI] [PubMed] [Google Scholar]
- Dwivedi G. R., Upadhyay H. C., Yadav D. K., Singh V., Srivastava S. K., Khan F., et al. (2014b). 4-hydroxy-α-tetralone and its derivative as Drug resistance reversal agents in multidrug resistant Escherichia coli. Chem. Biol. Drug Des. 83, 482–492. 10.1111/cbdd.12263 [DOI] [PubMed] [Google Scholar]
- Eicher T., Cha H. J., Seeger M. A., Brandstatter L., El-Delik J., Bohnert J. A., et al. (2012). Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. U.S.A. 109, 5687–5692. 10.1073/pnas.1114944109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner M., Bhattacharya S., Christiansen B., Gebel J., Goroncy-Bermes P., Hartemann P., et al. (2017). Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control. 12:Doc05. 10.3205/dgkh000290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmy A., Srinivasan A., Webber M. A. (2016). The relationship between bacterial multidrug efflux pumps and biofilm formation, in Efflux-Mediated Antimicrobial Resistance in Bacteria, edd Li X. Z., Elkins C. A., Zgurskaya H. I. (Adis; Cham: Springer International Publishing; ), 651–663. [Google Scholar]
- Fair R. J., Tor Y. (2014). Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 6, 25–64. 10.4137/PMC.S14459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L., Hancock R. (2012). Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681. 10.1128/CMR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiamegos Y. C., Kastritis P. L., Exarchou V., Han H., Bonvin A. M., Vervoort J., et al. (2011). Antimicrobial and efflux pump inhibitory activity of caffeoylquinic acids from Artemisia absinthium against gram-positive pathogenic bacteria. PLoS ONE 6:e18127. 10.1371/journal.pone.0018127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey M., Rahman M., Gibbons S., Piddock L. (2011). Medicinal plant extracts with efflux inhibitory activity against Gram negative bacteria. Int. J. Antimicrob. Agents 37, 145–151. 10.1016/j.ijantimicag.2010.10.027 [DOI] [PubMed] [Google Scholar]
- German N., Kaatz G. W., Kerns R. J. (2008). Synthesis and evaluation of PSSRI-based inhibitors of Staphylococcus aureus multidrug efflux pumps. Bioorg. Med. Chem. Lett. 18, 1368–1373. 10.1016/j.bmcl.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Gill E. E., Franco O. L., Hancock R. E. W. (2015). Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem. Biol. Drug Des. 85, 56–78. 10.1111/cbdd.12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J. E., Keshavan N. D. (2006). Messing with bacterial quorum sensing. Microbiol. Mol. Biol. Rev. 70, 859–875. 10.1128/MMBR.00002-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grkovic S., Brown M. H., Skurray R. A. (2002). Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66, 671–701. 10.1128/MMBR.66.4.671-701.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A., Zoete V., Michielin O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277. 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D., Singh A., Khan A. U. (2017). Nanoparticles as efflux pump and biofilm inhibitor to rejuvenate bactericidal effect of conventional antibiotics. Nanoscale Res. Lett. 12, 454. 10.1186/s11671-017-2222-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannula M., Hänninen M. L. (2008). Effect of putative efflux pump inhibitors and inducers on the antimicrobial susceptibility of Campylobacter jejuni and Campylobacter coli. J. Med. Microbiol. 57, 851–855. 10.1099/jmm.0.47823-0 [DOI] [PubMed] [Google Scholar]
- Hansen L. H., Johannesen E., Burmolle M., Sørensen A. H., Sørensen S. J. (2004). Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 48, 3332–3337. 10.1128/AAC.48.9.3332-3337.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Szewczyk P., Karyakin A., Evin M., Hong W. X. (2010). Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994. 10.1038/nature09408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F. (2007). Multiple molecular mechanisms for multidrug resistance transporters. Nature 446, 749–757. 10.1038/nature05630 [DOI] [PubMed] [Google Scholar]
