Abstract
From natural ecology1–4 to clinical therapy5–8, cells are often exposed to mixtures of multiple drugs. Two competing null models are used to predict the combined effect of drugs: response additivity (Bliss) and dosage additivity (Loewe)9–11. Here, noting that these models diverge with increased number of drugs, we contrast their predictions with growth measurements of four phylogenetically distant microorganisms including Escherichia coli, Staphylococcus aureus, Enterococcus faecalis and Saccharomyces cerevisiae, under combinations of up to ten different drugs. In all species, as the number of drugs increases, Bliss maintains accuracy while Loewe systematically loses its predictive power. The total dosage required for growth inhibition, which Loewe predicts should be fixed, steadily increases with the number of drugs, following a square-root scaling. This scaling is explained by an approximation to Bliss where, inspired by R. A. Fisher’s classical geometric model12, dosages of independent drugs add up as orthogonal vectors rather than linearly. This dose-orthogonality approximation provides results similar to Bliss, yet uses the dosage language as in Loewe and is hence easier to implement and intuit. The rejection of dosage additivity in favour of effect additivity and dosage orthogonality provides a framework for understanding how multiple drugs and stressors add up in nature and the clinic.
In both nature and the clinic, cells are often exposed to combinations of multiple stresses and drugs. In natural ecosystems, such as the soil, dozens of microbial species capable of producing different antimicrobial compounds coexist in very close proximity, thus exposing each other to a mixture of multiple stressors1–4. In clinical settings, drug combinations, aimed at reducing side effects and counteracting resistance13–17, are becoming increasingly important in treatment for infectious diseases and cancer5–8,18,19. It is therefore of wide importance to understand how cell growth is affected by combinations of a multitude of stressors, and thereby the general rules of high-dimensionality drug arithmetic.
When combined together, drugs can interact to synergize or antagonize each other’s effects relative to a null additive model. Synergy occurs when the combined effect of drugs is larger than expected based on their individual effects. Conversely, drugs can also antagonize each other, leading to a combined effect that is smaller than expected. These interactions are important in clinical settings as a way of increasing treatment potency and selectivity20–22 or slowing selection for resistance14,17,23. Importantly, the definition of both synergy and antagonism relies on comparing the effect of drug combinations with a null model of ‘additive expectation’6,24–27.
There are two primary models for the null effect of drug combinations6,28: the Bliss model10,29, which assumes response additivity; and the Loewe model9,11, which assumes dose additivity. According to Bliss, the combined effect of two drugs E1+2 is simply the sum of their individual effects30, E1+2 = E1+E2, where Ei = (g0 − gi)/g0 is the effect of drug i on the normalized growth rate g/g0 (Fig. 1; when effects are measured on the basis of total yield rather than growth rate, Bliss additivity becomes multiplicativity; Supplementary Note 1). In contrast, according to Loewe additivity, the effect of drugs in combination is determined not by the sum of their normalized effects, but rather by the sum of their normalized dosages, such that their combined effect is the same across all combinations that have the same total normalized dosage. Namely, according to Loewe, lines of equal combination effect in drug-dosage space (isoboles) are linear26 (Fig. 1). For example, if two drugs are additive with respect to Loewe, their 50% inhibition isobole is a straight line satisfying , where di is the dose of drug i and is the dose at which drug i alone causes 50% growth inhibition (IC50). Although conceptually different from one another, mechanistic support is available for both the Bliss and the Loewe models31 and there is no agreement on which model should generally be used32–35. Models that implement pairwise interaction data as well as higherorder interactions can improve multidrug predictions of either Bliss or Loewe21,36–39. Yet, regardless of pairwise interactions, it remains unknown which of these two null models best predicts the combined effect of multiple drugs.
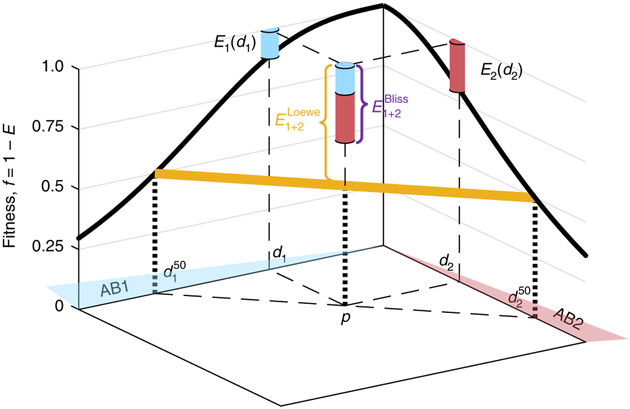
Fig. 1 |. Schematic depiction of effect additivity (Bliss) and dosage additivity (Loewe).
