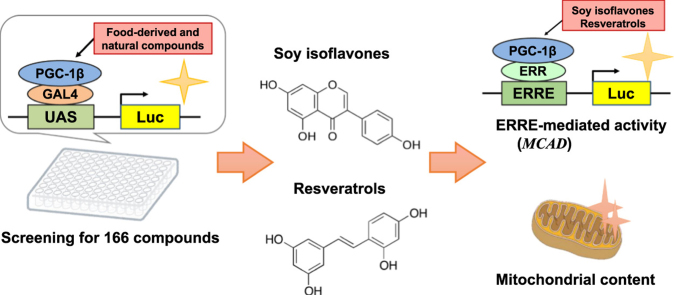
Abstract
PGC-1β is a transcriptional co-activator of nuclear receptors such as the estrogen receptor-related receptor (ERR). Transgenic overexpression of PGC-1β in mice increases energy expenditure and suppresses high-fat diet-induced obesity. In this study, we screened various food-derived and natural compounds using a reporter assay system to measure the transcriptional activity of PGC-1β. Soy-derived isoflavones, genistein and daidzein, and several resveratrols activated PGC-1β. Genistein, daidzein, and trans-oxyresveratrol activated ERR-responsive element-mediated reporter activity in the presence of PGC-1β. Stable overexpression of PGC-1β in C2C12 myoblasts increased the expression of medium-chain acyl-CoA dehydrogenase (MCAD), an important enzyme in fatty acid β-oxidation. Genistein and daidzein increased MCAD mRNA levels and mitochondrial content in PGC-1β-expressing C2C12 cells. These compounds activated ERR/PGC-1β complex-mediated gene expression, and our findings may be a practical foundation for developing functional foods targeting obesity.
Abbreviations: PGC-1β, Peroxisome proliferator activated receptor gamma coactivator-1β; ERR, estrogen receptor-related receptor; MCAD, medium-chain acyl-CoA dehydrogenase
Keywords: PGC-1β, Muscle cells, Isoflavone, ERR, Mitochondria
Graphical abstract
Highlights
-
•
We measured transcriptional activity of PGC-1β in the presence of various compounds.
-
•
Soy isoflavones and resveratrols increased the reporter activity of GAL4-PGC-1β.
-
•
They increased PGC-1β/ERR-target MCAD levels, and mitochondrial contents.
-
•
These findings may be a foundation for developing functional foods for anti-obesity.
1. Introduction
Peroxisome proliferator activated receptor gamma co-activator-1β (PGC-1β) and PGC-1α are transcriptional co-activators of nuclear receptors. PGC-1β has been cloned using a computer-based homology search based on similarity with PGC-1α by independent research groups including ours [1], [2]. PGC-1α co-activates transcriptional activity of multiple nuclear receptors, whereas PGC-1β preferentially co-activates estrogen receptor-related receptor (ERR) [2]. PGC-1α expression is induced by cold-exposure, exercise, and starvation in brown adipose tissue, skeletal muscle, and the liver, respectively [3]. By contrast, PGC-1β expression remains unaffected by external and endogenous stimuli. Therefore, it is possible that PGC-1β activity is not regulated by its expression but by other molecules such as the nuclear receptor ligands.
In mice, transgenic overexpression of PGC-1β in several tissues, including the skeletal muscle, increases the expression of medium-chain acyl-CoA dehydrogenase (MCAD), which is an ERR target and a pivotal enzyme in mitochondrial fatty acid β-oxidation [4], [5]. The transgenic mice are hyperphagic, have elevated energy expenditure, and are resistant to high-fat diet-induced obesity [2]. Therefore, PGC-1β is a promising target for developing functional foods to counter obesity.
In the present study, we screened for food-derived and natural compounds that activate PGC-1β. We used PGC-1β fused with a GAL4 DNA-binding domain, which allows the measurement of transcriptional activation of PGC-1β in the presence of various compounds in the culture medium.