- Hirakata Y., Srikumar R., Poole K., Gotoh N., Suematsu T., Kohno S., et al. (2002). Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196, 109–118. 10.1084/jem.20020005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs E. C., Yin X., Paul B. J., Astarita J. L., Storz G. (2012). Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 16696–16701. 10.1073/pnas.1210093109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler J. G., Christensen S. B., Slotved H. C., Rasmussen H. B., Gúzman A., Olsen C. E., et al. (2012). Novel inhibitory activity of the Staphylococcus aureus NorA efflux pump by a kaempferol rhamnoside isolated from Persea lingue Nees. J. Antimicrob. Chemother. 67, 1138–1144. 10.1093/jac/dks005 [DOI] [PubMed] [Google Scholar]
- Huda N., Lee E. W., Chen J., Morita Y., Kuroda T., Mizushima T., et al. (2003). Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 47, 2413–2417. 10.1128/AAC.47.8.2413-2417.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack D. L., Yang N. M., Saier M. H. (2001). The drug/metabolite transporter superfamily. Eur. J. Biochem. 68, 3620–3639. 10.1046/j.1432-1327.2001.02265.x [DOI] [PubMed] [Google Scholar]
- Jamshidi S., Sutton J. M., Rahman K. M. (2016). An overview of bacterial efflux pumps and computational approaches to study efflux pump inhibitors. Future Med. Chem. 8, 195–210. 10.4155/fmc.15.173 [DOI] [PubMed] [Google Scholar]
- Kaatz G. W., McAleese F., Seo S. M. (2005). Multidrug resistance in Staphylococcus aureu s due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49, 1857–1864. 10.1128/AAC.49.5.1857-1864.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarla P., Floyd J., Mukherjee M., Devireddy A. R., Inupakutika M. A., Ranweera I., et al. (2016). Inhibition of the multidrug efflux pump LmrS from Staphylococcus aureus by cumin spice Cuminum cyminum. Arch. Microbiol. 199, 465–474. 10.1007/s00203-016-1314-5 [DOI] [PubMed] [Google Scholar]
- Kalia N. P., Mahajan P., Mehra R., Nargotra A., Sharma J. P., Koul S., et al. (2012). Capsaicin, a novel inhibitor of the NorA efflux pump, reduces the intracellular invasion of Staphylococcus aureus. J. Antimicrob. Chemother. 67, 2401–2408. 10.1093/jac/dks232 [DOI] [PubMed] [Google Scholar]
- Kayser F. H., Benner E. J., Hoeprich P. D. (1970). Acquired and native resistance of Staphylococcus aureus to cephalexin and other beta-lactam antibiotics. Appl. Microbiol. 20, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesherwani M., Michael Gromiha M., Fukui K., Velmurugan D. (2017). Identification of novel natural inhibitor for NorM–a multidrug and toxic compound extrusion transporter–an insilico molecular modeling and simulation studies. J. Biomol. Struct. Dyn. 35, 58–77. 10.1080/07391102.2015.1132391 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Kim S. H., Jeon S. M., Park M. S., Rhie H. G., Lee B. K. (2008). Resistance to fluoroquinolones by the combination of target site mutations and enhanced expression of genes for efflux pumps in Shigella flexneri and Shigella sonnei strains isolated in Korea. Clin. Microbiol. Infec. 14, 760–765. 10.1111/j.1469-0691.2008.02033.x [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Nishino K., Yamaguchi A. (2001). Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644. 10.1128/JB.183.19.5639-5644.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T., Michea-Hamzehpour M., Henze U., Gotoh N., Curty L. K., Pechere J. C. (1997). Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23, 345–354. 10.1046/j.1365-2958.1997.2281594.x [DOI] [PubMed] [Google Scholar]
- Kourtesi C., Ball A., Huang Y., Jachak S., Vera D., Khondkar P., et al. (2013). Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol. J. 7, 34–52. 10.2174/1874285801307010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Shriram V., Mulla J. (2013). Antibiotic resistance reversal of multiple drug resistant bacteria using Piper longum fruit extract. J. Appl. Pharm. Sci. 3, 112–116. 10.7324/JAPS.2013.30322 [DOI] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist M., Hancock V., Klemm P. (2008). Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74, 7376–7382. 10.1128/AEM.01310-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. W., Huda M. N., Kuroda T., Mizushima T., Tsuchiya T. (2003). EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 47, 3733–3738. 10.1128/AAC.47.12.3733-3738.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennen R. M., Politz M. G., Kruziki M. A., Pfleger B. F. (2013). Identification of transport proteins involved in free fatty acid efflux in Escherichia coli. J. Bacteriol. 195, 135–144. 10.1128/JB.01477-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B. (2001). Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33, S124–S129. 10.1086/321837 [DOI] [PubMed] [Google Scholar]