Given fitness as a function of dosage of each of the individual drugs (fi = 1 − Ei, dose response curve, black solid line), Bliss and Loewe models predict the fitness f1 + 2 = 1 − E1+2 at any given point P = [d1,d2] in the drug concentration space. The Bliss prediction assumes additivity of normalized drug effects, , where E1 and E2 are the individual drug effects at their cognate concentrations (cyan and red piles, respectively). The Loewe model, on the other hand, assumes additivity of normalized drug dosage, such that the combined drug effect is fixed along linear lines of constant total normalized dosage (yellow, equals 50% in the example point P, and more generally is given by solving for , where is the concentration of drug i that leads to inhibition level E).
Here, measuring bacterial response to antibiotic combinations, we contrast the Bliss and Loewe models for an increasing number of drugs, where we show that expectations of these models increasingly diverge. We focus on four very different organisms: Escherichia coli as a model Gram-negative bacterium, Saccharomyces cerevisiae as a model eukaryote and the clinically important Gram-positive pathogens Staphylococcus aureus and Enterococcus faecalis. Quantifying their response to combinations of up to ten different drugs, we find that the Bliss model maintains accuracy with increased number of drugs, while the Loewe model loses its predictive power. Indeed, in contrast to Loewe, which predicts that the total drug dosage required for inhibition is constant, we find that total dosage increases monotonically with the number of drugs. Interestingly, our data show that this increase follows a square-root scaling, inspiring a simple model for orthogonality of dose additivity that follows a classical evolutionary optimization principle developed by R. A. Fisher12.
To contrast the Bliss and Loewe models, we calculated how their predictions scale with increased number of drugs. As a natural measure of the combined potency of multiple drugs, we considered the total drug dosage needed to achieve a given level of inhibition. Defining ‘total dosage’ (D) as the sum of the concentration of the individual drugs weighted by their corresponding IC50 values, , the ‘combination potency’ (D50) is the total dosage D that yields 50% growth inhibition. As the number of drugs N increases, Loewe additivity predicts that the combination potency remains fixed, . The prediction of Bliss, on the other hand, depends on the individual dose response curves of the different drugs. Assuming a Hill equation40 for the single-drug dose response , where h is the Hill coefficient, and equating the Bliss prediction of the combined effect E1..N = Σi=1..N Ei to 50%, yields the Bliss-predicted scaling of combination potency with the number of drugs: . Thus, while Loewe predicts that the total dosage required for inhibition is constant, Bliss predicts that it increases with the number of drugs (assuming Hill coefficient, h > 1). The two models can therefore best be contrasted by measuring the combined action of increased number of drugs.
Starting with E. coli, we considered ten mechanistically different antibiotics and measured their growth-inhibitory effect individually as well as in combinations with increased number of drugs. We chose antibiotics acting on a range of cellular functions, including cell wall synthesis, DNA replication, transcription and translation (Supplementary Table 1). Measuring optical density (OD) versus time of bacterial growth on gradients of each of the individual drugs, we determined the dose response curve g(di) for each of the drugs (Fig. 2a and Supplementary Fig. 1). These dose response curves are well fitted by Hill functions, defining the concentrations of 50% inhibition for each of the drugs in isolation (Supplementary Fig. 13a,b).
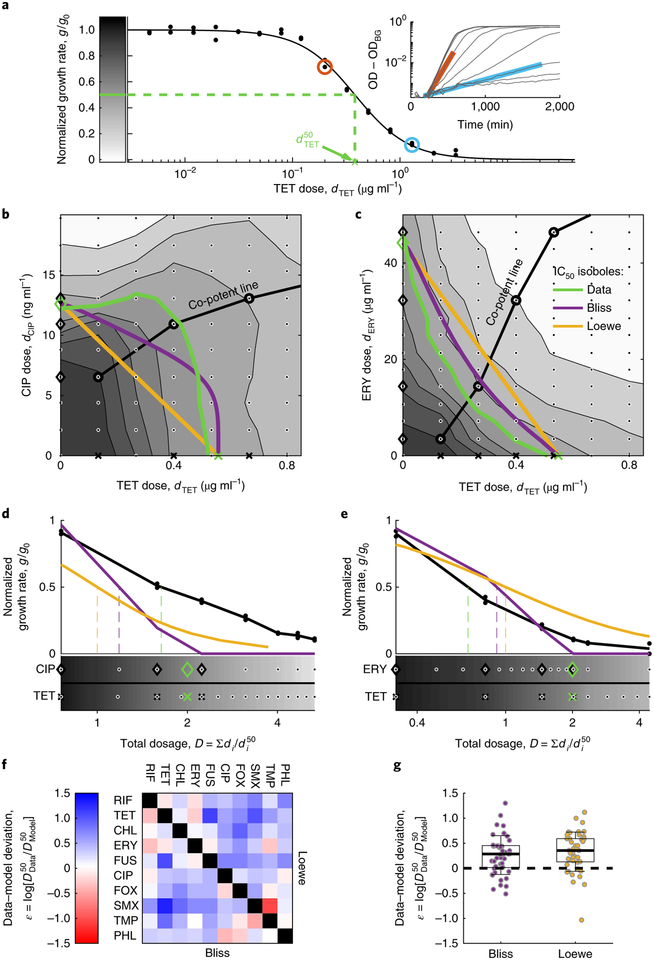
Fig. 2 |. Pairwise measurements do not resolve the Bliss and Loewe models of additivity.
