Abstract
Bronchopulmonary dysplasia in premature infants is a common and often severe lung disease with long-term sequelae. A genetic component is suspected but not fully defined. We performed an ancestry and genome-wide association study to identify variants, genes, and pathways associated with survival without bronchopulmonary dysplasia in 387 high-risk infants treated with inhaled nitric oxide in the Trial of Late Surfactant study. Global African genetic ancestry was associated with increased survival without bronchopulmonary dysplasia among infants of maternal self-reported Hispanic white race/ethnicity [odds ratio (OR) = 4.5, P = 0.01]. Admixture mapping found suggestive outcome associations with local African ancestry at chromosome bands 18q21 and 10q22 among infants of maternal self-reported African-American race/ethnicity. For all infants, the top individual variant identified was within the intron of NBL1, which is expressed in midtrimester lung and is an antagonist of bone morphogenetic proteins (rs372271081, OR = 0.17, P = 7.4 × 10−7). The protective allele of this variant was significantly associated with lower nitric oxide metabolites in the urine of non-Hispanic white infants (P = 0.006), supporting a role in the racial differential response to nitric oxide. Interrogating genes upregulated in bronchopulmonary dysplasia lungs indicated association with variants in CCL18, a cytokine associated with fibrosis and interstitial lung disease, and pathway analyses implicated variation in genes involved in immune/inflammatory processes in response to infection and mechanical ventilation. Our results suggest that genetic variation related to lung development, drug metabolism, and immune response contribute to individual and racial/ethnic differences in respiratory outcomes following inhaled nitric oxide treatment of high-risk premature infants.
Keywords: bronchopulmonary dysplasia, drug response, genetic ancestry, genome-wide association study, preterm infants
INTRODUCTION
Bronchopulmonary dysplasia (BPD) of premature infants is currently characterized by continuing requirement for supplemental oxygen and/or respiratory support at 36 wk postmenstrual age (PMA). BPD is the most common form of chronic lung disease in infants born prematurely and is associated with long-term respiratory morbidity, neurodevelopmental abnormalities, and death (33). The pathogenesis of BPD includes lung immaturity, with reduced pulmonary surfactant and low antioxidant and immune defenses, as well as exposure to insults of hyperoxia, barotrauma from ventilator support, and infections that damage lung epithelium and elicit inflammation. Sequelae of this injury are arrested lung development, fibrosis, and altered airway reactivity (7, 18, 24, 31, 33, 37, 47).
Therapeutic options for the prevention and treatment of BPD are limited and have not substantially affected the incidence of disease (reviewed in Refs. 26, 28). For example, vitamin A treatment evokes a modest reduction of BPD but is not in general use, and caffeine reduces oxygen use and is routinely used for prevention of apnea. Postnatal dexamethasone therapy improves respiratory status acutely and decreases the incidence of BPD. However, longer courses of this therapy are associated with neurodevelopmental abnormalities. Inhaled nitric oxide (iNO) is used off-label in preterm infants to prevent BPD, but the general efficacy of the drug has been brought into question (19).
The majority of studies evaluating the effectiveness of iNO have been performed in individuals with predominantly European ancestry (5). However, in the entire cohort of the Trial of Late Surfactant (TOLSURF; Ref. 60) and in a recent individual participant data meta-analysis across selected iNO trials (6), the incidence of BPD was significantly lower following treatment with iNO in infants of mothers who self-report as black/African-American ethnicity compared with those who self-report as non-Hispanic white. Coupled to observed differences in levels of urinary NO metabolites in black/African-American vs. non-Hispanic white infants (8), these results suggest that response to iNO in terms of preventing BPD varies between racial/ethnic groups.
Although both the intrauterine and postnatal environment play an important role in BPD, twin studies have estimated the heritability between 50 and 80% (13, 38), suggesting a genetic contribution as well (29). Genetic studies of BPD have identified several candidate genes and pathways through genome-wide association studies (GWAS; Refs. 4, 29, 43, 61) and exome sequencing (17, 39). However, none of the associations identified through GWAS has reached genome-wide significance, and replication of genetic associations has been problematic. This may, in part, be due to low statistical power given the relatively small sample size of each study (<1,000 preterm infants) combined with the absence of a single genetic risk factor of large effect. Similarly, disease heterogeneity, including the potential for differences in the genetic architecture of BPD between racial/ethnic groups, and the specific definition of BPD used may reduce statistical power (4). However, pathway and gene set enrichment analyses have identified candidates with high biological plausibility (4, 39).
In this study, we performed a GWAS for survival without BPD in preterm infants in TOLSURF, which included infants of maternal self-reported African-American, Hispanic, and non-Hispanic white race/ethnicity who all received iNO. We examine the effects of genetic variation at the level of individual variants, genes, and genetic pathways and test the hypothesis that genetic ancestry at both the genomic and local scale is associated with survival without BPD in admixed populations.
METHODS
Study approval.
Patient recruitment for the TOLSURF study was approved by the Institutional Review Boards at all participating sites, including the University of California, San Francisco.
Study subjects.
TOLSURF was a masked, randomized, sham-controlled trial conducted in 25 US hospitals (https://clinicaltrials.gov/: NCT01022580). The study was designed to assess the effect of late doses of surfactant on BPD at 36 wk PMA in infants of 23–28 wk gestation who required intubation and mechanical ventilation between 7 and 14 days of age (9). A total of 511 infants were enrolled, and all received iNO (Ikaria, Hampton, NJ) according to the protocol followed in the Nitric Oxide (to Prevent) Chronic Lung Disease (NO CLD) trial (10). BPD was assessed at 36 wk PMA by physiological testing as described (10). There was no statistical difference in BPD incidence between control and surfactant-treated groups at 36 wk, and the 2 groups were combined for this genetic study. Some infants were coenrolled in the multicenter, observational Prematurity and Respiratory Outcomes Program (PROP; Ref. 52).
Genotyping and quality control.
DNA was extracted from tracheal aspirate cells from 454 infants whose parents consented for DNA collection using cells from ≤5 tracheal aspirate collections per patient, obtained between postnatal days 7 and 21. DNA was isolated using an AutoGeneprep 965 instrument (AutoGen, Holliston, MA) by the manufacturer’s recommended standard protocol for human body fluids. In some cases, where protein contamination was evident, DNA was reprecipitated using 3 volumes of 100% ethanol and 3 M ammonium acetate at a 3:1 ratio after incubation at −80°C overnight. Samples were initially quantified by NanoDrop (Thermo Fisher Scientific, Waltham, MA) to assess purity [absorbance at 260 and 280 nm (A260/280)] followed by analysis using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) to assess DNA quantity more accurately. The values for DNA concentration (in ng/μl) ranged from 10 to 1,750, median 130; total DNA per patient (in ng) 130–4,200, median 1,600; A260/280, 1.32–1.91, median 1.77; 429 samples were of suitable quality and quantity for genotyping.
Genotyping was performed on the Affymetrix Axiom LAT 1 Array (World Array 4) that contained >800,000 single-nucleotide polymorphisms (SNPs) before quality control. SNPs were filtered based on call rates <95% and Hardy-Weinberg equilibrium P values <10−6 using PLINK (53). Subjects were evaluated for call rates, consistency between genetic and reported sex, autosomal heterozygosity, and cryptic relatedness/genetic identity using identical by descent/identical by state estimates in PLINK (53). In the case of multiples, one individual was selected at random to be included in the study.
Statistical analysis.
We inferred levels of African ancestry at both the genomic level (average across the genome) and the local level (at an individual locus). Genomic levels of African ancestry were evaluated using ADMIXTURE (3) in a quasisupervised analysis assuming three ancestral continental populations of origin (k = 3, African, European, and Native American). Windows were offset by a factor of 0.2, the cutoff for linkage was set to 0.1, and a constant recombination rate was set to 10−8/bp. The proportion of global African ancestry was compared between cases (BPD/death) vs. controls (survival without BPD) using logistic regression within infants of maternal self-reported African-American race/ethnicity and those of Hispanic ethnicity, adjusting for gestational age, sex, birth weight, and multiple gestation (yes/no). Local ancestry was inferred using LAMP-LD (11) in infants with maternal self-reported African-American race/ethnicity using a two-population model. Unrelated individuals from the International Haplotype Map (HapMap) Project of African ancestry [Yoruba in Ibadan, Nigeria (YRI)] and European ancestry [Utah residents with Northern and Western European ancestry (CEU)] were used as a reference to estimate global and local African and European ancestry.
Imputation of genetic variation from the Phase 3 1000 Genomes Project was performed using the Michigan Imputation Server (21), including ~79 million variants across all populations. Variants were then filtered for imputation quality scores >0.3. Genetic association testing for survival without BPD was performed at both genotyped and imputed SNPs using logistic regression, adjusting for global genetic ancestry, gestational age, sex, birth weight, and multiple gestation. Analyses were performed within each racial/ethnic group using PLINK (53) and then combined in a meta-analysis using METAL (64). Gene-based statistics were calculated using versatile gene-based association study (VEGAS; Ref. 41) using genotyped SNPs and intersected with a set of genes previously identified as being upregulated in BPD-dysregulated lungs (14). Pathway and gene set analyses were performed using canonical pathways in Ingenuity Pathway Analysis (IPA; Ref. 36) and Protein Analysis THrough Evolutionary Relationships (PANTHER; Ref. 46) and Molecular Signature Database (MSigDB; Ref. 56) using Genomic Regions Enrichment of Annotations Tool (GREAT) version 3.0.0 (45). With the use of GREAT, we assigned a foreground of gene coordinates with an association P > 0.05 for survival without BPD and a background of all gene coordinates for which a gene-based statistic was calculated (from VEGAS; Ref. 41).
Admixture mapping for local African ancestry was performed in infants with maternal self-reported African-American race/ethnicity using logistic regression. Similar to association testing on individual variants, we performed association testing for the number of haplotypes of African ancestry at each genotyped SNP (homologous to association testing for the number of copies of the minor allele). Identical to our GWAS, we adjusted for global genetic ancestry, gestational age, sex, birth weight, and multiple gestation. To account for multiple testing, we estimated the independent number of tests using the coda package in R and applied a Bonferroni correction.