2. Materials and methods
2.1. Screening compounds that increase GAL4-PGC-1β activity
Full-length PGC-1β cDNA was cloned into the pM vector (Clontech/Takara Bio, Shiga, Japan) to produce a fusion protein with the GAL4 DNA-binding domain [2]. HEK293T cells were cultured in 90-mm dishes and transfected at 90% confluence using Lipofectamine 2000 (Thermo Fisher Scientific Co., Waltham, MA, USA), according to the manufacturer's instructions. Luciferase reporter plasmid containing four copies of the GAL4 DNA-binding sequence (upstream activating sequence, UAS; 2.5 µg), pM-PGC-1β (2.5 µg), and phRL-TK luc (control Renilla luciferase; 0.2 µg) were transfected. Five hours after transfection, the cells were plated at a density of 1 × 105 cells per well in a 96-well plate in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Twenty-nine hours after transfection, the cells were treated with various commercially available compounds (Sigma-Aldrich Japan, Tokyo, Japan; final concentration, 10 µM). After 20 h, the cells were lysed and assayed for luciferase activity using the Dual-Glo Luciferase Assay kit (Promega Co., Madison, WI, USA). The activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity (internal control).
2.2. Transfection and luciferase assays
C2C12 cells were plated at a density of 1 × 105 cells per well in a 12-well plate containing DMEM supplemented with 10% FBS. A luciferase reporter plasmid containing ERR-responsive element (ERRE, 0.8 µg), expression plasmid (pCMX-PGC-1β or empty pCMX, 0.4 µg), and phRL-TK vector (25 ng, Promega Co., Madison, WI, USA), which acted as an internal control of transfection efficiency, was transfected into C2C12 cells using Lipofectamine 2000. ERRE was derived from the MCAD promoter [2]. Five hours after transfection, the cells were treated with compounds (final concentration, 10 µM). After 24 h, the cells were lysed and assayed for luciferase activity using the Dual-Glo Luciferase Assay kit (Promega). The activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity (internal control) and was expressed as an average of triplicate experiments [6].
2.3. Quantitative real-time RT-PCR analysis
Total RNA was isolated from cell homogenates using the TRIzol reagent (Thermo Fisher Scientific Inc., Tokyo, Japan). cDNA was synthesized using 500 ng of each RNA sample with ReverTra Ace (Toyobo, Tokyo, Japan). Gene expression was measured as described previously [6]. The primer sequences used were as follows: MCAD, forward 5′-GATCGCAATGGGTGCTTTTGATAGAA-3′ and reverse 5′-AGCTGATTGGCAATGTCTCCAGCAAA-3′; PGC-1β, forward 5’-AAGAACTTCAGACGTGAGAGCAGAG-3’ and reverse 5’-GCATGCCGGACGCTTG-3’; 36B4, forward 5′-GGCCCTGCACTCTCGCTTTC-3′ and reverse 5′-TGCCAGGACGCGCTTGT-3′.
2.4. Stable cell lines
PlatE packaging cells for retrovirus were cultured in 90-mm dishes and transfected at 70% confluence using Lipofectamine 2000 (Thermo Fisher Scientific), according to the manufacturer's instructions, using 2 µg of pMX-derived expression plasmid [6] either containing the PGC-1β cDNA or the vector on its own. Virus-containing supernatants were harvested 48 h after transfection and added to C2C12 cells, which were selected using 5 µg/mL of puromycin to eliminate uninfected cells. After drug selection, virally infected stable cells were cultured to confluence in DMEM containing 10% FBS, and the medium was changed every 2 days.
2.5. Mitochondrial staining
C2C12 cells stably overexpressing PGC-1β were plated at a density of 1 × 104 cells per well in a 24-well plate containing DMEM supplemented with 10% FBS. Twenty-four hours later, test compounds were added to the culture medium (final concentration, 10 µM). After 24 h, MitoTracker Red CMXRos, stains mitochondria in live cells and its accumulation is dependent upon membrane potential (Thermo Fisher Scientific) [7] was added to the medium (final concentration, 50 nM) and incubated for 20 min. The cells were observed using fluorescence microscopy. Quantification of images was performed using the ImageJ software (https://imagej.nih.gov/ij/docs/guide/index.html).
2.6. Statistical analyses
Statistical comparison was performed using the Student's two-tailed unpaired t-test and two-way analysis of variance, followed by Tukey's post-hoc test for more groups. Data were checked for normality and equal variances between the groups. P < 0.05 was considered significant.