a, Representative single-drug dose response curve showing the normalized growth rate g/g0 along a concentration gradient of TET (black dots, replicates), the Hill equation fit (, black line) and the IC50 (, green dashed line). Inset: growth rates g were calculated by fitting OD600 nm measurements over time (black) to exponential function, OD = OD0·2g·t + ODBG (cyan and red). b,c, Response surface showing growth rates (greyscale indicated in a) over a two-dimensional grid gradient (dots) of the antagonistic antibiotic pair TET–CIP (b) and the synergistic pair TET–ERY (c). The measured IC50 isoboles (green) are contrasted with Bliss (purple) and Loewe (orange) predictions. Indicated are the co-potent lines (circles), the corresponding co-potent single drugs (black crosses for TET, black diamonds for CIP and ERY) and the IC50 values (green symbols). d,e, Dose response along the co-potent line of the two drug mixtures (TET–CIP, d; TET–ERY, e) as a function of total dosage . Measured normalized growth rates of the combined drugs are contrasted with Bliss and Loewe predictions based on the single-drug data (shown below). All symbols correspond to those in b and c. f,g, Data-model deviations (, indicates the difference between measured () and predicted () combination potencies) for each of the two models are presented as an interaction matrix (f) and a box plot (g, box at first and third quartiles, whiskers at mean ± two standard deviations, black dashed line represents perfectly accurate prediction). There is no significant difference between the models in their predictions of measured pairwise potencies (45 different combinations, two-sided t-test, P = 0.42).
Moving to drug pairs, we measured their combination potency and compared it to Bliss and Loewe predictions. We first measured the full response surface across two-dimensional dose gradients for two drug pairs: tetracycline and ciprofloxacin (TET–CIP) and tetracycline and erythromycin (TET–ERY), representing well-known examples of antibiotic antagonism and synergy (Fig. 2b,c response surface and IC50 isoboles; Supplementary Fig. 12a)41,42. Using the growth measurements of the individual drug gradients Ei(di), we derived the response surface and the IC50 isobole predictions of Loewe (straight line connecting the points [, 0] and [0, ]) and Bliss (the set of all points [d1, d2] satisfying E1(d1) + E2(d2) = 50%, Methods). As expected, the measured IC50 isobole lies above these predictions for the TET–CIP pair (indicating antagonism) and below for the TET–ERY pair (indicating synergy). While these two-dimensional gradients allow clear definition of synergy and antagonism, they require many growth measurements and become combinatorically prohibitive in a high-dimensional multidrug space.
To effectively sample the concentration space of multiple drugs, we performed growth measurements along a ‘co-potent’ line38, a curve in concentration space where the individual drugs have equal potencies in isolation (E1(d1) = E2(d2) =.. = EN(dN), Fig. 2b–e). This co-potent line sampling method vastly reduces the dimensionality of the required measurements while guaranteeing that null models are evaluated in a region in drug concentration space where all drugs are active, rather than one in which the combined effect is dominated by a subset of drugs. Identifying the point P = (d1,d2,…,dN) on the co-potent line where growth is inhibited by 50% yields the combination potency, . This measured combination potency was contrasted with the expected potencies and , defined as the points along the co-potent line where the single-drug-based calculations of Bliss and Loewe predict 50% inhibition (Methods). The interaction between drugs was then defined as the deviation between the observed and predicted potencies of the combination , which captures the extent of antagonism (ε > 0) and synergy (ε > 0) (Fig. 2d,e).
Measuring combination effects for all drug pairs, we find that their joint potencies are similarly well predicted by both the Bliss and the Loewe models. For each of the 45 drug pairs, we measured their dose response along co-potent concentration gradient and determined their combination potency, (Methods, Fig. 2f and Supplementary Figs. 12b and 13a). Comparing these combination potencies with predictions of the Bliss and Loewe null models, we find that both positive (ε > 0, antagonism) and negative (ε > 0, synergism) deviations are prevalent with respect to either model (Fig. 2f,g). This prevalence of both antagonism and synergy among drug pairs overwhelms any small deviations between the two models (Fig. 2g; σ(εBliss) = 0.39; σ(εLoewe) = 0.39; < εBliss> − <εLoewe> = −0.066, t-test: P = 0.42). Further, clustering drugs on the basis of these pairwise interactions, defined with respect to either Bliss or Loewe, leads to similar grouping by mechanism of action (Supplementary Fig. 2; possible small advantage to Bliss in resolving fine functionality differences)43. The similarity of pairwise null predictions, the prevalence and magnitude of pairwise interactions with respect to both models, and their similar correlation with cellular function prohibit distinction of the Loewe and Bliss null models based on drug pairs.