Measures of NO metabolites (NOx), including nitrate, nitrite, and nitrosylated compounds, were made from the urine of 62 infants included in the current genetic study collected between 6 and 65 days postnatal age, both before and following administration of iNO at 2–20 ppm as previously described (8). Briefly, urine was collected for 4–8 h, and NOx were assayed according to Ref. 50 and normalized to creatinine to adjust for renal excretory function. NOx were measured off iNO and at three different doses of iNO (2, 5, and 10–20 ppm). Genetic association testing at a single SNP was performed using linear regression to test for a correlation between genotype and values of NOx at a dose of 5 ppm. Values of NOx at 5 ppm were selected for analysis because they are highly correlated to levels at 2 ppm and more closely resemble a normal distribution compared with 10–20 ppm.
For selected genes of interest, mRNA expression levels were obtained from a previous study that performed RNA sequencing on three specimens of human fetal lung of 23 wk gestational age (Gene Expression Omnibus acc. no. GSE83888; Ref. 12).
RESULTS
Following quality control, our study included a total of 795,465 genotyped SNPs and 387 unrelated infants; demographics by respiratory outcome are shown in Table 1 for 271 infants who died or had a diagnosis of BPD and 116 survivors without BPD. Overall, mean values for gestational age and birth weight were approximately 25 wk and 700 g, respectively, for this group of infants who still required intubation and ventilation between 7 and 14 days of age, representing a cohort at high risk for BPD. Within infants of maternal self-reported non-Hispanic white ethnicity (white), infants with BPD/death had a significantly higher respiratory severity score on study entry compared with survivors without BPD but had no significant difference in gestational age, birth weight, sex, and multiple gestations. Within infants of maternal self-reported black/African-American ethnicity (black/AA), infants with BPD/death had significantly lower gestational age, lower birth weight, and higher respiratory severity score compared with no BPD. These differences for infants with/without BPD are consistent with the known influence of immaturity and severity of early lung disease on BPD. No significant differences in clinical characteristics were observed between the two groups of maternal self-reported white Hispanic ethnicity (white Hispanic).
Table 1.
Baseline characteristics of participants from the TOLSURF study included in the GWAS
| Non-Hispanic White |
African American |
Hispanic White |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| BPD/Death | No BPD | P Value | BPD/Death | No BPD | P Value | BPD/Death | No BPD | P Value | |
| n | 136 | 41 | N/A | 82 | 51 | N/A | 28 | 14 | N/A |
| Gestational age, wk | 25.4 (1.3) | 25.2 (1.2) | 0.52 | 24.9 (1.0) | 25.4 (1.0) | 0.008 | 24.9 (1.3) | 25.5 (0.95) | 0.12 |
| Birth weight, g | 712 (182) | 750 (165) | 0.21 | 640 (147) | 704 (145) | 0.015 | 703 (155) | 740 (210) | 0.57 |
| %Male | 59.6 | 51.2 | 0.44 | 56.1 | 45.1 | 0.29 | 57.1 | 35.7 | 0.33 |
| %Multiple gestation | 15.4 | 22.0 | 0.46 | 9.76 | 9.80 | 1.0 | 3.57 | 14.3 | 0.53 |
| RSS at entry | 4.0 (2.1) | 3.1 (1.4) | 0.0008 | 4.0 (2.2) | 2.7 (0.94) | <0.0001 | 3.8 (2.8) | 3.3 (2.0) | 0.49 |
Data are means with standard deviations in parentheses. P values represent comparisons using a Student’s t-test for continuous measurements [gestational age, birth weight, and respiratory severity score (RSS) at entry] and a χ2-test for categorical (%male and %multiple gestation). Demographics are shown by maternal self-reported racial/ethnic group. BPD, bronchopulmonary dysplasia; GWAS, genome-wide association study; N/A, not applicable; TOLSURF, Trial of Late Surfactant.
Global ancestry and admixture mapping.
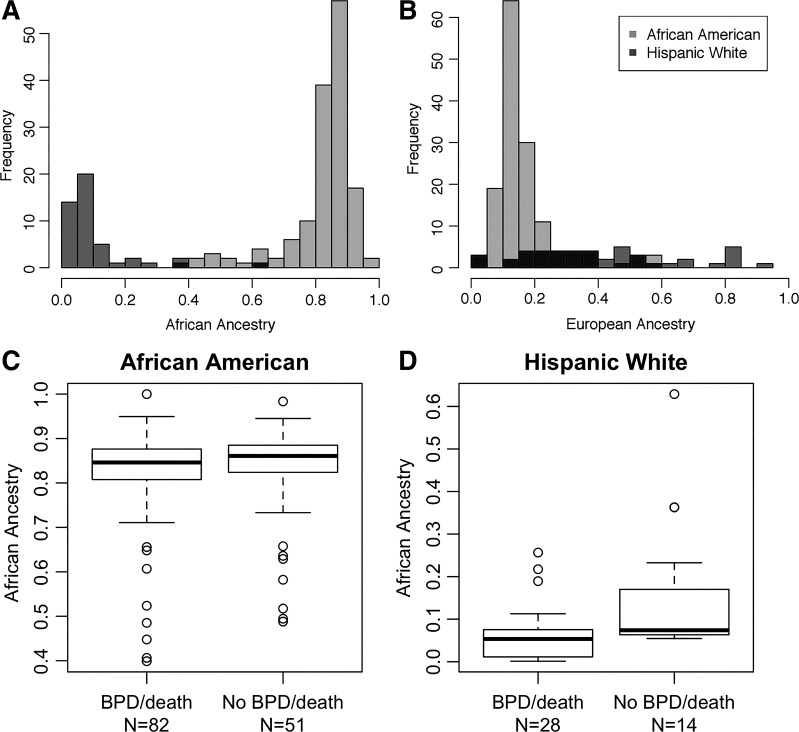
Individual proportions of African ancestry across the entire genome were consistent with expectations given maternal self-reported race/ethnicity (Fig. 1, A and B). Specifically, black/AA infants had a higher degree of genomic African ancestry [median = 85% (range = 40–100%)] compared with white Hispanic infants [median = 6.3% (range = 1.2–63%)].
Fig. 1.
Global ancestry proportions and survival without bronchopulmonary dysplasia (BPD). Proportions shown are of global African (A) and European (B) ancestry in preterm infants participating in the Trial of Late Surfactant (TOLSURF) study by maternal self-reported race/ethnicity. Global ancestry was inferred using ADMIXTURE. Box plots compare global African ancestry and survival without BPD in infants of maternal self-reported black/African-American race/ethnicity (C; logistic regression: P = 0.97, β = −0.015 ± 0.37) and Hispanic white race/ethnicity (D; P = 0.01, β = −1.5 ± 0.60).
Global African ancestry was not significantly different between infants with BPD/death compared with those surviving without BPD for black/AA infants adjusting for gestational age, sex, birth weight, and multiple gestation (β = −0.015, SE = 0.37, P = 0.97; Fig. 1C). However, genomic African ancestry was protective for BPD/death in white Hispanic infants adjusting for the same covariates (β = −1.5, SE = 0.6, P = 0.01; Fig. 1D). Results were similar when all covariates were excluded.
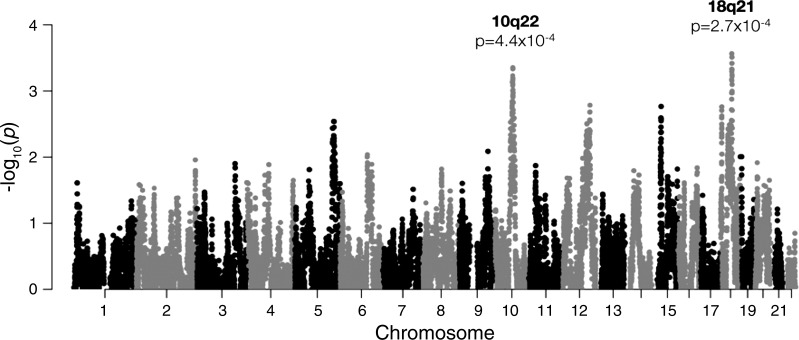
With the use of logistic regression, African ancestry was further examined in black/AA infants at individual loci (i.e., local ancestry or admixture mapping) to evaluate differences in African ancestry at specific regions of the genome between cases and controls. The strongest associations with local ancestry were observed at chromosome band 10q21, where African ancestry was protective for BPD/death [P = 4.4 × 10−4, odds ratio (OR) = 0.17], and 18q21, where African ancestry was risky for BPD/death (P = 2.7 × 10−4, OR = 4.6; Fig. 2). The estimated number of independent ancestry blocks was determined to be 478, and thus neither of the admixture mapping peaks was statistically significant following Bonferroni correction (α = 1.0 × 10−4).
Fig. 2.
Results of admixture mapping comparing local African ancestry and survival without bronchopulmonary dysplasia in 133 infants with maternal self-reported black/African-American race/ethnicity (82 cases, 51 controls). Top associations were observed at chromosome bands 10q21 (odds ratio = 0.17, P = 4.4 × 10−4) and 18q21 (odds ratio = 4.6, P = 2.7 × 10−4).
GWAS and gene-based comparisons.