- Li X-Z., Plésiat P., Nikaido H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Chen Y. M., Chen Y., Cheng Y. Y., Zhang L. H. (2016). RND efflux pump and its interrelationship with quorum sensing system. Hereditas (Beijing), 38, 894–901. 10.16288/j.yczz.16-139 [DOI] [PubMed] [Google Scholar]
- Limaverde P. W., Campina F. F., da Cunha F. A., Crispim F. D., Figueredo F. G., Lima L. F., et al. (2017). Inhibition of the TetK efflux-pump by the essential oil of Chenopodium ambrosioides L. and α-terpinene against Staphylococcus aureus IS-58. Food Chem. Toxicol. 109, 957–961. 10.1016/j.fct.2017.02.031 [DOI] [PubMed] [Google Scholar]
- Lin J., Michel L. O., Zhang Q. (2002). CmeABC functions as a multidrug effl ux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. 10.1128/AAC.46.7.2124-2131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister P. D., Wolter D. J., Hanson N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. 10.1128/CMR.00040-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlejohn T. G., Paulsen I. T., Gillespie M. T., Tennent J. M., Midgley M., Jones I. G., et al. (1992). Substrate specifi city and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 95, 259–265. 10.1111/j.1574-6968.1992.tb05376.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Hassan K. A., Ashwood H. E., Gamage H. K. A. H., Li L., Mabbutt B. C., et al. (2018). Regulation of the aceI multidrug efflux pump gene in Acinetobacter baumannii. J. Antimicrob. Chemother. 73, 1492–1500. 10.1093/jac/dky034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Warren M. S., Lee A., Galazzo J., Fronko R., Lee M., et al. (2001). Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45, 105–116. 10.1128/AAC.45.1.105-116.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Parsons S. M. (2010). Conformational propensities of peptides mimicking transmembrane helix 5 and motif C in wild-type and mutant vesicular acetylcholine transporters. ACS Chem. Neurosci. 1, 381–390. 10.1021/cn900033s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch A. S. (2006). Efflux systems in bacterial pathogens: an opportunity for therapeutic intervention? An industry view. Biochem. Pharmacol. 71, 949–956. 10.1016/j.bcp.2005.10.021 [DOI] [PubMed] [Google Scholar]
- Ma D., Alberti M., Lynch C., Nikaido H., Hearst J. E. (1996). The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19, 101–112. [DOI] [PubMed] [Google Scholar]
- Maisuria V., Hosseinidoust Z., Tufenkji N. (2015). Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Appl. Environ. Microbiol. 81, 3782–3792. 10.1128/AEM.00239-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiaterra G., Laudadio E., Cometti M., Mobbili G., Minnelli C., Massaccesi L., et al. (2017). Inhibitors of multidrug efflux pumps of Pseudomonas aeruginosa from natural sources: an in silico high-throughput virtual screening and in vitro validation. Med. Chem. Res. 26, 414–430. 10.1007/s00044-016-1761-1 [DOI] [Google Scholar]
- Marrer E., Schad K., Satoh A. T., Page M. G., Johnson M. M., Piddock L. J. (2006). Involvement of the putative ATP-dependent effl ux proteins PatA and PatB in fluoroquinolone resistance of a multidrug- resistant mutant of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 50, 685–693. 10.1128/AAC.50.2.685-693.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., Sánchez M. B., Martínez-Solano L., Hernandez A., Garmendia L., Fajardo A., et al. (2009). Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbial. Rev. 33, 430–449. 10.1111/j.1574-6976.2008.00157.x [DOI] [PubMed] [Google Scholar]
- Martins M., Dastidar S. G., Fanning S., Kristiansen J. E., Molnar J., Pagès J. M., et al. (2008). Potential role of non-antibiotics (helper compounds) in the treatment of multidrug- resistant Gram-negative infections: mechanisms for their direct and indirect activities. Int. J. Antimicrob. Agents 31, 198–208. 10.1016/j.ijantimicag.2007.10.025 [DOI] [PubMed] [Google Scholar]