However, for increasing number of drugs, we find that the combined effect is well predicted by Bliss, while the Loewe prediction systematically diverges. Given that predictions of the two models should diverge with increased number of drugs, we measured the combined effect of multiple combinations with a varying number of antibiotics. We chose 35 combinations of 3 to 10 antibiotics, including 8 randomly chosen sets from each combination size of N = 3, 5 and 7 drugs, all 10 sets of 9 drugs, and the whole 10-drug set (Fig. 3a,b; Supplementary Figs. 3a, 12c and 13b and Supplementary Tables 9 and 10). Following the procedure used for the drug pairs, we measured the combined effect of each multidrug set as a function of total dosage along co-potent lines and identified their combination potencies D50. Contrasting these measured potencies with the predicted potencies of Bliss and Loewe based on the single-drug measurements, we find that the Bliss model maintains good accuracy regardless of the number of drugs, while the accuracy of the Loewe model declines as the number of drugs increases (Fig. 3c, dots, and Supplementary Figs. 4 and 5). This rejection of Loewe in favour of Bliss is independent of the co-potency of the drug mixture (Supplementary Fig. 6) and also appears when using the multiplicative form of the Bliss model (more suitable for yield measurements, Supplementary Note 1; Supplementary Fig. 7), as well as when considering alternative derivation of growth rates from OD curves (Supplementary Fig. 1c)44. Our E. coli growth data therefore show that the effect of combinations of many diverse drugs is well predicted by Bliss, and not by Loewe.
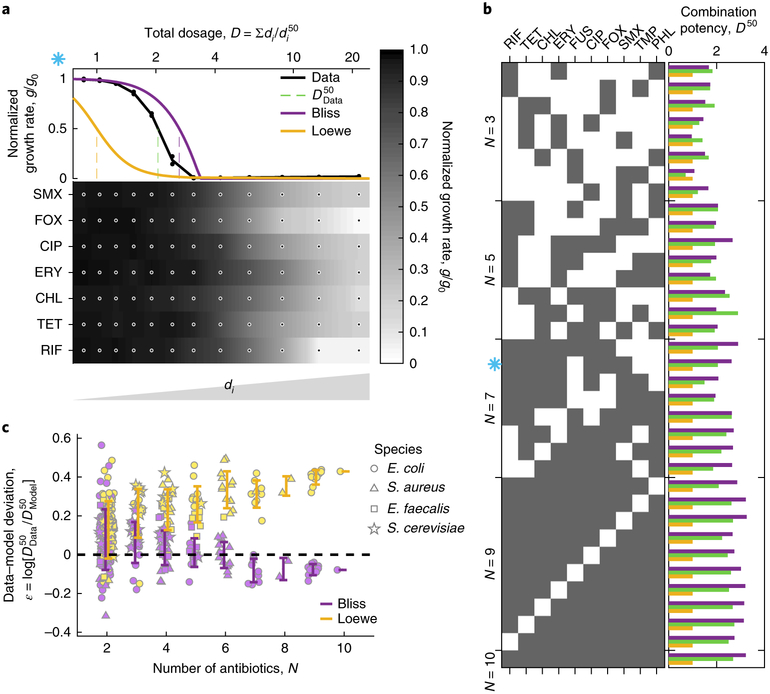
Fig. 3 |. The Loewe model of additivity loses its predictive power with increased number of combined drugs.
a, An example of dose response along the co-potent line of a mixture of seven drugs. The measured normalized growth rate calculated on the basis of duplicate measurements (dots) and the combined potency of the drug mixture are contrasted with Bliss and Loewe predictions calculated on the basis of the single-drug measurements (below, greyscale). b, The combination potency (, green) is contrasted with predictions of Bliss (, purple) and Loewe (, yellow) for 35 different combinations of 3 to 10 drugs (black squares). The blue asterisk matches the combination shown in a. c, The deviation of each of the models from the data is plotted for 172 different combinations as a function of the number of drugs (error bars represent one standard deviation of each combination size), showing that the Loewe predictions deviate from the data with increased number of drugs, while Bliss predictions remain accurate (the dashed line represents perfectly accurate prediction).
To test the generality of these findings, we applied our methodology to the Gram-positive pathogens E. faecalis and S. aureus as well as to the eukaryotic model of S. cerevisiae. For each of these organisms, we chose a repertoire of diverse drugs and measured their combined effect in random sets of increased number of drugs (up to eight in S. aureus, six in E. faecalis and five in S. cerevisiae; Supplementary Tables 1, 9 and 10 and Supplementary Figs. 3b–e, 12d–g and 13c–f). As in E. coli, both the Bliss and Loewe models provided comparable predictions for combinations of small number of drugs and when the number of drugs increases the fitness was well predicted by Bliss while Loewe’s prediction diverged (Fig. 3c and Supplementary Figs. 4 and 5). Furthermore, the power of Bliss over Loewe is also seen for combinations involving strongly synergistic drug pairs, treating the pair mixture as a single agent (TMP–SLF in S. aureus; Supplementary Fig. 8). We conclude that across species the Loewe model, predicting that the total dosage required for inhibition is fixed regardless of the number of drugs, can be rejected as a general predictor for the potency of diverse multidrug combinations.