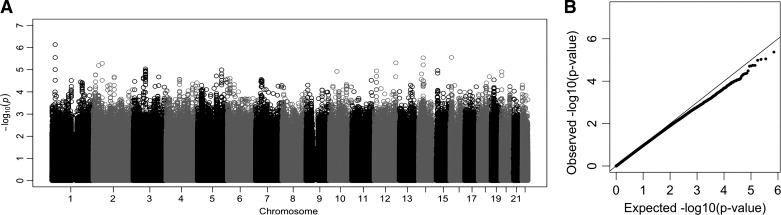
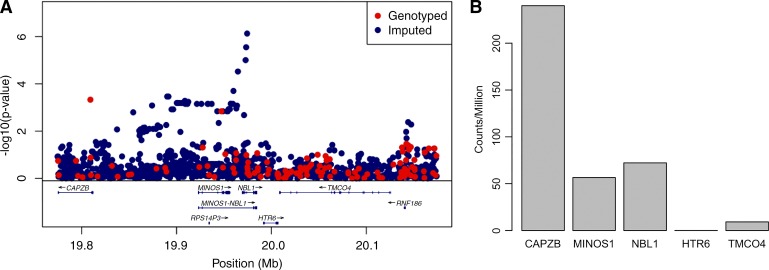
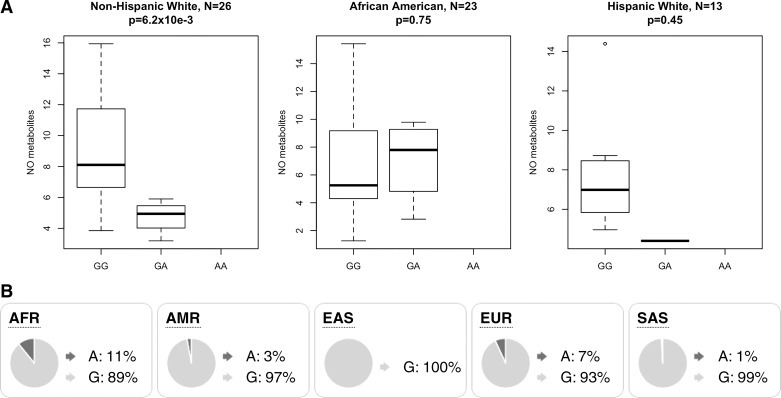
Following genotype imputation, which infers additional variants for each infant using whole genome sequences from the 1000 Genomes Project, we tested the entire cohort for an association with survival without BPD. We examined associations at 8.8 million individual variants, adjusting for global genetic ancestry, gestational age, sex, birth weight, and multiple gestations. No individual variant was genome-wide significantly associated with BPD (all P values >5 × 10−8, Fig. 3). However, the top association was observed at a variant within the intron of NBL1 (rs372271081; P = 7.4 × 10−7; Fig. 4A). The minor allele was protective for BPD (OR = 0.17) and showed a similar effect within each racial/ethnic group (Table 2). Local African ancestry at this locus was not significantly associated with BPD in black/AA infants (P = 0.24). NBL1 and two additional genes within the same region (CAPZB and MINOS1) were expressed in fetal lung at 23 wk gestation (Fig. 4B). Furthermore, the minor allele at rs372271081 was significantly associated with decreased urinary NOx in non-Hispanic white infants but was not significant in black/AA or white Hispanic infants (Table 3, Fig. 5A). Notably, the protective allele for BPD at rs372271081 is at a somewhat higher frequency in populations with African ancestry (Fig. 5B).
Fig. 3.
Manhattan plot (A) and quantile-quantile plot (B) showing the results of a weighted meta-analysis for survival without bronchopulmonary dysplasia (BPD) across 3 maternal self-reported racial/ethnic groups, including non-Hispanic white (136 BPD/death infants, 41 no BPD), black/African American (82 BPD/death infants, 51 no BPD), and Hispanic white (28 BPD/death infants, 14 no BPD).
Fig. 4.
A: LocusZoom plot of the region flanking the top association at rs372271081, an intronic variant of NBL1. Mb, megabase. B: expression of genes by RNA sequencing within this locus in fetal lung at 23 wk gestation.
Table 2.
Results of tests of association at rs372271081 for survival without BPD using logistic regression and urinary NO metabolites following iNO treatment using linear regression
| Survival without BPD |
Urinary NO Metabolites |
|||||||
|---|---|---|---|---|---|---|---|---|
| Population | Frequency in Cases, n | Frequency in Controls, n | Odds Ratio | P Value | n | β (SE) | 95% CI | P Value |
| Non-Hispanic white | 0.040 (136) | 0.12 (41) | 0.30 | 6.2 × 10−3 | 26 | −5.3 (1.7) | (−8.7, −1.9) | 6.2 × 10−3 |
| African American | 0.055 (82) | 0.19 (51) | 0.25 | 6.9 × 10−4 | 23 | 0.74 (2.3) | (−3.8, 5.2) | 0.75 |
| Hispanic white | 0.018 (28) | 0.11 (14) | 0.15 | 0.070 | 13 | −1.9 (2.3) | (−6.5, 2.7) | 0.45 |
Results are shown with respect to the minor allele (A), which trends as protective for bronchopulmonary dysplasia (BPD) in 3 populations and is significantly associated with lower urinary NO metabolites in infants of maternal non-Hispanic white race/ethnicity. β, Regression coefficient/effect size; CI, confidence interval; iNO, inhaled nitric oxide; SE, standard error.
Table 3.
Genetic variants associated with survival without BPD at P < 10−6 in a meta-analysis across 3 racial/ethnic groups
| Chr | Position (hg19) | SNP | Allele | Annotation | NHW OR | AA OR | HW OR | Meta OR | Meta P Value |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 19974397 | rs372271081 | A | Intron, NBL1 | 0.19 | 0.10 | 0.17 | 0.17 | 7.42 × 10−7 |
| 2 | 14648908 | rs10193074 | G | Intergenic | 0.26 | 0.25 | N/A | 0.26 | 4.17 × 10−6 |
| 2 | 33777089 | 2:33777089 | C | Intron, RASGRP3 | 0.39 | 0.17 | 0.22 | 0.28 | 6.41 × 10−6 |
| 2 | 54980799 | 2:54980799 | G | Intron, EML6 | 0.33 | 0.78 | 0.44 | 0.40 | 5.20 × 10−6 |
| 2 | 105035900 | rs4851694 | T | Intergenic | 3.8 | 8.7 | N/A | 4.3 | 5.92 × 10−6 |
| 2 | 105039687 | rs2889323 | C | Intergenic | 3.8 | 8.7 | N/A | 4.3 | 5.92 × 10−6 |
| 2 | 105091271 | rs6543256 | G | Intron, LOC150568 | 3.2 | 9.0 | N/A | 4.1 | 7.24 × 10−6 |
| 3 | 74073182 | rs1949931 | G | Intergenic | 0.38 | 0.082 | 0.44 | 0.39 | 9.25 × 10−6 |
| 10 | 134044152 | rs60417571 | T | Intron, STK32C | 0.19 | N/A | 0.27 | 0.21 | 3.06 × 10−6 |
| 12 | 131048872 | 12:131048872 | CTG | Intron, RIMBP2 | 0.44 | 0.11 | 0.41 | 0.39 | 4.91 × 10−6 |
| 14 | 47459909 | rs8016110 | A | Intron, MDGA2 | 2.9 | 43 | 2.5 | 3.5 | 2.92 × 10−6 |
| 16 | 8834085 | rs75055007 | A | Intron, ABAT | 0.30 | 0.061 | 0.28 | 0.26 | 2.79 × 10−6 |
For loci with multiple single-nucleotide polymorphisms (SNPs) at P < 10−6, only a single SNP with the smallest P value is included in the table. AA, African American [82 bronchopulmonary dysplasia (BPD)/death infants, 51 no BPD]; Chr, chromosome; HW, Hispanic white (28 BPD/death infants, 14 no BPD); Meta, meta-analysis (total 246 BPD/death infants, 106 no BPD); N/A, not applicable; NHW, non-Hispanic white (136 BPD/death infants, 41 no BPD); OR, odds ratio.
Fig. 5.
A: box plot showing levels of urinary NO metabolites by genotype at rs372271081 in preterm infants following treatment with inhaled nitric oxide at 5 ppm. B: frequency of rs372271081 in populations from the Phase 3 1000 Genomes Project. A and G, alleles; AFR, African; AMR, Admixed American; EAS, East Asian; EUR, European; SAS, South Asian.
To increase statistical power, we combined the results of association testing of individual variants within known genes to create a whole gene-based statistic. No individual gene was significantly associated with BPD following Bonferroni correction for 17,670 tests (the number of genes tested, α = 2.8 × 10−6; Table 4). However, by restricting our comparisons to 21 candidate genes for which expression is dysregulated in BPD lungs (14), variation in CCL18 was significantly associated with BPD after adjusting for a smaller number of tests in our hypothesis-driven approach (P = 0.0011). Local African ancestry at CCL18 was not significantly associated with BPD in black/AA infants (P = 0.76), suggesting that any causal variants are at the same frequency between European and African continental populations and that our association is not confounded by population structure. This gene is expressed at a low level in 23-wk human fetal lung (0.31 ± 0.10 counts per million). None of the genes implicated through exome sequencing from Li et al. (39) were significantly associated with BPD in our study (minimum P = 0.0018, ADCY8), nor did we find any significant associations with 11 NO-related candidate genes with reported associations with human disease (Table 5; minimum P = 0.18, KALRN).
Table 4.
Top genes associated with survival without BPD in a meta-analysis across racial/ethnic groups including 246 cases and 106 controls
| Chr | Gene | No. of SNPs | P Value |
|---|---|---|---|
| 5 | RICTOR | 17 | 4.5 × 10−5 |
| 4 | MED28 | 6 | 1.8 × 10−4 |
| 12 | IL23A | 8 | 2.2 × 10−4 |
| 19 | ZNF492 | 14 | 2.5 × 10−4 |
Gene-based statistics were calculated using versatile gene-based association study (VEGAS). None of the genes was statistically significant following Bonferroni correction for the total number of genes examined (n = 17,671). BPD, bronchopulmonary dysplasia; Chr, chromosome; SNPs, single-nucleotide polymorphisms.
Table 5.