3. Results and discussion
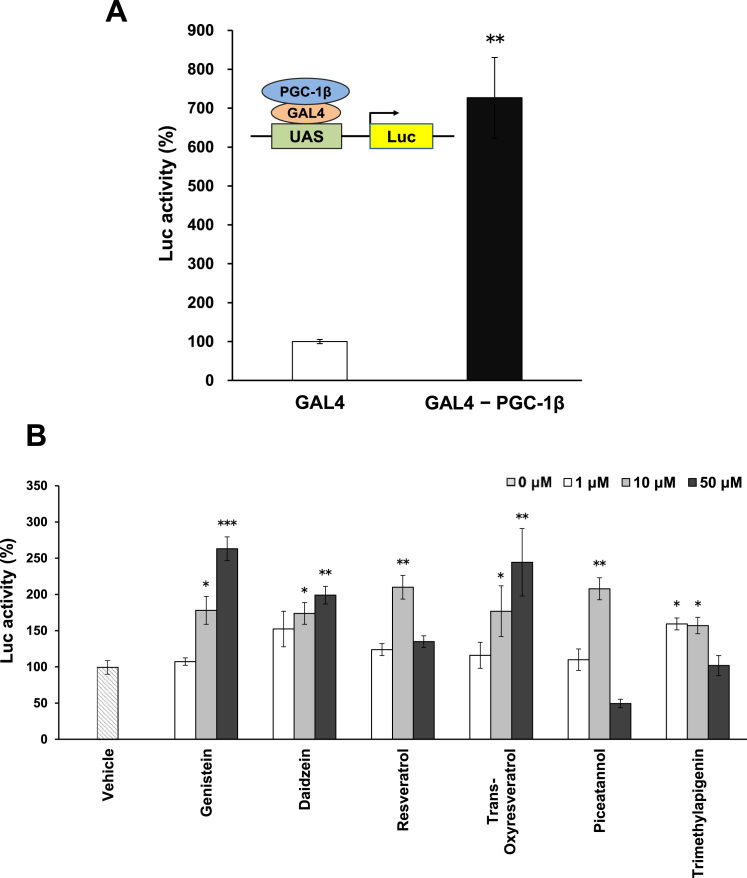
PGC-1β is a transcriptional co-activator that does not bind to promoter DNA directly, but interacts with transcription factors such as nuclear receptors. To evaluate the transcriptional activity of PGC-1β, we made a fusion protein of PGC-1β with GAL4 DNA-binding domain [2]. Compared with GAL4 alone, GAL4-PGC-1β showed a markedly higher reporter luciferase activity (Fig. 1A). We used this system and added various food-derived and natural compounds to the culture medium and examined their effects on PGC-1β-mediated transcriptional activity.
Fig. 1.
Establishment of a reporter system to measure the transcriptional activity of PGC-1β A) HEK293T cells were transfected with a reporter luciferase gene containing four copies of a GAL4-biding site (UAS) in the presence of GAL4 DNA-binding domain-fusion construct of full-length PGC-1β. Mean values from triplicate experiments are shown as fold induction, where the activity in the presence of GAL4 DNA-binding domain alone (without PGC-1β) serves as the reference value (set as 100). A schematic representation of the reporter plasmid is shown. B) Different concentrations (1, 10, and 50 µM) of the indicated compounds were added to the culture medium, and reporter luciferase activity was measured. Mean values are shown as fold induction, where the reference value is from GAL4-PGC-1β in the presence of vehicle alone. Each value represents mean ± SE (N = 3); ***P < 0.001, **P < 0.01, *P < 0.05.
We used 166 commercially available compounds (final concentration, 10 µM), examined the reporter activity, and compared it with that of the vehicle. The screening results are shown in [8]. Of the 166 compounds, 23 increased the reporter activity by more than 1.5-fold. To confirm this, we used different concentrations (1, 10, and 50 µM) of the 23 compounds and observed their effect on GAL-PGC-1β. Compounds that significantly increased reporter activity at any concentration are shown in Fig. 1B. Chemical structures of the compounds are shown in Supplemental Fig. 1. Compounds that increased reporter activity were genistein and daidzein, which are soy isoflavones, trans-oxyresveratrol, trans-resveratrol, and piceatannol, which are resveratrols, and trimethylapigenin, which is a flavone [9]. Because multiple soy isoflavones and resveratrols increased reporter activity, they were likely to be valid compounds. Genistein, daidzein, and trans-oxyresveratrol increased the reporter activity in a dose-dependent manner (Fig. 1B). Therefore, we used these three compounds for further analysis.