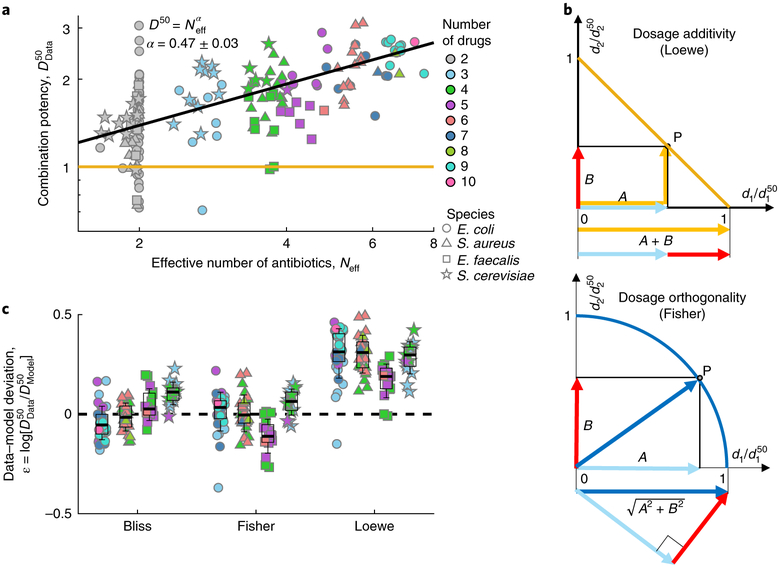
Next, we tested how the potency of drug combinations, namely the total dosage required for inhibition, varies with the number of drugs. To account for any slight experimental deviations from the ideal co-potent line, we use a natural entropy-like definition of an effective number of drugs Neff that is based on the uniformity of the individual drug effects (Neff equals N if all drugs have the same effect; is slightly smaller than N when these effects are uneven; and converges to 1 at the extreme case when a single drug dominates; see definition of Neff in Fig. 4 caption). Contrary to the Loewe prediction, we find that the total dosage required for inhibition increases with the effective number of drugs (Fig. 4a and Supplementary Fig. 8a). Moreover, this inhibitory total dosage seems to obey a simple scaling law: it increases as the square root of the effective number of drugs (D50 = (Neff)α, least-square fit yields: α = 0.47 ± 0.03).
Fig. 4 |. A square-root scaling law of inhibitory total dosage with effective number of drugs is explained by a simple dosage-orthogonality model.
a, The combination potency, of all 172 different drug combinations is plotted as a function of effective number of drugs (Neff=exp[− Σipi log(pi)], where pi = Ei/ΣjEj, and Ei(di) are the single-drug individual effects at their cognate concentrations; colours represent the actual number of drugs, N). In contrast to Loewe, which assumes that the total dosage required for inhibition is fixed (yellow line), the total dosage increases as the square root of the effective number of drugs (black line, fit of D50 = (Neff)α yields α = 0.47 ± 0.03, 0.95 confidence interval). b, The square-root scaling is explained by a Fisher-inspired dose-orthogonality model, which assumes that for small perturbations the dosages of independent drugs should be added as orthogonal vectors rather than linearly as in Loewe. Hence, isoboles of X% inhibition are spherical surfaces defined by (Fisher, bottom, circles in two-drug space, blue line) instead of linear surfaces (Loewe, top, straight lines in two-drug space, yellow line). c, Data-model deviation of all the combinations of more than two drugs for each of the three models (box at median and first and third quartiles, whiskers at mean ± two standard deviations, black dashed line represents perfectly accurate prediction). The data strongly reject Loewe and are instead consistent with both Fisher and Bliss (96 data points, two sided t-test: Loewe, P < 10−40; Bliss, P = 0.61; Fisher, P = 0.8).
The square-root scaling of the inhibitory dosage with number of drugs can be explained by an approximation to Bliss, inspired by the classical optimization principle of Fisher’s geometric model of adaptation12. Fisher’s model describes the fitness f in a space of N independent orthogonal phenotypes and assumes that it declines as a function of the Euclidean distance from an optimal point (, where and xi are phenotypic distances from the optimal point). For a given fitness value, the phenotypic distances xi therefore decline as and their sum, , increases as . The analogy of drug dosages with Fisher’s phenotypes, and more generally the analogy of drug and mutations, explains the square-root scaling of total inhibitory dosage with number of drugs and underscores that drug concentrations should be summed not linearly by simple addition as in Loewe, but rather as the geometric sum of orthogonal vectors (Fig. 4b; of course, orthogonality is an idealization from which drug combinations can deviate due to synergy or antagonism, or when similar drugs act along the same axis). This Fisher-inspired ‘dose-orthogonality’ model can also be derived as a second-order approximation of Bliss additivity at the limit of small dosages, emphasizing the dependency of this approximation on optimality of the growth rate at the origin (Supplementary Note 2; for Hill coefficients close to 1, as common in the drug combinations applied to E. faecalis, Bliss prediction can be closer to Loewe than to Fisher). Indeed, we find that even strongly interacting drug pairs assume more circular isoboles for small fitness effects (Fig. 2b,c and Supplementary Fig. 10) and that the square-root scaling becomes more accurate at a lower level of inhibition (Supplementary Fig. 9b). Similarly to Bliss and in contrast to Loewe, combination potency predictions of this dosage-orthogonality model (derived by intersecting the co-potent lines with spherical IC50 isoboles , Methods) are consistent with the drug combination measurements (Fig. 4c). Yet, unlike Bliss these predictions do not require fine measurements of the minute individual drug effects Ei(di), but rather depend on the more robust measurements of their individual IC50 dosages, . While it allows the use of dose language like Loewe, the dose-orthogonality model provides an intuitive and robust approximation of Bliss’s response additivity (Supplementary Table 2), which well predicts the potency of drug combinations and explains the square-root scaling law of potency with number of drugs.