- Masi M., Refregiers M., Pos K. M., Pages J. M. (2017). Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2:17001. 10.1038/nmicrobiol.2017.1 [DOI] [PubMed] [Google Scholar]
- Maurya A., Dwivedi G., Darokar M., Srivastava S. (2013). Antibacterial and synergy of clavine alkaloid lysergol and its derivatives against nalidixic acid-resistant Escherichia coli. Chem. Biol. Drug Des. 81, 484–490. 10.1111/cbdd.12103 [DOI] [PubMed] [Google Scholar]
- May T., Ito A., Okabe S. (2009). Induction of multidrug resistance mechanism in Escherichia coli biofilms by interplay between tetracycline and ampicillin resistance genes. Antimicrob. Agents Chemother. 53, 4628–4639. 10.1128/AAC.00454-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár J., Engi H., Hohmann J., Molnár P., Deli J. (2010). Reversal of multidrug resistance by natural substances from plants. Curr. Topics Medi. Chem. 10, 1757–1768. 10.2174/156802610792928103 [DOI] [PubMed] [Google Scholar]
- Moore J. D., Gerdt J. P., Eibergen N. R., Blackwell H. E. (2014). Active efflux influences the potency of quorum sensing inhibitors in Pseudomonas aeruginosa. Chembiochem 15, 435–442. 10.1002/cbic.201300701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand B., Mühlemann K. (2007). Heteroresistance to penicillin in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 104, 14098–14103. 10.1073/pnas.0702377104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Kodama K., Shiota S., Mine T., Kataoka A., Mizushima T., et al. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782. 10.1128/AAC.42.7.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 16, 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta S. S., Cluzel P., Aldana M. (2015). Adaptive resistance in bacteria requires epigenetic inheritance, genetic noise, and cost of efflux pumps. PLoS ONE 10:e0118464. 10.1371/journal.pone.0118464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukanganyama S., Chirisa E., Hazra B. (2012). Antimycobacterial activity of diospyrin and its derivatives against Mycobacterium aurum. Res. Pharm. 2, 1–13. [Google Scholar]
- Nakashima R., Sakurai K., Yamasaki S., Hayashi K., Nagata C., Hoshino K., Onodera Y., Nishino K., et al. (2013). Structural basis for the inhibition of bacterial multidrug exporters. Nature 500, 102–106. 10.1038/nature12300 [DOI] [PubMed] [Google Scholar]
- Napier B. A., Band V., Burd E. M., Weiss D. S. (2014). Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob. Agents Chemother. 58, 5594–5597. 10.1128/AAC.02432-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D., Cragg G. (2012). Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335. 10.1021/np200906s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S. T., Kwasny S. M., Ding X., Cardinale S. C., McCarthy C. T., Kim H-S., et al. (2015). Structure–activity relationships of a novel pyranopyridine series of Gram negative bacterial efflux pump inhibitors. Bioorg. Med. Chem. 23, 2024–2034. 10.1016/j.bmc.2015.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Li Y., Guan J., Zhao J., Cui J., Wang R., et al. (2016). Effects of efflux pump inhibitors on colistin resistance in multidrug resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 60, 3215–3218. 10.1128/AAC.00248-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R. J., Sen S., Choo Y. J., Beltrao P., Zietek M., Chaba R., et al. (2011). Phenotypic landscape of a bacterial cell. Cell 144, 143–156. 10.1016/j.cell.2010.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Takatsuka Y. (2009). Mechanisms of RND multidrug efflux pumps. Biochem. Biophys. Acta 1794, 769–781. 10.1016/j.bbapap.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino K., Latifi T., Groisman E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141. 10.1111/j.1365-2958.2005.04940.x [DOI] [PubMed] [Google Scholar]