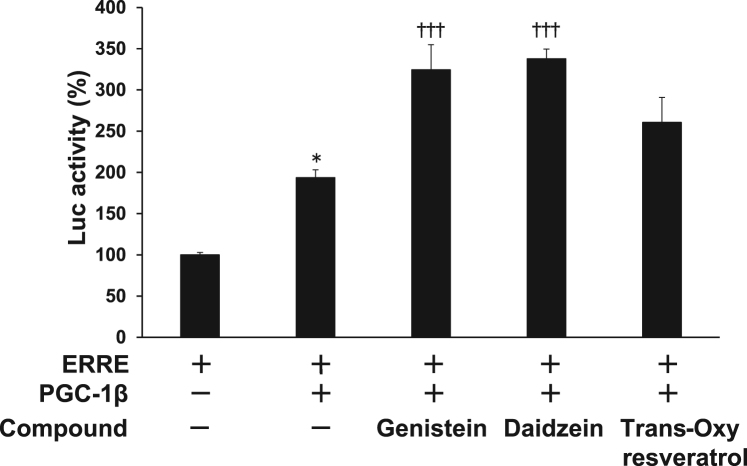
PGC-1β preferentially co-activates ERR-mediated transcription [2]. Therefore, we examined the effect of genistein, daidzein, and trans-oxyresveratrol on ERRE-mediated transcription via PGC-1β. We used C2C12 myoblast cells, which express endogenous ERR [6]. PGC-1β-expression vector increased ERRE-mediated luciferase reporter activity by two-fold (Fig. 2). In the presence of PGC-1β, genistein and daidzein significantly increased ERRE-luciferase activity, and trans-oxyresveratrol also marginally increased ERRE-luciferase activity (Fig. 2).
Fig. 2.
Effect of genistein, daidzein, and trans-oxyresveratrol on the reporter containing ERREs in the presence of PGC-1β The effect of genistein, daidzein, and trans-oxyresveratrol (10 µM) on ERRE-mediated reporter activity was measured in the presence of PGC-1β expression vector. The reporter construct contained an ERRE, derived from the MCAD gene, and the luciferase reporter gene. Each value represents mean ± SE (N = 3); *P < 0.05: vs ERRE alone, ††† P < 0.001: vs GAL4-PGC-1β plus ERR (vehicle).
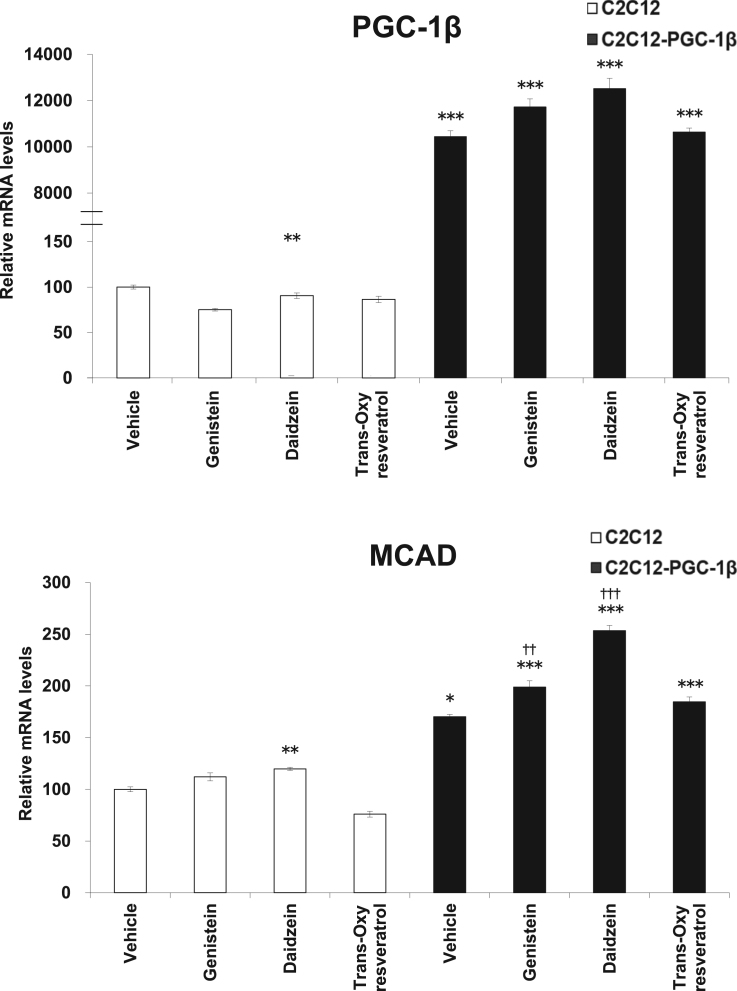
Next, we stably overexpressed PGC-1β in C2C12 myoblasts and examined the effect of the genistein, daidzein, and trans-oxyresveratrol on the expression of ERR-target gene MCAD. In C2C12 myoblasts (mock, without PGC-1β overexpression), daidzein treatment significantly increased MCAD mRNA levels (Fig. 3). The overexpression of PGC-1β mRNA was confirmed by quantitative real-time PCR (Fig. 3). Stable overexpression of PGC-1β increased MCAD mRNA levels. Moreover, genistein and daidzein treatment in C2C12-PGC-1β cells further increased MCAD mRNA expression significantly compared with that of vehicle-treatment in C2C12-PGC-1β cells. Trans-oxyresveratrol marginally increased MCAD mRNA levels in C2C12-PGC-1β cells (Fig. 3).