List of NO-related candidate genes/variants previously associated with disease
| Gene | Variant | Disease (Measurement) (Ref.) |
|---|---|---|
| NOS2 | rs944722 | Radiation lung injury (lung function) (58) |
| Infant RSV-related respiratory morbidity (22) | ||
| rs2274894, rs7215373 | Tuberculosis susceptibility (59) | |
| rs3794767 | Malaria susceptibility (blood Plasmodium/NO) (57) | |
| NOS3 | rs1799983, rs2070744 | Coronary artery disease (54, 67) |
| G894T | Essential hypertension (40) | |
| Hypoxic ischemic encephalopathy (65) | ||
| −922 G>A, −786 T>C | Ischemic stroke susceptibility (30) | |
| GUCY1A3 | A680T | Pulmonary hypertension (63) |
| LYRM9 | rs3751972 | Asthma (FeNO*) (58) |
| GSDMB | rs8069176 | Asthma (FeNO*) (58) |
| GSR | rs2253409 | Lupus (NO production) (55) |
| KALRN | rs9289231 | Coronary artery disease (15) |
| TSNAX-DISC1 | rs821722 | Nicotine dependence (25) |
| PON1 | Q192R | Coronary artery disease (49) |
| IFNGR1 | rs1327474 | Tuberculosis susceptibility (59) |
| PDE5 | G1142T | Congestive heart failure response to inhaled NO (20) |
A, C, G, and T, alleles; RSV, respiratory syncytial virus.
FeNO, fractional concentration of nitric oxide in exhaled air.
Pathway analysis.
Pathway analysis can be a powerful means to identify an enrichment of genes with marginal signals of association on their own but which function in a similar biological pathway. Pathway analysis was performed using GREAT (45) on 1,024 genes with gene-based P values <0.05 compared with 17,640 genes as a background. A total of 5 pathways/gene sets were identified with a false discovery rate <0.05 from the PANTHER and MSigDB databases; this group contains 2 pathways related to cancers, 1 related to immune function, 1 related to methylation marks, and 1 implicated in experimental lung injury (Table 6). The pathway of highest statistical significance (P = 5 × 10−12) was “Genes within amplicon 1q21 identified in a copy number alterations study of 191 breast tumor samples.” Eight of the eleven genes in this pathway are expressed in human fetal lung at 23 wk GA, and none is regulated by glucocorticoids, which enhance fetal lung maturity. Biological functions of potential relevance to lung development, injury, and repair for these genes include tyrosine kinase receptor signaling pathway (EFNA4, RUSC1, SHC1, ADAM15), developmental processes (EFNA4, RUSC1, ZBTB7B, PBXIP1, SHC1, ADAM15, PYGO2) including angiogenesis (SHC1), NF-κB signaling (RUSC1, ZBTB7B), and sex steroid receptor signaling (PBXIP1, SHC1).
Table 6.
PANTHER and MSigDB pathways showing a significant enrichment of genes associated with survival without BPD at P < 0.05
| Pathway/Gene Set | P Value | FDR | Fold Enrichment | No. of Genes |
|---|---|---|---|---|
| PANTHER | ||||
| Toll receptor signaling pathway | 2.3 × 10−4 | 0.035 | 3.1 | 13 |
| MSigDB | ||||
| Genes within amplicon 1q21 identified in a copy number alterations study of 191 breast tumor samples | 1.5 × 10−15 | 5.1 × 10−12 | 8.6 | 21 |
| Genes with low-CpG-density promoters bearing H3K4me3 marks in embryonic fibroblasts | 4.1 × 10−7 | 6.8 × 10−4 | 2.5 | 35 |
| Nearest neighbors of TAL1, based on the close agreement of their expression profiles with that of TAL1 in pediatric T cell acute lymphoblastic leukemia | 6.7 × 10−6 | 0.0076 | 5.0 | 11 |
| Genes upregulated in lung tissue on LPS aspiration with mechanical ventilation | 2.4 × 10−5 | 0.020 | 2.2 | 32 |
Foreground set of 1,024 genes with association P value <0.05 was compared with a background set of 17,640 genes using Genomic Regions Enrichment of Annotations Tool (GREAT). Gene-based association P values were calculated using versatile gene-based association study (VEGAS). BPD, bronchopulmonary dysplasia; FDR, false discovery rate; MSigDB, Molecular Signature Database; PANTHER, Protein Analysis THrough Evolutionary Relationships.
Pathway analysis using IPA of 181 genes with gene-based P values <0.01 of 209 canonical pathways identified 2 with a significant enrichment of genes following a Bonferroni correction: agranulocyte adhesion and diapedesis (P = 3.06 × 10−5) and granulocyte adhesion and diapedesis (P = 1.22 × 10−4); genes in these 2 pathways are identical except for MYL9 (Table 7). With the exception of CLDN17, all genes identified in these pathways are expressed in human fetal lung (12).
Table 7.
Canonical pathways from Ingenuity Pathway Analysis with a significant enrichment of genes with association P < 0.01 for survival without BPD
| Canonical Pathway | No. of Genes (%) | Genes in Pathway with P < 0.01 | P Value |
|---|---|---|---|
| Agranulocyte adhesion and diapedesis | 9/181 (4.8) | CCL3, CCL4, CCL17, CCL18, CCL22, CLDN17, CX3CL1, MYL9, RDX | 3.06 × 10−5 |
| Granulocyte adhesion and diapedesis | 8/181 (4.4) | CCL3, CCL4, CCL17, CCL18, CCL22, CLDN17, CX3CL1, RDX | 1.22 × 10−4 |
Statistical significance was determined using a Bonferroni adjustment for 209 canonical pathways tested (α = 2.39 × 10−4). BPD, bronchopulmonary dysplasia.
DISCUSSION
Unique aspects of our study are the patient population and rigorous assignment of BPD. All infants in TOLSURF were <28 wk gestation and were intubated at 7–14 days, representing infants with severe early respiratory failure and high risk for BPD as reflected by the occurrence of BPD/death in 68.5% of the total population (9). In addition, infants were enrolled from 25 different US sites, providing both racial/ethnic and geographic diversity. The diagnosis of BPD was assigned on a physiological basis using an oxygen/flow reduction challenge to establish a requirement for respiratory support. Thus it is possible that some of our findings may be restricted to extremely premature infants with severe early respiratory disease. Other characteristics of our cohort, which may limit generalization of some of our findings, are exposure to late surfactant treatment in approximately half of the infants and the use of iNO for 3 wk in all infants. Although surfactant therapy transiently improved respiratory status, it did not affect outcome at 36 wk PMA. iNO therapy likely influenced overall outcome in infants of self-identified black/African-American women but not non-Hispanic white women (6), and thus we examined NO metabolism as it relates to genetic associations with BPD in our study.
Higher genomic levels of African ancestry were associated with better respiratory outcome in iNO-treated infants with maternal self-reported Hispanic white race/ethnicity but not for infants with maternal self-reported black/African-American race/ethnicity. Although the protective effect of African genomic ancestry in white Hispanic infants requires independent replication, our results suggest that the protective effect of African ancestry may be saturated at lower levels of ancestry than are present in the majority of black/African-American infants in the study. In other words, there is an increase in the protective effect of African ancestry between the range of 1.2 and 63% (that observed in white Hispanic infants in the study) but not between the range of 40 and 100% (that observed in black/African-American infants in the study). This may reflect a polygenic basis for the beneficial effect of iNO whereby similar protection from BPD can be conferred by a variety of different variants of African ancestry in genes of key NO-related pathways. Alternatively, there may be differential gene-environment interactions between racial/ethnic groups. For example, the protective effect of African ancestry may only be relevant under specific environmental exposures that vary by racial/ethnic group.
Genetic ancestry does not form a direct causal relationship but rather indicates differences in the underlying patterns of genetic variation in infants with/without BPD that differ by continental origin. If only small proportions of African ancestry are required for a protective effect, this would suggest a highly polygenic contribution to BPD distributed throughout the genome. Admixture mapping in black/AA infants identified two suggestive, but not statistically significant, peaks, at 10q22 and 18q21, whereby African ancestry was associated with a decreased and increased risk of death/BPD, respectively. Therefore, admixture mapping further supports the hypothesis that the effect of ancestry is not limited to a single locus of large effect. It is possible the relationship between ancestry and BPD is restricted to infants receiving iNO; this is supported by prior studies that identified racial differences in endogenous NO levels or metabolism in infants (8) and adults (32, 34, 35, 44).
In our agnostic scan including ~9 million genotyped and imputed variants, no individual variant was genome-wide significantly associated with survival without BPD. This was not unexpected, given our small sample size, and is consistent with prior GWAS that similarly failed to identify individual variants with large effects at genome-wide significance levels (4, 29, 43, 61). However, numerous biologically plausible genes have been implicated with top GWAS variants identified at suggestive levels of significance, including ADARB2 (4), CRP (43), SPOCK2 (29), and an intergenic region on chromosome 18 (61). We were unable to replicate any of the top findings from prior GWAS, but note that our current study has many unique characteristics as outlined above.
The biggest limitation of our study is the sample size, given that any one genetic variant is likely to have a small effect. Small/modest sample size is a common limitation to genetic studies of preterm infants, and thus there is a need to integrate additional biological measurements to increase power of discovery. For example, after implicating variants around CRP at suggestive levels of statistical significance, Mahlman et al. (43) identified CRP protein as being significantly elevated in infants that went on to develop BPD at 36 wk. Ambalavanan et al. (4) queried gene expression in BPD lungs and validated suggestive associations identified through GWAS at variants in CD44 and the miR-219 pathway. By taking a similar integrative approach, we found our top BPD-associated variant, rs372271081, was significantly associated with differences in NOx in white infants, whereby the protective allele for BPD was associated with decreased levels of NOx in the urine following treatment of iNO (but not before treatment). This finding lends supports for a true genetic association with BPD at rs372271081 and indicates the association is likely specific to iNO treatment and an effect through differential drug metabolism. However, we found no significant association between genotype and NO metabolites in black/AA or Hispanic white infants, which may reflect the limited statistical power given the smaller sample sizes, the lower frequency of the allele, varying patterns of linkage disequilibrium, and/or the presence of genetic and environmental interactions.
rs372271081 Lies within an intron of neuroblastoma 1, DAN family BMP antagonist (NBL1), which is a highly plausible candidate gene for contributing to BPD susceptibility via differential response to iNO. Numerous studies in mice indicate that the BMP pathway is important for lung development, including branching morphogenesis in early gestation and distal lung epithelial cell differentiation, alveolization, and vasculogenesis in late gestation (16, 23, 62). The transforming growth factor-β-BMP signaling pathway is disrupted by hyperoxia (1), which is known to play a role in the development of BPD (1, 2). In humans, disrupted BMP signaling has been implicated in the pathogenesis of heritable pulmonary arterial hypertension and hereditary hemorrhagic telangiectasis (27, 48). Finally, in addition to ligand inhibition of BMP, DAN family members are known to modulate Wnt and VEGF signaling pathways that have a role in lung development and injury/repair (48). Overall, there is strong biological plausibility for a role of genetic variation in NBL1 and respiratory outcome in iNO-treated infants based on 1) the critical role of BMP signaling in lung development and disease, 2) the mediation of BMP action via NO, 3) the expression of NBL1 and BMPs in human fetal lung (35), and 4) the racial differences in BPD and NO metabolism (12).