Across diverse taxons, our measurements reject the Loewe model of dosage additivity for predicting the effect of combination of multiple diverse drugs, favouring the Bliss effect additivity and motivating a simple model of dosage-orthogonality. In contrast to Loewe additivity, which predicts that the total dosage required for inhibition is fixed, we find that the total inhibitory dosage increases with the number of drugs, following a square-root scaling law. This general reduction in potency with increased number of drugs could be important for understanding ecological environments where bacteria are exposed to a multitude of drugs and stresses and any one toxin does not typically work on its own but rather combined within a complex soup of natural inhibitors. While the results could also be important in the clinical settings where multiple drugs are combined, any such implications will require extending the conceptual approach and methodology and to consider the killing regime, the impact on multispecies communities and the complexity that stems from transient effects introduced by the pharmacokinetics and pharmacodynamics of each of the individual drugs. The square-root scaling supports a model for drug additivity where dosages of independent drugs add up orthogonally rather than linearly. This dosage-orthogonality model provides an approximation to Bliss, yet it uses dosage arithmetic that allows a more robust implementation and simple intuition. It will be interesting to explore the generality of these results and the limit on the number of orthogonal pharmacological axes as more antibiotics and stresses are added, as well as beyond the minimal inhibitory concentration and in more complex systems such as in cancer therapy. Throughout such clinical systems and natural ecologies, our findings provide a uniform framework for understanding the null arithmetic of many-drug combinations.
Methods
Strains and media.
Experiments were performed with: E. coli strain MC4100 in M9 media (Na2HPO4 6 g/l−1, Na2HPO4 3 g l−1, NaCl 0.5 g l−1, NH4Cl 1 g l−1, glucose 2 g l−1, Casamino acids 1 g l−1, thiamine 0.34 g l−1, MgSO4 1 mM, CaCl2 0.1 mM); S aureus sp. Rosenbach (ATCC 29213 - Wichita) in Mueller–Hinton broth; E. faecalis ATCC 49757 in brain heart infusion broth; and S. cerevisiae BY4741 Euroscarf in YPD broth (yeast extract 1%, peptone 2%, glucose 2%). Antibiotics were added as indicated (Sigma Aldrich).
Growth rate assay.
The data represent seven different experimental set-ups: three for E. coli: two-dimensional gradient of ERY–TET and CIP–TET (Fig. 2b–e and Supplementary Figs. 9 and 12a), all 45 pairwise combinations (Fig. 2f,g and Supplementary Figs. 12b and 13a), and 35 combinations of order higher than two (Figs. 3 and 4, dots, and Supplementary Figs. 12c and 13b); two for S. aureus: 22 combinations not involving beta-lactam antibiotics, and 21 combinations involving beta-lactam antibiotics (Figs. 3c and 4, triangles, and Supplementary Figs. 12d,e and 13c,d); one for E. faecalis, composed of 23 combinations (Figs. 3c and 4, squares, and Supplementary Figs. 12f and 13e); and one for S. cerevisiae, composed of 26 combinations (Figs. 3c and 4, stars; Supplementary Figs. 12g and 13e). In each of these experiments, an inoculum of 104 cells was inoculated into 150 μl of media in a Nunc 96-well flat-bottom microplate. Antibiotics were added into the wells as indicated using a D300e digital dispenser (Tecan), which dispenses a nanolitre-scale volume of each antibiotic. Each concentration combination was performed in duplicate wells. Multiple untreated control wells (no antibiotics) were designated on each plate (2–6% of the wells in each experiment). To avoid systematic positional effects across the plates, the wells chosen for each drug combination on the plates were randomized. The plates were incubated at 30 °C with shaking (Liconic orbital shaker STX44), and OD600 nm was measured at least every 25 min using a Tecan robotic system and the Infinite M200 Pro reader. To enhance uniformity, the plate orientations in the shaker were rotated 180° following every measurement.
Growth rate analysis.