Fig. 3.
Effect of genistein, daidzein, and trans-oxyresveratrol on PGC-1β-stably overexpressed C2C12 cells in relationship to the expression of the MCAD gene. PGC-1β was stably overexpressed in C2C12 cells (C2C12-PGC-1β). Compared with C2C12 without exogenous PGC-1β, MCAD mRNA levels increased in C2C12-PGC-1β cells. The C2C12-PGC-1β cells were treated with genistein, daidzein, and trans-oxyresveratrol (final concentration 10 µM), for 24 h. Genistein and daidzein significantly increased MCAD mRNA levels. Each value represents mean ± SE (N = 4); ***P < 0.001, **P < 0.01: vs C2C12 (vehicle), ††† P < 0.001, †† P < 0.01: vs C2C12-PGC-1β (vehicle).
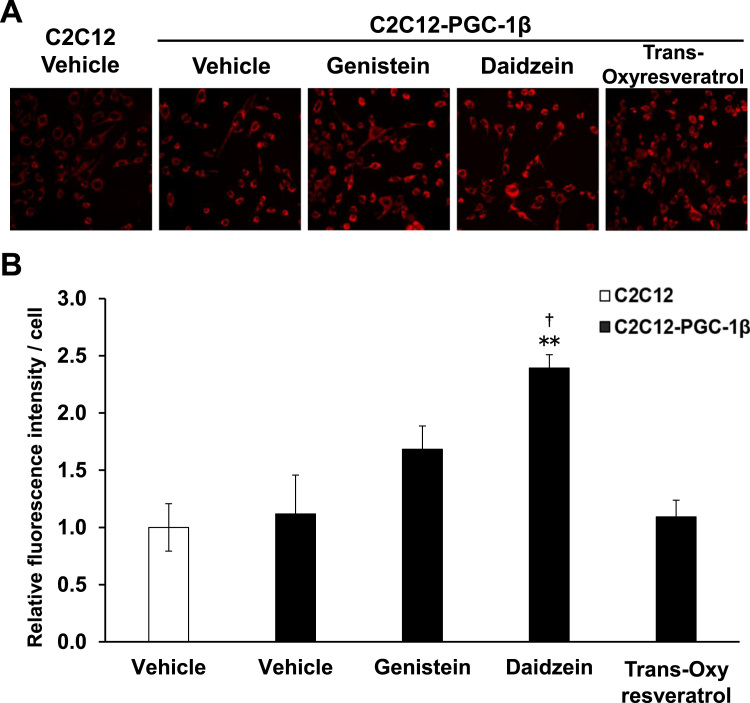
PGC-1β increases mitochondrial content [10]. Stable overexpression of PGC-1β in C2C12 myoblasts increased mitochondrial staining, as assessed using MitoTracker Red (increased strength of the fluorescence signal). Genistein and daidzein treatment further enhanced the fluorescence signal compared with that of vehicle treatment in C2C12-PGC-1β cells, which suggested increased mitochondrial content (Fig. 4).
Fig. 4.