Although NBL1 has not been specifically implicated in prior GWAS/exome sequencing studies, genes involved in lung development are strong candidates for a role in BPD, which only occurs in immature lungs. For example, a common variant in SPOCK2, an extracellular matrix protein, was implicated in BPD through GWAS and found to be upregulated during lung alveolar development and after exposure to hyperoxia in rats (29). Furthermore, pathway analyses have implicated other genes involved in pulmonary structure and functions (39). Replication and both laboratory and functional validation are necessary to confirm a causal relationship of variants in NBL1 and BPD in infants treated with iNO. Currently, there are no other cohorts of premature infants treated with iNO with DNA samples available for validation studies.
We further performed hypothesis-driven tests of association with BPD using a set of 21 genes that are dysregulated in BPD lungs (14) and 11 genes in the nitric oxide pathway that are reported to have variants associated with disease (Table 5). First, we hypothesized that genes showing differential expression in BPD-dysregulated vs. control lungs may contain variants that contribute to survival without BPD. We found a significant association with genetic variation in a single gene, CCL18, a cytokine involved in the immune response that promotes collagen production in lung fibroblasts (42) and is associated with pulmonary fibrosis and interstitial lung diseases in adults (51, 66). Inflammation is known to be important in the pathogenesis of BPD, and anti-inflammatory therapy (dexamethasone) suppresses a variety of inflammatory mediators, including CCL18, and reduces BPD (12, 26). Second, because all infants in the study received iNO, we hypothesized that variation in genes in the NO pathway may contribute to differential response to iNO treatment as indicated by survival without BPD. However, no individual variant or candidate gene (based on known association with human disease) was significantly associated with survival without BPD following correction for multiple tests.
Nonetheless, because exposure to iNO appears to influence the differential rates of BPD between racial/ethnic groups (6, 60), we hypothesized that genetic variants that contribute to BPD may act through differential response to iNO. In support of this, the protective allele for BPD at rs372271081 is significantly associated with decreased NOx and is more common in populations with African ancestry. Several studies indicate reduced bioavailability of NO in African Americans vs. Caucasians, likely, in part, due to increased oxidation of NO. In laboratory studies, release of NO from umbilical venous endothelial cells was substantially lower in African-American vs. Caucasian infants (34, 44). Levels of urinary NOx are lower in African-American and Hispanic premature infants vs. Caucasian infants regardless of iNO treatment, reflecting baseline differences in NO metabolism and thus bioavailability (8). In adults, African Americans are known to have increased frequency of hypertension and cardiovascular disease, and a NO-targeted medication (isosorbide dinitrates and hydralazine) is indicated therapy for heart failure specifically in African Americans (i.e., a racially directed therapy; Refs. 32, 35). However, further studies are needed to evaluate the contribution of rs372271081 to racial/ethnic differences in NO bioavailability and differential response to iNO.
Pathway analyses identified pathways and sets of genes that were significantly enriched for genes with association P values <0.05. Across IPA, PANTHER, and MSigDB data sets, a common theme that emerged was genes involved in immune function, including granulocyte and agranulocyte adhesion and diapedesis from IPA canonical pathways, Toll receptor signaling pathway from PANTHER, and genes upregulated in response to LPS exposure and mechanical ventilation from MSigDB. These results suggest that variation in immune response, including recruitment of leukocytes and lymphocytes, contributes to survival without BPD.
Overall, our results for this cohort of iNO-treated, high-risk infants suggest that genomic African ancestry is protective for BPD and that an intronic variant in NBL1 may contribute to BPD via differential activity of the transforming growth factor-β-BMP pathway and production/metabolism of NO. Furthermore, we implicated variation in genes involved in the immune response, including CCL18, as contributing to differences in respiratory outcomes of preterm infants.
GRANTS
This study was funded by grants from the National Institutes of Health (5-U01-HL-094338M, 1-R01-MD-010443, R21-ES-24844, R01-HL-117004, U54-MD-009523, R01-HL-128439, and U01-HL-101798) and an Edward A. Dickson Emeritus Professorship Award (to P. L. Ballard). Ikaria, Inc./Mallinckrodt Pharmaceuticals funded the genetic analyses, including support for supplies, technical effort, and statistical analyses. In addition, Ikaria, Inc. and ONY, Inc. provided drugs for the conduct of the parent trial, but neither company had input into study design, data analysis, data interpretation, or manuscript preparation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.G.T., P.L.B., R.L.K., E.G.B., and R.A.B. conceived and designed research; D.G.T., P.L.B., R.L.K., C.E., E.G.B., and R.A.B. performed experiments; D.G.T., P.L.B., R.L.K., S.S.O., S.H., D.H., C.E., and R.A.B. analyzed data; D.G.T., P.L.B., R.L.K., and R.A.B. interpreted results of experiments; D.G.T. prepared figures; D.G.T. and P.L.B. drafted manuscript; D.G.T., P.L.B., R.L.K., E.G.B., and R.A.B. edited and revised manuscript; D.G.T., P.L.B., R.L.K., S.S.O., S.H., D.H., C.E., E.G.B., and R.A.B. approved final version of manuscript.
ACKNOWLEDGMENTS
Preprint is available at https://doi.org/10.1101/263814.
We thank the TOLSURF Study Investigators, study coordinators, physicians, nurses, respiratory therapists, and families who participated in the TOLSURF study.
TOLSURF Study Investigators: Robin H. Steinhorn, Department of Pediatrics, Children’s National Medical Center, Washington, DC; Catherine M. Bendel, Department of Pediatrics, University of Minnesota, Minneapolis, MN; Ellen M. Bendel-Stenzel, Department of Pediatrics, Children’s Hospital and Clinics of Minnesota, St. Paul and Minneapolis, MN; Sherry E. Courtney, Department of Pediatrics, University of Arkansas for Medical Sciences, Little Rock, AR; Ramasubbareddy Dhanireddy, Department of Pediatrics, University of Tennessee Health Science Center, Memphis, TN; Frances R. Koch, Department of Pediatrics, Medical University of South Carolina, Charleston, SC; Mark L. Hudak, Department of Pediatrics, University of Florida College of Medicine, Jacksonville, FL; Dennis E. Mayock, Department of Pediatrics, University of Washington, Seattle, WA; Rajan Wadhawan, Department of Pediatrics, Florida Hospital for Children, Orlando, FL; Nicolas F. Porta, Department of Pediatrics, Northwestern University Feinberg School of Medicine, Chicago, IL; Victor J. McKay, Department of Pediatrics, All Children’s Hospital, St. Petersburg, FL; Jeffrey D. Merrill, Children’s Hospital Oakland Research Institute, Oakland, CA; Eric C. Eichenwald; Department of Pediatrics, Children’s Hospital of Philadelphia, Philadelphia, PA; William E. Truog, Department of Pediatrics, Children’s Mercy Hospital, Kansas City, MO; Mark C. Mammel, Children’s Hospital and Clinics of Minnesota, St. Paul, MN; Elizabeth E. Rogers, Department of Pediatrics, University of California, San Francisco, CA; Rita M. Ryan, Department of Pediatrics, Medical University of South Carolina, Charleston, SC; David J. Durand, Children’s Hospital Oakland Research Institute, Oakland, CA; T. Michael O’Shea, Department of Pediatrics, Wake Forest School of Medicine Winston-Salem, NC; and Dennis M. Black, Department of Epidemiology and Biostatistics, University of California, San Francisco, CA.