Data analysis was performed using MATLAB. The OD600 nm measurements were averaged using a running window of two data points. The log phase of each of these curves was determined using an algorithm based on the sensitivity analysis method (‘tornado diagram’, Supplementary Fig. 1), and OD data in this constant exponential growth region were fitted to OD = OD0·2gt + ODBG (in some of the wells, the log-phase period was mis-determined, and were cured manually; all fits are indicated in Supplementary Fig. 12). From these fitted parameters, we obtained for each well the background ODBG and the growth rate g. Fitness for each drug mixture was then calculated as normalized growth rates g/g0, where g is the average growth rate in duplicate wells and g0 is the median growth rate of all untreated wells in the experiment. All calculated normalized growth rates are available in Supplementary Table 10. While OD is commonly used as a measure of microbial density, the relation between OD and cell number depends on cell size and morphology, which can be affected by the antibiotics. As the growth rate is defined as the logarithmic derivative of the OD, it is independent of this effect of antibiotic on cell size, as long as these effects are constant throughout the fitted region. Therefore, we measure the growth rate in the regime of steady exponential growth long after the addition of the antibiotic.
Determining drug concentrations for co-potent combinations.
To achieve co-potent concentrations for the different drugs, we first measured growth rates on gradients of each of the individual drugs and identified the IC20 and IC80 for each of the drugs, and . Then, we designed the drug gradients such that the individual drugs have matching effects: , where di(k) is the dosage of drug i at dilution number k, is the dilution factor (chosen to have three dilutions between the IC20 and IC80), is the highest concentration (strong enough to inhibit growth) and k varies from 1 to 15 such that the last concentration is low enough to have negligible effect (all drug concentrations are provided in Supplementary Tables 3–8). For example: 7.3 ng ml−1 and 53.1 ng ml−1 of TET inhibit the growth of S. aureus by 20% and 80%, respectively; hence, wTET = (53/7.3)1/3 = 1.93. At the same time, 35 ng ml−1 and 152 ng ml−1 ERY inhibit the growth by 20% and 80%, respectively; hence, wERY = (152/35)1/3 = 1.64. On the co-potent line, we mix ERY and TET (Supplementary Fig. 3b) with the following concentrations TET with Ery (‘O’ in Supplementary Table 5), 199 ng ml−1 TET with 405 ng ml−1 ERY (‘N’), …, 0.52 ng ml−1 TET with 2.93 ng ml−1 ERY (‘E’).
Calculating model predictions for Bliss, Loewe and Fisher.
The models predict fitness across drug concentration space f(d1,d2,…,dN) based on the single-drug dose response curves (fi(di), Hill fitted, Supplementary Fig. 12). For any given level of inhibition E, we define in each of the models the N − 1-dimensional isobolic surface of all points (d1d2,…,dN) in concentration space predicted to have fitness f = 1−E. For Bliss, this isobolic surface is the collection of points satisfying Σi (1 − fi (di)) = 1 – E. For Loewe, the isobolic surface is predicted to be linear, satisfying , where is the concentration of drug i that inhibits growth by E, as defined by . For Fisher, this isobolic surface is an N - 1-dimensional sphere defined by . Substituting E = 50%, these calculations define the IC50 isoboles (Fig. 2b,c). The intersections of the co-potent lines with the predicted isobolic surfaces for ranges of values of E define the predicted fitness along the co-potent line (Figs. 2 and 3, Supplementary Fig. 3 and Supplementary Table 11). In particular, the intersections of the co-potent lines with the E = 50% isobolic surfaces define the predicted potencies , and (Figs. 2–4, Supplementary Fig. 3 and Supplementary Table 11).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability.
MATLAB codes to calculate growth rate and model predictions are available on the laboratory website at https://kishony.net.technion.ac.il/resources/; any additional codes are available from the corresponding author.