- Norgan A. P., Freese J. M., Tuin P. M., Cunningham S. A., Jeraldo P. R., Patel R. (2016). Carbapenem- and colistin-resistant Enterobacter cloacae from Delta, Colorado, in 2015. Antimicrob. Agents Chemother. 60, 3141–3144. 10.1128/AAC.03055-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa W., Minato Y., Dodan H., Onishi M., Tsuchiya T., Kuroda T. (2015). Characterization of MATE-type multidrug efflux pumps from Klebsiella pneumoniae MGH78578. PLoS ONE 10:e0121619. 10.1371/journal.pone.0121619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohene-Agyei T., Mowla R., Rahman T., Venter H. (2014). Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. Microbiologyopen 3, 885–896. 10.1002/mbo3.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada U., Yamashita E., Neuberger A., Morimoto M., van Veen H. W., Murakami S. (2017). Crystal structure of tripartite-type ABC transporter MacB from Acinetobacter baumannii. Nat. Commun, 8, 1336. 10.1038/s41467-017-01399-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T. J., Kwasny S. M., Kim H. S., Nguyen S. T., Houseweart C., D'Souza S., et al. (2014). Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 58, 722–733. 10.1128/AAC.01866-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T. J., Nguyen S. T. (2015). Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 6:421. 10.3389/fmicb.2015.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès J. M., Amaral L. (2009). Mechanisms of drug efflux and strategies to combat them: challenging the efflux pump of Gram-negative bacteria. Biochim. Biophys. Acta, Proteins Proteomics 1794, 826–833. 10.1016/j.bbapap.2008.12.011 [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Piddock L. J. (2006). Multidrug-resistance efflux pumps-Not just for resistance. Nat. Rev. Microbiol. 4, 629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- Piddock L. J. (2014). Understanding the basis of antibiotic resistance: a platform for drug discovery. Microbiology 160, 2366–2373. 10.1099/mic.0.082412-0 [DOI] [PubMed] [Google Scholar]
- Ponnusamy K., Ramasamy M., Savarimuthu I., Paulraj M. G. (2010). Indirubin potentiates ciprofloxacin activity in the NorA efflux pump of Staphylococcus aureus. Scand. J. Infect. Dis. 42, 500–505. 10.3109/00365541003713630 [DOI] [PubMed] [Google Scholar]
- Poole K. (2004). Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10, 12–26. 10.1111/j.1469-0691.2004.00763.x [DOI] [PubMed] [Google Scholar]
- Poole K. (2005). Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56, 20–51. 10.1093/jac/dki171 [DOI] [PubMed] [Google Scholar]
- Poole K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162–176. 10.1080/07853890701195262 [DOI] [PubMed] [Google Scholar]
- Poole K., Gotoh N., Tsujimoto H., Zhao Q., Wada A., Yamasaki T., et al. (1996). Overexpression of the mexC- mexD- oprJ effl ux operon in nfxB-type multi drug resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724. 10.1046/j.1365-2958.1996.281397.x [DOI] [PubMed] [Google Scholar]
- Prasch S., Bucar F. (2015). Plant derived inhibitors of bacterial efflux pumps: an update. Phytochem. Rev. 14:961 10.1007/s11101-015-9436-y [DOI] [Google Scholar]
- Purssell A., Poole K. (2013). Functional characterization of the NfxB repressor of the mexCD -oprJ multidrug efflux operon of Pseudomonas aeruginosa. Microbiology 159, 2058–2073. 10.1099/mic.0.069286-0 [DOI] [PubMed] [Google Scholar]
- Putman M., van Veen H., Konings W. (2000). Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693. 10.1128/MMBR.64.4.672-693.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman T., Yarnall B., Doyle D. A. (2017). Efflux drug transporters at the forefront of antimicrobial resistance. Eur. Biophys. J. 46, 647–653. 10.1007/s00249-017-1238-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamohan G., Srinivasan V. B., Gebreyes W. A. (2010). Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J. Antimicrob. Chemother. 65, 228–232. 10.1093/jac/dkp427 [DOI] [PubMed] [Google Scholar]
- Ramaswamy V. K., Cacciotto P., Malloci G., Vargiu A. V., Ruggerone P. (2017). Computational modelling of efflux pumps and their inhibitors. Essays Biochem. 61, 141–156. 10.1042/EBC20160065 [DOI] [PubMed] [Google Scholar]