Effect of genistein, daidzein, and trans-oxyresveratrol on the mitochondrial staining of C2C12-PGC-1β cells A) Control C2C12 cells (without exogenous PGC-1β) and C2C12 cells stably expressing PGC-1β were used (as in Fig. 3). MitoTracker (final concentration 50 nM) was added to the culture medium, and fluorescence was observed with the same exposure time. Compared with control C2C12 cells, C2C12-PGC-1β cells had a marginally increased MitoTracker signal. C2C12-PGC-1β cells were treated with genistein and daidzein, which further enhanced the MitoTracker signal. Typical microscopy images are shown. B) Quantification of images. Relative fluorescence intensity per cells are shown (N = 3). **P < 0.001: vs C2C12 (vehicle), † P < 0.05: vs C2C12-PGC-1β (vehicle).
Our screening of food-derived and natural compounds showed that the soy-derived isoflavones, genistein and daidzein, were PGC-1β-transcriptional activators. These compounds also increased ERRE-mediated PGC-1β transcriptional activity and ERR-target MCAD gene expression and mitochondrial content, which are targets of PGC-1β. Genistein and daidzein functionally enhanced PGC-1β-mediated cellular events. PGC-1β co-activates ERR [2], and genistein and daidzein are reported to be agonists of ERRs [11], which is consistent with our current finding that genistein and daidzein activated PGC-1β-mediated gene expression. PGC-1β likely interacts with endogenous ERR and is activated by isoflavones. Indeed, in the presence of the ERR expression vector, the reporter luciferase activity derived from GAL4-PGC-1β increased. Genistein and daidzein further enhanced ERR/GAL4-PGC-1β reporter activity (Supplemental Fig. 2).
We also found that resveratrol activated PGC-1β, albeit to a lesser extent than the isoflavones. PGC-1α, a homologue of PGC-1β, is activated by resveratrol via the activation of Sirt1. Sirt1 is a deacetylase, and deacetylation of PGC-1α stimulates its transcriptional activity [12]. Examining the amino acid sequences of PGC-1β and PGC-1α revealed that several lysine residues known to be regulated by acetylation in PGC-1α were also present in PGC-1β (Supplemental Fig. 3). Although acetylation-based regulation has not been reported in PGC-1β, resveratrol and Sirt1 may regulate PGC-1β as well as PGC-1α. For trimethylapigenin (Fig. 1B) whose role in PGC-1β regulation remains unclear, further studies are needed.
In our previous in vivo study, transgenic overexpression of PGC-1β in mice increased energy expenditure and prevented obesity. Other epidemiological studies have shown that people consuming soy tend to be lean (i.e., having a low body mass index) [13]. Therefore, soy isoflavones and resveratrols may be useful to prevent obesity as functional foods or supplements.
Acknowledgments
This study is supported by grants-in-aid for scientific research (KAKENHI) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT, Tokyo). This study is also supported by the Council for Science, Technology, and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), and “Technologies for creating next-generation agriculture, forestry and fisheries” (funding agency: Bio-oriented Technology Research Advancement Institution, NARO). This study is also supported by The Public Foundation of Elizabeth Arnold-Fuji, and Japan Dairy Association (J-milk). The funders had no role in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.11.009.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.11.009.
Appendix A. Transparency document
Supplementary material.
Appendix A. Supplementary material
Supplemental Fig. 1 Diagrammatic representation of the chemical structures of the screened compounds Structures of genistein, daidzein, resveratrol, trans-oxyresveratrol, piceatannol, and trimethylapigenin are shown.
Supplemental Fig. 2. Effect of genistein and daidzein on the reporter in the presence of GAL4-PGC-1β and ERR. HEK293T cells were transfected with a reporter luciferase gene containing four copies of a GAL4-biding site (UAS) in the presence of GAL4 DNA-binding domain-fusion construct of full-length PGC-1β. Values are shown as fold induction, where the activity in the presence of GAL4 DNA-binding domain alone (without PGC-1β) serves as the reference value (set as 100). The expression vector of ERR (pCMX-ERRγ) was also used for transfection. A schematic representation of the reporter plasmid is shown. 10 µM (final concentration) of genistein or daidzein were added to the culture medium, and reporter luciferase activity was measured. Each value represents mean ± SE (N = 3); ###P < 0.001, with or without ERR, ***P < 0.001: vs GAL4-PGC-1β (vehicle), ††† P < 0.001: vs GAL4-PGC-1β plus ERR (vehicle).