Supplementary Material
Acknowledgements
We thank U. Alon and I. Katzir for thoughtful discussions and suggestions, Y. Arava for strains, and members of the Kishony lab, especially M. Datta, E. Tamar, I. Yelin and N. Yin, for experimental support and comments on the manuscript. This work was supported in part by the US National Institutes of Health grant R01-GM081617, the Israeli Centers of Research Excellence I-CORE Program ISF grant 152/11 and the European Research Council FP7 ERC Grant 281891 (to R.K.).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-018-0252-1.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The data sets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
- 1.Ueda K et al. Wide distribution of interspecific stimulatory events on antibiotic production and sporulation among Streptomyces species. J. Antibiot 53, 979–982 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Clardy J, Fischbach MA & Currie CR The natural history of antibiotics. Curr. Biol 19, R437–R441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vetsigian K, Jajoo R & Kishony R Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol 9, e1001184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chait R, Vetsigian K & Kishony R What counters antibiotic resistance in nature? Nat. Chem. Biol 8, 2–5 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Blumberg HM et al. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med 167, 603–662 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Keith CT, Borisy AA & Stockwell BR Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov 4, 71–78 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald JB, Schoeberl B, Nielsen UB & Sorger PK Systems biology and combination therapy in the quest for clinical efficacy. Nat. Chem. Biol 2, 458–466 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Palmer AC & Sorger PK Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 171, 1678–1691. e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loewe S & Muischnek H Über kombinationswirkungen. Archivf. experiment. Pathol. u. Pharmakol 114, 313–326 (1926). [Google Scholar]
- 10.Bliss CI The toxicity of poisons applied jointly. Ann. Appl. Biol 26, 585–615 (1939). [Google Scholar]
- 11.Loewe S The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3, 285–290 (1953). [PubMed] [Google Scholar]
- 12.Fisher RA The Genetical Theory of Natural Selection (Clarendon Press, Oxford, 1930). [Google Scholar]
- 13.Chait R, Craney A & Kishony R Antibiotic interactions that select against resistance. Nature 446, 668–671 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Michel J-B et al. Drug interactions modulate the potential for evolution of resistance. Proc. Natl Acad. Sci. USA 105, 14918–14923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamovic L & Sommer MOA Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med 5, 204ra132 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Pál C, Papp B & Lázár V Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol 23, 401–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baym M, Stone LK & Kishony R Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann GR, Lehár J & Keith CT Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today 12, 34–42 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Bayat Mokhtari R et al. Combination therapy in combating cancer. Oncotarget 8, 38022–38043 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehár J et al. Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol 27, 659–666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cokol M et al. Systematic exploration of synergistic drug pairs. Mol. Syst. Biol 7, 544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrasekaran S et al. Chemogenomics and orthology-based design of antibiotic combination therapies. Mol. Syst. Biol 12, 872 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegreness M, Shoresh N, Damian D, Hartl D & Kishony R Accelerated evolution of resistance in multidrug environments. Proc. Natl Acad. Sci. USA 105, 13977–13981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou TC & Talalay P Quantitative analysis of dose–effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul 22, 27–55 (1984). [DOI] [PubMed] [Google Scholar]
- 25.Greco WR, Bravo G & Parsons JC The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev 47, 331–385 (1995). [PubMed] [Google Scholar]
- 26.Berenbaum MC The expected effect of a combination of agents: the general solution. J. Theor. Biol 114, 413–431 (1985). [DOI] [PubMed] [Google Scholar]
- 27.Tang J, Wennerberg K & Aittokallio T What is synergy? The Saariselkä agreement revisited. Front. Pharmacol 6, 181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeh PJ, Hegreness MJ, Aiden AP & Kishony R Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol 7, 460–466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao W et al. A new bliss independence model to analyze drug combination data. J. Biomol. Screen 19, 817–821 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Boucher AN & Tam VH Mathematical formulation of additivity for antimicrobial agents. Diagn. Microbiol. Infect. Dis 55, 319–325 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Baeder DY, Yu G, Hozé N, Rolff J & Regoes RR Antimicrobial combinations: Bliss independence and Loewe additivity derived from mechanistic multi-hit models. Phil. Trans. R. Soc. B 371, 20150294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foucquier J & Guedj M Analysis of drug combinations: current methodological landscape. Pharmacol. Res. Perspect 3, e00149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drescher K & Boedeker W Assessment of the combined effects of substances: the relationship between concentration addition and independent action. Biometrics 51, 716–730 (1995). [Google Scholar]
- 34.Goldoni M & Johansson C A mathematical approach to study combined effects of toxicants in vitro: evaluation of the Bliss independence criterion and the Loewe additivity model. Toxicol. In Vitro 21, 759–769 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Lee SI Drug interaction: focusing on response surface models. Korean J. Anesthesiol 58, 421–434 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood K, Nishida S, Sontag ED & Cluzel P Mechanism-independent method for predicting response to multidrug combinations in bacteria. Proc. Natl Acad. Sci. USA 109, 12254–12259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmer A, Katzir I, Dekel E, Mayo AE & Alon U Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc. Natl Acad. Sci. USA 113, 10442–10447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cokol M, Kuru N, Bicak E, Larkins-Ford J & Aldridge BB Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci. Adv 3, e1701881 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tekin E et al. Prevalence and patterns of higher-order interactions in Escherichia coli. NPJ Syst. Biol. Appl 4, 31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner JG Kinetics of pharmacologic response I. Proposed relationships between response and drug concentration in the intact animal and man. J. Theor. Biol 20, 173–201 (1968). [DOI] [PubMed] [Google Scholar]
- 41.Bollenbach T, Quan S, Chait R & Kishony R Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139, 707–718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.How SJ, Hobson D, Hart CA & Webster RE An in-vitro investigation of synergy and antagonism between antimicrobials against Chlamydia trachomatis. J. Antimicrob. Chemother 15, 533–538 (1985). [DOI] [PubMed] [Google Scholar]
- 43.Yeh P, Tschumi AI & Kishony R Functional classification of drugs by properties of their pairwise interactions. Nat. Genet 38, 489–494 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Kahm M, Hasenbrink G, Lichtenberg-Fraté H, Ludwig J & Kschischo M grofit: fitting biological growth curves with R. J. Stat. Softw 33, 1–21 (2010).20808728 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.