- Ramón-García S., Mick V., Dainese E., Martin C., Thompson C. J., De Rossi E., et al. (2012). Functional and genetic characterization of the Tap efflux pump in Mycobacterium bovis BCG. Antimicrob. Agents Chemother. 56, 2074–2083. 10.1128/AAC.05946-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A., Liu Y., Michaelis M. L., Himes R. H., Georg G. I., Audus K. L. (2005). Chemical modification of paclitaxel (Taxol) reduces P-glycoprotein interactions and increases permeation across the blood-brain barrier in vitro and in situ. J. Med. Chem. 48, 832–838. 10.1021/jm040114b [DOI] [PubMed] [Google Scholar]
- Roca I., Marti S., Espinal P., Martinez P., Gibert I., Vila J. (2009). CraA: an MFS efflux pump associated with chloramphenicol resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 4013–4014. 10.1128/AAC.00584-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L., Villellas C., Bailo R., Viveiros M., Ainsa J. A. (2013). Role of the Mmr efflux pump in drug resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 57, 751– 757. 10.1128/AAC.01482-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Pahwa S., Nandanwar H., Jachak S. (2012). Phenylpropanoids of Alpinia galanga as efflux pump inhibitors in Mycobacterium smegmatis mc2 155. Fitoterapia 83, 1248–1255. 10.1016/j.fitote.2012.06.008 [DOI] [PubMed] [Google Scholar]
- Roy S. K., Kumari N., Pahwa S., Agrahari U. C., Bhutani K. K., Jachak S. M., et al. (2013). NorA efflux pump inhibitory activity of coumarins from Mesua ferrea. Fitoterapia 90, 140–150. 10.1016/j.fitote.2013.07.015 [DOI] [PubMed] [Google Scholar]
- Ruzin A., Keeney D., Bradford P. A. (2007). AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Antimicrob. Chemother. 59, 1001–1004. 10.1093/jac/dkm058 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Piccioni M., Felicetti T., De Marco S., Manfroni G., Pagiotti R., et al. (2017). Investigation on the effect of known potent S. aureus NorA efflux pump inhibitors on the staphylococcal biofilm formation. RSC Adv. 7, 37007–37014. 10.1039/C7RA03859C [DOI] [Google Scholar]
- Saier M. H., Paulsen I. T., Sliwinski M. K., Pao S. S., Skurray R. A. (1998). Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12, 265–274. 10.1096/fasebj.12.3.265 [DOI] [PubMed] [Google Scholar]
- Santajit S., Indrawattana N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res. Int. 2016:2475067. 10.1155/2016/2475067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler B. D., Kaatz G. W. (2016). Multidrug efflux pumps of Gram-positive bacteria. Drug Resist. Updat. 27, 1–13. 10.1016/j.drup.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Schuldiner S. (2009). EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim. Biophys. Acta 1794, 748–762. 10.1016/j.bbapap.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Sharma S., Kumar M., Sharma S., Nargotra A., Koul S., Khan I. A. (2010). Piperine as an inhibitor of Rv1258c, a putative multidrug efflux pump of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 1694–1701. 10.1093/jac/dkq186 [DOI] [PubMed] [Google Scholar]
- Shigemura K., Osawa K., Kato A., Tokimatsu I., Arakawa S., Shirakawa T., et al. (2015). Association of overexpression of efflux pump genes with antibiotic resistance in Pseudomonas aeruginosa strains clinically isolated from urinary tract infection patients. J. Antibiot. 68, 568. 10.1038/ja.2015.34 [DOI] [PubMed] [Google Scholar]
- Shin J., Prabhakaran V. S., Kim K. S. (2018). The multi-faceted potential of plant-derived metabolites as antimicrobial agents against multidrug-resistant pathogens. Microb. Pathog. 116, 209–214. 10.1016/j.micpath.2018.01.043 [DOI] [PubMed] [Google Scholar]
- Shiu W., Malkinson J., Rahman M., Curry J., Stapleton P., Gunaratnam M., et al. (2013). A new plant-derived antibacterial is an inhibitor of efflux pumps in Staphylococcus aureus. Int. J. Antimicrob. Agents 42, 513–518. 10.1016/j.ijantimicag.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Kahne D., Walker S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriyong T., Srimanote P., Chusri S., Yingyongnarongkul B. E., Suaisom C., Tipmanee V., et al. (2017). Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 17:405. 10.1186/s12906-017-1913-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler G., Kincses A., Gajdacs M., Amaral L. (2017). New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules 22:468. 