In addition to the authors and other TOLSURF Study Investigators, the following members of the TOLSURF Study Group participated in this study. University of California, San Francisco, CA: Suzanne Hamilton Strong, RN; Jill Immamura-Ching, RN; Margaret Orfanos-Villalobos, RN; Cassandra Williams, RN; and Lisa Palermo, MA. Alta Bates Summit Medical Center, Berkeley, CA, and UCSF Benioff Children’s Hospital Oakland, Oakland, CA: Dolia Horton, RRT; Loretta Pacello, RCP; and April Willard, RN. Children’s Mercy Hospital, Kansas City, MO: Cheryl Gauldin, RN; Anne Holmes, RN; Patrice Johnson, RRT; and Kerrie Meinert, RRT. Women and Children’s Hospital of Buffalo, Buffalo, NY: Anne Marie Reynolds, MD; Janine Lucie, NNP; Patrick Conway; Michael Sacilowski; Michael Leadersdorff, RRT; Pam Orbank, RRT; and Karen Wynn, NNP. Anne and Robert H. Lurie Children’s Hospital/Northwestern University, Chicago, IL: Maria deUngria, MD; Janine Yasmin Khan, MD; Karin Hamann, RN; Molly Schau, RN; Brad Hopkins, RRT; and James Jenson, RRT. Texas Children’s Hospital, Houston, TX: Carmen Garcia, RN. Stony Brook University Hospital, Stony Brook, NY: Aruna Parekh, MD; Jila Shariff, MD; Rose McGovern, RN; Jeff Adelman, RRT; Adrienne Combs, RN; and Mary Tjersland, RRT. University of Washington, Seattle, WA: Elizabeth Howland; Susan Walker, RN; Jim Longoria, RRT; and Holly Meo, RRT. University of Texas Health Science Center, Houston, TX: Amir Khan, MD, Georgia McDavid, RN; Katrina Burson, RN, BSN; Richard Hinojosa, BSRT, RRT; Christopher Johnson, MBA, RRT; Karen Martin, RN, BSN; Sarah Martin, RN, BSN; Shawna Rogers, RN, BSN; and Sharon Wright, MT. University of Florida College of Medicine, Jacksonville, UF Health Shands Hospital, and Wolfson Children’s Hospital, Jacksonville, FL: Kimberly Barnette, RRT; Amanda Kellum, RRT; Michelle Burcke, RN; Christie Hayes, RRT; Stephanie Chadwick, RN; Danielle Howard, RN; Carla Kennedy, RRT; and Renee Prince, RN. Wake Forest School of Medicine and Forsyth Medical Center, Winston Salem, NC: Beatrice Stefanescu, MD; Kelly Warden, RN; Patty Brown, RN; Jennifer Griffin, RRT; and Laura Conley, RRT. University of Minnesota Amplatz Children’s Hospital, Minneapolis, MN: Michael Georgieff, MD; Bridget Davern; Marla Mills, RN; and Sharon Ritter, RRT. Medical University of South Carolina, Charleston, SC: Carol Wagner, MD; Deanna Fanning, RN; and Jimmy Roberson, RRT. Children’s Hospitals and Clinics of Minnesota, St. Paul, MN: Andrea Lampland, MD; Pat Meyers, RRT; and Angela Brey, RRT. Children’s Hospitals and Clinics of Minnesota, Minneapolis, MN: Neil Mulrooney, MD; Cathy Worwa, RRT; Pam Dixon, RN, ANM; Gerald Ebert, RRT-NPS; Cathy Hejl, RRT; Molly Maxwell, RT; and Kristin McCullough, RN. University of Tennessee Health Science Center, Memphis, TN: Mohammed T. El Abiad, MD; Ajay Talati, MD; Sheila Dempsey, RN; Kathy Gammage, RRT, MBA; Gayle Gower, RN; Kathy James, RRT; and Pam LeNoue, RN. All Children’s Hospital, St. Petersburg, FL: Suzi Bell, DNP; Dawn Bruton, RN, BSN, CCRP; Michelle Beaulieu, DNP; and Richard Williams, RRT. Florida Hospital for Children, Orlando, FL: Robin Barron-Nelson, RN; and Shane Taylor, RRT. Arkansas Children’s Hospital and University of Arkansas Medical Sciences, Little Rock, AR: Carol Sikes, RN; Gary Lowe, RRT; and Betty Proffitt, RRT.
Clinical Coordinating Center: University of California, San Francisco, Department of Pediatrics: Cheryl Chapin; Hart Horneman; Karin Hamann, RN; Susan Kelley, RRT; Karin Knowles; and Nancy Newton, RN, MS.
Data Coordinating Center: University of California, San Francisco, Department of Epidemiology and Biostatistics: Eric Vittinghoff, PhD; Jean Hietpas; Laurie Denton; and Lucy Wu.
Data Safety Monitoring Board: Cincinnati Children’s Hospital Medical Center, Cincinnati, OH: Allan Jobe, MD (Chair 2009–2010). UH Rainbow Babies and Children’s Hospital, Cleveland, OH: Avroy Fanaroff, MD (Chair 2010–2016). EMMES Corporation, Rockville, MD: Traci Clemons, PhD. Boston University School of Public Health, Boston, MA: Leonard Glantz, JD. Wake Forest School of Medicine, Winston-Salem, NC: David Reboussin, PhD. Stanford University, Stanford, CA: Krisa Van Meurs, MD (2009–2010). Johns Hopkins University, Baltimore, MD: Marilee Allen, MD (2010–2016). Women and Infants Hospital, Providence, RI: Betty Vohr, MD.
Clinical Steering Committee: Department of Pediatrics, UCSF Benioff Children’s Hospital San Francisco, San Francisco, CA: Roberta Ballard, MD; Philip Ballard, MD, PhD; Roberta Keller, MD; Elizabeth Rogers, MD; and Nancy Newton, MS, RN. University of California, San Francisco, Department of Epidemiology and Biostatistics: Dennis Black, PhD. National Heart, Lung, and Blood Institute (NIH/NHLBI): Carol Blaisdell, MD. UCSF Benioff Children’s Hospital Oakland, Oakland, CA: David Durand, MD; Jeffrey Merrill, MD; and Jeanette Asselin, MS, RRT. University of Texas Health Science Center, Houston, TX: Eric Eichenwald, MD. Children’s Hospital and Clinics of Minnesota, St. Paul, MN: Mark Mammel, MD. Medical University of South Carolina, Charleston, SC: Rita Ryan, MD. Children’s Mercy Hospital, Kansas City, MO: William Truog, MD.
REFERENCES
- 1.Alejandre-Alcázar MA, Kwapiszewska G, Reiss I, Amarie OV, Marsh LM, Sevilla-Pérez J, Wygrecka M, Eul B, Köbrich S, Hesse M, Schermuly RT, Seeger W, Eickelberg O, Morty RE. Hyperoxia modulates TGF-β/BMP signaling in a mouse model of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol 292: L537–L549, 2007. doi: 10.1152/ajplung.00050.2006. [DOI] [PubMed] [Google Scholar]
- 2.Alejandre-Alcázar MA, Shalamanov PD, Amarie OV, Sevilla-Pérez J, Seeger W, Eickelberg O, Morty RE. Temporal and spatial regulation of bone morphogenetic protein signaling in late lung development. Dev Dyn 236: 2825–2835, 2007. doi: 10.1002/dvdy.21293. [DOI] [PubMed] [Google Scholar]
- 3.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664, 2009. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambalavanan N, Cotten CM, Page GP, Carlo WA, Murray JC, Bhattacharya S, Mariani TJ, Cuna AC, Faye-Petersen OM, Kelly D, Higgins RD; Genomics and Cytokine Subcommittees of the Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network . Integrated genomic analyses in bronchopulmonary dysplasia. J Pediatr 166: 531–537.e13, 2015. doi: 10.1016/j.jpeds.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Askie LM, Ballard RA, Cutter GR, Dani C, Elbourne D, Field D, Hascoet JM, Hibbs AM, Kinsella JP, Mercier JC, Rich W, Schreiber MD, Wongsiridej PS, Subhedar NV, Van Meurs KP, Voysey M, Barrington K, Ehrenkranz RA, Finer NN; Meta-analysis of Preterm Patients on Inhaled Nitric Oxide Collaboration . Inhaled nitric oxide in preterm infants: an individual-patient data meta-analysis of randomized trials. Pediatrics 128: 729–739, 2011. doi: 10.1542/peds.2010-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Askie LM, Davies LC, Schreiber MD, Hibbs AM, Ballard PL, Ballard RA. Race effects of inhaled nitric oxide in preterm infants: an individual participant data meta-analysis. J Pediatr 193: 34–39.e2, 2018. doi: 10.1016/j.jpeds.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Balinotti JE, Chakr VC, Tiller C, Kimmel R, Coates C, Kisling J, Yu Z, Nguyen J, Tepper RS. Growth of lung parenchyma in infants and toddlers with chronic lung disease of infancy. Am J Respir Crit Care Med 181: 1093–1097, 2010. doi: 10.1164/rccm.200908-1190OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballard PL, Keller RL, Black DM, Durand DJ, Merrill JD, Eichenwald EC, Truog WE, Mammel MC, Steinhorn R, Ryan RM, Courtney SE, Horneman H, Ballard RA; Investigators of TOLSURF Pilot and TOLSURF . Inhaled nitric oxide increases urinary nitric oxide metabolites and cyclic guanosine monophosphate in premature infants: relationship to pulmonary outcome. Am J Perinatol 32: 225–232, 2015. doi: 10.1055/s-0034-1382255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballard RA, Keller RL, Black DM, Ballard PL, Merrill JD, Eichenwald EC, Truog WE, Mammel MC, Steinhorn RH, Rogers EE, Ryan RM, Durand DJ, Asselin JM, Bendel CM, Bendel-Stenzel EM, Courtney SE, Dhanireddy R, Hudak ML, Koch FR, Mayock DE, McKay VJ, O’Shea TM, Porta NF, Wadhawan R, Palermo L, Strong SH, Immamura-Ching J, Orfanos-Villalobos M, Williams C, Horton D, Pacello L, Willard A, Gauldin C, Holmes A, Johnson P, Meinert K, Reynolds AM, Lucie J, Conway P, Sacilowski M, Leadersdorff M, Orbank P, Wynn K, deUngria M, Khan J, Hamann K, Schau M, Hopkins B, Jenson J, Garcia C, Shariff J, McGovern R, Adelman J, Combs A, Tjersland M, Walker S, Howland E, Longoria J, Meo H, McDavid G, Burson K, Hinojosa R, Johnson C, Miller K, Rogers S, Wright S, Barnette K, Kellum A, Burke M, Hayes C, Chadwick S, Howard D, Kennedy C, Prince R, Stefanescu B, Helderman J, Warden K, Brown P, Griffin J, Conley L, Georgieff M, Davern B, Mills M, Ritter S, Wagner C, Fanning D, Roberson J, Lampland A, Meyers P, Brey A, Worwa C, Dixon P, Ebert G, Hejl C, Maxwell M, McCullough K, El Abiad MT, Talati A, Dempsey S, Gammage K, Gower G, James K, LeNoue P, Bell S, Bruton D, Beaulieu M, Williams R, Barron-Nelson R, Taylor S, Sikes NC, Lowe G, Proffitt B, Chapin C, Horneman H, Hamann K, Kelley S, Vittinghoff E, Hietpas J, Denton L, Wu L, Jobe A, Fanaroff A, Clemons T, Glantz L, Reboussin D, Van Meurs K, Allen M, Vohr B, Ballard R, Ballard P, Blaisdell C, Durand D, Black D, Eichenwald E, Keller R, Mammel M, Merrill J, Rogers E, Ryan R, Truog W, Asselin J, Newton N; TOLSURF Study Group . Randomized trial of late surfactant treatment in ventilated preterm infants receiving inhaled nitric oxide. J Pediatr 168: 23–29.e4, 2016. doi: 10.1016/j.jpeds.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballard RA, Truog WE, Cnaan A, Martin RJ, Ballard PL, Merrill JD, Walsh MC, Durand DJ, Mayock DE, Eichenwald EC, Null DR, Hudak ML, Puri AR, Golombek SG, Courtney SE, Stewart DL, Welty SE, Phibbs RH, Hibbs AM, Luan X, Wadlinger SR, Asselin JM, Coburn CE; NO CLD Study Group . Inhaled nitric oxide in preterm infants undergoing mechanical ventilation. N Engl J Med 355: 343–353, 2006. doi: 10.1056/NEJMoa061088. [DOI] [PubMed] [Google Scholar]