Supplemental Fig. 3 Comparison of the amino acid sequences of PGC-1β and PGC-1α Conserved lysine residues regulated by Sirt1-mediated deacetylation in PGC-1α (Rodgers et al. [12]) are indicated in bold-face and marked off with a red box.
References
- 1.Lin J., Puigserver P., Donovan J., Tarr P., Spiegelman B.M. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J. Biol. Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 2.Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., Kawada T., Miyoshi M., Ezaki O., Kakizuka A. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. USA. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamei Y., Hatazawa Y., Yoshimura R., Miura S., PGC-1α, Stimuli-Inducible A. Nuclear receptor coactivator: structural features and role in skeletal muscle metabolism gene regulation. Biomed. Sci. 2015;1:6–9. [Google Scholar]
- 4.Sladek R., Bader J.A., Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol. Cell. Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vega R.B., Kelly D.P. A role for estrogen-related receptor alpha in the control of mitochondrial fatty acid beta-oxidation during brown adipocyte differentiation. J. Biol. Chem. 1997;272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- 6.Hatazawa Y., Qian K., Gong D.W., Kamei Y. PGC-1alpha regulates alanine metabolism in muscle cells. PLoS One. 2018;13:e0190904. doi: 10.1371/journal.pone.0190904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chazotte B. Labeling mitochondria with MitoTracker dyes. Cold Spring Harb. Protoc. 2011;2011:990–992. doi: 10.1101/pdb.prot5648. [DOI] [PubMed] [Google Scholar]
- 8.Nakai S., Uchitomi R., Matsuda R., Onishi T., Hatazawa Y., Kamei Y. Screening of food components that enhance transcriptional activity of PGC1-beta. Data Brief. 2018 doi: 10.1016/j.dib.2019.103814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean M., Murphy B.T., Burdette J.E. Phytosteroids beyond estrogens: regulators of reproductive and endocrine function in natural products. Mol. Cell. Endocrinol. 2017;442:98–105. doi: 10.1016/j.mce.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meirhaeghe A., Crowley V., Lenaghan C., Lelliott C., Green K., Stewart A., Hart K., Schinner S., Sethi J.K., Yeo G., Brand M.D., Cortright R.N., O'Rahilly S., Montague C., Vidal-Puig A.J. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suetsugi M., Su L., Karlsberg K., Yuan Y.C., Chen S. Flavone and isoflavone phytoestrogens are agonists of estrogen-related receptors. Mol. Cancer Res.: MCR. 2003;1:981–991. [PubMed] [Google Scholar]
- 12.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 13.Maskarinec G., Aylward A.G., Erber E., Takata Y., Kolonel L.N. Soy intake is related to a lower body mass index in adult women. Eur. J. Nutr. 2008;47:138–144. doi: 10.1007/s00394-008-0707-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplemental Fig. 1 Diagrammatic representation of the chemical structures of the screened compounds Structures of genistein, daidzein, resveratrol, trans-oxyresveratrol, piceatannol, and trimethylapigenin are shown.
Supplemental Fig. 2. Effect of genistein and daidzein on the reporter in the presence of GAL4-PGC-1β and ERR. HEK293T cells were transfected with a reporter luciferase gene containing four copies of a GAL4-biding site (UAS) in the presence of GAL4 DNA-binding domain-fusion construct of full-length PGC-1β. Values are shown as fold induction, where the activity in the presence of GAL4 DNA-binding domain alone (without PGC-1β) serves as the reference value (set as 100). The expression vector of ERR (pCMX-ERRγ) was also used for transfection. A schematic representation of the reporter plasmid is shown. 10 µM (final concentration) of genistein or daidzein were added to the culture medium, and reporter luciferase activity was measured. Each value represents mean ± SE (N = 3); ###P < 0.001, with or without ERR, ***P < 0.001: vs GAL4-PGC-1β (vehicle), ††† P < 0.001: vs GAL4-PGC-1β plus ERR (vehicle).
Supplemental Fig. 3 Comparison of the amino acid sequences of PGC-1β and PGC-1α Conserved lysine residues regulated by Sirt1-mediated deacetylation in PGC-1α (Rodgers et al. [12]) are indicated in bold-face and marked off with a red box.