10.3390/molecules22030468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V. B., Rajamohan G. (2013). KpnEF, a new member of the Klebsiella pneumonia cell envelope stress response regulon, is an SMR-type efflux pump involved in broadspectrum antimicrobial resistance. Antimicrob. Agents Chemother. 57, 4449–4462. 10.1128/AAC.02284-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V. B., Singh B. B., Priyadarshi N., Chauhan N. K., Rajamohan G. (2014). Role of novel multidrug efflux pump involved in drug resistance in Klebsiella pneumoniae. PLoS ONE 9:e96288. 10.1371/journal.pone.0096288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavri M., Piddock L., Gibbons S. (2007). Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 59, 1247–1260. 10.1093/jac/dkl460 [DOI] [PubMed] [Google Scholar]
- Su X. Z., Chen J., Mizushima T., Kuroda T., Tsuchiya T. (2005). AbeM, an H+-coupled Acinetobacter baumannii multidrug efflux pump belonging to the MATE family of transporters. Antimicrob. Agents Chemother. 49, 4362–4364. 10.1128/AAC.49.10.4362-4364.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Deng Z., Yan A. (2014). Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. 10.1016/j.bbrc.2014.05.090 [DOI] [PubMed] [Google Scholar]
- Suresh M., Nithya N., Jayasree P. R., Vimal K. P., Kumar P. M. (2018). Mutational analyses of regulatory genes, mexR, nalC, nalD and mexZ of mexAB-oprM and mexXY operons, in efflux pump hyperexpressing multidrug-resistant clinical isolates of Pseudomonas aeruginosa. World J. Microbiol. Biotechnol. 34, 83. 10.1007/s11274-018-2465-0 [DOI] [PubMed] [Google Scholar]
- Suriyanarayanan B., Sarojini S. R. (2015). Docking analysis insights quercetin can be a non-antibiotic adjuvant by inhibiting Mmr drug efflux pump in Mycobacterium sp. and its homologue EmrE in Escherichia coli. J. Biomol. Struct. Dyn. 33, 1819–1834. 10.1080/07391102.2014.974211 [DOI] [PubMed] [Google Scholar]
- Swick M. C., Morgan-Linnell S. K., Carlson K. M., Zechiedrich L. (2011). Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrob. Agents Chemother. 55, 921–924. 10.1128/AAC.00996-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait-Kamradt A., Clancy J., Cronan M., Dib-Hajj F., Wondrack L., Yuan W., et al. (1997). mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41, 2251–2255. 10.1128/AAC.41.10.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telke A. A., Olaitan A. O., Morand S., Rolain J-M. (2017). soxRS indices colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J. Antimicrob. Chemother. 72, 2715–2721. 10.1093/jac/dkx215 [DOI] [PubMed] [Google Scholar]
- Tikhonova E. B., Yamada Y., Zgurskaya H. I. (2011). Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem. Biol. 18, 454–463. 10.1016/j.chembiol.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargiu A. V., Ramaswamy V. K., Malvacio I., Malloci G., Kleinekathöfer U., Ruggerone P. (2018). Water-mediated interactions enable smooth substrate transport in a bacterial efflux pump. Biochim. Biophys. Acta Gen. Sub. 1862, 836–845. 10.1016/j.bbagen.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Venter H., Mowla R., Ohene-Agyei T., Ma S. (2015). RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6:377 10.3389/fmicb.2015.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P., Tiwari M., Tiwari V. (2017). In silico high-throughput virtual screening and molecular dynamics simulation study to identify inhibitor for AdeABC efflux pump of Acinetobacter baumannii. J. Biomol. Struct. Dyn. 36, 1182–1194. 10.1080/07391102.2017.1317025 [DOI] [PubMed] [Google Scholar]
- Vimberg V., Lenart J., Janata J., Balikova Novotna G. (2015). ClpP-independent function of ClpX interferes with telithromycin resistance conferred by Msr(A) in Staphylococcus aureus. Antimicrob. Agents Chemother. 59, 3611–3614. 10.1128/AAC.04367-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viveiros M., Jesus A., Brito M., Leandro C., Martins M., Ordway D., et al. (2005). Inducement and reversal of tetracycline resistance in Escherichia coli K-12 and expression of proton gradient-dependent multidrug efflux pump genes. Antimicrob. Agents Chemother. 49, 3578–3582. 10.1128/AAC.49.8.3578-3582.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A., Hoyle C., Palmer S., O'Reilly J., Griffith J. (2001). Prokaryote multidrug efflux proteins of the major facilitator superfamily: amplified expression, purification and characterisation. J. Mol. Microbiol. Biotechnol. 3, 193–200. [PubMed] [Google Scholar]