- 11.Baran Y, Pasaniuc B, Sankararaman S, Torgerson DG, Gignoux C, Eng C, Rodriguez-Cintron W, Chapela R, Ford JG, Avila PC, Rodriguez-Santana J, Burchard EG, Halperin E. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 28: 1359–1367, 2012. doi: 10.1093/bioinformatics/bts144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrette AM, Roberts JK, Chapin C, Egan EA, Segal MR, Oses-Prieto JA, Chand S, Burlingame AL, Ballard PL. Antiinflammatory effects of budesonide in human fetal lung. Am J Respir Cell Mol Biol 55: 623–632, 2016. doi: 10.1165/rcmb.2016-0068OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhandari V, Bizzarro MJ, Shetty A, Zhong X, Page GP, Zhang H, Ment LR, Gruen JR; Neonatal Genetics Study Group . Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics 117: 1901–1906, 2006. doi: 10.1542/peds.2005-1414. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharya S, Go D, Krenitsky DL, Huyck HL, Solleti SK, Lunger VA, Metlay L, Srisuma S, Wert SE, Mariani TJ, Pryhuber GS. Genome-wide transcriptional profiling reveals connective tissue mast cell accumulation in bronchopulmonary dysplasia. Am J Respir Crit Care Med 186: 349–358, 2012. doi: 10.1164/rccm.201203-0406OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boroumand M, Ziaee S, Zarghami N, Anvari MS, Cheraghi S, Abbasi SH, Jalali A, Pourgholi L. The Kalirin gene rs9289231 polymorphism as a novel predisposing marker for coronary artery disease. Lab Med 45: 302–308, 2014. doi: 10.1309/LMLS813ZDPHRFLUU. [DOI] [PubMed] [Google Scholar]
- 16.Bragg AD, Moses HL, Serra R. Signaling to the epithelium is not sufficient to mediate all of the effects of transforming growth factor β and bone morphogenetic protein 4 on murine embryonic lung development. Mech Dev 109: 13–26, 2001. doi: 10.1016/S0925-4773(01)00508-1. [DOI] [PubMed] [Google Scholar]
- 17.Carrera P, Di Resta C, Volonteri C, Castiglioni E, Bonfiglio S, Lazarevic D, Cittaro D, Stupka E, Ferrari M, Somaschini M, Magaldi R, Rinaldi M, Maffei G, Stronati M, Tzialla C, Borghesi A, Tagliabue P, Fedeli T, Citterio M, Mosca F, Colnaghi M, Lavizzari A, Agosti M, Francescato G, Pomero G, Dalmazzo C, Boldrini A, Scaramuzzo R, Bertino E, Borgione S, Martano C, Carnielli V, Nobile S, Auriemma A, Bellan C, Carrera G, Zambetti C, Pucello R, Palatta S; BPD and Genetics Study Group . Exome sequencing and pathway analysis for identification of genetic variability relevant for bronchopulmonary dysplasia (BPD) in preterm newborns: a pilot study. Clin Chim Acta 451: 39–45, 2015. doi: 10.1016/j.cca.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Coalson JJ. Pathology of new bronchopulmonary dysplasia. Semin Neonatol 8: 73–81, 2003. doi: 10.1016/S1084-2756(02)00193-8. [DOI] [PubMed] [Google Scholar]
- 19.Cole FS, Alleyne C, Barks JD, Boyle RJ, Carroll JL, Dokken D, Edwards WH, Georgieff M, Gregory K, Johnston MV, Kramer M, Mitchell C, Neu J, Pursley DM, Robinson WM, Rowitch DH. NIH Consensus Development Conference statement: inhaled nitric-oxide therapy for premature infants. Pediatrics 127: 363–369, 2011. doi: 10.1542/peds.2010-3507. [DOI] [PubMed] [Google Scholar]
- 20.Damy T, Lesault PF, Guendouz S, Eddahibi S, Tu L, Marcos E, Guellich A, Dubois-Randé JL, Teiger E, Hittinger L, Adnot S. Pulmonary hemodynamic responses to inhaled NO in chronic heart failure depend on PDE5 G(-1142)T polymorphism. Pulm Circ 1: 377–382, 2011. doi: 10.4103/2045-8932.87303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nat Genet 48: 1284–1287, 2016. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drysdale SB, Prendergast M, Alcazar M, Wilson T, Smith M, Zuckerman M, Broughton S, Rafferty GF, Johnston SL, Hodemaekers HM, Janssen R, Bont L, Greenough A. Genetic predisposition of RSV infection-related respiratory morbidity in preterm infants. Eur J Pediatr 173: 905–912, 2014. doi: 10.1007/s00431-014-2263-0. [DOI] [PubMed] [Google Scholar]
- 23.Eblaghie MC, Reedy M, Oliver T, Mishina Y, Hogan BL. Evidence that autocrine signaling through Bmpr1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev Biol 291: 67–82, 2006. doi: 10.1016/j.ydbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Ehrenkranz RA, Walsh MC, Vohr BR, Jobe AH, Wright LL, Fanaroff AA, Wrage LA, Poole K; National Institutes of Child Health and Human Development Neonatal Research Network . Validation of the National Institutes of Health consensus definition of bronchopulmonary dysplasia. Pediatrics 116: 1353–1360, 2005. doi: 10.1542/peds.2005-0249. [DOI] [PubMed] [Google Scholar]
- 25.Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, Zhao H, Farrer LA. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry 77: 493–503, 2015. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gien J, Kinsella JP. Pathogenesis and treatment of bronchopulmonary dysplasia. Curr Opin Pediatr 23: 305–313, 2011. doi: 10.1097/MOP.0b013e328346577f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goumans MJ, Zwijsen A, Ten Dijke P, Bailly S. Bone morphogenetic proteins in vascular homeostasis and disease. Cold Spring Harb Perspect Biol 10: a031989, 2018. doi: 10.1101/cshperspect.a031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenough A, Ahmed N. Perinatal prevention of bronchopulmonary dysplasia. J Perinat Med 41: 119–126, 2013. doi: 10.1515/jpm-2012-0084. [DOI] [PubMed] [Google Scholar]
- 29.Hadchouel A, Durrmeyer X, Bouzigon E, Incitti R, Huusko J, Jarreau PH, Lenclen R, Demenais F, Franco-Montoya ML, Layouni I, Patkai J, Bourbon J, Hallman M, Danan C, Delacourt C. Identification of SPOCK2 as a susceptibility gene for bronchopulmonary dysplasia. Am J Respir Crit Care Med 184: 1164–1170, 2011. doi: 10.1164/rccm.201103-0548OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howard TD, Giles WH, Xu J, Wozniak MA, Malarcher AM, Lange LA, Macko RF, Basehore MJ, Meyers DA, Cole JW, Kittner SJ. Promoter polymorphisms in the nitric oxide synthase 3 gene are associated with ischemic stroke susceptibility in young black women. Stroke 36: 1848–1851, 2005. doi: 10.1161/01.STR.0000177978.97428.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Husain AN, Siddiqui NH, Stocker JT. Pathology of arrested acinar development in postsurfactant bronchopulmonary dysplasia. Hum Pathol 29: 710–717, 1998. doi: 10.1016/S0046-8177(98)90280-5. [DOI] [PubMed] [Google Scholar]
- 32.Ishizawar D, Yancy C. Racial differences in heart failure therapeutics. Heart Fail Clin 6: 65–74, 2010. doi: 10.1016/j.hfc.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Jobe AH, Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med 163: 1723–1729, 2001. doi: 10.1164/ajrccm.163.7.2011060. [DOI] [PubMed] [Google Scholar]
- 34.Kalinowski L, Dobrucki IT, Malinski T. Race-specific differences in endothelial function: predisposition of African Americans to vascular diseases. Circulation 109: 2511–2517, 2004. doi: 10.1161/01.CIR.0000129087.81352.7A. [DOI] [PubMed] [Google Scholar]
- 35.Khan BV, Rahman ST, Haque T, Merchant N, Bhaheetharan S, Harris J 3rd, Umar K, Wahi J, Ferdinand KC. Vascular effects of nebivolol added to hydrochlorothiazide in African Americans with hypertension and echocardiographic evidence of diastolic dysfunction: the NASAA study. J Cardiovasc Pharmacol Ther 17: 291–297, 2012. doi: 10.1177/1074248412436607. [DOI] [PubMed] [Google Scholar]
- 36.Krämer A, Green J, Pollard J Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530, 2014. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laughon MM, Langer JC, Bose CL, Smith PB, Ambalavanan N, Kennedy KA, Stoll BJ, Buchter S, Laptook AR, Ehrenkranz RA, Cotten CM, Wilson-Costello DE, Shankaran S, Van Meurs KP, Davis AS, Gantz MG, Finer NN, Yoder BA, Faix RG, Carlo WA, Schibler KR, Newman NS, Rich W, Das A, Higgins RD, Walsh MC; Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network . Prediction of bronchopulmonary dysplasia by postnatal age in extremely premature infants. Am J Respir Crit Care Med 183: 1715–1722, 2011. doi: 10.1164/rccm.201101-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavoie PM, Pham C, Jang KL. Heritability of bronchopulmonary dysplasia, defined according to the consensus statement of the national institutes of health. Pediatrics 122: 479–485, 2008. doi: 10.1542/peds.2007-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Yu KH, Oehlert J, Jeliffe-Pawlowski LL, Gould JB, Stevenson DK, Snyder M, Shaw GM, O’Brodovich HM. Exome sequencing of neonatal blood spots and the identification of genes implicated in bronchopulmonary dysplasia. Am J Respir Crit Care Med 192: 589–596, 2015. doi: 10.1164/rccm.201501-0168OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Wang L, Liu Y, Wang Z, Li M, Zhang B, Wang H, Liu K, Wen S. The association between endothelial nitric oxide synthase gene G894T polymorphism and hypertension in Han Chinese: a case-control study and an updated meta-analysis. Ann Hum Biol 42: 185–195, 2015. doi: 10.3109/03014460.2014.911958. [DOI] [PubMed] [Google Scholar]
- 41.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S; AMFS Investigators . A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87: 139–145, 2010. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luzina IG, Tsymbalyuk N, Choi J, Hasday JD, Atamas SP. CCL18-stimulated upregulation of collagen production in lung fibroblasts requires Sp1 signaling and basal Smad3 activity. J Cell Physiol 206: 221–228, 2006. doi: 10.1002/jcp.20452. [DOI] [PubMed] [Google Scholar]
- 43.Mahlman M, Karjalainen MK, Huusko JM, Andersson S, Kari MA, Tammela OK, Sankilampi U, Lehtonen L, Marttila RH, Bassler D, Poets CF, Lacaze-Masmonteil T, Danan C, Delacourt C, Palotie A, Muglia LJ, Lavoie PM, Hadchouel A, Rämet M, Hallman M. Genome-wide association study of bronchopulmonary dysplasia: a potential role for variants near the CRP gene. Sci Rep 7: 9271, 2017. doi: 10.1038/s41598-017-08977-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason RP, Kalinowski L, Jacob RF, Jacoby AM, Malinski T. Nebivolol reduces nitroxidative stress and restores nitric oxide bioavailability in endothelium of black Americans. Circulation 112: 3795–3801, 2005. doi: 10.1161/CIRCULATIONAHA.105.556233. [DOI] [PubMed] [Google Scholar]
- 45.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501, 2010. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res 44: D336–D342, 2016. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Natarajan G, Pappas A, Shankaran S, Kendrick DE, Das A, Higgins RD, Laptook AR, Bell EF, Stoll BJ, Newman N, Hale EC, Bara R, Walsh MC. Outcomes of extremely low birth weight infants with bronchopulmonary dysplasia: impact of the physiologic definition. Early Hum Dev 88: 509–515, 2012. doi: 10.1016/j.earlhumdev.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolan K, Thompson TB. The DAN family: modulators of TGF-β signaling and beyond. Protein Sci 23: 999–1012, 2014. doi: 10.1002/pro.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park KW, Park JJ, Kang J, Jeon KH, Kang SH, Han JK, Lee SE, Yang HM, Lee HY, Kang HJ, Koo BK, Oh BH, Park YB, Kim HS. Paraoxonase 1 gene polymorphism does not affect clopidogrel response variability but is associated with clinical outcome after PCI. PLoS One 8: e52779, 2013. doi: 10.1371/journal.pone.0052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posencheg MA, Gow AJ, Truog WE, Ballard RA, Cnaan A, Golombek SG, Ballard PL; NO CLD Investigators . Inhaled nitric oxide in premature infants: effect on tracheal aspirate and plasma nitric oxide metabolites. J Perinatol 30: 275–280, 2010. doi: 10.1038/jp.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasse A, Pechkovsky DV, Toews GB, Jungraithmayr W, Kollert F, Goldmann T, Vollmer E, Müller-Quernheim J, Zissel G. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am J Respir Crit Care Med 173: 781–792, 2006. doi: 10.1164/rccm.200509-1518OC. [DOI] [PubMed] [Google Scholar]
- 52.Pryhuber GS, Maitre NL, Ballard RA, Cifelli D, Davis SD, Ellenberg JH, Greenberg JM, Kemp J, Mariani TJ, Panitch H, Ren C, Shaw P, Taussig LM, Hamvas A; Prematurity and Respiratory Outcomes Program Investigators . Prematurity and respiratory outcomes program (PROP): study protocol of a prospective multicenter study of respiratory outcomes of preterm infants in the United States. BMC Pediatr 15: 37, 2015. doi: 10.1186/s12887-015-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575, 2007. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rai H, Parveen F, Kumar S, Kapoor A, Sinha N. Association of endothelial nitric oxide synthase gene polymorphisms with coronary artery disease: an updated meta-analysis and systematic review. PLoS One 9: e113363, 2014. doi: 10.1371/journal.pone.0113363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramos PS, Oates JC, Kamen DL, Williams AH, Gaffney PM, Kelly JA, Kaufman KM, Kimberly RP, Niewold TB, Jacob CO, Tsao BP, Alarcón GS, Brown EE, Edberg JC, Petri MA, Ramsey-Goldman R, Reveille JD, Vilá LM, James JA, Guthridge JM, Merrill JT, Boackle SA, Freedman BI, Scofield RH, Stevens AM, Vyse TJ, Criswell LA, Moser KL, Alarcón-Riquelme ME, Langefeld CD, Harley JB, Gilkeson GS. Variable association of reactive intermediate genes with systemic lupus erythematosus in populations with different African ancestry. J Rheumatol 40: 842–849, 2013. doi: 10.3899/jrheum.120989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trovoada MJ, Martins M, Ben Mansour R, Sambo MR, Fernandes AB, Antunes Gonçalves L, Borja A, Moya R, Almeida P, Costa J, Marques I, Macedo MP, Coutinho A, Narum DL, Penha-Gonçalves C. NOS2 variants reveal a dual genetic control of nitric oxide levels, susceptibility to Plasmodium infection, and cerebral malaria. Infect Immun 82: 1287–1295, 2014. doi: 10.1128/IAI.01070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Valk RJ, Duijts L, Timpson NJ, Salam MT, Standl M, Curtin JA, Genuneit J, Kerhof M, Kreiner-Møller E, Cáceres A, Gref A, Liang LL, Taal HR, Bouzigon E, Demenais F, Nadif R, Ober C, Thompson EE, Estrada K, Hofman A, Uitterlinden AG, van Duijn C, Rivadeneira F, Li X, Eckel SP, Berhane K, Gauderman WJ, Granell R, Evans DM, St Pourcain B, McArdle W, Kemp JP, Smith GD, Tiesler CM, Flexeder C, Simpson A, Murray CS, Fuchs O, Postma DS, Bønnelykke K, Torrent M, Andersson M, Sleiman P, Hakonarson H, Cookson WO, Moffatt MF, Paternoster L, Melén E, Sunyer J, Bisgaard H, Koppelman GH, Ege M, Custovic A, Heinrich J, Gilliland FD, Henderson AJ, Jaddoe VW, de Jongste JC; EArly Genetics & Lifecourse Epidemiology (EAGLE) Consortium . Fraction of exhaled nitric oxide values in childhood are associated with 17q11.2-q12 and 17q12-q21 variants. J Allergy Clin Immunol 134: 46–55, 2014. doi: 10.1016/j.jaci.2013.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Velez DR, Hulme WF, Myers JL, Weinberg JB, Levesque MC, Stryjewski ME, Abbate E, Estevan R, Patillo SG, Gilbert JR, Hamilton CD, Scott WK. NOS2A, TLR4, and IFNGR1 interactions influence pulmonary tuberculosis susceptibility in African-Americans. Hum Genet 126: 643–653, 2009. doi: 10.1007/s00439-009-0713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wai KC, Kohn MA, Ballard RA, Truog WE, Black DM, Asselin JM, Ballard PL, Rogers EE, Keller RL; Trial of Late Surfactant (TOLSURF) Study Group . Early cumulative supplemental oxygen predicts bronchopulmonary dysplasia in high risk extremely low gestational age newborns. J Pediatr 177: 97–102.e2, 2016. doi: 10.1016/j.jpeds.2016.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, St Julien KR, Stevenson DK, Hoffmann TJ, Witte JS, Lazzeroni LC, Krasnow MA, Quaintance CC, Oehlert JW, Jelliffe-Pawlowski LL, Gould JB, Shaw GM, O’Brodovich HM. A genome-wide association study (GWAS) for bronchopulmonary dysplasia. Pediatrics 132: 290–297, 2013. doi: 10.1542/peds.2013-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Warburton D, Bellusci S. The molecular genetics of lung morphogenesis and injury repair. Paediatr Respir Rev 5, Suppl A: S283–S287, 2004. doi: 10.1016/S1526-0542(04)90052-8. [DOI] [PubMed] [Google Scholar]
- 63.Wilkins MR, Aldashev AA, Wharton J, Rhodes CJ, Vandrovcova J, Kasperaviciute D, Bhosle SG, Mueller M, Geschka S, Rison S, Kojonazarov B, Morrell NW, Neidhardt I, Surmeli NB, Aitman TJ, Stasch JP, Behrends S, Marletta MA. α1-A680T variant in GUCY1A3 as a candidate conferring protection from pulmonary hypertension among Kyrgyz highlanders. Circ Cardiovasc Genet 7: 920–929, 2014. doi: 10.1161/CIRCGENETICS.114.000763. [DOI] [PubMed] [Google Scholar]
- 64.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Y, Zhu Z, Fang X, Yin L, Liu Y, Xu S, Li A. The association between NOS3 gene polymorphisms and hypoxic-ischemic encephalopathy susceptibility and symptoms in Chinese Han population. BioMed Res Int 2016: 1957374, 2016. doi: 10.1155/2016/1957374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Kaminski N. Biomarkers in idiopathic pulmonary fibrosis. Curr Opin Pulm Med 18: 441–446, 2012. doi: 10.1097/MCP.0b013e328356d03c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao GL, Li QJ, Lu HY. Association between NOS3 genetic variants and coronary artery disease in the Han population. Genet Mol Res 15: gmr8044, 2016. doi: 10.4238/gmr.15028044. [DOI] [PubMed] [Google Scholar]