- Warner D. M., Levy S. B. (2010). Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): rob-dependent CAMP induction of the marRAB operon. Microbiol. 156, 570–578. 10.1099/mic.0.033415-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T. (1963). Infective heredity of multiple drug resistance in bacteria. Bacteriol. Rev. 27, 87–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch A., Awah C. U., Jing S., van Veen H. W., Venter H. (2010). Promiscuous partnering and independent activity of MexB, the multidrug transporter protein from Pseudomonas aeruginosa. Biochem. J. 430, 355–364. 10.1042/BJ20091860 [DOI] [PubMed] [Google Scholar]
- Wendlandt S., Kadlec K., Fessler A. T., Schwarz S. (2015). Identification of ABC transporter genes conferring combined pleuromutilin-lincosamide-streptogramin A resistance in bovine methicillin- resistant Staphylococcus aureus and coagulase-negative staphylococci. Vet. Microbiol. 177, 353–358. 10.1016/j.vetmic.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Willers C., Wentzel J. F., du Plessis L. H., Gouws C., Hamman J. H. (2016). Efflux as a mechanism of antimicrobial drug resistance in clinical relevant microorganisms: the role of efflux inhibitors. Expert Opin. Ther. Targets 21, 23–36. 10.1080/14728222.2017.1265105 [DOI] [PubMed] [Google Scholar]
- World Economic Forum (2013). Global Risks, Eighth Edition Ed. 2013. 28–33. Available online at: http://www3.weforum.org/docs/WEF_GlobalRisks_Report_2013.pdf (Accessed March 15, 2018).
- World Health Organization (2014). Antimicrobial Resistance. Available online at: http://www.searo.who.int/thailand/factsheets/fs0023/en/ (Accessed October 22, 2018). [Google Scholar]
- Wright G. D. (2016). Antibiotic adjuvants: Rescuing antibiotics from resistance. Trends Microbiol. 24, 862–871. 10.1016/j.tim.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Yaffe D., Radestock S., Shuster Y., Forrest L. R., Schuldiner S. (2013). Identification of molecular hinge points mediating alternating access in the vesicular monoamine transporter VMAT2. Proc. Natl. Acad. Sci. U.S.A. 110, E1332–1341. 10.1073/pnas.1220497110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Lopez C. R., Zechiedrich E. L. (2006). Quorum sensing and multidrug transporters in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 2386–2391. 10.1073/pnas.0502890102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. K., Low L. Y., Yap P. S. X., Yusoff K., Mai C. W., Lai K. S., et al. (2018). Plant-Derived Antimicrobials: Insights into Mitigation of Antimicrobial Resistance. Rec. Nat. Prod. 12, 295–316. 10.25135/rnp.41.17.09.058 [DOI] [Google Scholar]
- Yasufuku T., Shigemura K., Shirakawa T., Matsumoto M., Nakano Y., Tanaka K., et al. (2011). Correlation of overexpression of efflux pump genes with antibiotic resistance in Escherichia coli strains clinically isolated from urinary tract infection patients. J. Clin. Microbiol. 49, 189–194. 10.1128/JCM.00827-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Bogaki M., Nakamura S., Ubukata K., Konno M. (1990). Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172, 6942–6949. 10.1128/jb.172.12.6942-6949.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B., Wang H., Zou L., Zhang A., Yang X., Guan Z. (2010). Evaluation and target validation of indole derivatives as inhibitors of the AcrAB-TolC efflux pump. Biosci. Biotechnol. Biochem. 74, 2237–2241. 10.1271/bbb.100433 [DOI] [PubMed] [Google Scholar]
- Zgurskaya H. I., Lopez C. A., Gnanakaran S. (2015). Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 1, 512–522. 10.1021/acsinfecdis.5b00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun Y., Wang Y., Lu M., He J., Liu J., et al. (2014). Non-antibiotic agent ginsenoside 20(S)-Rh2 enhanced the antibacterial effects of ciprofloxacin in vitro and in vivo as a potential NorA inhibitor. Eur. J. Pharmacol. 740, 277–284. 10.1016/j.ejphar.2014.07.020 [DOI] [PubMed] [Google Scholar]